Abstract
Purpose
To assess exome data for pre-emptive pharmacogenetic screening for 203 clinically-relevant pharmacogenetic variant positions from the Pharmacogenomics Knowledgebase and Clinical Pharmacogenetics Implementation Consortium and identify copy number variants (CNVs) in CYP2D6.
Methods
We examined the coverage and genotype quality of 203 pharmacogenetic variant positions in 973 exomes vs. 5 genomes vs. 5 genotyping chip datasets. Then we determined the agreement of exome and chip genotypes by evaluating concordance in a three-way comparison of exome, genome and chip-based genotyping at 1,929 variant positions in 5 individuals. Finally, we evaluated the utility of exomes for detecting CYP2D6 CNVs.
Results
For 5 individuals examined for 203 pharmacogenetic variants (5 × 203 = 1,015), 998/1,015 were identified by genome, 849/1,015 by exome and 295/1,015 by genotyping chip. Thirty-six pharmacogenetic star allele variants with moderate to strong CPIC therapeutic recommendations were identified in 973 exomes. Exomes had high (98%) genotype concordance with chip-based genotyping. CYP2D6 CNVs were identified in 57/973 exomes.
Conclusions
Exomes outperformed the current chip-based assay in detecting more important pharmacogenetic variant positions and CYP2D6 CNVs for preemptive pharmacogenetic screening. Tools should be developed to derive pharmacogenetic variants from exomes.
Keywords: massively parallel sequencing, pharmacogenetic screening
INTRODUCTION
One of the goals of precision medicine is to use pharmacogenomics to optimize treatment efficacy and minimize adverse drug reactions. Barriers to the implementation of pharmacogenomics-guided therapy include the turn-around time in obtaining a pharmacogenetic (PGx) result1 and the clinical utility of returning PGx variants.2 One recommendation to avoid treatment delays is to implement preemptive PGx testing.1 Current PGx testing using array-based genotyping platforms (e.g., Affymetrix DMET Plus {Drug Metabolizing Enzymes and Transporters array}) screens for a predefined set of PGx variants.3,4 Genomic testing platforms such as exome sequencing (ES) or genome sequencing (GS),5 also called massively-parallel sequencing (MPS), have potentially wider utility than the aforementioned genotyping platforms and this begs the question of whether MPS sequence data could be used for pre-emptive PGx testing. Part of the larger challenge for the field of medical genomics is to identify all potential uses of sequencing so that the cost of these assays can be amortized across multiple applications, thus decreasing the effective cost of the test. Prior studies with small sample size showed high ES genotype concordance rate with other platforms (99.6% with MiSeq and 98.9% with iPLEX ADME PGx panel)6 and variable (60–80%) ES coverage of DMET Plus PGx variant positions depending on the capture kit used.7 An extensive analysis with a larger data set was needed to assess the capability of ES in detecting clinically relevant PGx variants. We set out to assess MPS concordance and coverage of annotated PGx variants compared to a current genotyping platform to determine if MPS could serve as a genotyping source for pre-emptive PGx testing.
MATERIAL AND METHODS
Participants
This study was performed at the NIH Clinical Center as part of the ClinSeq® project and included 973 participants enrolled between 45 and 65 years of age who were consented for baseline clinical tests, ES and/or GS, return of genetic results, and iterative phenotyping based on an individual’s genetic variants.8,9 The National Human Genome Research Institute IRB reviewed and approved this study. See Supplementary Methods online.
