Abstract
Objectives
To utilize a non-biased assay of circulating tumour cells (CTCs) in prostate cancer (PCa) patients in order to identify non-traditional CTC phenotypes potentially excluded by conventional detection methods reliant upon antigen and/or sized based enrichment.
Patients and Methods
41 metastatic castration resistant prostate cancer (mCRPC) patients and 20 healthy volunteers were analysed on the Epic CTC Platform, via high throughput imaging of DAPI expression and CD45/cytokeratin (CK) immunofluorescence (IF) in all circulating nucleated cells plated on glass slides. IF for androgen receptor [AR] expression, and FISH for PTEN and ERG confirmed PCa origin of CTCs.
Results
Traditional (t) CTCs (CD45−/CK+/morphologically distinct) were identified in 100% mCRPC patients. Using the above markers, we identified non-traditional CTCs in mCRPC patients, including CK− and apoptotic CTCs. Small CTCs (≤WBC size) were identified in 98% of mCRPC patients. Total, traditional and non-traditional CTCs were significantly increased in deceased vs. living patients at 18 months; however only non-traditional CTCs associated with overall survival. Traditional and total CTC counts by the Epic platform in the mCRPC cohort were also significantly correlated with CTC counts by the CellSearch system.
Conclusions
Heterogeneous non-traditional CTC populations that may be missed by other approaches are frequent in mCRPC; characterization of non-traditional CTCs may provide additional prognostic or predictive information.
Keywords: Circulating tumour cells, liquid biopsy, metastatic castration resistant prostate cancer, ERG, PTEN
INTRODUCTION
Dissemination of cancer cells from primary sites into circulation and seeding of metastases is the underlying cause of mortality for most non-hematopoietic malignancies. Detection and enumeration of circulating tumour cells (CTCs) is associated with decreased progression-free survival and overall survival for a variety of tumours, including prostate cancer (PCa) [1–3]. Based on these findings, CTC enumeration could be a valuable tool for disease response and progression monitoring. Additionally, CTCs can also be characterized for predictive biomarkers. Given that CTCs circulate at very low concentrations (10−6–10−7) [4], detection in millilitres of blood requires extremely sensitive and specific methods. Numerous technologies have been developed to this end [5, 6], with the majority requiring an enrichment step based on differences in various physical (size and/or density) or biological parameters (surface marker expression) to distinguish CTCs from surrounding hematopoietic cells. For example, the CellSearch® Circulating Tumor Cell System, the only FDA cleared method for CTC detection [7], requires cells to contain a DAPI-intact nucleus, lack expression of the hematopoietic marker CD45, express epithelial cell adhesion molecule (EpCAM) and cytokeratin (CK), and have a diameter larger than 4 μm [7]. Enrichment technologies however will potentially fail to capture non-traditional tumour cells lacking the selected characteristic.
Indeed, recent reports utilizing non-EpCAM or size based selection techniques have underscored this point, revealing PCa CTCs with non-traditional phenotypes, describing heterogeneity in size [8], epithelial marker expression [9], cell integrity [10], and proliferation rate [11]; both between and within individual patients. CTCs isolated from men with metastatic PCa have also shown evidence of epithelial-mesenchymal transition (EMT), via detection of EMT related transcription factors, expression of N-cadherin and vimentin, and loss of CK and E-cadherin expression [9]. However, the degree and scope of this heterogeneity within PCa CTC populations is incompletely described, as most approaches are only able to assess heterogeneity in a single parameter.
Numerous studies have identified a relationship between CTC counts and disease progression or overall survival in metastatic castrate-resistant prostate cancer (mCRPC) [2, 3]. These studies have been based on single-parametric enrichment technologies which likely miss non-traditional CTC phenotypes that may have prognostic or therapeutic significance. Here, we utilize a non-biased assay technique (the Epic CTC platform) that retains all nucleated cells and interrogates them in a multi-parametric fashion, analysing size, shape, DAPI staining and immunofluorescence (IF) antigen characterization (typically CD45 and CK expression). Concurrently, we evaluated both traditional (CD45−, CK+, morphologically distinct) and candidate non-traditional CTCs for PCa-specific molecular aberrations, including androgen receptor (AR) expression by IF, and PTEN deletion and ERG rearrangement by fluorescence in situ hybridization (FISH) [12–15]. Through this approach, we demonstrate that patients with mCRPC harbour a variety of non-traditional CTC phenotypes.
PATIENTS AND METHODS
Sample Collection and Handling
Whole blood samples were obtained from 41 unique patients, required to have histologically or biochemically confirmed prostate adenocarcinoma and planned for initiation of a new treatment for progressive mCRPC in the presence of castrate levels of serum testosterone (<50 ng per decilitre [1.73 nmol per litre]), consistent with Prostate Cancer Clinical Trials Working Group 2 (PCWG2) guidelines. After collection at the Royal Marsden, Sutton, UK, samples were shipped to Epic Sciences (San Diego, CA, USA) at ambient temperature. Sample collection for this study was approved by The Royal Marsden (London, UK) Research Ethics Committee (REC 04/Q0801/6) and by The Royal Marsden Committee for Clinical Research. Additionally, 21 samples were obtained from 20 consenting healthy adults by The Institute of Cancer Research (London, U.K.) or Epic Sciences (San Diego, CA, USA) and processed in the same manner as patient samples. All samples were collected with informed consent. Patient and healthy volunteer demographics are summarized in Table 1. The median blood sample transit time for patient samples was 32 hours with a range of 28–78 hours. Additional draws from select patients were evaluated as needed for FISH.
Table 1.
Demographic and clinical characteristics of patients and healthy volunteers at the time of study inclusion
| CRPC patients | n=41 |
|---|---|
| Age (yr) | |
| Median | 70 |
| Range | 40–82 |
| Serum PSA (ng/mL) | |
| Median | 90 |
| Range | 2–2532 |
| LDH (U/L) | |
| Median | 170 |
| Range | 112–678 |
| Albumin (g/L) | |
| Median | 35 |
| Range | 24–42 |
| Haemoglobin (g/dL) | |
| Median | 12.2 |
| Range | 8.3–15.5 |
| Sites of metastases | |
| Bone | 39 (95%) |
| Nodal | 28 (68%) |
| Visceral | 13 (32%) |
| ECOG performance score | |
| 0 | 6 (15%) |
| 1 | 33 (80%) |
| 2 | 2 (5%) |
| Previous CRPC therapies (n) | |
| Median | 3 |
| Range | 1–5 |
| Previous therapies | |
| Bicalutamide | 41 (100%) |
| Docetaxel | 26 (63%) |
| Cabazitaxel | 4 (10%) |
| Abiraterone (AA) | 14 (34%) |
| Enzalutamide (E) | 3 (7%) |
| Investigational agents | 13 (32%) |
|
| |
| Healthy Volunteers | n=20 |
|
| |
| Age (yr) | |
| Median | 33.5 |
| Range | 21–53 |
| Sex | |
| Male | 10 |
| Female | 10 |
CRPC = castration resistant prostate cancer
Blood sample preparation and storage
Upon sample receipt, red blood cells were lysed and nucleated cells dispensed onto glass microscope slides according to methods previously described [16–18]. Up to 12 slides were prepared from each blood sample at 3×106 cells/slide. Slides were then stored at −80°C (stable for longer than one year, unpublished data). The number of slides created from each individual sample was determined by the volume of blood received and the white blood cell (WBC) count.
CTC identification and protein characterization on the Epic platform
For CTC analysis, two slides from each patient sample were thawed, then stained by IF to distinguish CTCs from WBCs as described previously [16–18]. In addition to DAPI, CD45, and CK, an additional antibody targeting the N-terminal region of AR (clone D6F11, Cell Signalling Technology, Danvers, MA, USA) was utilized, capable of detecting full length and LBD-truncated variants of the protein. Stained slides were imaged on a high-speed fluorescent imaging system. Captured images were analysed by an automated algorithm that characterizes each cell by more than 90 parameters, including protein expression and morphology to distinguish CTCs from normal nucleated cells. All CTC candidates were then reviewed by trained technicians, and CK+/CD45− cells with intact, DAPI+ nuclei exhibiting tumour-associated morphologies were classified as traditional CTCs. The trained technicians were blinded as to whether the sample was from a cancer patient or healthy donor. Candidate CTCs that did not meet the criteria for traditional CTC, as described in the Results, were identified as CD45−/CK− cells with abnormal morphology or apoptotic CTCs (CD45− with characteristic nuclear fragmentation or condensation). In a separate subsequent analysis, cell morphological characteristics, including cell area, were determined for each CTC by Epic proprietary software. Analysis was reviewed by a Board certified Anatomic Pathologist with experience in genitourinary pathology and molecular characterization of mCRPC (S.A.T.) to confirm cancer origin.
