Abstract
Morphological plasticity in response to environmental cues in migrating cancer cells requires F-actin cytoskeletal rearrangements. Conserved formin family proteins play critical roles in cell shape, tumor cell motility, invasion and metastasis, in part, through assembly of non-branched actin filaments. Diaphanous-related formin-2 (mDia2/Diaph3/Drf3/Dia) regulates mesenchymal-to-amoeboid morphological conversions and non-apoptotic blebbing in tumor cells by interacting with its inhibitor diaphanous-interacting protein (DIP), and disrupting cortical F-actin assembly and bundling. F-actin disruption is initiated by a CXCL12-dependent mechanism. Downstream CXCL12 signaling partners inducing mDia2-dependent amoeboid conversions remain enigmatic. We found in MDA-MB-231 tumor cells CXCL12 induces DIP and mDia2 interaction in blebs, and engages its receptor CXCR4 to induce RhoA-dependent blebbing. mDia2 and CXCR4 associate in blebs upon CXCL12 stimulation. Both CXCR4 and RhoA are required for CXCL12-induced blebbing. Neither CXCR7 nor other Rho GTPases that activate mDia2 are required for CXCL12-induced blebbing. The Rho Guanine Nucleotide Exchange Factor (GEF) Net1 is required for CXCL12-driven RhoA activation and subsequent blebbing. These results reveal CXCL12 signaling, through CXCR4, directs a Net1/RhoA/mDia-dependent signaling hub to drive cytoskeleton rearrangements to regulate morphological plasticity in tumor cells. These signaling hubs may be conserved during normal and cancer cells responding to chemotactic cues.
Keywords: formin, cytoskeleton, amoeboid, CXCL12
1. Introduction
Morphological plasticity during cell motility relies on regulation of F-actin cytoskeletal networks. Formin family proteins include mammalian Diaphanous-related (mDia/Drf/Diaph/Dia) formins (mDia1–3) [1–3]. mDias are highly-conserved actin nucleators that elongate and, in some cases, bundle F-actin filaments underlying protrusive cytoskeleton structures including filopodia, lamellipodia, and ruffles [2, 4]. mDia-based regulation of F-actin dynamics is critical to maintenance of the cortical F-actin cytoskeleton underlying cell contraction, amoeboid bleb formation, and cell motility [4–6]. mDias are regulated through autoinhibition. They are activated when Rho GTPases bind the mDia GTPase-binding domain (GBD), releasing the autoinhibited conformation and allowing protein effector binding [1].
mDia2 functional inhibition occurs upon association with Diaphanous-interacting protein (DIP). DIP (SPIN90/NCKIPSD/WASp-interacting SH3 protein (murine) (WISH)) [7–9] has a DIP leucine-rich region (LRR) that binds mDia2 FH2 domains directly and inhibits FH2-mediated F-actin filament elongation and bundling [10]. This disrupts the cortical actin cytoskeleton. DIP-mediated mDia2 inhibition converts elongated, mesenchymal cancer cells to rounded amoeboid morphologies enriched in non-apoptotic membrane blebs which undergo expansion/retraction cycles, typically without detachment [10, 11]. Amoeboid morphological shifts can underlie migration programs that are typically independent of integrins or proteolytic extracellular matrix degradation [12]. The mesenchymal-to-amoeboid transition (MAT) may allow cancer cells to evade therapeutics targeting matrix metalloproteinase (MMP)-driven motility [13, 14] or adapt to specific environmental cues.
The chemokine CXCL12 promotes both DIP:mDia2 axis assembly and subsequent non-apoptotic amoeboid blebbing [10, 11]. In breast, prostate and lung cancers, CXCL12 promotes metastasis to distal sites that express high chemokine levels [15, 16]. CXCL12 has two receptors- CXCR4 and CXCR7. CXCR4 is expressed uniformly in both normal breast and breast cancer cells; Only metastatic cells show receptor activation and propagation of associated downstream signaling (i.e., Erk, Ca2+ mobilization, cAMP production) upon CXCL12 stimulation [17]. CXCR4 is required for invasion of the metastatic, mesenchymal breast cancer cell line MDA-MB-231 [18]. CXCL12, via CXCR4 interaction, is implicated in amoeboid migration of zebrafish primordial germ cells [19]. Spatial/temporal regulation of RhoA GTPase activation is required for CXCL12/CXCR4 signaling [20]. The foregoing findings suggest that CXCL12 drives amoeboid transitions in tumor cells via CXCR4 interaction. Is CXCR4 engagement linked to Rho-directed mDia2-dependent amoeboid morphological transitions in breast tumor cells?
Rho GTPases, including RhoA-G, Cdc42, and Rac, regulate mDia activity, cellular transformation, tumor development, and progression [21, 22]. Rho GTPase activation regulates amoeboid blebbing, and is mediated by various extracellular signals [23]. RhoA promotes amoeboid transitions in tumor cells through RhoA, ROCK and myosin light chain [24–27]. Rho GTPases are regulated via GEFs and GTPase-activating proteins (GAPs), which modulate GTPase activity. RhoGEFs are required for Rho GTPases to cycle between GTP- and GDP-bound states [28–31]. GTP binding to GTPase proteins activates downstream signaling.
Neuroepithelioma transforming gene-1 (Net1) is a RhoGEF implicated in human glioma, and breast, gastric, and hepatocellular carcinoma progression [31–35]. In glioma and hepatocellular carcinoma, Net1 overexpression correlates with poor patient prognosis [34]. Net1A drives amoeboid cell motility (ACM) and invasion in MDA-MB-231s [36]. Is regulation of Net1-mediated RhoA activation linked to CXCL12/mDia2/DIP mechanisms driving amoeboid morphologies in breast tumor cells?
