Abstract
Interoperability across data sets is a key challenge for quantitative histopathological imaging. There is a need for an ontology that can support effective merging of pathological image data with associated clinical and demographic data. To foster organized, cross-disciplinary, information-driven collaborations in the pathological imaging field, we propose to develop an ontology to represent imaging data and methods used in pathological imaging and analysis, and call it Quantitative Histopathological Imaging Ontology – QHIO. We apply QHIO to breast cancer hot-spot detection with the goal of enhancing reliability of detection by promoting the sharing of data between image analysts.
Keywords: histopathology imaging, image analysis, hot spot, ontology, breast cancer
Graphical abstract
Introduction
Interoperability across data sets is a key challenge for quantitative histopathological imaging (QHI). Interoperability describes the extent to which systems and devices can share data and interpret the data that is shared. The ideal is for multiple systems to be able to use and interpret each other’s data in just the same way that they can use and interpret their own data. Limited interoperability between imaging data systems is a major obstacle to coherent multi-institutional collaboration, in digital pathology as in many other areas.
Interoperability, according to the HIMSS Dictionary, may be seen on three different levels.1 At the foundational level interoperability describes the capability for simple data exchange from one information system to another, without any requirement for the receiving information technology system to be able to interpret the data that it receives. Interoperability at the structural level refers to the capability for data exchange in which the format and organization of the data is preserved unaltered. Here interoperability relates to the syntax of the data exchanged. The highest level of semantic interoperability is achieved, according to the HIMSS Dictionary, when data systems can take advantage “of both the structuring of the data exchange and the codification of the data including vocabulary so that the receiving information technology systems can interpret the data.”
Many strategies to achieve semantic interoperability nowadays require the use of controlled vocabularies which provide single annotations or tags to be used to address the problems which arise when multiple coding systems use different codes describe the same entities in reality. “Ontologies” improve on controlled vocabularies by using links and logical definitions to connect terms in a rich network of well-defined relationships. With the advance of pathological imaging technology and of associated software for the processing of pathological images, the need arises for an ontology which can support effective merging of pathological image data with associated clinical and demographic data that have already been described using existing controlled vocabularies such as SNOMED-CT, the NCI Thesaurus, or the ontologies such as the Cell Ontology constituting the OBO Foundry [1].
To this end we are constructing a Quantitative Histopathological Image Ontology (QHIO) incorporating terms representing the different types and subtypes of pathological images, imaging processes and techniquesand computational algorithms. In addition the ontology will incorporate formal definitions of these terms and specify formally the relations that hold between entities of the corresponding types. Because QHIO will itself follow the principles of the OBO Foundry, the data resulting from the use of QHIO terms in annotations will be in a form that allows integration with other commonly used ontologies in the biomedical domain.
The result will allow us to leverage the opportunities brought by new imaging platforms and algorithms to create an environment in which the clinical imaging data, and clinician and algorithmically created annotations deriving from different communities of clinicians and scientists, can be combined and analyzed as a single whole.
The Problem of Reproducibility of Image Analysis
A further urgent factor in contemporary research is the issue of reproducibility of clinical and scientific findings. The reproducibility and validation of large-scale, cross-institutional imaging research is limited by the fact that there is lacking any common structured framework for describing images and the results of their analysis. Here, too, we believe, ontologies can play a role by providing controlled vocabularies which can be used to describe in standardized ways the steps taken to achieve particular results [2]. Currently, pathology image data is collected in local “silos” using in-house protocols, and is processed using proprietary algorithms developed in isolation. Typically the software itself may be unavailable to downstream image consumers, and even when it is available, there is rarely information about the sets of parameters necessary to run the software. Even the type of outputs of these algorithms, for example, a new potential prognostic feature, is not open to discovery by third party software. For these and a series of related reasons digital histopathology is difficult to reproduce and validate. While reproducibility and validation are important goals in their own right, even more value can be gained by combining and building upon image data and software methods across institutions.
Quantitative Histopathological Imaging Ontology (QHIO)
To foster organized, cross-disciplinary, information-driven collaborations in the pathological imaging field, we propose to develop an ontology to represent imaging data and methods used in pathological imaging and analysis with an initial focus on enabling effective communication and collaboration between developers of algorithms for analyzing histopathology images. Our ontology is modeled after the Gene Ontology (GO) [3] and follows the principles of the Open Biomedical Ontologies (OBO) Foundry [4], which are now used by some dozens of ontology-driven research efforts in clinical and translational science. An essential aspect of our work is the reuse of existing ontology terms wherever possible, thereby increasing the potential for interoperability as well as reducing the effort involved in defining new terms [5]. Hereafter when we refer to QHIO we mean the aggregate of terms we define, together with those terms that have been reused (imported) from other ontologies [5].