CTC analysis via CellSearch Assay
CTC isolation and enumeration were carried out using the CellSearch™ system (Janssen Diagnostics, Raritan, NJ, USA) according to the manufacturer’s instructions. Blood samples were drawn into CellSave™ tubes (Janssen Diagnostics) and samples were kept at room temperature and processed within 72 h of collection. To calculate the CTC count, 7.5 ml of blood was enriched immunomagnetically using anti-EpCAM antibodies, followed by fluorescent labelling and individual capture using a four-colour semiautomated fluorescent microscope. The images were then presented to trained operators, who selected cells that met the definition of CTC. Criteria used to define a CTC include round to oval morphology, size >5 μm, a visible nucleus (4′,6-diamidino-2-phenylindole positive), positive staining for cytokeratins 8,18 and/or 19 (phycoerythrin) and negative staining for CD45 (allophycocyanin). Results were expressed as the number of cells per 7.5 ml of blood.
CTC FISH analysis
Following CTC IF analysis, a subset of slides with a sufficient number of CTCs was tested for PTEN loss or ERG rearrangements by fluorescence in situ hybridization (FISH). Coverslips were removed and slides were hybridized using a two-color probe solution targeting either PTEN and chromosome 10 centromere (CC10) DNA sequences or regions flanking 5′ and 3′ ERG (Cymogen Dx, New Windsor, NY, USA). After processing, slides were counterstained with DAPI and mounted with an anti-fade mounting medium. Epic software was used to relocate CTCs for scoring. A minimum of 20 WBCs on each slide were also scored as internal controls.
PTEN loss was defined by a cell containing fewer PTEN signals than CC10 signals or only one of each signal (loss of chromosome 10). Cells with no PTEN signals and at least one CC10 signal were classified as homozygous PTEN loss (HO). Heterozygous PTEN loss (HE) cells contained at least one PTEN signal and either more CC10 signals than PTEN or one PTEN signal and one CC10 signal. Cells with a 1:1 ratio of PTEN:CC10 and at least two of each signal were considered PTEN non-deleted. ERG rearrangements can present as a split of the 5′ and 3′ FISH signals (representing ERG fusions though insertion, inversion or translocation) or as a deletion of the 5′ FISH signal (representing TMPRSS2:ERG fusions through deletion of the intervening region on chromosome 21). Hence, cells were considered ERG-rearranged if a separation of at least one pair of 5′ and 3′ ERG signals (by a distance of at least 2 signal diameters) or a deletion of at least one 5′ ERG signal was observed. Cells in which all 3′ ERG signals had a corresponding 5′ ERG signal within two signal diameters were considered ERG non-rearranged.
Statistical Analysis
Correlations/associations of traditional and non-traditional CTC counts and clinicopathological parameters were assessed by Spearman rank correlation, two-sided Mann-Whitney tests, Kruskal-Wallis tests or Kaplan-Meier analysis using MedCalc v 15.6 (MedCalc Software bvba, Belgium). Area under the curve (AUC) from receiver operator characteristic (ROC) curves was determined using MedCalc v 15.6 for traditional and non-traditional CTC counts for predicting mCRPC vs. healthy volunteer status. Youden index CTC/mL cutoff, value, and CTC cutoffs with sensitivity and specificity at 100% specificity and sensitivity, respectively, were determined as part of the ROC analysis. Kaplan-Meier analysis was performed by stratifying patients per CTC type into those with ≤ vs. > median CTC count. For all tests, p values < 0.05 were considered significant.
RESULTS
Detection of Traditional CTCs in mCRPC patients
A total of 41 blood samples from 41 mCRPC patients and 21 blood samples from 20 healthy volunteers were analysed. Patient and healthy volunteer demographics are shown in Table 1. An average of 11 slides per patient was plated (range 4 to 16) from an average blood volume of 7.0 mL per patient (range 3.68–7.83 mL). Traditional CTCs were defined (described previously [16–18]) as cells with an intact, DAPI-positive nucleus lacking features of apoptosis, absence of CD45 staining (CD45−) and positive CK staining (CK+) (Figure 1). Additionally, traditional CTCs were required to have characteristic cytomorphologic features consistent with malignancy (including nucleomegaly, nuclear membrane irregularity, eccentric cytoplasmic distribution, and polygonal/elongated cell shapes) [16–19]. Using this traditional CTC definition, 41/41 (100%) mCRPC patients had detectable traditional CTCs (median 5/mL, range 1–121) and 22/41 (54%) had >4/mL.
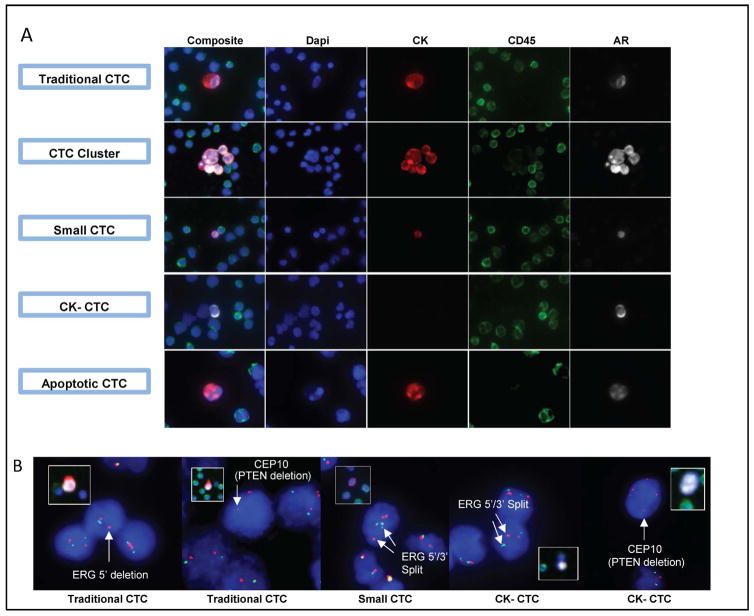
Figure 1. Patients with metastatic castration resistant prostate cancer (mCRPC) harbour traditional and non-traditional CTCs with prostate cancer specific molecular alterations.
A. Representative immunofluorescence (IF) image (100× magnification) examples of traditional and non-traditional CTCs in mCRPC patient samples (positive androgen receptor (AR) staining by IF is shown for each CTC). Traditional (DAPI+/CD45−/cytokeratin [CK]+/abnormal morphology), cluster (two or more adjacent traditional CTCs), small (DAPI+/CD45−/CK+/small cellular area), CK− (DAPI+/CD45−/CK−/AR+/abnormal morphology), and apoptotic (DAPI+/CD45−/CK+/nuclear disintegration/abnormal morphology) CTCs from mCRPC patient samples are shown. B. Example photomicrographs (400× magnification) of ERG and PTEN molecular alterations detected by fluorescence in situ hybridization (FISH) in traditional and non-traditional CTCs from mCRPC patient samples are shown (inset images (100× magnification) show CTC prior to FISH). PTEN deletions are indicated by loss of the PTEN locus green signal) in the presence of chromosome 10 centromeric signal (CEP 10, red). ERG rearrangements can be identified by 5′ deletion (resulting in loss of 5′ green signal; far left panel) or by split 5′/3′ signals (centre and second from right panel).
When two or more adjacent traditional CTCs were identified, they were classified as CTC clusters. CTC clusters were detected in 7/41 (17%) patients (median 0/mL, range 0–6) (Figure 1). Of 20 healthy volunteers tested, none had >4 events in 1mL meeting the definition for a traditional CTC and five (25%) had 1–4/mL (median 0/mL, range 0–4/mL, see Figure 2 for representative events in healthy volunteers). CTC clusters were not detected in healthy volunteer samples. Figure 3 shows the frequency of traditional CTCs and CTC clusters in patient samples compared to healthy volunteer samples.