Selection of clinically-relevant pharmacogenetic variants for comparison
We identified 50 Pharmacogenomics Knowledgebase (PharmGKB) level 1A and 1B PGx variants (https://www.pharmgkb.org/) and 154 Clinical Pharmacogenetics Implementation Consortium (CPIC) (http://www.pharmgkb.org/page/cpic) variants from 40 gene-drug pairs with level A evidence (2 promoter variants were located at the same genomic position) for a total of 203 PGx variant positions. We evaluated coverage of these 203 PGx variant positions from 973 exomes, 5 genomes and 5 chip data. Three HLA-B variants (HLA-B*52:01:01, HLA-B*57:01:01, HLA-B*58:01:01) were excluded as they were not amenable to genotyping by the chip, ES or GS. MPS genotype concordance was determined by comparing 5 individuals with ES, GS and DMET Plus genotypes (hereafter referred to as the chip). The chip has been previously shown to have high genotype concordance (91 to 99%) compared to 6 orthogonal genotyping platforms.10 We selected CYP2D6 for CNV analysis because 1–2% of individuals carry more than two functional copies may have an ultrarapid metabolizer phenotype that can lead to codeine toxicity.11
Laboratory Methods
See Supplementary Methods online.
RESULTS
Detection of 203 CPIC/PharmGKB variant positions by exome vs. genome vs. chip
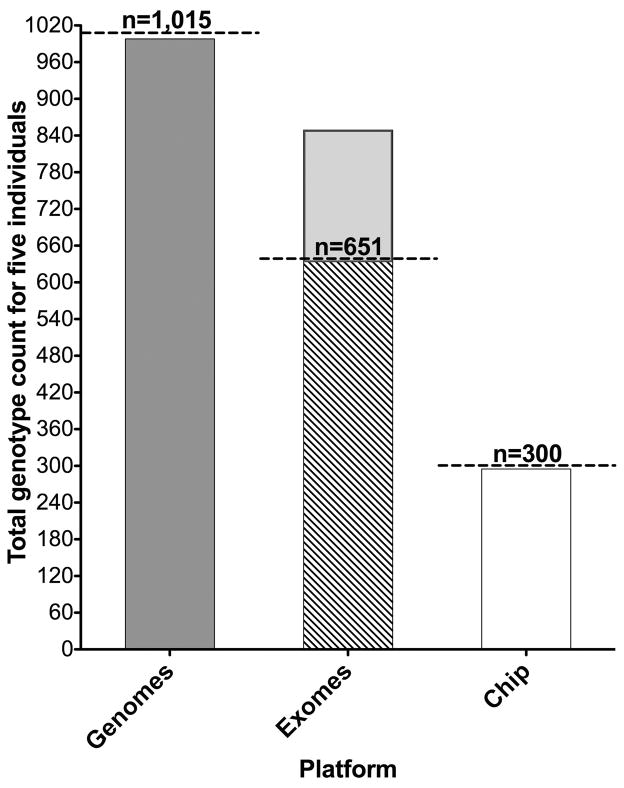
Five individuals were examined for 203-curated variants (132 coding, 71 noncoding positions) by ES, GS and chip-based testing. One would ideally like to detect a total of 1,015 genotypes (203×5). A total genotype count regardless of genotype quality from five individuals is shown in figure 1. GS detected 998/1,015 genotypes (657/660 {coding}, 341/351 {noncoding}). In the coding positions, 129/132 positions were covered in 5 individuals and 3/132 covered in four individuals. In the noncoding positions, 63/71 positions were covered in five, 5/71 covered in four and 3/71 covered in two individuals. For ES, 117/203 positions were targeted by two capture kits for five individuals (Agilent38Mb n=2, TruSeqV2 n=3), 12/203 targeted only by Agilent38Mb and 14/203 targeted only by TruseqV2. The expected total genotype count is 651 ({117×5}+{12×2}+{14×3}). The targeted genotype detection rate was 647/651. Of the positions targeted by both capture kits, 114/117 variant positions were covered in five individuals, 2/117 covered in four individuals and 1/117 covered in three individuals. All 26 positions targeted by only one of the capture kits had complete coverage. The total ES genotype count was 849 (647 {targeted} + 202 {off-target}). The chip targeted 46/132 coding and 14/71 noncoding positions and the targeted detection rate was 225/230 (coding) and 70/70 (noncoding). (Figure 1, Table S1–S3 online). In-house cost per genotypable site was $43.77 ($8710/199 positions) for genomes and $4.79 ($810/169 positions) for exomes and $9.31 for chip ($549/59 positions). These figures may not reflect clinical costs.