We examined roles for CXCR4-CXCL12 signaling in inducing Rho-dependent mDia2:DIP interaction, amoeboid morphological switching, and non-apoptotic blebbing in MDA-MB-231s. DIP and mDia2 interact with CXCR4 within blebs. CXCL12 activates RhoA, an essential feature of CXCL12-driven blebbing. CXCL12-mediated RhoA/mDia2 activation and subsequent blebbing requires Net1. Our data suggest CXCL12 drives modulation of the Net1/RhoA/mDia2-directed F-actin cytoskeleton in cancer cells undergoing morphological switching to amoeboid phenotypes with non-apoptotic blebs.
2. Materials and Methods
2.1 Cell Culture, Antibodies, and Transfection
MDA-MB-231 and HEK293 cells (ATCC) were maintained in DMEM (Gibco) supplemented with 10% FBS (v/v), 100 units/ml penicillin and 100mg/ml streptomycin at 37°C with 5% CO2.
RhoA, Net1, PDZ, LARG, p115, Ect2 (Santa Cruz); RhoC (Cell Signaling); CXCR4, NCKIPSD, tubulin, and CXCR7 (AbCam); CXCR4 (Sigma); and mDia2 (Proteintech) antibodies were used at 1:100–200 dilutions for immunoprecipitation (IP), western blotting, pull-down assays and immunofluorescence (IF).
MDA-MB-231s were transfected with Lipofectamine LTX/Plus reagent (Invitrogen) per manufacturer’s specifications. For siRNA transfections, Dharmafect ON-TARGETplus SMARTpools (Thermoscientific) against human NCKIPSD, DRF3, CXCR7, CXCR4 or GAPDH were used at 100nM with Dharmafect-1 reagent. Stable GEF knockdown cells were generated using lentiviral shRNA (Thermoscientific) (Supplemental Table 1) selected with 35µg/ml puromycin. Control cells were generated using empty pLKO1 or shGFPscr vector.
CFP-RhoA V14, CFP-RhoA V14I41A, CFP-RhoB V14, CFP-RhoB V14I41A, CFP-RhoC V14 and CFP-RhoC V14I41A were kind gifts from Dr. Art Alberts (Van Andel Institute, Grand Rapids, MI). Whole cell and IP lysates were prepared, and IPs were performed as described [8].
2.2 Immunofluorescence (IF)
Cells imaged in 2D were fixed with 4% paraformaldehyde in PBS (PFA/PBS), permeabilized with 0.2% Triton X-100 and incubated with primary antibody overnight at 4°C. Cells were incubated with secondary antibody (Alexa 488, 546, or 647) and phalloidin (Molecular Probes) for 3h at 37°C. Coverslips were mounted with fluoromount-G (Southern Biotech).
For 3D IF, cells were mixed with 2mg/ml Type-I collagen (BD Biosciences), gelled for 1h at 37°C and incubated 24h. Cells were fixed, permeabilized and incubated with antibodies as above. Gels were covered with fluoromount-G.
Images were acquired using a TCS SP5 multiphoton laser scanning confocal microscope (Leica Microsystems) with 458, 488, 514, 561 and 633nm laser lines in a sequential manner. Z-stack images were acquired using 0.5µm optical sections, creating 3D reconstructions for image analysis using MetaMorph software.
2.3 Percentage blebbing cells, C3 treatment/rescue
Cells were treated for indicated times and imaged using an EVOS digital inverted microscope (Life Technologies). Percent blebbing was determined by counting blebbing cells amongst total cells in a 20× field of view using an Olympus 20X LWD FL/PH 0.40NA objective. Three fields of view were quantified per sample with >30 cells quantified per experiment. The experiment was repeated thrice. Cells for representative images were fixed and phalloidin-stained prior to mounting.
Transfected cells were treated with 0.5µg/ml C3 transferase (C3) (Cytoskeleton) or vehicle (H20) for 5h, followed by 25ng/ml CXCL12. After 60m, cells were imaged and percent blebbing was determined as above. Three fields of view were quantified per sample with >30 cells quantified per experiment. The experiment was repeated thrice.
2.4 GTPase Activity Assays
GTPase activity was assessed with pull-down assays as before [37] using 50µg of purified GST-tagged Rhotekin Rho-binding domain (RBD) (Addgene) or GST-tagged PAK binding domain (PBD) (Addgene) bound to glutathione-sepharose beads. Bound proteins were resolved on SDS-PAGE gels. Densitometry was performed using ImageJ 1.47v [38]. The experiment was replicated thrice.
2.5 Statistical Analysis
One-tailed student’s t-test evaluated statistical significance with a 95% confidence interval. A p value ≤0.05 was considered statistically significant.