We intend to continue to design and use QHIO to promote long-term interoperability of data across pathology imaging. We will cultivate wide community dissemination through development of model collections that include sample image data, example software that can compute useful information about them as well as the annotated output of this software. Both the model collections and the ontology will be released into the public domain, again following the model of the GO [4].
The workflow in histopathology-based clinical testing extends from biopsy collection, to slide production and analysis, to the assignment of a diagnostic category, to the creation of a predictive statement for an individual patient based on that diagnostic category. All of these activities combine to support the choice of a treatment plan by a clinician for a particular patient.
At the same time these activities cover a variety of different domains and thus they raise multiple questions that will impact interoperability. Some of these questions include: How are demographic, anatomical and relevant health information to be gathered and communicated? How are annotations for histology image data created and what do they identify? What information will be needed to enable productive communication between image analysis researchers and clinicians? Answers to these questions will influence the success and quality of interoperability. Our long-term goal is to demonstrate that an ontology can enable interoperability in the pathology image domain. To achieve this goal, we are in the process of curating an annotated dataset and developing parallel sets of algorithms for quantifying breast cancer histopathology. We will attempt to combine the results of these algorithms, run on the same dataset, as a test, while developing and testing QHIO.
QHIO’s overarching goal is to foster interoperability of computationally derived image data. We need to emphasize that this study is not designed so much to help routine clinical diagnosis; rather it is designed to create an enriched data resource in which various kinds of clinical and translational software approaches can be tested out. It can also be used to explore the role image data may play in clinical decision support systems and to test hypotheses regarding correlations between image features and patient outcomes, or between image features and other features catalogued by OBO Foundry ontologies.
Motivating examples
We present two general activities (i.e. validation and combining multiple analyses to make diagnosis), and give a more detailed use case (i.e. hot spot detection) that motivates our effort.
Activity 1: Validation
The issue of interoperability is a major concern in validation. For example, two groups working on feature extraction may be investigating different but overlapping features, but currently have no way to develop a comparison tool to use their extracted features on the same dataset and thus produce quantitative results in a manner that can be easily communicated. This is important because such comparison is not only important for the qualitative validation of subject matter experts’ prognoses but also for timely identification of divergence of results flowing from use of independently developed algorithms targeting the same object.
Activity 2: Combining multiple analyses to make diagnoses
Another scenario where QHIO might be useful is in the making of diagnoses on the basis of data derived from multiple analyses. Several different algorithms can be run on the same images to analyse different aspects (e.g. the amount of stroma and mitotic count). A diagnostic program needs to check whether the available data have the specific features it needs: Relative amounts of cells by type, quantity of mitotic cells and potentially a subgroup of those within certain regions. It is important that we can establish effectively that these features are among those found in several different analyses while the algorithm determines the case, retrieves the relevant information, and computes the diagnostic in a robust manner.
Use case: Hot spot detection
Ki67, a nuclear marker expressed in all phases of the cell cycle except G0 [6], has been widely used in pathology to assess proliferation within multiple neoplasms. “Hot Spots”, are areas with prevalent Ki67 staining with the highest number of positively staining nuclei within the invasive component. In the literature, there is no uniform approach to scoring hot spots. Many studies have specifically targeted hot spots in determining the Ki67 index while others have done an overall assessment of Ki67 with the incorporation of the hot spots in the overall index. Clearly, studies addressing outcome with detailed hot spot scoring are warranted. However in order to do better we need to develop an automatic hot spot detection method that can minimize the intra- and inter-pathologist’s variability in identification of hot spots.