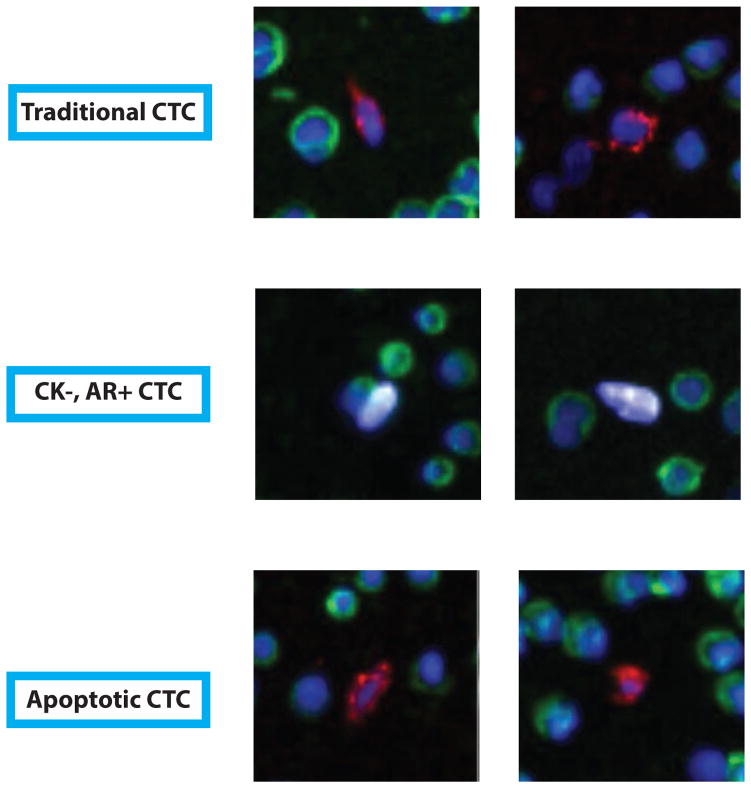
Figure 2. CTC-like cells detected in healthy volunteers.
Representative immunofluorescence (IF) examples (100× magnification) of traditional and non-traditional CTC-like cells in healthy volunteer samples. Traditional (DAPI+/CD45−/cytokeratin [CK]+/abnormal morphology), CK− (DAPI+/CD45−/CK−/AR+/abnormal morphology), and apoptotic (DAPI+/CD45−/CK+/nuclear disintegration/abnormal morphology) CTC-like cells from healthy volunteers are shown. Blue = DAPI, green = CD45, Red = CK, White = AR.
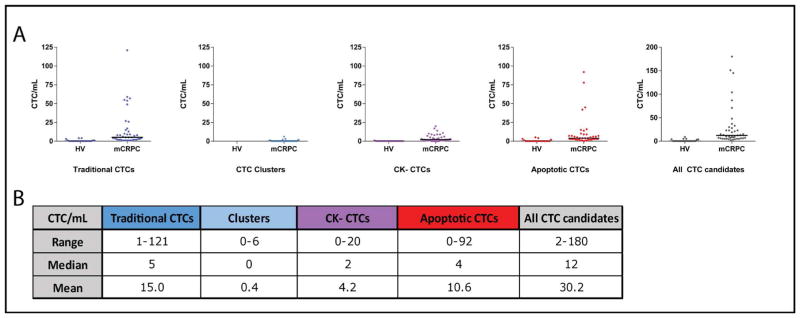
Figure 3. CTC incidence in mCRPC patient samples compared to healthy volunteers (HV).
A. The number of traditional CTCs, CTC clusters, CK− CTCs, apoptotic CTCs and all CTC candidates (traditional, clusters, CK−, and apoptotic CTCs)/mL identified in HV and mCRPC patient samples are plotted. Black lines indicate the median CTCs/mL. B. Range, median, and mean CTCs/mL in mCRPC patient samples for all categories are given.
ROC curve analysis of traditional CTC counts, CTC clusters (as well as other non-traditional CTCs as described below) to discriminate healthy volunteers from mCRPC patients—although not the intended use of the assay— was performed for each traditional and non-traditional CTC type. The area under the curve (AUC) for prediction of CRPC status for traditional CTCs (CTCs/mL), CTC clusters, CK− CTCs, apoptotic CTCs and all CTCs was 0.93 (Youden index 0.71 at >0 CTCs/mL), 0.59 (Youden index 0.17 at >0 CTCs/mL), 0.91 (Youden index 0.78 at >0 CTCs/mL), 0.91 (Youden index 0.74 at >4 CTCs/mL) and 0.96 (Youden index 0.76 at >4 CTCs/mL), as shown in Table 2..
Table 2.
Receiver operator characteristic curve (ROC) analysis of circulating tumor cell (CTC) counts for discriminating healthy volunteers and patients with castration resistant prostate cancer (CRPC)
| CTC type | ROC | Youden index | 100% Sensitivity | 100% Specificity | ||||
|---|---|---|---|---|---|---|---|---|
| AUC | 95% CI | Value | CTC/mL cutoff | CTC/mL cutoff | Specificity | CTC/mL cutoff | Sensitivity | |
| All CTCs | 0.96 | (0.88–0.99) | 0.76 | >4 | >0 | 71% | >9 | 56% |
| Traditional CTCs | 0.93 | (0.84–0.98) | 0.71 | >0 | >0 | 71% | >4 | 54% |
| Apoptotic CTCs | 0.91 | (0.81–0.97) | 0.74 | >0 | N/A | N/A | >5 | 39% |
| CK− CTCs | 0.91 | (0.81–0.97) | 0.78 | >0 | N/A | N/A | >1 | 63% |
| CTC Clusters | 0.59 | (0.45–0.71) | 0.17 | >0 | N/A | N/A | >0 | 17% |
ROC curves were generated from CTC/mL counts for indicated CTC types (All CTCs is the sum of the other 4 types) for predicting CRPC status in 21 healthy volunteer samples (from 20 patients) and 41 CRPC samples. Cell size was not included in this analysis (all categories may include small nuclear sized CTCs). Area under the curve (AUC) and 95% confidence intervals (CI) are given, along with Youden index values and CTC/mL cutoffs. CTC/mL cutoffs for 100% sensitivity and specificity are given, along with corresponding specificity and sensitivity.
Phenotypic and Molecular Characterization of Traditional CTCs
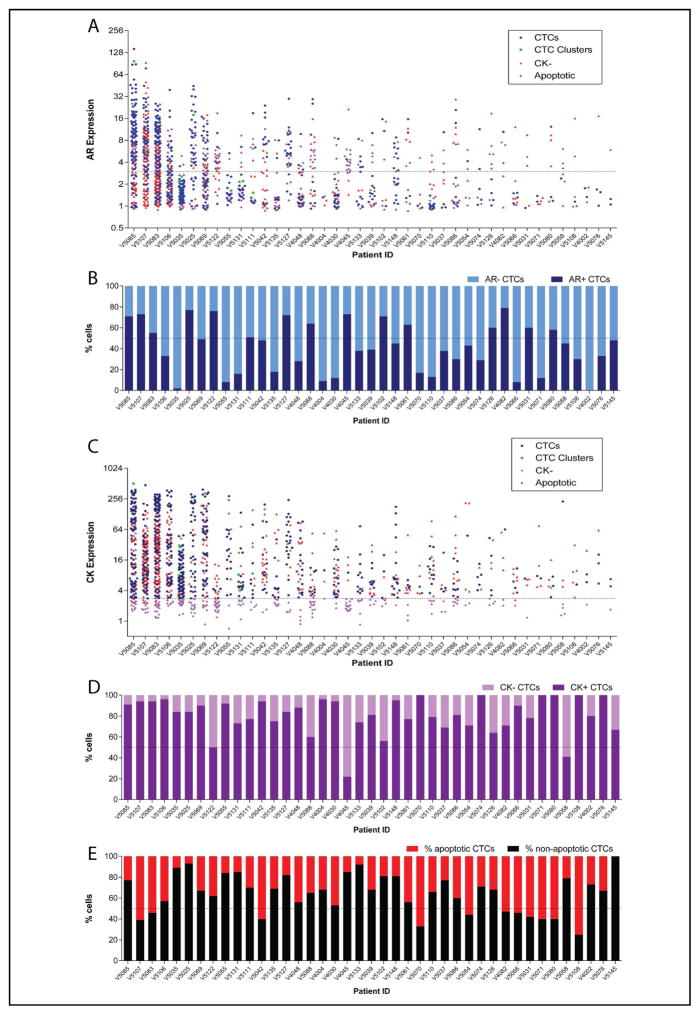
AR over-expression, loss of PTEN, and ERG rearrangements (most commonly resulting in TMPRSS2:ERG gene fusions) are common molecular events in mCRPC and can be used to confirm captured cells are CTCs, especially ERG rearrangements that are PCa specific [12–15]. AR expression was evaluated in all mCRPC patients and select healthy volunteers. To generate a relative AR expression value for each CTC, Epic’s software normalized AR expression in CTCs to approximately 1 million surrounding CD45+ WBCs present on the same slide, resulting in CTCs with AR expression values ≥3.0 being considered positive. Across the 41 mCRPC patients, 28 (68%) had at least one AR-positive traditional CTC/mL (range 0–96) (Figure 4A). The proportion of AR+ to AR− traditional CTCs was variable (Figure 4B). Of six healthy volunteer samples tested, none had >1 AR+ CTC/mL.