3. Results
3.1 CXCL12 causes amoeboid conversions and mDia2:DIP interaction
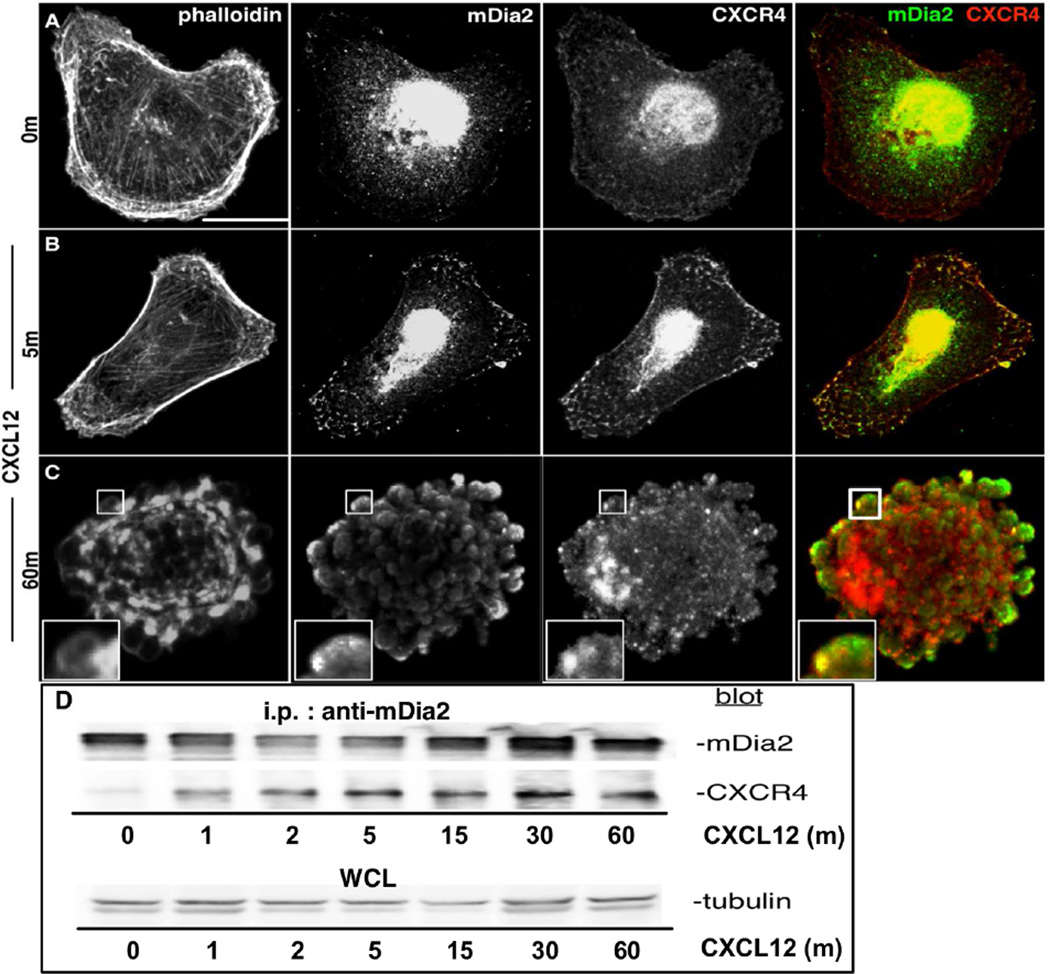
We previously showed CXCL12 initiates mDia2-dependent non-apoptotic blebbing in MDA-MB-231 cells in 2D matrices and within 3D collagen gels, and causes cells to adopt an elongated phenotype[11]. This resembled phenotypes observed upon treatment with a functional mDia FH2 domain inhibitor, SMIFH2. CXCL12 and its receptors CXCR4 and CXCR7 regulate not only amoeboid blebbing in developing zebrafish [19], but also cancer cell motility [15, 16, 39–41]. To assess whether CXCL12 induced association between mDia2 and CXCR4, MDA-MB-231s were CXCL12-stimulated and stained for F-actin, mDia2, and CXCR4. Unstimulated cells retained mesenchymal morphologies, with no mDia2 and CXCR4 co-localization (Fig. 1A). Five minutes after CXCL12 treatment, robust mDia2 and CXCR4 vesicular and plasma membrane co-localization was observed. While cells retained a primarily mesenchymal morphology at 5m, CXCL12 induced interaction between mDia2 (green) and CXCR4 (red) at the cell periphery (yellow in overlay) (Fig. 1B). CXCL12 initiated blebbing within an hour of treatment [11], with mDia2 and CXCR4 distinctly localizing in blebs at the cell periphery (Fig. 1C). To validate kinetics of mDia2:CXCR4 interaction, we co-IP’d mDia2 and CXCR4using HEK293 cells, which easily transfect relative to MDA-MB-231s. HEK293s overexpressing mDia2 and CXCR4 were stimulated with CXCL12 through 60m. A small amount of mDia2:CXCR4 complex was seen in absence of stimulation. More robust association occurred by 1m post-stimulation and was sustained through 60m (Fig. 1D). Thus, CXCL12 has an important role in spatial and temporal regulation of mDia2/CXCR4 interaction during amoeboid morphological shifts.
Figure 1. CXCL12 induces CXCR4:mDia2 complexes.
A–C. MDA-MB-231s were CXCL12-stimulated, and stained for F-actin, mDia2 and CXCR4. Cells were imaged using a 63× oil objective. Bar=15µm. D. HEK293s expressing FLAG-mDia2 and CXCR4 were stimulated with 25ng/ml CXCL12. Lysates were immunoprecipitated for mDia2 and blotted for mDia2 or CXCR4. Whole cell lysates blotted for tubulin serve as loading controls.