In previous work, we developed a hot spot tool [7] for breast cancer as Ki67 has shown promise as a prognostic marker and predictor of responsiveness to chemotherapy or endocrine therapy. The method first segments Ki67 positive pixels using a previously-developed Visually Meaningful Segmentation (VMS) method [8]. VMS generates an image-dependent filter, which in turn generates a density map from the segmented image. The smoothness of the density image simplifies the detection of local maxima, which directly correspond to the hot spots in the image. The method was tested on 23 different regions of interest extracted from 10 breast cancer Ki67 slide images. To determine intra-reader variability, each image was annotated twice for hot spots by a board-certified pathologist with a two-week interval between the two readings. A computer-generated hot spot was considered true-positive if it agreed with either of the two annotation sets provided by the pathologist. While intra-reader variability was 57%, our method correctly detected hot spots with 81% precision. In order to run this tool at multiple institutions, some interoperability issues need to be overcome. For instance, we need a way to identify the stain type as Ki67. Additionally, several terms need to be defined and communicated, for example, positive and negative stain, hot spot boundary, image magnification, etc. both to the algorithm as well as the pathologist who will use this system.
Methods
In our paper “Biomedical imaging ontologies: A survey and proposal for future work” [9] we surveyed the state of the art as concerns the development and application of controlled vocabularies and ontologies for digital pathology [10, 11]. We also laid out a plan for building QHIO and for using QHIO in promoting the sharing of pathology image data and associated algorithm and algorithm output information. We have begun to execute this plan by
creating a prototype of QHIO and
applying it to breast cancer hot-spot detection with the goal of enhancing reliability of detection by promoting the sharing of data between image analysts.
Our goal here is to demonstrate that QHIO can be used for pathology data sharing, and that it can serve as a starting point for further development toward realizing our longer term goals of advancing interoperability of histopathological imaging systems and reproducibility of histopathological imaging assays. The current version of QHIO is available at https://github.com/ontodev/QHIO.
A turning point in the development of ontologies and their extensive use in biology and biomedicine was the building and application of the Gene Ontology (GO) [3]. That work showed the benefits of tagging sequence data obtained from both humans and multiple model organism species with a single set of species-neutral terms. The success of the GO created a situation in which many biomedical subdisciplines saw a need to develop ontologies of their own, often in uncoordinated fashion with resultant tendencies to forking and redundancy. To counteract these tendencies a group of researchers developing ontologies centered on the GO established, in 2004, the Open Biomedical Ontology (OBO) Foundry initiative, promulgating a set of principles for ontology development which have been tested in practice and refined in light of the lessons learned by the many groups who have sought to apply them in their work. It is these principles which we have used also to guide our work on QHIO. Chief among them is a commitment by the developers of each ontology to ensure interoperability with its neighbouring ontologies.
The Ontology for Biomedical Investigations
One neighbour to QHIO is the Ontology for Biomedical Investigations (OBI), an OBO Foundry ontology with over 2500 terms for describing biomedical investigations, including core terms such as investigation, assay, planning, protocol, specimenand conclusion based on data.
Currently, OBI contains a small number of terms for medical imaging and pathology, including terms such as imaging assay, staining, feature extraction, and pathologist role. We are developing QHIO as an extension of OBI, with the immediate advantage of interoperability between QHIO and the larger OBO ecosystem. QHIO will be subject to the same processes of ongoing review and maintenance and be able to draw on the deep expertise that the OBI community has developed over several years.
OBI’s hierarchy of terms can be used to represent investigations either in great detail or in broad strokes, as appropriate. For example, OBI can be used to annotate a given body of experiment records along dimensions such as: funding agency, antibodies, staining methods, statistical algorithms used, and so forth. In this way OBI enables powerful querying across experimental data, made still more powerful through cross-linkage to other OBO Foundry ontologies such as the Gene Ontology the Cell Ontology (CL) [12], the Foundational Model of Anatomy (FMA) [13], the Human Disease Ontology [14], Chemical Entities of Biological Interest (ChEBI) ontology [15], as well as to external ontology resources including – most importantly for our purposes – the National Cancer Institute Thesaurus (NCIT). This enables other researchers to find experimental data that have been annotated with OBI because they know which terms to look for when searching. It also enables other researchers to understand how the data was acquired, since OBI enables provenance information pertaining not only to the persons, organizations, places and times of generation of the data, but also the experimental and data processing methods used.
A number of initiatives are under way to map these and other OBO Foundry ontologies to major clinical vocabularies (especially SNOMED CT and ICD) in order to allow them to be used for querying and analytical purposes in tandem with Electronic Health Record and other clinical data [16]. We will take this work further in the specific field of breast cancer research, taking account of the set of common data elements (CDEs) recommended by the NCI's Early Detection Research Network (EDRN) for the treatment of breast cancer biomarker data. We will explore applications of our work to the creation of CDEs for cancer imaging biomarkers, drawing also on the preliminary work performed within the framework of the Quantitative Imaging Biomarker Ontology (QIBO) project [17].