Figure 4. AR and CK expression by immunofluorescence varies across traditional and non-traditional CTCs in mCRPC patient samples.
AR (A&B) and CK (C&D) intensity distribution across traditional and non-traditional CTCs per patient. The dashed lines at 3 (A) and 2.8 units (C) indicate the cut-off for AR and CK positivity, respectively. Charts B and D show the percentage of all CTCs/mL (traditional and non-traditional) with or without B) AR or D) CK expression per patient. E. The percentage of apoptotic and non-apoptotic CTCs/mL per patient.
To further establish the prostatic origin of CTCs in our cohort, we also assessed PTEN and ERG status by FISH, using surrounding WBCs as internal controls. To determine assay specificity, we assessed PTEN and ERG status of WBCs from patient slides tested by FISH. Of 120 patient WBCs evaluated for PTEN FISH, 3 (2.5%) and 0 (0.0%) cells were detected with PTEN heterozygous (HE) and homozygous (HO) loss, respectively, resulting in 97.5–100% specificity of PTEN FISH on the Epic platform. Of 220 patient WBCs evaluated with ERG FISH, 10 (4.5%) and 0 (0.0%) cells had a split signal or loss of the 5′ probe confirming 95.5–100% specificity.
After IF staining for CTC and AR detection on the Epic platform, CTCs from select mCRPC patients were then re-located and evaluated for PTEN and/or ERG FISH status. PTEN deletions (HE and/or HO) were detected in traditional CTCs from 4 of 5 patients evaluated (Table 3). Likewise, ERG rearrangements (translocation and/or deletion) were identified in traditional CTCs from 5 of 6 patients evaluated (Table 3). Representative images of traditional CTCs with AR expression and/or PTEN deletion or ERG rearrangement are shown in Figure 1. Among slides from healthy volunteers, none had sufficient detectable CTCs to perform PTEN or ERG assessment.
Table 3.
FISH characterization of prostate cancer specific molecular alterations (PTEN deletions top; ERG rearrangements bottom) in phenotypically diverse CTCs in patients with mCRPC
| Patient | Cell Type | PTEN evaluable cells | AR+ cells | PTEN heterozygous cells | PTEN homozygous cells | 1PTEN deletion cells |
|---|---|---|---|---|---|---|
| (n) | (%) | (n) | (n) | (%) | ||
| V4004* | Traditional | 7 | 14% | 2 | 0 | 29% |
| CK− | 0 | N/A | N/A | N/A | N/A | |
| Small | 0 | N/A | N/A | N/A | N/A | |
|
| ||||||
| V5035 | Traditional | 32 | 0% | 1 | 0 | 3% |
| CK− | 4 | 25% | 0 | 0 | 0% | |
| Small | 1 | 0% | 0 | 0 | 0% | |
|
| ||||||
| V5042* | Traditional | 11 | 36% | 0 | 4 | 37% |
| CK− | 9 | 44% | 1 | 4 | 56% | |
| Small | 2 | 0% | 0 | 0 | 0% | |
|
| ||||||
| V5131 | Traditional | 12 | 0% | 1 | 1 | 17% |
| CK− | 5 | 0% | 0 | 0 | 0% | |
| Small | 0 | N/A | N/A | N/A | N/A | |
|
| ||||||
| V5055 | Traditional | 13 | 0% | 0 | 0 | 0% |
| CK− | 0 | N/A | N/A | N/A | N/A | |
| Small | 0 | N/A | N/A | N/A | N/A | |
| Patient | Cell Type | ERG evaluable cells | AR+ cells | ERG 5′ deletion cells | ERG 5′/3′ split cells | 2ERG rearranged cells |
|---|---|---|---|---|---|---|
| (n) | (%) | (n) | (n) | (%) | ||
| V4048 | Traditional | 11 | 0%** | 0 | 0 | 0% |
| CK− | 0 | N/A | N/A | N/A | N/A | |
| Small | 0 | N/A | N/A | N/A | N/A | |
|
| ||||||
| V5025* | Traditional | 73 | 73% | 6 | 12 | 25% |
| CK− | 10 | 90% | 0 | 0 | 0% | |
| Small | 23 | 48% | 2 | 4 | 26% | |
|
| ||||||
| V5083 | Traditional | 35 | 63% | 4 | 7 | 31% |
| CK− | 2 | 100% | 0 | 1 | 50% | |
| Small | 3 | 0% | 0 | 0 | 0% | |
|
| ||||||
| V5085* | Traditional | 64 | 94% | 0 | 4 | 6% |
| CK− | 6 | 83% | 0 | 1 | 17% | |
| Small | 0 | N/A | N/A | N/A | N/A | |
|
| ||||||
| V5106 | Traditional | 9 | 56% | 0 | 3 | 33% |
| CK− | 1 | 100% | 0 | 1 | 100% | |
| Small | 0 | N/A | N/A | N/A | N/A | |
|
| ||||||
| 5131* | Traditional | 21 | 0% | 0 | 3 | 14% |
| CK− | 7 | 0% | 0 | 0 | N/A | |
| Small | 1 | 100% | 0 | 0 | N/A | |
FISH analysis for prostate cancer specific alterations was performed on selected patients. The percentage of CTCs (stratified by CTC type) with androgen receptor (AR) positivity by immunofluorescence (IF) is given for patients assessed for PTEN loss (top panel) or ERG rearrangement (bottom) by FISH. False positive PTEN HE and HO rate in white blood cells (WBCs) 2.5% (3/120 WBCs) and 0% (0/120 WBCs), respectively. False positive ERG 5′ deletion and 5′/3′ split rate 0% (0/220 WBCs) and 4.5% (10/220 WBCs), respectively.
Including both HE and HO cells.
Both 5′ deletion and 5′/3′ split cells.
Additional slides beyond the standard 2 slides were used for FISH evaluation.
3 cells were unevaluable for AR expression.
PTEN FISH was performed on 5 representative patients with CK− CTCs and we were able to evaluate 18 CK− CTCs from 3 patients. Of 3 patients with evaluable CK− CTCs, PTEN deletions were identified in 1 patient, with 5/9 (56%) CK− CTCs showing PTEN deletion. PTEN deletions were also detected in 4/11 (36%) traditional CTCs from this patient. ERG FISH was performed on 6 representative patients with CK− CTCs and we were able to evaluate 26 CK− CTCs from 5 patients. Of 5 patients with evaluable CK− CTCs, ERG rearrangements were identified in 3 patients, with 3/26 (12%) CK− CTCs showing ERG rearrangement. ERG rearrangements were also detected in 18/108 (17%) traditional CTCs from these 3 patients.
