Abstract
The processing of amyloid precursor protein (APP) to amyloid beta (Aβ) is of great interest to the Alzheimer’s disease (AD) field. Decades of research define how APP is altered to form Aβ, and how Aβ generates oligomers, protofibrils, and fibrils. Numerous signaling pathways and changes in cell physiology are known to influence APP processing. Existing data additionally indicate a relationship exists between mitochondria, bioenergetics, and APP processing. Here, we review data that address whether mitochondrial function and bioenergetics modify APP processing and Aβ production.
Introduction
Aβ associates with and is believed by many to cause Alzheimer’s disease (AD), and thus is central to major therapeutic initiatives. Well before the advent of positron emission tomography (PET) – based amyloid imaging, Aβ plaques were known to deposit within brains of cognitively normal individuals (Bennett et al., 2006; Dickson et al., 1992; Price and Morris, 1999). PET-based Aβ imaging in living subjects advanced the field by enabling longitudinal measurements of Aβ plaque accumulation. These studies showed brain Aβ fibril deposition increases prior to the onset of cognitive decline, and profoundly slows during the clinically active stages (Burns and Swerdlow, 2013; Jack et al., 2013). Therefore, plaque deposition may precede cognitive decline by 1–2 decades and Aβ fibril accumulation essentially plateaus before overt clinical symptoms manifest. Why fibrillary Aβ accumulation begins, accelerates, and decelerates to the point that little additional accumulation occurs prior to symptom onset is unknown.
Studies depicting connections between mitochondrial dysfunction and Aβ suggest potential leads to these questions. Amyloid Precursor Protein (APP) and Aβ reportedly co-localize with mitochondria (Devi et al., 2006; Hansson Petersen et al., 2008). Aβ inhibits respiratory chain function, and Aβ toxicity appears reduced in cells that lack functional respiratory chains (reviewed in (Swerdlow, 2012)). Altering mitochondrial function also changes APP processing, and can increase or decrease the production of amyloidogenic derivatives (Canevari et al., 1999; Casley et al., 2002a; Gabuzda et al., 1994; Gasparini et al., 1997; Khan et al., 2000; Leuner et al., 2012; Onyango et al., 2010; Pereira et al., 1998; Webster et al., 1998). Here, we review evidence for the relationship between mitochondria, bioenergetics, and APP processing.
I. Amyloid Precursor Protein
In humans the APP gene is located on chromosome 21. Alternative splicing generates 8–11 APP protein isoforms of varying amino acid length (305, 639, 677, 695, 696, 714, 733, 746, 751,752, 770) (Matsui et al., 2007; Sandbrink et al., 1996). It is important to note APP splicing may vary between species (i.e. rodent versus human), and there may be additional unidentified splice variants. Of the currently known isoforms, APP 751 and 770 are expressed in glial cells, while APP 695 is the predominate form expressed in neurons (Matsui et al., 2007). APP is a type I transmembrane protein which is trafficked through both secretory and endocytic pathways.
Cellular localization of APP is dynamic. In the secretory pathway APP moves from the endoplasmic reticulum (ER) to the plasma membrane (Haass et al., 2012). During this process APP is post-translationally modified (phosphorylation, tyrosine sulphation, and N- or O- linked glycosylation) (Bhattacharyya et al., 2013; Lee et al., 2003; Pahlsson et al., 1992). APP cleaved at the plasma membrane is internalized (this requires a YENPTY motif), where it is endocytosed by either endosomes or lysosomes (Lai et al., 1995). Data from overexpression studies suggest the majority of APP is trafficked to the trans-Golgi network, while only 10% arrives at the plasma membrane (Haass et al., 2012). As mentioned above, the YENPTY domain is required for APP endocytosis. Mutations in this domain inhibit amyloidogenic APP processing (La Rosa et al., 2015; Perez et al., 1999). This domain contains a cytosolic adaptor in which proteins that contain a phosphotyrosine binding domain bind to APP. These adaptor proteins include but are not limited to, Fe65, Dab1, Mint 1, Mint 2, Mint 3, and JNK (Haass et al., 2012; Thinakaran and Koo, 2008).