What will QHIO achieve?
Currently whole slide pathology images are annotated using vendor-supplied, freely available software programs. The resultant annotations in these programs typically employ natural language phrases, sometimes using machine-readable formats such as XML. They are limited to small controlled vocabularies of terms for colors and basic geometrical shapes (e.g. lines, circles, free-form figures), and use unstructured text fields to describe features of interest (e.g. ‘necrotic region’, ‘necrosis’, etc.). Annotations created by members of given communities using given vendor software are in almost every case incompatible with those created by other communities using other software. This locks valuable data into information silos.
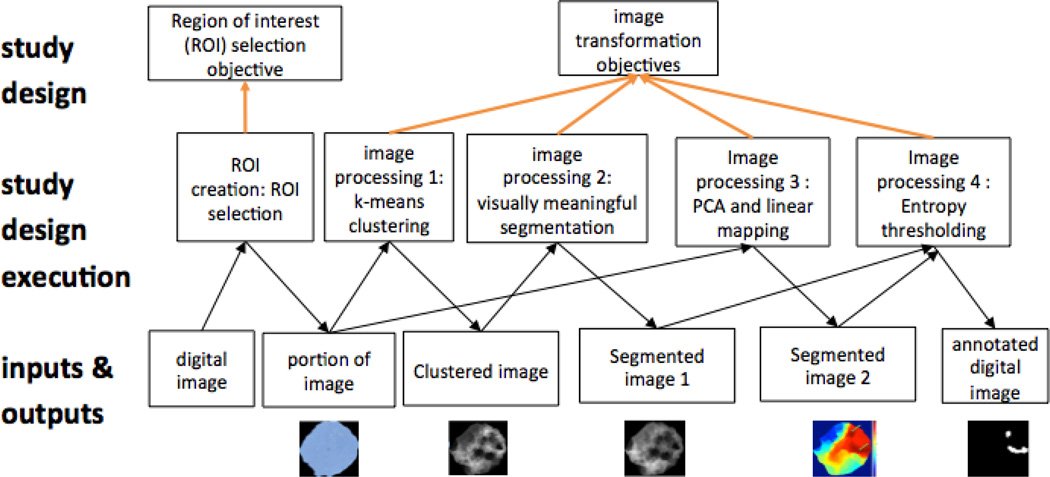
The challenges of sharing algorithms and image features are even greater than those of sharing image annotations because no standards currently exist to describe algorithms and image features. To rectify this problem QHIO will include standard terms not merely for algorithm and feature types and attributes but also for all other entities involved in each stage of the pathological imaging and analysis workflow (Figure 1). These will include:
Input: type (e.g. image), preparation processes such as staining, slide characteristics, magnification, resolution, the meaning and typical range of parameters and other annotations
Parameters: size of the filter window sizes, the number of iterations, etc.
Output: type (e.g. image, measurement), the meaning and typical range of parameters, statistical details (e.g. accuracy, false positive rate), statistical evaluation methodology (e.g. ROC) and the methods/software required
Execution: operating system (e.g. Windows 7), software environment (e.g. Matlab), required resources (e.g. RAM, storage), expected time to run the algorithm.
Figure 1.
From specimen to hot spot detection. Overview of types and relationships in QHIO. Types are taken from OBI. More specific types from QHIO noted after colon.
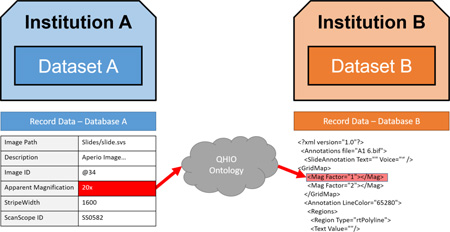
In addition there is a need to improve the existing annotation scheme by using QHIO in tandem with standard linked data formats that generalize current approaches based on XML, by applying QHIO and associated software to make image and algorithm annotations cleanly interoperable between sites (Figure 2).
Figure 2.