Small CTCs were evaluated in select patient samples by FISH for ERG and PTEN aberrations. ERG FISH was performed on 6 patients with small CTCs and we were able to evaluate 27 small CTCs from 3 patients. Of these, ERG rearrangements were detected in 1 patient in 6/23 (26%) small CTCs. ERG rearrangements were also detected in 18/73 (25%) traditional CTCs (non-small) from this patient. The size of small CTCs precluded interpretation of PTEN deletions in all but 3 small CTCs in 2 patients, which were wild type. Examples of small CTCs harbouring AR positivity by IF or ERG alterations by FISH are shown in Figure 1. Importantly, these results demonstrate the utility of this assay to characterize CTC populations with cancer-specific markers, verifying the prostatic origin of traditional CTCs in this cohort.
Detection of Non-traditional CTCs in mCRPC patients
As described in detail below, we identified two categories of cells with non-traditional phenotypes (in addition to CTC clusters) that were potential CTCs: cells with weak or no CK expression (CK− CTCs), and cells with degenerative changes and nuclear disintegration consistent with apoptosis (apoptotic CTCs). All patients harboured circulating cells with these non-traditional phenotypes (median 6/mL, range 1–101). Four of 20 (20%) healthy volunteers had circulating cells that met these expanded CTC criteria (median 0/mL, range 0–5) and 3/20 (15%) had cells with AR positivity (median 0/mL per patient, range 0–4). Figure 3 shows the frequency of non-traditional CTCs in patient samples compared to healthy volunteer samples. For all non-traditional CTCs, AR expression was evaluated by IF and showed a wide variability similar to traditional CTCs (Figure 4A). Representative images of non-traditional CTCs, including those with PCa specific molecular alterations, are shown in Figure 1.
Cytokeratin-negative CTCs
CK intensity in CD45− circulating cells with abnormal morphology varied widely across patients with mCRPC (Figure 4C). As with AR expression measurements, relative CK expression values for every CTC were generated by Epic’s software, normalizing CK expression in CTCs to approximately 1 million surrounding CD45+ WBCs present on the same slide. CTCs with CK expression values ≥2.8 were considered positive (signal ≥ 2.8 fold higher than background WBCs). Hence, we defined non-traditional CK− CTCs as CD45− circulating cells with morphological distinction and/or AR positivity, but CK intensity less than 2.8. Such CK− CTCs were identified in 34/41 (83%) mCRPC patients (median 2 CK− CTC/mL, range 0–20). Amongst patients with CK− CTCs, 26 (76%) had at least 1 AR+ CK− CTC/mL (range 0–20). The proportion of CK+/CK− CTCs varied across the cohort, with two patients demonstrating a CTC population that was predominantly CK− (Figure 4D). Of healthy volunteers evaluated, none had >1 CD45−/CK− cell/mL. Both ERG rearrangements and PTEN deletions were detected in CK− CTCs from mCRPC patients who harboured these same alterations in traditional CTCs (Table 3).
Apoptotic and Small CTCs
CD45−/CK+ cells with nuclear fragmentation or condensation characteristic of apoptosis were also observed across the mCRPC cohort. These cells, which we classified as non-traditional apoptotic CTCs, were detected in 40/41 (98%) of mCRPC patients (median 4/mL, range 0–92) as shown in Figure 3. Of the 40 patients with apoptotic CTCs, 31 (78%) had at least 1 AR+ apoptotic CTC/mL (range 0–60). Similar to the other categories of non-traditional CTCs, the frequency of apoptotic CTCs varied among mCRPC patients, with 11/41 (27%) patients exhibiting CTC populations composed primarily of apoptotic CTCs (Figure 4E). Due to nuclear fragmentation or disintegration, apoptotic CTCs were not amenable to FISH analysis.
We then utilized Epic software to objectively measure the total cell area of each CTC to elucidate the subpopulations of CTCs in patient samples that were similar in size to (or smaller than) WBCs, that could potentially be missed by size or density enrichment strategies for CTC isolation. Approximately 300 patient WBCs were used to generate the median and interquartile ranges for WBC area (75 μm2 and 64–90 μm2, respectively). We therefore used a cell area ≤ 90 μm2 to define a small CTC (Figure 5A). By this approach, cell area of traditional and non-traditional CTC subtypes was evaluated for 40/41 patient samples (1 sample was not evaluable by this method and omitted from analysis). Apoptotic CTCs were omitted from analysis because of poor segmentation due to fragmented cell morphology. We observed a wide range of CTC sizes within individual patients and across the mCRPC patient cohort using this objective assessment, with small CTCs detectable in 39/40 (97.5%) mCRPC patients. CTCs detected in mCRPC patients had a median cell area of 99.0 μm2 (range 28.9–1631.0). The proportion of small CTCs (CTCs with cell area ≤ 90 μm2) is shown for each patient in Figure 5B, with 17/40 (42.5%) patients exhibiting CTC populations composed predominantly of small CTCs. Small CTCs were evaluated in select patient samples by FISH for ERG and PTEN aberrations (Table 3). Examples of small CTCs harbouring AR positivity by IF or ERG alterations by FISH are shown in Figure 1.
Figure 5. CTC size varies greatly across patients with mCRPC.
A. Cellular area (μm2) per CTC as calculated by Epic software for each patient is plotted. White blood cell (WBC) size frequency distribution curve was generated by measuring the cellular area of approximately 300 normal WBCs (right side). The blue dashed line represents the median WBC area (75 μm2). Red and green dashed lines indicate WBC size cut-offs equal to the 25th (64 μm2) and 75th (90 μm2) percentile, respectively. Traditional CTCs (blue), CTC clusters (green) and CK− CTCs (purple) are indicated according to the legend. Apoptotic CTCs were omitted from analysis due to poor segmentation of fragmented nuclei in these cells. B. The percentage of all CTCs for each patient with area greater than (non-Small CTC; green) or less than the WBC 75th percentile (Small CTC; red) is plotted. *Sample 5037 was not evaluable and is not included in panel A.
Clinical significance of CTC types
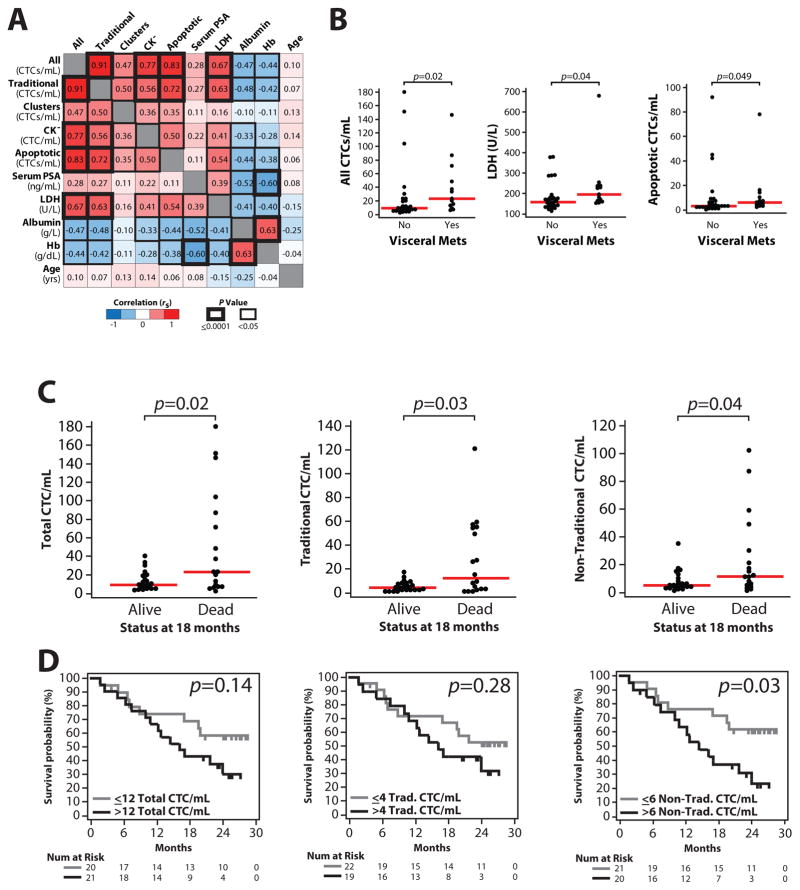
Although the main focus of our work was to define the morphologic and phenotypic range of CTCs in patients with mCRPC using an unbiased approach complemented by molecular characterization, we also assessed associations between traditional and non-traditional CTC types and clinicopathological parameters (serum prostate-specific antigen (PSA), lactate dehydrogenase (LDH), albumin and hemoglobin levels, as well as prior treatment regimens and presence of visceral metastasis). Importantly, as shown in Figure 6A, this analysis demonstrated that counts of all CTC types detected by the Epic platform (traditional, CTC clusters, and each non-traditional CTC type, as well as the sum of all CTCs) are significantly correlated with each other, ranging from Spearman rank correlation (rs) = 0.35 for apoptotic CTC and CTC cluster counts (p=0.03), to rs=0.72 for traditional CTC and apoptotic CTC counts (p<0.0001). Of interest, there was no significant correlation between any CTC type count and serum PSA levels. However, counts of CTC types other than CTC clusters were significantly associated with high LDH and low albumin, two important prognostic parameters. Likewise, both all CTC (sum of traditional CTCs, CTC clusters, apoptotic CTCs and CK− CTCs) and apoptotic CTC counts were significantly associated with the presence of visceral metastasis Figure 6B. We did not identify a statistically significant difference between any CTC type count and prior treatment regimen.