3.2 CXCL12-mediated blebbing requires CXCR4 and DIP expression
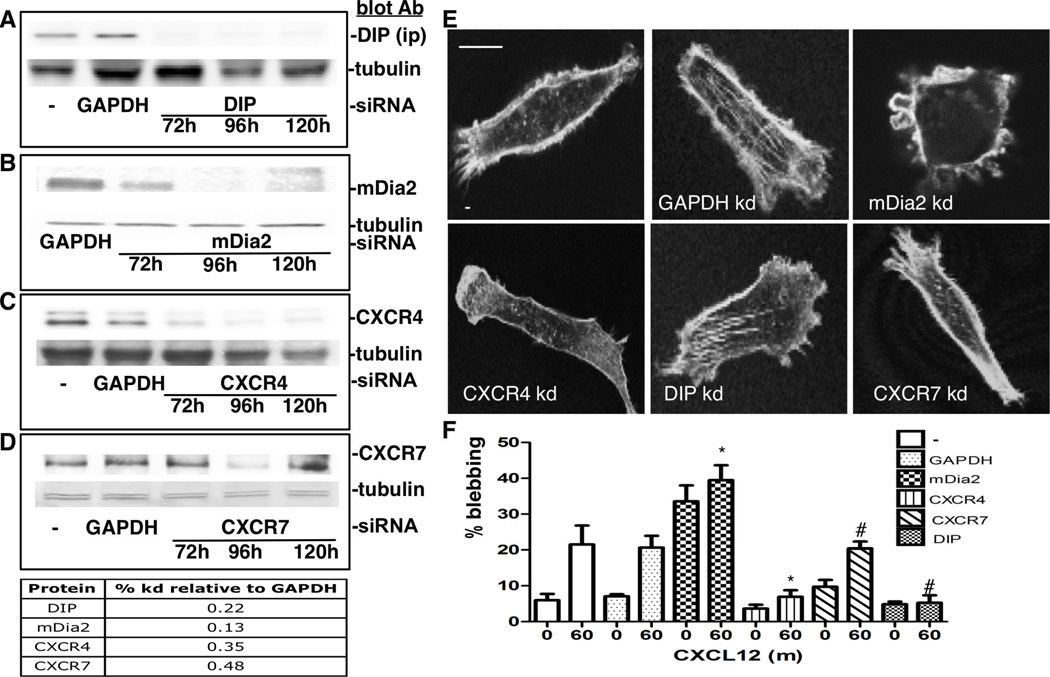
To determine whether CXCL12-driven amoeboid morphological shifts require CXCR4 or CXCR7 expression, or the mDia2 negative regulator DIP, we used siRNA to deplete MDA-MB-231s of GAPDH (control), DIP, mDia2, CXCR4, or CXCR7 (Fig. 2A–D). Knockdown was evaluated 72, 96, and 120h after transfection; the 96h time-point was used for experiments as it achieved robust knockdown (Fig. 2A–D). Cells were CXCL12-treated for 30–60m (30m, not shown). Cells were stained for F-actin at time zero (T0) (Fig 2E). Percentage of blebbing cells was calculated for live and fixed samples treated for 0 and 60m. Both untransfected and GAPDH siRNA-transfected cells showed significant blebbing increases after 1h of CXCL12 treatment (Fig. 2F). mDia2 knockdown induced a robust blebbing morphology that was not significantly altered by CXCL12. DIP depletion suppressed CXCL12-driven blebbing [10, 11]. CXCR4 depletion reduced blebbing in the CXCL12 presence or absence. CXCR7 depletion did not affect CXCL12-induced blebbing, consistent with CXCR4 being the primary receptor for CXCL12-driven blebbing.
Figure 2. CXCR4 is required in CXCL12-mediated blebbing.
A–D. MDA-MB-231s were depleted of GAPDH (control), DIP, mDia2, CXCR4 or CXCR7 using siRNA-mediated knockdown through 120h. Lysates were immunoprecipitated for DIP (A) or whole cell lysates (B-D) were blotted against respective proteins. Tubulin acts as a loading control. Densitometry quantified knockdown relative to GAPDH. E. siRNA-depleted cells stained at T0 for F-actin. Cells were imaged using a 63× oil objective. Bar=10 µm. F. siRNA-depleted cells were stimulated with 25ng/ml CXCL12. Percentage blebbing cells was determined 30 (not shown)-60m after stimulation. A representative experiment performed in triplicate is shown, where at least 30 cells are counted per condition. *p<0.05 vs. untreated T0, #p<0.05 untreated vs. T60.
3.3 CXCL12-activated RhoA regulates membrane blebbing
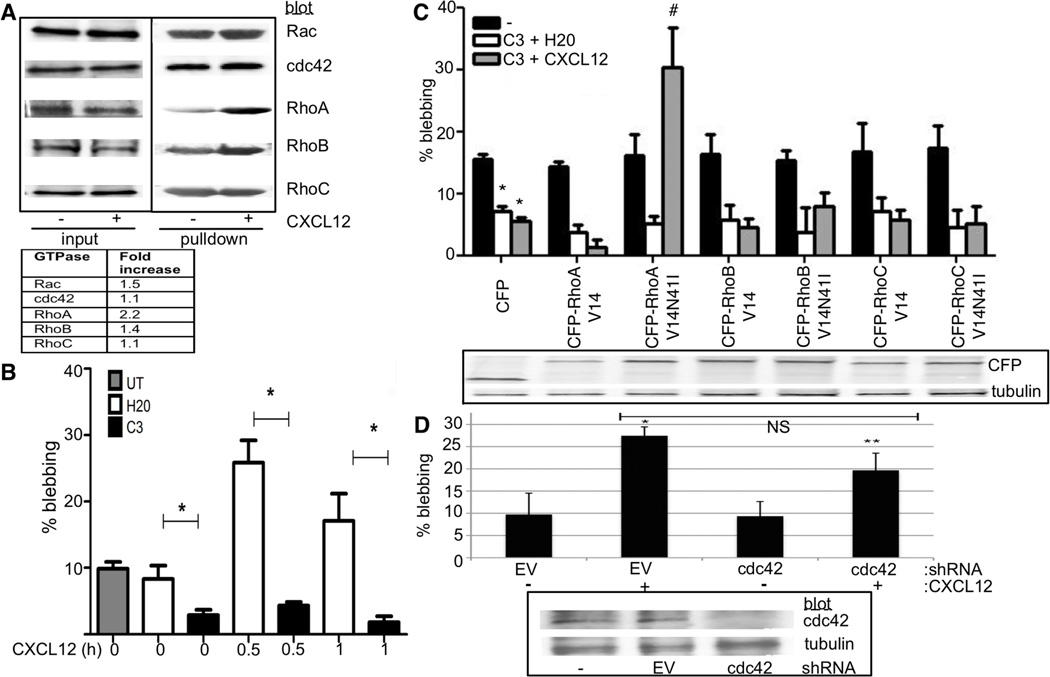
GTP-bound Rho GTPase binding releases mDias from autoinhibition [42, 43]. RhoA may drive CXCL12-mediated activation of mDia2 and subsequent blebbing because CXCL12 induces Rho-directed blebbing in migrating zebrafish germline cells [19]. RhoA is involved in amoeboid transitions [44]. To determine whether activated RhoA and other GTPases are required for CXCL12-induced mDia2-dependent amoeboid blebbing, we evaluated Rho GTPase activation through pull-downs using either the PBD or RBD that specifically interact with GTP-bound Rac, Cdc42, or RhoA-C. Each GTPase had modestly increased GTP-binding in response to CXCL12, but RhoA saw robust increases with CXCL12 (2.2-fold increase, compared to untreated, Fig. 3A).