Sample workflow represented through QHIO and OBI terms. (PCA = Principal Component Analysis)
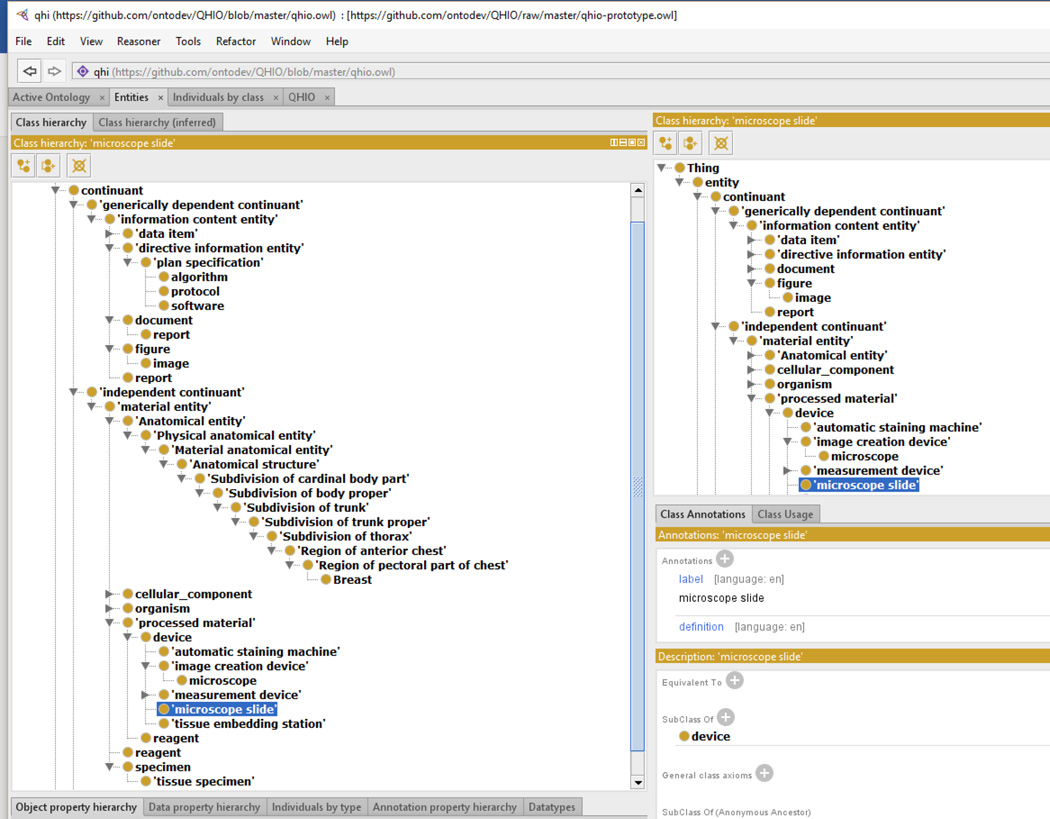
For this study, we have identified hot spot detection algorithm-related terms (Table 1). These terms as well as ontological terms necessary to represent images were included in QHIO prototype (Figure 3).
Table 1.
Hot spot detection algorithm related terms, existing OBO superclass and URI, and upper level term. Information entity is from the IAO, disorder from OGMS, and rest of the terms are from BFO.
| Term needed | mid-level parent | mid-level URI |
high-level parent | |
|---|---|---|---|---|
| 1 | color space transformation | data transformation | OBI_0200000 | process |
| 2 | image segmentation | biological feature identification | OBI_0000015 | process |
| 3 | object classification | class discovery data transformation | OBI_0200175 | process |
| 4 | clustering | class discovery data transformation | OBI_0200175 | process |
| 5 | pixel | unit | UO_0000000 | information entity |
| 6 | image markup | symbol | IAO_0000028 | information entity |
| 7 | image annotation | textual entity | IAO_0000300 | information entity |
| 8 | histological slide scanning | image creation | OBI_0001007 | process |
| 9 | histological slide scanner | image creation device | OBI_0000398 | material entity |
| 10 | image region | part_of some image | IAO_0000101 | information entity |
| 11 | histological mounting | histological sample preparation | OBI_0000341 | process |
| 12 | ischemia | ischemia | DOID_326 | disorder |
| 13 | tissue dehydration | material processing | OBI_0000094 | process |
| 14 | histological sectioning | histological sample preparation | OBI_0000341 | process |
| 15 | optical magnification | ratio device setting | OBI_0000654 | quality |
| 16 | microns per pixel | unit label | UO_0000000 | information entity |
| 17 | image annotation creation | documenting | IAO_0000572 | process |
| 18 | histological slide | microscope slide | OBI_0400170 | material entity |
| 19 | Ki67 stain | cytological stain role | OBI_0000026 | material entity |
| 20 | ki67 | antigen KI-67 | PR_000010425 | material entity |
| 21 | hot spot | cellular feature identification | OBI_0000219 | information entity |
| 22 | histological section | tissue specimen | OBI_0001479 | material entity |
| 23 | mitosis count | substance unit | UO_0000006 | information entity |
| 24 | Ki67 percentage | concentration unit | UO_0000051 | information entity |
Figure 3.