APP Processing Pathways
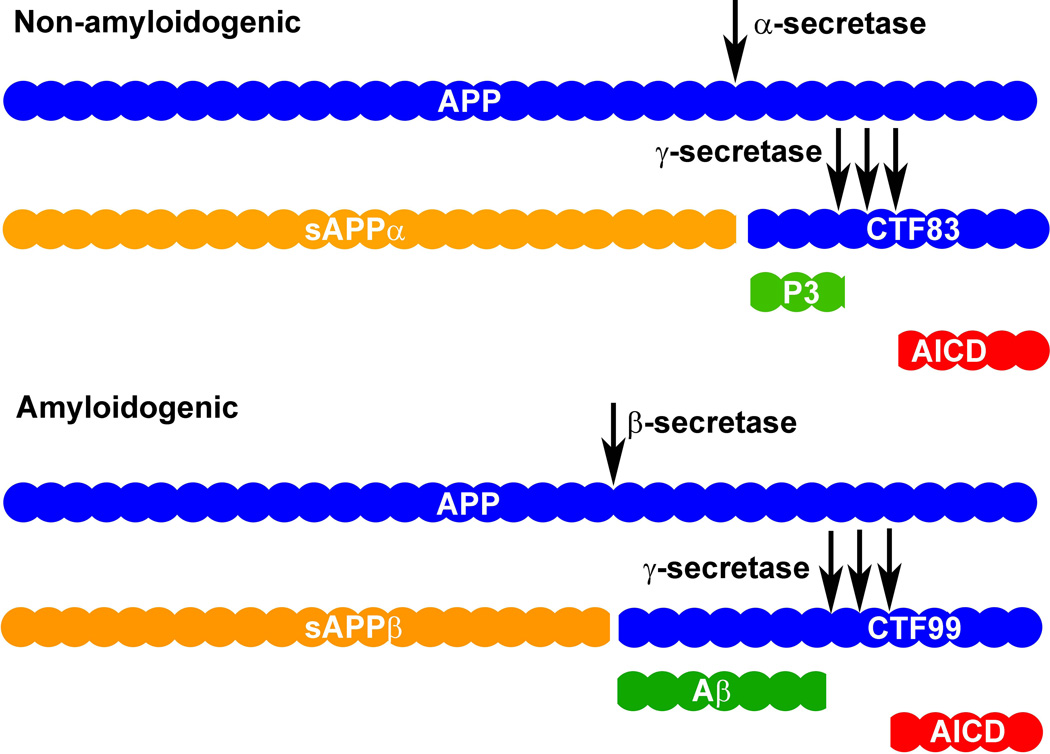
APP processing is dependent on secretase enzymes, which yield products that are secreted into the extracellular space or which remain within or associated with the cell. APP processing is generally divided into two pathways, non-amyloidogenic and amyloidogenic. The non-amyloidogenic pathway begins with α-secretase-mediated cleavage of APP at amino acid 687 (in the APP 770 isoform) which releases the ectodomain, soluble APP α (sAPPα), into the extracellular space. As a result, a C-terminal fragment of APP that is 83 amino acids in length (CTF83) remains embedded in the plasma membrane. Cleavage of CTF83 by γ-secretase releases a small p3 fragment into the extracellular space and the APP intracellular domain (AICD) into the cytoplasm (Figure 1). Conversely, amyloidogenic processing begins with β-secretase-mediated APP cleavage at amino acid 671 (in the APP 770 isoform). As a result, a smaller ectodomain, soluble APP β (sAPPβ), is released into the extracellular space. A larger APP C-terminal fragment that is 99 amino acids in length (CTF99) remains embedded in the plasma membrane. Finally, cleavage of CTF99 by γ-secretase releases Aβ into the extracellular space and the AICD into the cytoplasm (Figure 1). Numerous in depth reviews of APP processing pathways are available (Haass et al., 2012; Zhang et al., 2011).
Figure 1. APP Processing Pathways.
The non-amyloidogenic pathway begins with cleavage of full-length APP (membrane bound) by α-secretase, yielding sAPPα (ectodomain; released to extracellular space) and CTF83 (membrane bound). CTF83 is then cleaved by γ-secretase (three consecutive cleavage sites) to generate p3 (released to extracellular space) and AICD (released to cytoplasm). The amyloidogenic pathway begins with cleavage of full-length APP (membrane bound) by β-secretase, yielding sAPPβ (ectodomain; released to extracellular space) and CTF99 (membrane bound). CTF99 is then cleaved by γ-secretase (three consecutive cleavage sites) to generate Aβ (released to extracellular space) and AICD (released to cytoplasm).