Figure 6. Associations of CTC/mL counts and clinicopathological parameters.
A. Spearman rank correlation (rs) matrix of CTC/mL counts (All=summed traditional, clusters, CK− and apoptotic) and clinicopathological parameters for the 41 patients with mCRPC in our cohort. Correlations are given and indicated according to the colour scale in the legend). Statistical significance of each comparison is indicated by the cell border thickness according to the legend. B. Significant (p<0.05, Mann-Whitney test) associations between CTC counts (and clinicopathological parameters) with the presence/absence of visceral metastases are shown. C. All, traditional and non-traditional CTC counts stratified by patient status (dead vs. alive) at 18 months are plotted (40 of 41 patients evaluable at that time point). p values from Mann-Whitney tests are shown. D. Kaplan-Meier analysis for all, traditional, and non-traditional CTC counts for overall survival time for all 41 patients. For each CTC type, patients were stratified by having > or ≤ median CTC count/mL. Log-rank test p values are shown. Plots for each non-traditional CTC type for C&D are shown in Figure S2.
Next, we assessed correlations between CTC counts by the CellSearch Circulating Tumor Cell System and the Epic platform in the mCRPC cohort. Importantly, as shown in Table 4, we observed strong correlation between traditional CTC counts by CellSearch and traditional CTCs by the Epic platform in the mCRPC patients (Pearson’s correlation r = 0.78, p<0.0001). Likewise, we also observed significant correlations between CellSearch CTC counts and apoptotic (Pearson’s correlation r = 0.91, p<0.0001) or CK− CTC counts (r = 0.39, p=0.02) by the Epic platform (Table 4). No significant correlation was observed between CellSearch CTC counts and CTC cluster counts (r = −0.07, p=0.68) by the Epic platform, although CTC clusters were infrequently observed in our cohort. Importantly, we identified a strong, significant correlation between CellSearch CTC counts and total CTC counts by the Epic Platform (r = 0.89, p<0.0001, Table 4).
Table 4.
CTC counts by Epic vs. CellSearch platforms in mCRPC patients
| Patient | CellSearch | Epic CTC counts | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| CTCs/7 mL | Traditional CTCs/mL | CTC Clusters/mL | CK-CTCs/mL | Apoptotic CTCs/mL | All CTCs/mL | |
| V4002 | NA | 2 | 0 | 1 | 1 | 4 |
| V4004 | 2 | 9 | 3 | 1 | 6 | 19 |
| V4030 | NA | 7 | 0 | 1 | 7 | 15 |
| V4045 | NA | 1 | 0 | 10 | 2 | 13 |
| V4048 | 12 | 9 | 0 | 2 | 9 | 20 |
| V4082 | 5 | 1 | 0 | 2 | 3 | 6 |
| V5025 | 157 | 54 | 1 | 11 | 5 | 71 |
| V5031 | NA | 1 | 0 | 1 | 3 | 5 |
| V5035 | 1 | 57 | 6 | 14 | 10 | 87 |
| V5037 | 0 | 4 | 0 | 3 | 2 | 9 |
| V5039 | 27 | 6 | 0 | 2 | 4 | 12 |
| V5042 | 285 | 8 | 0 | 1 | 14 | 23 |
| V5054 | 11 | 1 | 0 | 2 | 4 | 7 |
| V5055 | 52 | 26 | 2 | 3 | 6 | 37 |
| V5058 | 1 | 1 | 0 | 3 | 1 | 5 |
| V5061 | 6 | 3 | 0 | 2 | 4 | 9 |
| V5066 | NA | 2 | 0 | 1 | 3 | 6 |
| V5069 | 70 | 27 | 0 | 5 | 16 | 48 |
| V5070 | 1 | 3 | 0 | 0 | 6 | 9 |
| V5071 | 6 | 2 | 0 | 0 | 3 | 5 |
| V5074 | 18 | 5 | 0 | 0 | 2 | 7 |
| V5076 | 1 | 2 | 0 | 0 | 1 | 3 |
| V5080 | 3 | 2 | 0 | 0 | 3 | 5 |
| V5083 | 646 | 59 | 0 | 9 | 78 | 146 |
| V5085 | 563 | 121 | 0 | 17 | 42 | 180 |
| V5086 | 26 | 3 | 0 | 1 | 3 | 7 |
| V5088 | 0 | 5 | 0 | 8 | 7 | 20 |
| V5102 | 5 | 4 | 0 | 5 | 2 | 11 |
| V5106 | 448 | 55 | 0 | 4 | 45 | 104 |
| V5107 | 568 | 49 | 1 | 9 | 92 | 151 |
| V5108 | 1 | 1 | 0 | 0 | 3 | 4 |
| V5110 | 0 | 4 | 0 | 2 | 3 | 9 |
| V5111 | 0 | 13 | 1 | 7 | 9 | 30 |
| V5122 | 0 | 5 | 0 | 20 | 15 | 40 |
| V5126 | 5 | 2 | 0 | 2 | 2 | 6 |
| V5127 | 95 | 15 | 0 | 4 | 4 | 23 |
| V5131 | 0 | 17 | 2 | 9 | 5 | 33 |
| V5133 | 12 | 8 | 0 | 3 | 1 | 13 |
| V5135 | 2 | 10 | 0 | 6 | 7 | 23 |
| V5145 | 34 | 1 | 0 | 1 | 0 | 2 |
| V5148 | 21 | 8 | 0 | 0 | 2 | 10 |
| Correlation with CellSearch: | 0.78 | −0.07 | 0.39 | 0.91 | 0.89 | |
| p value: | <0.0001 | 0.68 | 0.02 | <0.0001 | <0.0001 | |
Circulating tumour cell (CTC) counts for 41 patients with metastatic castration resistant prostate cancer (mCRPC) as determined by the CellSearch and Epic platforms are given. For Epic determined CTC counts, counts for traditional CTCs, non-traditional CTCs (CTC clusters, apoptotic CTCs and CK− CTCs) and all CTCs (sum of traditional and non-traditional CTCs) are given. NA = not available. Pearson correlation and p values of CellSearch counts with each Epic CTC category across the cohort are given below the table.