An example representation in the QHIO prototype generated using Protégé
Discussion and Conclusion
Experience shows that – even with the use of traditional Delphi techniques [18] – current image annotation formats and current ways of creating annotations (in many cases on the basis of free text or locally developed codes) leave much of the data accessible only at the site where it was created. Such data is both difficult to interpret and understand outside that context, and is in practice thus undiscoverable by external researchers. Our future work will be about bridging that gap by building an ontology and supporting software for better data sharing.
The potential value of QHIO is its ability to aggregate data from multiple sources that can then be subject to analysis for research purposes. Establishing which images are compatible with a chosen algorithm and which might be pooled in an aggregate analysis certainly could benefit researchers, and the provision of information of the types in the QHIO would be very valuable for this purpose, whether embedded in the image metadata or in the database from which they are derived. Using a subset of the common dataset, ontology development experts need to continue the development of QHIO. QHIO must be applied to convert the annotated image data into a set of interoperable, standardized data, supported by tools that allow for rich search and analysis, and automated matching of algorithms to appropriate input data.
Ontologies serve as crucial aids to human communication, and are supported by a range of technologies that aid machine communication, search, and analysis [19]. We will use linked data standards developed by the World Wide Web Consortium (W3C) and associated technologies to build, maintain and use QHIO, and to process, store, and query annotations on images and algorithms. In particular the W3C standard Web Ontology Language (OWL) [20] will serve as the foundational technology for QHIO, as it is for all OBO ontologies. OWL brings both the ability to define and organize the classes in the ontology and the ability to instantiate those classes in the case of particular workflows or studies. OWL builds on the Resource Description Framework (RDF), which can be queried using the SPARQL Query Language. These and other software resources used in our project are based on open source tools and libraries. We have already demonstrated the use of OWL in QHIO, and used it to model the application of a hot-spot detection algorithm.
QHIO will also build upon existing work done by the Open Microscopy Environment project (http://www.openmicroscopy.org/site/support/ome-model/). For example, OME already deals very effectively with the details of various file formats. QHIO and OME are complementary efforts. One future research direction is to establish tight integration between OME and QHIO, although this may require changes to both OME and QHIO. Additionally, we need to supply resources for keeping track of the image artifact itself, of how it changes as different sorts of annotations are added.
We will use QHIO and linked data tools to develop an automated workflow for converting existing image annotations in our common dataset to linked data annotations using QHIO terms and terms from associated ontologies and structured resources referencing clinical data. The converted data will be loaded into an RDF database where it can be shared and queried using SPARQL. It is at this point the benefits of using OBO Foundry ontologies become increasingly clear:
Every ontology term belongs to a hierarchy of more-and-less general terms, allowing for queries at many levels of specificity;
Ontology terms are defined through logical axioms that provide further links across the network of data, such as parthood, aboutness, adjacency, inclusion and other relations;
The logic of OWL allows for automated reasoning software to infer further links in the network from existing links and logical axioms, facilitating error checking and discovery of new knowledge;
Pathology imaging data can be easily integrated with data from other medical and scientific domains;
The technology that is involved is now widely dispersed through the biomedical research community through the sustained support not merely of the W3C [20] but also of the National Cancer Institute, the International Health Terminology Standards Development Organization (IHTSDO, now responsible for SNOMED – CT) and by other bodies.
Highlights.
The Quantitative Histopathological Imaging Ontology (QHIO) is proposed.
QHIO facilitates interoperability between histopathology datasets and algorithms.
By enforcing data compatibility, QHIO enables large-scale collaborative studies.
Researchers can easily find data and algorithms to suit their experimental needs.