The α-secretase is a membrane bound zinc metalloproteinase consisting of the A Disintegrin And Metalloprotease (ADAM) family members ADAM9, ADAM10, ADAM19, and Tumor Necrosis Factor Converting Enzyme (TACE)/ADAM17 (Asai et al., 2003; Buxbaum et al., 1998; Lammich et al., 1999). ADAM10 appears to be the dominant α-secretase in neurons (Kuhn et al., 2010). Other substrates for α-secretase include NOTCH receptors, tumor necrosis factor α (TNFα), epidermal growth factor (EGF) receptor ligands, cadherins, and interleukin 6 (IL6) receptor (Haass et al., 2012). Cleavage of APP by α-secretase is dependent on both the α helical conformation of the cleavage site and the distance between the bond undergoing hydrolysis and the plasma membrane (Sisodia, 1992). Details about α-secretase function, structure, and regulation have been reviewed elsewhere (Allinson et al., 2003; Lichtenthaler, 2011)
The β-secretase 1 (BACE1) is required for APP cleavage and is rate limiting for the generation of Aβ from APP. Knockout of BACE1, but not BACE2, completely blocks the generation of Aβ (Cai et al., 2001; Haass et al., 2012). Beyond APP BACE1 also cleaves myelin, voltage dependent sodium channels, platelet selecting glycoprotein ligand 1, type II α 2,6 sialytransferase, and interleukin like receptor type II (Haass et al., 2012). BACE1 is largely membrane bound with highest activity in an acidic pH environment (Haass et al., 1993a; Haass et al., 1993b; Selkoe et al., 1996). BACE1 structure, function, and cell trafficking are reviewed in depth elsewhere (Cole and Vassar, 2007).
The γ-secretase, an aspartyl protease, is comprised of four subunits. These include Presenilin 1 (PS1), Presenilin 2 (PS2), nicastrin, anterior pharynx defective (APH-1), and PS enhancer (PEN2) (Francis et al., 2002). PS1 and PS2 form the catalytic domain of γ-secretase while APH-1 may act as a stabilizer, PEN2 acts as a regulator/enhancer of activity, and nicastrin serves as a substrate receptor (Bolduc et al., 2016; Dries and Yu, 2008; Holmes et al., 2014). The cleavage event catalyzed by γ-secretase does not occur at a single site as there are three cleavage sites on APP separated by three amino acids; ε, γ, and δ. γ-secretase assembly, trafficking, and substrates have been reviewed elsewhere (De Strooper et al., 2012; Dries and Yu, 2008; Haapasalo and Kovacs, 2011).
The location of APP processing is still unclear. Data support APP processing at the plasma membrane, ER, trans-Golgi network, endosome vesicles, mitochondria, and lipid rafts (Choy et al., 2012; Devi and Ohno, 2012; Di Paolo and Kim, 2011; Haass et al., 2012; Hartmann et al., 1997; Hicks et al., 2012; Placido et al., 2014). Further uncertainty exists in the field regarding the existence of intracellular Aβ (Wirths et al., 2012), as some have argued this could represent an artifact of overexpressing a mutant human transgene in mice (Oakley et al., 2006; Youmans et al., 2012). It is important to note, though, that studies depicting intracellular Aβ have also been performed in post-mortem human brains (Aho et al., 2010; Glabe, 2001; Gyure et al., 2001; Hashimoto et al., 2010; Takahashi et al., 2002).
The hypothesis that amyloidogenic APP processing occurs within endosomes is supported by discrete yet convincing studies (Koo and Squazzo, 1994). For example, endosomes contain the optimum pH for BACE1 activity, APP and BACE1 interact within the endosomal pathway, and APP CTFs are processed in endosomes. Furthermore, inhibition of endosomal signaling prevents amyloidogenic APP processing (Haass et al., 1993b; Haass et al., 2012; Selkoe et al., 1996). APP trafficking/recruitment to endosomes of the trans-Golgi network are both sites at which Aβ has been shown to be generated (Choy et al., 2012; Edgar et al., 2015; Morel et al., 2013).