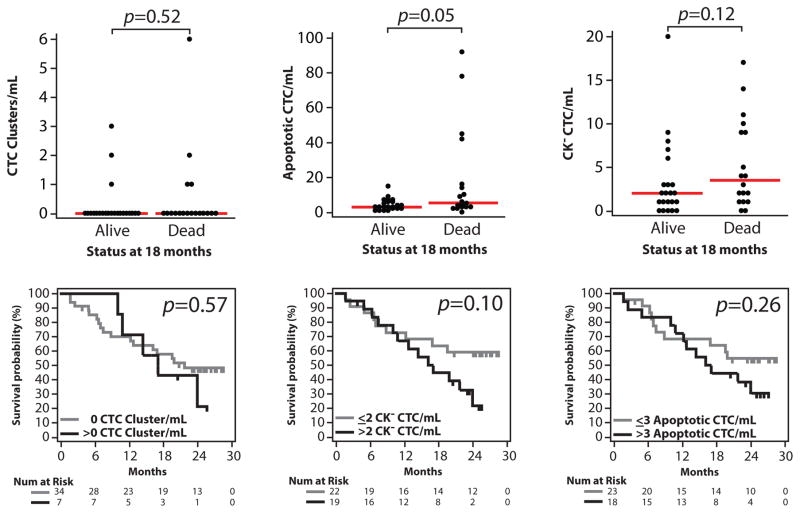
Lastly, to provide preliminary insight into associations with clinical outcome, we determined CTC counts by the Epic Platform in mCRPC patients who were alive or dead after 18 months (40 of 41 patients with sufficient follow-up). As shown in Figure 6C, total, traditional and non-traditional (clusters, apoptotic and CK−) CTC counts were significantly higher in patients dead versus alive at 18 months (Mann-Whitney test, p=0.02, p=0.03 and p=0.04, respectively). By Kaplan-Meier analysis on all 41 patients, the number of non-traditional CTCs was significantly associated with overall survival (, > vs. ≤ median CTC count, p=0.03, Fig 4D). Results for individual non-traditional CTC types are shown in Figure 7. Of interest, there were 14 patients with <5 CTCs detected by the CellSearch assay, of which 4 were deceased at 18 months; each of these deceased patients demonstrated non-traditional CTC cells as detected by the Epic platform.
Figure 7. Associations of non-traditional CTC/mL counts in patients with mCRPC and overall survival.
A. Individual non-traditional CTC type counts (CTC clusters, CK− CTCs and apoptotic CTCs) stratified by patient status (dead vs. alive) at 18 months are plotted (40 of 41 patients evaluable at that time point). p values from Mann-Whitney tests are shown. D. Kaplan-Meier analysis for CTC clusters, CK− CTCs and apoptotic CTCs and overall survival time for all 41 patients. For each CTC type, patients were stratified by having > or ≤ median CTC count/mL. Log-rank test p values are shown.
DISCUSSION
To characterize the phenotypic diversity of CTCs in mCRPC, we used the Epic platform to characterize all nucleated cells from whole blood samples [20]. Using DAPI, CD45, CK and AR IF, we identified and morphologically characterized cells with cancer-associated features, irrespective of whether they met the traditional CTC definition. We then used FISH and slide scanning with single-cell resolution to confirm that sub-populations of non-traditional CTCs were PCa in origin. We detected non-traditional CTC in all patients, including CTC clusters, small, apoptotic and/or CK− CTCs. We also observed intra-patient heterogeneity of AR and CK expression and PTEN and ERG status consistent with a heterogeneous model of advanced disease, as has been observed in recent studies of CRPC assessing AR expression in CellSearch-isolated CTCs, metastatic tissues assessed by whole genome sequencing, and circulating cell free DNA assessed using targeted next generation sequencing [21–23].
Although non-traditional CTC counts correlated with traditional CTC counts, substantial variation in the proportions of traditional to non-traditional CTCs were observed. Importantly, some CTCs we identified could be missed by CTC detection platforms using antigen capture (i.e. EpCAM or CK) or size selection (such as filtration). In keeping with this idea, Chen et al., recently reported that EpCAM captured CTCs (enriched using NanoVelcro Chips) in patients with mCRPC showed variable nuclear size, and patients with visceral metastases specifically showed elevated very small nuclear sized CTCs [8]. Our results suggest that in addition to nuclear size variability, CTCs in mCRPC patients show considerable heterogeneity in nuclear organization, and CK and AR expression.
CK+/CD45− cells were identified in a small minority of healthy volunteer samples at low frequencies, with the majority demonstrating low CK and AR expression. Given no universally established reference range for the definition of CTC, nor a recognized pan-CTC marker, it is difficult to discern the relevance of these cells in healthy volunteers. CTC-like cells can be detected in the blood of patients with benign conditions as well as in those with early stage disease [24–26], and have been reported in labelled healthy volunteers [27]. The ideal healthy volunteer or non-cancer control population would be matched in age and demographics, with confirmed absence of malignancy. While these caveats are impediments for utilizing any CTC platform for the primary diagnosis of prostate cancer, the increased sensitivity and specificity of the Epic platform for CTC detection could be suited for acquiring prognostic and predictive information in mCRPC patients.
Detection of significant non-traditional CTC populations in mCRPC patients is consistent with substantial evidence supporting their existence and clinical relevance. For example, CTCs postulated to have undergone epithelial to mesenchymal transition (EMT), and displaying reduced/no EpCAM or CK expression, have been identified by multiple platforms using both cell lines and patient samples [27–33]. These cells may be enriched for multipotent cancer stem cells, with increased self-renewal and metastasis forming capacity [34, 35]. Although prior studies demonstrate substantial heterogeneity of PTEN deletion status in CTCs and CRPC tissue foci in a given patient [12, 36], ERG rearrangements have generally been identified as a clonal alteration (when present) in both CTCs and mCRPC tissues [12, 21, 37–39]. Possible explanations for these apparently discrepant results include increased sensitivity for traditional and non-traditional CTCs (identified here) that may represent multiple clonal populations when compared to our previous CellSearch based study showing homogeneous ERG status in traditional CTCs [12]. Also, our recent study on circulating cell-free DNA in patients with mCRPC supported the existence of both ERG rearranged and wildtype clones in the same patient during disease progression [23]. This contrasts with the observation that multiple CRPC foci at autopsy in an individual patient nearly always show homogeneous ERG status. Further studies are required to interrogate this difference, which we hypothesize could be informed by more comprehensive genetic analysis of individual traditional and non-traditional CTC populations.
Our cohort represents a single institution mCRPC cohort and the relatively small size in this non-uniformly treated initial study limits the ability to draw robust conclusions on associations between non-traditional CTCs and clinical outcome. Likewise, additional studies are needed to characterize the metastatic potential of distinct CTC classes. Importantly, evaluation of these distinct CTC populations described here in larger, well-defined cohorts to inform on their prognostic and predictive utility in mCRPC is now warranted. Here, using pathologic and molecular biological approaches, we have described expanded categories of CTC phenotypes in patients with mCRPC and demonstrate that considerable heterogeneity exists within these categories. These expanded CTC subtypes may provide novel prognostic and predictive information for patients with mCRPC, as well as other advanced cancers. Intriguing preliminary clinical associations between mCRPC outcomes and various CTC subpopulations will need to be assessed in additional ongoing and planned studies to expand on our study.
Acknowledgments
This work was in-part supported by funding from the Movember GAP CTC programme, National Institute for Health Research to The Royal Marsden/Institute of Cancer Research Biomedical Research Centre and National Institutes of Health (R01 CA183857 to S.A.T.). G.A. is a Cancer Research UK clinician scientist. We thank the participating men and their families who suffered from metastatic prostate cancer and nonetheless gave the gift of participation so that others might benefit. Authorship contributions to the manuscript are as follows: A.S.M. and R.F. wrote the manuscript; ASM, RF, JSdB, RD, SAT, and GA conceived of the study; R.F., Z.Z., P.F., S.S., J.M., J.S.dB. and G.A. provided samples; A.S.M., R.F. R.K., M.L., R.G., J.L., A.J., N.B., D.M., J.M., R.D., S.A.T., G.A. carried out experiments and analysed the data. All authors contributed to the editing and approval of the manuscript prior to submission. S.A.T. and G.A. are the guarantors of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis
Footnotes
Disclosures: The University of Michigan has been issued a patent on the detection of ETS gene fusions in prostate cancer, on which S.A.T. is listed as a co-inventor. The University of Michigan licensed the diagnostic field of use to Gen-Probe, Inc., who has sublicensed some rights to Ventana/Roche. S.A.T. serves as a consultant to, and has received honoraria from, Ventana/Roche, Astellas, Medivation and Abbvie, and is a co-founder of Strata Oncology. R.K., M.L., R.G, J.L., A.J., N.B., D.M. and R.D. are employees of Epic Sciences. R.F., Z.Z., P.F., S.S., J.M., J.S.dB. and G.A. are employees of The Institute of Cancer Research that has developed abiraterone and therefore has a commercial interest in this agent. G.A. is on the ICR list of rewards to inventors for abiraterone. J.S.d.B. has received consulting fees and travel support from Amgen, Astellas, AstraZeneca, Boehringer Ingelheim, Bristol-Myers Squibb, Dendreon, Enzon, Exelixis, Genentech, GlaxoSmithKline, Medivation, Merck, Novartis, Pfizer, Roche, Sanofi-Aventis, Supergen and Takeda, and grant support from AstraZeneca and Genentech. G.A. has received honoraria, consulting fees or travel support from Astellas, Medivation, Janssen, Millennium Pharmaceuticals, Ipsen, Ventana and Sanofi-Aventis, and grant support from Janssen, AstraZeneca and Arno.