Designed with OBO principles, QHIO integrates with existing biomedical ontologies.
Acknowledgments
Funding
The project described was supported in part by Awards Number U24CA199374 (PIs: Gurcan, Madabushi, Martel) from the National Cancer Institute and UL1TR001412 (co-PI: Smith) from National Center for Advancing Translational Sciences (NCATS). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute, National Center for Advancing Translational Sciences (NCATS), or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
HIMSS Dictionary of Healthcare Information Technology Terms, Acronyms and Organizations, 2nd Edition, 2010, Appendix B
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflict of interest.
References
- 1.Diehl AD, Meehan TF, Bradford YM, Brush MH, Dahdul WM, Dougall DS, et al. The Cell Ontology 2016: enhanced content, modularization, and ontology interoperability. J Biomed Semantics. 2016;7:44. doi: 10.1186/s13326-016-0088-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinkman RR, Courtot M, Derom D, Fostel J, He Y, Lord PW, et al. Modeling biomedical experimental processes with OBI. J. Biomedical Semantics. 2010;1:S7. doi: 10.1186/2041-1480-1-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nature biotechnology. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Courtot M, Gibson F, Lister AL, Malone J, Schober D, Brinkman RR, et al. MIREOT: The minimum information to reference an external ontology term. Applied Ontology. 2011;6:23–33. [Google Scholar]
- 6.Gerdes J, Lemke H, Baisch H, Wacker H-H, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. The Journal of Immunology. 1984;133:1710–1715. [PubMed] [Google Scholar]
- 7.Niazi MKK, Downs-Kelly E, Gurcan MN. Hot spot detection for breast cancer in Ki-67 stained slides: image dependent filtering approach. SPIE Medical Imaging. 2014:904106-904106-8. [Google Scholar]
- 8.Niazi M, Beamer G, Gurcan MN. Detecting and characterizing cellular responses to Mycobacterium tuberculosis from histology slides. Cytometry Part A. 2014;85:151–161. doi: 10.1002/cyto.a.22424. [DOI] [PubMed] [Google Scholar]
- 9.Smith B, Arabandi S, Brochhausen M, Calhoun M, Ciccarese P, Doyle S, et al. Biomedical imaging ontologies: A survey and proposal for future work. Journal of pathology informatics. 2015;6 doi: 10.4103/2153-3539.159214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adebayo S, McLeod K, Tudose I, Osumi-Sutherland D, Burdett T, Baldock R, et al. PhenoImageShare: an image annotation and query infrastructure. J Biomed Semantics. 2016;7:35. doi: 10.1186/s13326-016-0072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galton A, Landini G, Randell D, Fouad S. Ontological Levels in Histological Imaging. Formal Ontology in Information Systems: Proceedings of the 9th International Conference (FOIS 2016) 2016:271.
- 12.Meehan TF, Masci AM, Abdulla A, Cowell LG, Blake JA, Mungall CJ, et al. Logical development of the cell ontology. BMC bioinformatics. 2011;12:6. doi: 10.1186/1471-2105-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. Journal of biomedical informatics. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Schriml LM, Arze C, Nadendla S, Chang Y-WW, Mazaitis M, Felix V, et al. Disease Ontology: a backbone for disease semantic integration. Nucleic acids research. 2012;40:D940–D946. doi: 10.1093/nar/gkr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hastings J, de Matos P, Dekker A, Ennis M, Harsha B, Kale N, et al. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic acids research. 2013;41:D456–D463. doi: 10.1093/nar/gks1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson PN. Computational Phenotype Analysis in Human Medicine. Phenomics. 2014:8. [Google Scholar]
- 17.Buckler AJ, Ouellette M, Danagoulian J, Wernsing G, Liu TT, Savig E, et al. Quantitative Imaging Biomarker Ontology (QIBO) for Knowledge Representation of Biomedical Imaging Biomarkers. Journal of digital imaging. 2013;26:630–641. doi: 10.1007/s10278-013-9599-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.NickBowles. The Delphi technique. Nursing Standard. 1999;13:32–36. doi: 10.7748/ns1999.07.13.45.32.c2650. 1999/07/28. [DOI] [PubMed] [Google Scholar]
- 19.Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, et al. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic acids research. 2011;39:W541–W545. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.OWL 2 Web Ontology Language Document Overview. 2012 Available: http://www.w3.org/TR/owl2-overview/