Figure 1.
Genotype count of 5 genomes, 5 exomes, and 5 chip data at 203 pharmacogenetic variant positions in 5 individuals
Total genotype count for five individuals at 203 pharmacogenetic variant positions regardless of genotype quality from genome, exome and chip. Total genotype count is 1,015 for genomes, 651-targeted genotypes for exomes and 300-targeted genotypes for the chip (represented by the horizontal dashed line). Genomes detected 998/1,015, exomes detected 849/1,015 ({647 targeted represented by diagonal striped area} and {202 off-target represented by light grey area}) and chip detected 295/1,1015.
Chip, Affymetrix DMET Plus (Drug Metabolizing and Transporters array).
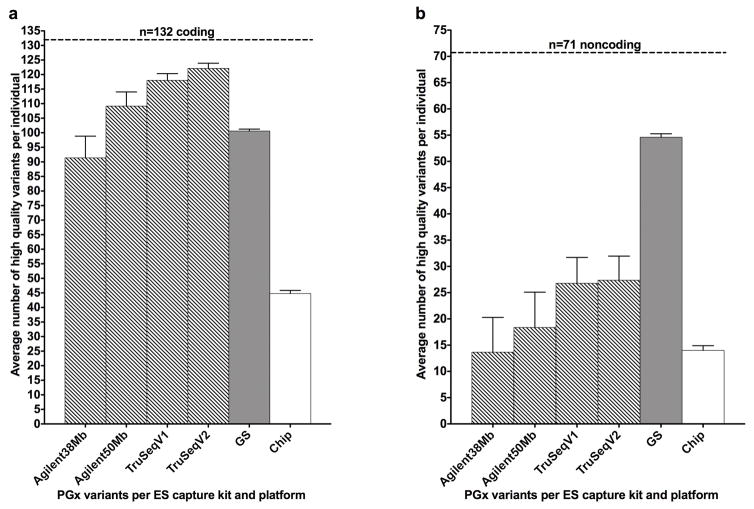
We next examined the detection rate of high quality genotypes (GQ ≥50) per individual at 203 positions. We included 973 exomes captured with four kits (Agilent38Mb, Agilent50Mb, TruSeqV1, TruSeqV2). ES, GS and chip data were grouped by coding vs. noncoding variants (intergenic, intronic, promoter, or 3′ untranslated region). GS detected an average of 101 and ES detected an average of 120 genotypes per individual at coding positions. At noncoding positions, GS detected an average of 55 and ES detected 27 genotypes per individual (Figure 2a, 2b {ES average based on TruSeqV1 and V2 data}). ES coverage was the highest in coding regions and the TruSeqV2 kit had the highest average (122), while the chip captured 45 genotypes per individual (Figure 2a). ES coverage in non-coding regions was low. Among the 71 noncoding positions, TruSeqV1/V2 had the highest average (27) while the Agilent38Mb kit and the chip had the lowest average of 14 genotypes per individual (Figure 2b). GS coverage was outperformed by the Agilent50Mb, TruSeqV1/V2 kits in coding regions (Figure 2a). (Table S4 online).
Figure 2.
a. Exome capture kits vs. genome vs. chip coverage of 132 coding pharmacogenetic variant positions
b. Exome capture kits vs. genome vs. chip coverage of 71 noncoding pharmacogenetic variant positions
Total number of variant positions represented by the horizontal dashed line. Bar graphs shows the average number of high quality variants per individual by four exome capture kits (Agilent 38Mb (n=393), Agilent 50Mb (n=318), Illumina TruSeqV1 (n=147), Illumina TruSeqV2 (n=115)) versus genome sequence (n=5) versus chip data (n=5). The top of the bars indicates the average number of high quality (GQ score equal to or greater than 50) variant(s) detected per individual for exomes, genomes and chip. The whiskers above the bars represent the SEM. See Table S4 online for mean, SEM and N.