References
- 1.Miyamoto DT, Sequist LV, Lee RJ. Circulating tumour cells-monitoring treatment response in prostate cancer. Nat Rev Clin Oncol. 2014 Jul;11:401–12. doi: 10.1038/nrclinonc.2014.82. [DOI] [PubMed] [Google Scholar]
- 2.Diamond E, Lee GY, Akhtar NH, et al. Isolation and characterization of circulating tumor cells in prostate cancer. Front Oncol. 2012;2:131. doi: 10.3389/fonc.2012.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong AJ, Eisenberger MA, Halabi S, et al. Biomarkers in the management and treatment of men with metastatic castration-resistant prostate cancer. Eur Urol. 2012 Mar;61:549–59. doi: 10.1016/j.eururo.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Cremoux P, Extra JM, Denis MG, et al. Detection of MUC1-expressing mammary carcinoma cells in the peripheral blood of breast cancer patients by real-time polymerase chain reaction. Clin Cancer Res. 2000 Aug;6:3117–22. [PubMed] [Google Scholar]
- 5.Krebs MG, Metcalf RL, Carter L, Brady G, Blackhall FH, Dive C. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol. 2014 Mar;11:129–44. doi: 10.1038/nrclinonc.2013.253. [DOI] [PubMed] [Google Scholar]
- 6.Hong B, Zu Y. Detecting circulating tumor cells: current challenges and new trends. Theranostics. 2013;3:377–94. doi: 10.7150/thno.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 8.Chen JF, Ho H, Lichterman J, et al. Subclassification of prostate cancer circulating tumor cells by nuclear size reveals very small nuclear circulating tumor cells in patients with visceral metastases. Cancer. 2015 May 14; doi: 10.1002/cncr.29455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armstrong AJ, Marengo MS, Oltean S, et al. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res. 2011;9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larson CJ, Moreno JG, Pienta KJ, et al. Apoptosis of circulating tumor cells in prostate cancer patients. Cytometry A. 2004 Nov;62:46–53. doi: 10.1002/cyto.a.20073. [DOI] [PubMed] [Google Scholar]
- 11.Stott SL, Lee RJ, Nagrath S, et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med. 2010 Mar 31;2:25ra3. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Attard G, Swennenhuis JF, Olmos D, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–8. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 13.Beltran H, Rubin MA. New strategies in prostate cancer: translating genomics into the clinic. Clin Cancer Res. 2013 Feb 1;19:517–23. doi: 10.1158/1078-0432.CCR-12-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenner JC, Chinnaiyan AM, Tomlins SA. ETS Fusion Genes in Prostate Cancer. In: Tindall DJ, editor. Prostate Cancer: Biochemistry, Molecular Biology and Genetics, Vol. Protein Reviews. New York Springer; New York: 2013. pp. 139–83. [Google Scholar]
- 15.Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012 Jul 12;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh HB, Marrinucci D, Bethel K, et al. High speed detection of circulating tumor cells. Biosens Bioelectron. 2006 Apr 15;21:1893–9. doi: 10.1016/j.bios.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 17.Marrinucci D, Bethel K, Bruce RH, et al. Case study of the morphologic variation of circulating tumor cells. Hum Pathol. 2007 Mar;38:514–9. doi: 10.1016/j.humpath.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 18.Marrinucci D, Bethel K, Kolatkar A, et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol. 2012 Feb;9:016003. doi: 10.1088/1478-3975/9/1/016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marrinucci D, Bethel K, Luttgen M, Bruce RH, Nieva J, Kuhn P. Circulating tumor cells from well-differentiated lung adenocarcinoma retain cytomorphologic features of primary tumor type. Arch Pathol Lab Med. 2009 Sep;133:1468–71. doi: 10.1043/1543-2165-133.9.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werner SL, Graf RP, Landers M, et al. Analytical Validation and Capabilities of the Epic CTC Platform: Enrichment-Free Circulating Tumour Cell Detection and Characterization. J Circ Biomark. 2015:4. doi: 10.5772/60725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gundem G, Van Loo P, Kremeyer B, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015 Apr 16;520:353–7. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crespo M, van Dalum G, Ferraldeschi R, et al. Androgen receptor expression in circulating tumour cells from castration-resistant prostate cancer patients treated with novel endocrine agents. Br J Cancer. 2015;112( Suppl):1166–74. doi: 10.1038/bjc.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carreira S, Romanel A, Goodall J, et al. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med. 2014 Sep 17;6:254ra125. doi: 10.1126/scitranslmed.3009448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller MC, Doyle GV, Terstappen LW. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J Oncol. 2010;2010:617421. doi: 10.1155/2010/617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pantel K, Deneve E, Nocca D, et al. Circulating epithelial cells in patients with benign colon diseases. Clinical chemistry. 2012 May;58:936–40. doi: 10.1373/clinchem.2011.175570. [DOI] [PubMed] [Google Scholar]
- 26.Reyal F, Valet F, de Cremoux P, et al. Circulating tumor cell detection and transcriptomic profiles in early breast cancer patients. Annals of oncology : official journal of the European Society for Medical Oncology/ESMO. 2011 Jun;22:1458–9. doi: 10.1093/annonc/mdr144. [DOI] [PubMed] [Google Scholar]
- 27.Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013 Apr 3;5:179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pecot CV, Bischoff FZ, Mayer JA, et al. A novel platform for detection of CK+ and CK− CTCs. Cancer Discov. 2011 Dec;1:580–6. doi: 10.1158/2159-8290.CD-11-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe M, Uehara Y, Yamashita N, et al. Multicolor detection of rare tumor cells in blood using a novel flow cytometry-based system. Cytometry A. 2013 Dec 10; doi: 10.1002/cyto.a.22422. [DOI] [PubMed] [Google Scholar]
- 30.Giordano A, Gao H, Anfossi S, et al. Epithelial-mesenchymal transition and stem cell markers in patients with HER2-positive metastatic breast cancer. Mol Cancer Ther. 2012 Nov;11:2526–34. doi: 10.1158/1535-7163.MCT-12-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu M, Bardia A, Wittner BS, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science (New York, NY. 2013 Feb 1;339:580–4. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorges TM, Tinhofer I, Drosch M, et al. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC cancer. 2012;12:178. doi: 10.1186/1471-2407-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Ridgway LD, Wetzel MD, et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med. 2013 Apr 10;5:180ra48. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009 Jun;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Seminars in cancer biology. 2012 Oct;22:396–403. doi: 10.1016/j.semcancer.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferraldeschi R, Nava Rodrigues D, Riisnaes R, et al. PTEN Protein Loss and Clinical Outcome from Castration-resistant Prostate Cancer Treated with Abiraterone Acetate. Eur Urol. 2015 Nov 4;67:795–802. doi: 10.1016/j.eururo.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Udager AM, Shi Y, Tomlins SA, et al. Frequent discordance between ERG gene rearrangement and ERG protein expression in a rapid autopsy cohort of patients with lethal, metastatic, castration-resistant prostate cancer. Prostate. 2014 Sep;74:1199–208. doi: 10.1002/pros.22836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mehra R, Tomlins SA, Yu J, et al. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer Res. 2008 May 15;68:3584–90. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W, Laitinen S, Khan S, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009 May;15:559–65. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]