Figure 3. Requirement for RhoA in response to CXCL12.
A. MDA-MB-231s were stimulated with 25ng/mL CXCL12 for 15m. GTPase activation was analyzed via GTPase pull-down assay. Fold-changes relative to 10% lysate input were calculated by densitometry. B. MDA-MB-231s were treated with C3 for 5h. Post-inhibition cells were treated with CXCL12 for 30–60m and percentage blebbing was determined using a 20× objective. * p<0.001 vs. respective H2O time-point. C. MDA-MB-231s transfected with CFP-fused plasmids and treated with (white and gray bars) or without (black bar) C3 were stimulated with 25ng/ml CXCL12. Percentage blebbing quantified after 1h. *p<0.001 vs. CFP T0. #p<0.001 vs. CFP C3 + CXCL12; *p<0.01 vs. CFP untreated cells. Expression was confirmed (lower). Tubulin acts as a lysate loading control. D. MDA-MB-231s expressing empty vector (EV) or cdc42 shRNA were assessed for CXCL12-driven blebbing as above. *p<0.02 vs. EV T0; **p<0.04 vs. cdc42 kd T0. NS=not significant. Blot validated cdc42 knockdown (lower). Shown is a representative experiment of three, with at least 30 cells counted per field for B, C, D.
While RhoA activity was enhanced by CXCL12, GTPase activity may or may not have been required for CXCL12-induced blebbing and amoeboid morphological transitions. To address this, we used C3, a potent pan-Rho inhibitor targeting RhoA-C through ADP-ribosylation on asparagine 41 in the GTPase effector-binding domain. MDA-MB-231s were C3-treated for 5h and then stimulated with CXCL12 or vehicle (Fig. 3C). MDA-MB-231s bleb ~10% of the time, without stimulation. CXCL12 treatment increases blebbing to 27–40%. This remained unchanged in untreated and H20-treated cells. C3-mediated RhoA-C inhibition markedly blocked CXCL12-driven MDA-MB-231 blebbing at both 30 and 60m. C3 targets RhoA-C. Does one of them play a more important role? RhoA had the greatest activity increase following chemokine stimulation, but that does not mean it is required.
To evaluate specificity of Rho GTPases involved in the CXCL12-driven blebbing mechanism, we performed C3 rescues using activated Rho mutants that are C3 resistant. The N41I Rho point mutant renders active GTPase resistant to C3 [45]. MDA-MB-231s were transfected with CFP-tagged constitutively-active RhoA-C V14 (expression confirmed in Fig. 3C). At T0, ~15% of transfected cells blebbed (Fig. 3C, black bars). C3 caused significant decreases in all samples lacking N41I mutation. Of plasmids encoding N41I C3-resistant V14-activated Rho, CXCL12-mediated blebbing is recovered after C3 treatment only with expression of C3-resistant RhoA (CFP-RhoA V14 N41I), and not C3-resistant activated RhoB or C (Fig 3C, gray, white bars). To confirm cdc42 requirement in the CXCL12-driven blebbing mechanism, we stably expressed cdc42 shRNA in MDA-MB-231s. Depletion was confirmed and blebbing determined in response to CXCL12 (Fig 3D). Cdc42 depletion failed to inactivate CXCL12-driven blebbing. Of those GTPases assessed, only RhoA was involved in the CXCL12-driven blebbing mechanism.