3′UTR, 3 prime untranslated region; Chip, Affymetrix DMET Plus (Drug Metabolizing and Transporters array); CPIC, Clinical Pharmacogenetics Implementation Consortium; ES, exome sequence; GQ, genotype quality; GS, genome sequence; Mb, megabase; N, number of individuals tested per platform; PGx, pharmacogenetic; PharmGKB, Pharmacogenomics Knowledgebase; SEM, standard error of the mean.
Detection of CPIC and PharmGKB pharmacogenetic variants and rare loss-of-function variants in known pharmacogenes
ES identified 36 star (*) allele variants with CPIC recommendations for change in therapy including individuals homozygous for CYP2C19 *2 (n=18), TPMT *3B, *3C (n=5),12 SLCO1B1 *5 (n=21)13 and individuals heterozygous for DPYD*13 (n=2) and rs67376798 (n=6) (Table S5 online).14 Twenty individuals with rare, loss-of-function and eight with splice variants were identified in eight known pharmacogenes (Table S6 online).
Genotype concordance between exomes, genomes and genotyping chip
The chip had 1,929 unique variant positions and identified 9,598 genotypes for the five samples tested.
Of 8,040 genotype calls made by chip-ES, 7,258 homozygous/hemizygous and 639 heterozygous calls were concordant and 143 (1.8%) calls were discordant. Of the chip-ES discordant calls, the chip called 89/143 heterozygous and 54/143 homozygous and 83/143 of the discordant calls had ES GQ <50 and 77/83 are non-coding. For discordant calls with ES GQ ≥50, 57/60 were concordant in ES-GS (12/57 heterozygous and 45/57 homozygous) (Table S7, S8 online).
Of 9,543 genotype calls made by chip-GS, 8,411 homozygous/hemizygous and 1,029 heterozygous calls were concordant and 103 (1.1%) were discordant. Of the discordant chip-GS calls, the chip called 19/103 heterozygous and 84/103 homozygous/hemizygous and 29/103 had GS GQ <50 and 74/103 had GS GQ ≥50. Over 2/3 (20/29) were discordant coding calls. Among the discordant calls with GS GQ ≥50, 52/74 were concordant between ES-GS (12/52 heterozygous, 40/52 homozygous) (Table S7, S8 online).
Of 8,013 genotype calls made by ES-GS, 7,267 homozygous/hemizygous and 649 heterozygous were concordant and 97 (1.2%) were discordant. Of the discordant ES-GS calls, the chip called 78/97 heterozygous and 19/97 homozygous and 80/97 had ES GQ <50 and 73/80 were noncoding. The majority (76/97) of the discordant ES-GS calls were concordant between chip-GS (75/76 heterozygous {GS GQ ≥50}, 1/76 homozygous {GS GQ <50}). A few (17/97) of the discordant ES-GS calls had ES GQ ≥50 and 14/17 were concordant between chip-ES (2/14 heterozygous, 12/14 homozygous) (Table S7, S8 online).
Detection and validation of CYP2D6 CNVs using eXome hidden Markov model
CYP2D6 CNVs were detected in 57/973 exomes (duplication n=39, deletion n=18) (Table S9 online). XHMM quality scores (QS) ranged from 38–99. Seven individuals with the highest XHMM QS of 99 (duplication n=6, deletion n=1) were selected for validation with real-time quantitative PCR (qPCR) and all samples were confirmed (Table S10 online). An additional 19 samples with XHMM QS ranging from 38–99 (duplication n=17, deletion n=2) were selected for a second round of validation with qPCR and all were confirmed (Table S10). Of the 26 samples tested, 11 samples showed agreement across all CNV regions, nine samples were inconclusive (XHMM does not make CNV calls in noncoding regions) and six samples (168397, 136439, 181872, 181608, 185076, 196659) showed breakpoint discrepancies between the XHMM predictions and the qPCR result. This was not a surprising finding, as their XHMM Q_exact scores ({confidence measure of the predicted CNV breakpoint) were low, ranging from 4–18 (data not shown). Nine samples (142307, 175100, 187383, 140601, 190031, 190871, 194883, 131340, 167715) with predicted whole gene duplication showed a normal copy number with the 5′ probe (Table S10). This may be due to the CYP2D6 duplication not extending into the 5′ region. High sequence identity (96.9%) of CYP2D6 and the CYP2D7P pseudogene (NM_000106.5, NR_002570.2 respectively) can result in CYP2D6-2D7P hybrid genes.15 For these nine individuals, ES data analysis did not find paired-end reads mapping to both CYP2D6-CYP2D7P. Our paired-end reads (89 bp) and inserts (180 bp) are short, thus the absence of detecting paired-end reads mapping to CYP2D6-CYP2D7P does not rule out the presence of fusion/hybrid genes.