Lipid rafts bring APP and BACE1 together, providing an interaction interface for amyloidogenic APP processing. Lowering cholesterol levels in N2a neuroblastoma cells reduces Aβ production. It is hypothesized that APP within lipid rafts undergoes amyloidogenic processing, while APP not found within lipid rafts is subjected to non-amyloidogenic processing (Ehehalt et al., 2003). γ-secretase may also exist within lipid rafts (Urano et al., 2005). Overall, cholesterol and lipid metabolism may play a pivotal role in determining where and how APP is processed (Di Paolo and Kim, 2011; Hicks et al., 2012).
APP function
Overall, the function of APP is unclear. It has been assumed to relate to several processes, including cell adhesion and trophic support. APP has been implicated in cell adhesion due to its interaction with laminin and collagen and it co-localizes with integrins, specifically within neuronal axons that are sites of cell adhesion (Yamazaki et al., 1997; Young-Pearse et al., 2008). APP was also found to support cell growth. In fibroblasts, genetic deletion of APP led to slow cell growth and division. This consequence was rescued by restoring sAPPα levels (Saitoh et al., 1989). Furthermore, sAPPα may stimulate the differentiation of neuronal stem cells into an astrocytic phenotype (Caille et al., 2004). sAPPβ may have a similar function to sAPPα, but may be less potent (Chasseigneaux et al., 2011). Data also support an autocrine/paracrine function for APP. The structure of APP may also provide some insight into its possible function. APP contains metal binding domains (for copper and zinc), extracellular matrix components (such as heparin, collagen, and laminin), and a protease inhibitor domain (Kunitz, only in isoforms 751 and 770) (Dawkins and Small, 2014). Comparisons of data depicting functions of full length APP, sAPPα, and sAPPβ can be found elsewhere (Chasseigneaux and Allinquant, 2012).
Independent functions for smaller APP processing fragments have also been reported. While the APP ectodomains may be pivotal for cell growth and adhesion, the intracellular domains may play different roles. For example the AICD, when bound to its cofactor, Fe65, can regulate gene expression (Muller et al., 2008). Most studies focus on how and why Aβ forms aggregates, however limited studies suggest Aβ may also have a physiological function. Inhibition of Aβ production (which also limits sAPPβ production) induces neuronal cell death (Pearson and Peers, 2006; Plant et al., 2003). Further, data support a role for Aβ as an antimicrobial protein that protects distinct organisms (worms and mice) from microbial infections (Kumar et al., 2016; Soscia et al., 2010).
Substantial data support a role for APP and its fragments (including the AICD and Aβ) in modulating mitochondrial function. As we will discuss below, studies completed over several decades indicate a relationship exists between APP processing, APP (and its fragments), mitochondria, and bioenergetics. While Aβ and APP influence mitochondrial function and bioenergetic pathways, the converse is also true; mitochondrial function and bioenergetics modulate APP processing and trafficking pathways.
II. APP, Aβ, and Mitochondria
APP is known to localize to mitochondria, where it appears to impede protein import. In particular, APP arrests and forms a “super-complex” with the translocase of the outer mitochondrial membrane 40 (TOMM40) and the translocase of the inner mitochondrial membrane 23 (TIM23). Data suggest this transmembrane arrest of APP inhibits the import of nuclear-encoded mitochondrial proteins (Anandatheerthavarada et al., 2003; Devi et al., 2006). Observations of this interaction have been observed both in vivo and in vitro. This transmembrane arrest of APP induces mitochondrial dysfunction, including disruption of mitochondrial ATP synthesis, mitochondrial membrane potential, and reductions in cytochrome oxidase (COX) activity (Anandatheerthavarada et al., 2003). APP localization to mitochondria is dependent on three positively charged amino acid residues, as mutation of these residues to neutral amino acids prevents mitochondrial APP accumulation (Anandatheerthavarada et al., 2003).
More recent evidence suggests a 12 amino acid stretch located within the C-terminal domain of APP is important for its mitochondrial localization. Deletion of this 12 amino acid stretch (which contains a transmembrane domain) decreased APP mitochondrial localization, while also inducing mitochondrial dysfunction. In vitro, this APP deletion mutant led to decreased cell viability, increased ROS, decreased ATP, and depolarized mitochondrial membrane potential (Wang et al., 2016). More studies are warranted to understand the consequences of mitochondrial localized APP.