3.4 Net1-mediated RhoA activation is essential for CXCL12-driven blebbing
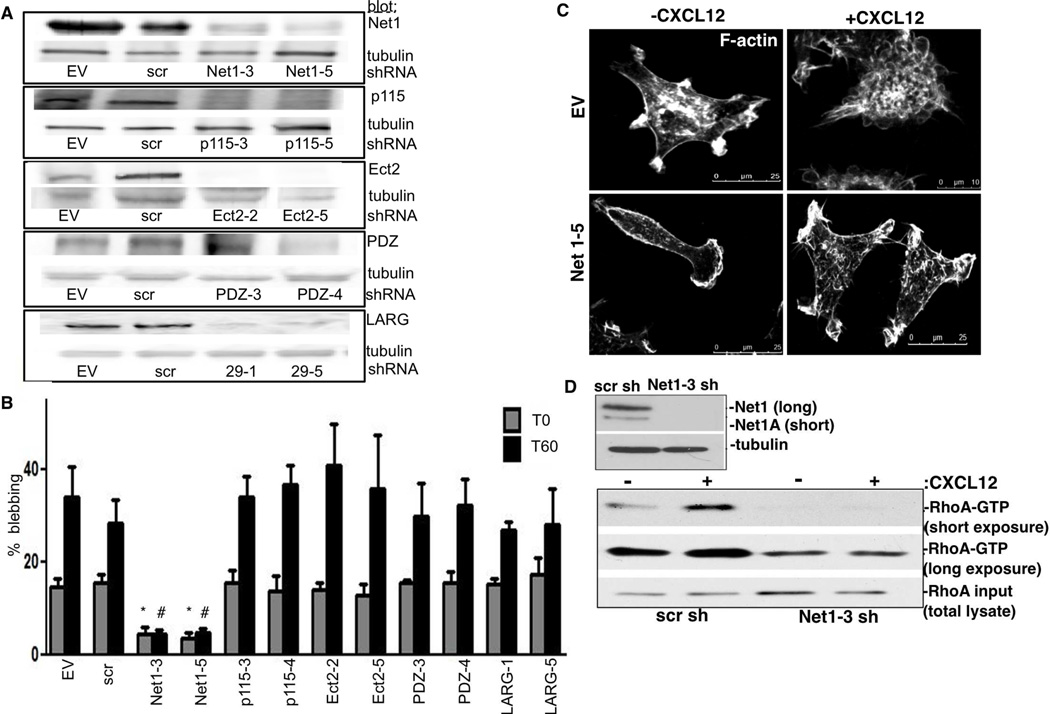
CXCL12/CXCR4 induces formation of the RhoA-directed mDia2:DIP complex, leading to non-apoptotic membrane blebbing and amoeboid morphological conversions. We sought to identify upstream GEFs responsible for RhoA activation. We generated stable knockdown cell lines for five GEFs, empty vector and non-targeting scrambled vector controls. We focused on p115, Net1, Ect2, PDZ RhoGEF, and LARG because they interact with RhoA and potentially with either formins or CXCR4 [20, 32, 46] (Fig. 4A). Only Net1 loss significantly decreased blebbing in CXCL12 presence/absence (Fig. 4B). Unlike control cells, Net1-depleted cells failed to bleb and maintained an elongated, polarized morphology in response to CXCL12 (Fig. 4C). These data agree with a recent study indicating a role for Net1A in amoeboid transitions in tumor cells [35].
Figure 4. Net1 loss inhibits CXCL12-driven blebbing.
A. Stable knockdowns were generated using shRNA against Net1, p115, Ect2, PDZ and LARG, scrambled (scr) and EV controls. Tubulin was used as a loading control. B. Cells were stimulated with 25ng/mL CXCL12. Percentage blebbing was determined at T0 and T60. *p<0.02 vs. EV T0; #p<0.02 vs. EV T60. The experiment was repeated three times, and in triplicate. At least 30 cells were counted per field. C–F. EV and Net1–5 knockdown cells were CXCL12-stimulated for 60m. F-actin was visualized using a 60× objective. D. Stable MDA-MB-231s expressing scr or Net1–3 shRNA were confirmed by blotting (upper). Tubulin acts as a loading control. GTPase activation in response to 60m CXCL12-treatment was analyzed via Rhotekin-RBD pull-down and RhoA blotting with short and long exposure. Equal protein input (10%) was validated by blotting for total cellular RhoA.
Finally, we evaluated whether Net1 depletion inhibited CXCL12-induced RhoA activation. Net1 (and Net1A) depletion was confirmed (Fig. 4D), and Rhotekin RBD pull-downs performed. Net1-depleted cells failed to activate RhoA (Fig. 4D), while control MDA-MB-231s activated RhoA in response to CXCL12.
4. Discussion
Cancer cells often alter their morphology and behavior in response to environmental cues. One mechanism that cancer cells utilize to adapt to different extracellular environments is switching to amoeboid morphologies, with hallmark dynamic non-apoptotic blebs. The DIP:mDia2 signaling node promotes a CXCL12-driven bleb-enriched amoeboid morphological switch in migrating human breast tumor cells [11]. CXCL12 induced interaction between mDia2 and its negative regulator DIP. Here we analyze the downstream mechanism of CXCL12-induced non-apoptotic membrane blebbing. CXCR4 is the primary receptor driving membrane blebbing, as CXCR7 knockdown did not inhibit CXCL12-induced blebbing. CXCR4 associates with mDia2 upon CXCL12 stimulation. RhoA is necessary and sufficient to drive the CXCL12-mediated blebbing mechanism. Finally, Net1 RhoGEF mediates CXCL12-directed RhoA activation and is required for CXCL12-directed blebbing in MDA-MB-231s.
Our data suggest an additional novel level of regulation of mDia2 to drive migratory plasticity in response to environmental cues. Net1 is an essential downstream effector of CXCL12/CXCR4 signaling. Net1 may activate RhoA, which is free to bind the mDia2 GBD. This releases mDia2 autoinhibition, promoting DIP interaction to drive amoeboid conversions. Interestingly, PDZ-GEF was shown to be important to CXCL12/CXCR4-driven, RhoA-dependent MDA-MB-231 motility [20]. However, that study did not discriminate amoeboid from mesenchymal cell motility. In our cells, PDZ depletion did not alter blebbing in the presence of CXCL12 (Fig. 4B). p115, Ect2, or LARG loss also failed to disrupt blebbing, indicating that Net1 is a critical and specific signaling partner in CXCL12-driven amoeboid conversions.
Both Net1 and its related isoform Net1A have roles in MDA-MB-231 migration in response to either FBS or LPA gradients. Yet, Net1A was suggested to specifically impact cell invasion through amoeboid conversions [31]. Like Net1A, Net1 influenced pMLC2 levels. Further, Net1 overexpression caused amoeboid conversions in a subset of cells. Net1 knockdown and amoeboid conversion quantification during cellular invasion was not determined. Our studies show a role for Net1 in driving amoeboid conversions and non-apoptotic blebbing through the Rho:mDia2:DIP signaling node. The shRNA sequences used target both Net1 and Net1A isoforms (Figure 4A). In the future, we will determine contributions of individual isoforms to CXCL12-driven amoeboid conversions and subsequent motility programs.