DISCUSSION
Adoption of PGx-guided therapy has been limited due to insufficient data to support clinical utility and cost effectiveness, knowledge gaps in pharmacogenomics and the inherent delay engendered by PGx testing. We propose leveraging existing MPS data by extracting PGx variants pre-emptively based on two premises. The first is that thousands of patients are currently undergoing clinical ES and GS and these data comprise a valuable resource for pharmacogenomics. The second is that the extraction of PGx variants from ES/GS data is part of a larger effort to maximize the utility of ES/GS testing results. Studies have demonstrated how ES data can be used to extract variants for the secondary screening of susceptibility to cancer, malignant hyperthermia, cardiomyopathy, cardiac dysrhythmias, and aortic dissection.16–19 We assessed the capability of MPS for pre-emptive PGx testing by comparing the coverage of 203 important PGx variants in 973 exomes to a widely used PGx chip.
ES and GS had several advantages over chip-based testing. The genome-wide coverage of ES and GS allowed coverage of more PharmGKB class 1A, 1B and CPIC gene-drug level A variants than the genotyping chip and identify both known and as yet to be discovered PGx variants in one test.
CYP2D6 is a good example for exploring the ability of ES to interrogate CNVs. XHMM detected complete and partial CYP2D6 deletions and duplications while the chip only detects deletions.
Limitations of this study include 399/973 of the ES sequences were generated with the Agilent38Mb capture kit which accounted for the majority of the NC in the ES data, thus decreasing the coverage of some variant positions. The use of four capture kits provided us an opportunity to assess variance in capture kit coverage.
Our results showed high exome genotype concordance rate and higher coverage with the TruSeq capture kits (using GQ ≥50) are consistent with findings from recent studies evaluating exome capability for pharmacogenomics screening.6,7 An updated array targeting PharmGKB level 1A, 1B and CPIC level A variants may be a more cost efficient initial screen than exomes, however, panel testing and enhanced exome capture with additional targets in noncoding regions20 will require periodic updating of the test platform and repeat testing of subjects for future discoveries. Although our results showed that exomes can be used to extract PGx variants, we are not advocating ordering an exome primarily for pharmacogenomics screening as our analyses did not answer the question of whether there is clinical utility and validity of using MPS for pre-emptive PGx screening for these variants.
We have demonstrated the utility of MPS data for the detection of single PGx variants and CYP2D6 CNVs. Currently, no tools are available to extract and annotate PGx variants from MPS data. We conclude that tools should be developed to extract PGx variants from existing ES and GS data for research and potential future use.
Supplementary Material
Acknowledgments
The authors are grateful for the contributions of the staff at the NIH Intramural Sequencing Center, NIH Clinical Center, and the ClinSeq® study participants. This study was funded by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
Footnotes
Supplementary material is linked to the online version of the paper at http://www.nature.com/gim.
DISCLOSURE:
Drs. Ng, Hong, Singh, Johnston, and Mullikin declare no conflict of interests. Dr. Biesecker is an uncompensated advisor to the Illumina Corp, receives royalties from Genentech, Inc, and receives honoraria for Editing from Wiley-Blackwell, Inc.
References
- 1.Weitzel KW, Elsey AR, Langaee TY, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet. 2014;166C(1):56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janssens AC, Evans JP. Returning pharmacogenetic secondary findings from genome sequencing: let’s not put the cart before the horse. Genet Med. 2015;17(11):854–856. doi: 10.1038/gim.2015.59. [DOI] [PubMed] [Google Scholar]
- 3.Daly TM, Dumaual CM, Miao X, et al. Multiplex assay for comprehensive genotyping of genes involved in drug metabolism, excretion, and transport. Clin Chem. 2007;53(7):1222–1230. doi: 10.1373/clinchem.2007.086348. [DOI] [PubMed] [Google Scholar]
- 4.Sissung TM, English BC, Venzon D, Figg WD, Deeken JF. Clinical pharmacology and pharmacogenetics in a genomics era: the DMET platform. Pharmacogenomics. 2010;11(1):89–103. doi: 10.2217/pgs.09.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;370(25):2418–2425. doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- 6.Chua EW, Cree SL, Ton KN, et al. Cross-Comparison of Exome Analysis, Next-Generation Sequencing of Amplicons, and the iPLEX((R)) ADME PGx Panel for Pharmacogenomic Profiling. Front Pharmacol. 2016;7:1. doi: 10.3389/fphar.2016.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Londin ER, Clark P, Sponziello M, Kricka LJ, Fortina P, Park JY. Performance of exome sequencing for pharmacogenomics. Per Med. 2014;12(2):109–115. doi: 10.2217/PME.14.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biesecker LG. Hypothesis-generating research and predictive medicine. Genome Res. 2013;23(7):1051–1053. doi: 10.1101/gr.157826.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biesecker LG, Mullikin JC, Facio FM, et al. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome research. 2009;19(9):1665–1674. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez CA, Smith C, Yang W, et al. Concordance of DMET plus genotyping results with those of orthogonal genotyping methods. Clin Pharmacol Ther. 2012;92(3):360–365. doi: 10.1038/clpt.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther. 2014;95(4):376–382. doi: 10.1038/clpt.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Relling MV, Gardner EE, Sandborn WJ, et al. Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin Pharmacol Ther. 2013;93(4):324–325. doi: 10.1038/clpt.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramsey LB, Johnson SG, Caudle KE, et al. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin Pharmacol Ther. 2014;96(4):423–428. doi: 10.1038/clpt.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caudle KE, Thorn CF, Klein TE, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing. Clin Pharmacol Ther. 2013;94(6):640–645. doi: 10.1038/clpt.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Black JL, 3rd, Walker DL, O’Kane DJ, Harmandayan M. Frequency of undetected CYP2D6 hybrid genes in clinical samples: impact on phenotype prediction. Drug Metab Dispos. 2012;40(1):111–119. doi: 10.1124/dmd.111.040832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnston JJ, Rubinstein WS, Facio FM, et al. Secondary Variants in Individuals Undergoing Exome Sequencing: Screening of 572 Individuals Identifies High-Penetrance Mutations in Cancer-Susceptibility Genes. Am J Hum Genet. 2012 doi: 10.1016/j.ajhg.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonsalves SG, Ng D, Johnston JJ, et al. Using exome data to identify malignant hyperthermia susceptibility mutations. Anesthesiology. 2013;119(5):1043–1053. doi: 10.1097/ALN.0b013e3182a8a8e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng D, Johnston JJ, Teer JK, et al. Interpreting secondary cardiac disease variants in an exome cohort. Circ Cardiovasc Genet. 2013;6(4):337–346. doi: 10.1161/CIRCGENETICS.113.000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorschner MO, Amendola LM, Turner EH, et al. Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am J Hum Genet. 2013;93(4):631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patwardhan A, Harris J, Leng N, et al. Achieving high-sensitivity for clinical applications using augmented exome sequencing. Genome Med. 2015;7(1):71. doi: 10.1186/s13073-015-0197-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.