CXCL12 induced blebbing in ~27–40% of MDA-MB-231s, versus ~10% in CXCL12 absencea. Heterogeneic receptor expression and/or activation may drive blebbing kinetics/responsiveness. An ~1.5 log range in receptor expression was seen in MDA-MB-231s [17]., while receptor activation is variable within a non-clonal MDA-MB-231 cell population [17]. Differences in G protein coupling drove differences in CXCL12 responsiveness amongst normal and metastatic breast cell lines. Third, spatial and temporal regulation of downstream CXCR4 signaling components may dictate responsiveness, including regulation of activated Net1/RhoA and mDia2. mDia2 and CXCR4 are coupled in stimulated cells (Fig. 1), yet all mDia2 did not interact with CXCR4. The interaction between DIP and mDia2, which drives amoeboid blebbing, is punctate and not uniform throughout cells [11], indicating spatial restriction of signaling nodes driving this mechanism. There may be a critical threshold of association of receptor and mDia2 signaling machinery needed to tip cells towards amoeboid conversion.
In summary, tumor cells utilize adaptive cues, such as CXCL12, present in the local microenvironment to alter their morphology and escape the primary tumor. A novel mechanism by which CXCL12 induces initial morphological plasticity, an early step driving ACM in tumor cells, is through engaging CXCR4 and activating a Net1/RhoA/mDia2 complex. This may allow interaction with negative regulators of mDia2 F-actin dynamics (DIP), driving amoeboid morphological switching and non-apoptotic blebbing. Our results set the stage for a broader understanding of molecular cues and coordinating signaling hubs that drive morphological interconversion through changes in F-actin dynamics.
Supplementary Material
Acknowledgments
We thank the late Dr. Art Alberts for reagents, fruitful discussions and off-site lab meetings (mountain biking) throughout the years, and Peterson Grimsby and Dr. Alan Goodridge for critical reading/manuscript editing. This work was supported by National Cancer Institute R01CA151632 (KME, AN-K), Ohio Cancer Research Associates (RG-M) and University of Toledo Foundation (KME, AN-K, RG-M).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Evangelista M, Zigmond S, Boone C. Formins: signaling effectors for assembly and polarization of actin filaments. J Cell Sci. 2003;116:2603–2611. doi: 10.1242/jcs.00611. [DOI] [PubMed] [Google Scholar]
- 2.Wallar BJ, Alberts AS. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 2003;13:435–446. doi: 10.1016/s0962-8924(03)00153-3. [DOI] [PubMed] [Google Scholar]
- 3.Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci. 2013;126:1–7. doi: 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baarlink C, Brandt D, Grosse R. SnapShot: Formins. Cell. 2010;142:172, 172, e171. doi: 10.1016/j.cell.2010.06.030. [DOI] [PubMed] [Google Scholar]
- 5.Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis. 2009;26:273–287. doi: 10.1007/s10585-008-9174-2. [DOI] [PubMed] [Google Scholar]
- 6.Nurnberg A, Kitzing T, Grosse R. Nucleating actin for invasion. Nat Rev Cancer. 2011;11:177–187. doi: 10.1038/nrc3003. Epub 2011 Feb 2010. [DOI] [PubMed] [Google Scholar]
- 7.Fukumi-Tominaga T, Mori Y, Matsuura A, et al. DIP/WISH-deficient mice reveal Diaand N-WASP-interacting protein as a regulator of cytoskeletal dynamics in embryonic fibroblasts. Genes Cells. 2009;14:1197–1207. doi: 10.1111/j.1365-2443.2009.01345.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee S, Lee K, Hwang S, et al. SPIN90/WISH interacts with PSD-95 and regulates dendritic spinogenesis via an N-WASP-independent mechanism. EMBO J. 2006;25:4983–4995. doi: 10.1038/sj.emboj.7601349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim CS, Park ES, Kim DJ, et al. SPIN90 (SH3 protein interacting with Nck, 90 kDa), an adaptor protein that is developmentally regulated during cardiac myocyte differentiation. J Biol Chem. 2001;276:12871–12878. doi: 10.1074/jbc.M009411200. [DOI] [PubMed] [Google Scholar]
- 10.Eisenmann KM, Harris ES, Kitchen SM, et al. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr Biol. 2007;17:579–591. doi: 10.1016/j.cub.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 11.Wyse MM, Lei J, Nestor-Kalinoski AL, et al. Dia-Interacting Protein (DIP) Imposes Migratory Plasticity in mDia2-Dependent Tumor Cells in Three-Dimensional Matrices. PLoS One. 2012;7:e45085. doi: 10.1371/journal.pone.0045085. Epub 42012 Sep 45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf K, Mazo I, Leung H, et al. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 14.Overall CM, Lopez-Otin C. Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nat Rev Cancer. 2002;2:657–672. doi: 10.1038/nrc884. [DOI] [PubMed] [Google Scholar]
- 15.Teicher BA, Fricker SP. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 2010;16:2927–2931. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 16.Muller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 17.Holland JD, Kochetkova M, Akekawatchai C, et al. Differential functional activation of chemokine receptor CXCR4 is mediated by G proteins in breast cancer cells. Cancer Res. 2006;66:4117–4124. doi: 10.1158/0008-5472.CAN-05-1631. [DOI] [PubMed] [Google Scholar]
- 18.Liang Z, Yoon Y, Votaw J, et al. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 2005;65:967–971. [PMC free article] [PubMed] [Google Scholar]
- 19.Blaser H, Reichman-Fried M, Castanon I, et al. Migration of zebrafish primordial germ cells: a role for myosin contraction and cytoplasmic flow. Dev Cell. 2006;11:613–627. doi: 10.1016/j.devcel.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 20.Struckhoff AP, Rana MK, Kher SS, et al. PDZ-RhoGEF is essential for CXCR4-driven breast tumor cell motility through spatial regulation of RhoA. J Cell Sci. 2013;126:4514–4526. doi: 10.1242/jcs.132381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 22.Lozano E, Betson M, Braga VM. Tumor progression: Small GTPases and loss of cell-cell adhesion. Bioessays. 2003;25:452–463. doi: 10.1002/bies.10262. [DOI] [PubMed] [Google Scholar]
- 23.Fackler OT, Grosse R. Cell motility through plasma membrane blebbing. J Cell Biol. 2008;181:879–884. doi: 10.1083/jcb.200802081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Struckhoff AP, Rana MK, Worthylake RA. RhoA can lead the way in tumor cell invasion and metastasis. Front Biosci. 2011;16:1915–1926. doi: 10.2741/3830. [DOI] [PubMed] [Google Scholar]
- 25.Sahai E. Illuminating the metastatic process. Nat Rev Cancer. 2007;7:737–749. doi: 10.1038/nrc2229. [DOI] [PubMed] [Google Scholar]
- 26.Gadea G, de Toledo M, Anguille C, et al. Loss of p53 promotes RhoA-ROCK-dependent cell migration and invasion in 3D matrices. J Cell Biol. 2007;178:23–30. doi: 10.1083/jcb.200701120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettee KM, Dvorak KM, Nestor-Kalinoski AL, et al. An mDia2/ROCK Signaling Axis Regulates Invasive Egress from Epithelial Ovarian Cancer Spheroids. PLoS One. 2014;9:e90371. doi: 10.1371/journal.pone.0090371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sit ST, Manser E. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci. 2011;124:679–683. doi: 10.1242/jcs.064964. [DOI] [PubMed] [Google Scholar]
- 29.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 30.Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene. 2013 doi: 10.1038/onc.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carr HS, Morris CA, Menon S, et al. Rac1 controls the subcellular localization of the Rho guanine nucleotide exchange factor Net1A to regulate focal adhesion formation and cell spreading. Mol Cell Biol. 2013;33:622–634. doi: 10.1128/MCB.00980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Mata R, Dubash AD, Sharek L, et al. The nuclear RhoA exchange factor Net1 interacts with proteins of the Dlg family, affects their localization, and influences their tumor suppressor activity. Mol Cell Biol. 2007;27:8683–8697. doi: 10.1128/MCB.00157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen SQ, Li K, Zhu N, et al. Expression and clinical significance of NET-1 and PCNA in hepatocellular carcinoma. Med Oncol. 2008;25:341–345. doi: 10.1007/s12032-008-9042-6. [DOI] [PubMed] [Google Scholar]
- 34.Tu Y, Lu J, Fu J, et al. Over-expression of neuroepithelial-transforming protein 1 confers poor prognosis of patients with gliomas. Jpn J Clin Oncol. 2010;40:388–394. doi: 10.1093/jjco/hyp186. [DOI] [PubMed] [Google Scholar]
- 35.Menon S, Oh W, Carr HS, et al. Rho GTPase-independent regulation of mitotic progression by the RhoGEF Net1. Mol Biol Cell. 2013;24:2655–2667. doi: 10.1091/mbc.E13-01-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carr HS, Zuo Y, Oh W, et al. Regulation of focal adhesion kinase activation, breast cancer cell motility, and amoeboid invasion by the RhoA guanine nucleotide exchange factor Net1. Mol Cell Biol. 2013;33:2773–2786. doi: 10.1128/MCB.00175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubash AD, Guilluy C, Srougi MC, et al. The small GTPase RhoA localizes to the nucleus and is activated by Net1 and DNA damage signals. PLoS One. 2011;6:e17380. doi: 10.1371/journal.pone.0017380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hinton CV, Avraham S, Avraham HK. Role of the CXCR4/CXCL12 signaling axis in breast cancer metastasis to the brain. Clin Exp Metastasis. 2010;27:97–105. doi: 10.1007/s10585-008-9210-2. [DOI] [PubMed] [Google Scholar]
- 40.Hawkins OE, Richmond A. The dynamic yin-yang interaction of CXCR4 and CXCR7 in breast cancer metastasis. Breast Cancer Res. 2012;14:103. doi: 10.1186/bcr3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hernandez L, Magalhaes MA, Coniglio SJ, et al. Opposing roles of CXCR4 and CXCR7 in breast cancer metastasis. Breast Cancer Res. 2011;13:R128. doi: 10.1186/bcr3074. Epub 2011 Dec 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorelik R, Yang C, Kameswaran V, et al. Mechanisms of plasma membrane targeting of formin mDia2 through its amino terminal domains. Mol Biol Cell. 2011;22:189–201. doi: 10.1091/mbc.E10-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staus DP, Taylor JM, Mack CP. Enhancement of mDia2 activity by Rho-kinasedependent phosphorylation of the diaphanous autoregulatory domain. Biochem J. 2011;439:57–65. doi: 10.1042/BJ20101700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 45.Wilde C, Genth H, Aktories K, et al. Recognition of RhoA by Clostridium botulinum C3 exoenzyme. J Biol Chem. 2000;275:16478–16483. doi: 10.1074/jbc.M910362199. [DOI] [PubMed] [Google Scholar]
- 46.Goulimari P, Knieling H, Engel U, et al. LARG and mDia1 link Galpha12/13 to cell polarity and microtubule dynamics. Mol Biol Cell. 2008;19:30–40. doi: 10.1091/mbc.E06-11-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.