In addition to mitochondrial APP localization, mitochondria also reportedly contain a functional γ-secretase. The mitochondrial γ-secretase cleaves the CTF83 of APP (of which has been shown to associate with the mitochondrial outer membrane-in addition to full-length APP protein). The topology of the CTF83 of APP is likely to generate the AICD and Aβ within the mitochondria (Pavlov et al., 2011). This is important because Aβ is found within mitochondria where it interacts with mitochondrial proteins and alters mitochondrial function (Du et al., 2008; Lustbader et al., 2004). Data support the accumulation of APP C-terminal fragments and full-length APP within mitochondria, particularly within the inner membrane and matrix of 5X Familial Alzheimer’s disease (5XFAD; mice expressing 5 human transgenes including 3 mutations in APP and 2 mutations in PS1) mice. Heterozygous genetic depletion of BACE1 prevented the mitochondrial accumulation of both CTF99 and full length APP protein in an age-dependent manner (i.e. at 6 months of age, but not 12 months of age). This partial deletion of BACE1 also rescued mitochondrial dysfunction within this model as illustrated by reduced oxidative damage and preserved complex II activity (Devi and Ohno, 2012).
Mitochondrial-localized APP is also cleaved by the serine protease HtrA2. This cleavage event generates a C-terminal fragment that is 161 amino acids in length (CTF161). CTF161 was found accumulated within the cytosol, under physiological conditions in vitro and in vivo. HtrA2 processing of mitochondrial-localized APP is hypothesized to prevent the accumulation of full-length APP and downstream mitochondrial dysfunction (Park et al., 2006).
Overall, the mechanism by which Aβ accumulates within mitochondria is hypothesized to occur via several plausible mechanisms. These include Aβ/ABAD interaction, local APP processing within mitochondria, or aberrant intracellular Aβ accumulation due to increased Aβ production or failed Aβ degradation. The effect of Aβ on mitochondrial function are discussed below.
Aβ and Mitochondria
In numerous model systems Aβ impairs mitochondrial function. In neuroblastoma cells APP overexpression lead to elevated Aβ1–40 with reductions in cellular respiration, ATP levels, and COX activity; complex III activity, though, was elevated (Rhein et al., 2009). In PC12 cells, exogenous Aβ depolarizes the mitochondrial membrane potential and decreases activities of the mitochondrial electron transport chain complexes I, III, and IV while also reducing oxygen consumption (Pereira et al., 1998). Shorter fragments, such as Aβ25–35, reduce cellular ATP production, levels of antioxidants (glutathione, GSH), mitochondrial membrane potential, and electron transport chain activities (CI-IV) in primary cortical neurons (Casley et al., 2002b). However, in isolated rat brain mitochondria Aβ25–35 reduced the activity of COX but did not affect complex I, II, or III activities (Canevari et al., 1999; Casley et al., 2002a). Aβ25–35 impairs anterograde transport of mitochondria to synapses in mouse hippocampal neurons (Calkins and Reddy, 2011). Aβ may alter mitochondrial dynamics by interacting with a mitochondrial fission protein, dynamin related protein 1 (DRP1) (Manczak et al., 2011). Both oligomeric and monomeric forms of Aβ were shown to bind to DRP1. Additional further mechanisms have also been suggested.
In transgenic mice and brains from AD subjects, Aβ interacts with Aβ binding alcohol dehydrogenase (ABAD), an interaction which inhibits the binding of NAD+ (Lustbader et al., 2004). Neurons from mice transgenic for mutant APP and wild-type ABAD display elevated levels of apoptosis and reactive oxygen species (ROS) production, with reduced COX activity. Whole brain preparations from these mice revealed decreased glucose utilization, lower COX activity, and reduced ATP levels (Takuma et al., 2005). Inhibiting the interaction between ABAD and Aβ improves mitochondrial function and reduces Aβ accumulation (Yao et al., 2011).
Aβ toxicity appears in some studies to be mediated through its effects on the mitochondrial respiratory chain. In N2a neuroblastoma cells Aβ25–35 increased caspase activation, ROS production, and cytochrome c release. However, in ρ0 cells that lack a functional respiratory chain due to an absence of mitochondrial DNA (mtDNA), Aβ25–35 failed to induce ROS production, caspase activation, or cytochrome c release (Cardoso et al., 2002). This indicates the damaging effects of Aβ work through mitochondria, particularly through direct or indirect interactions with the respiratory chain.
Other APP fragments, including the AICD, also alter mitochondrial function. For example, overexpression of the AICD and its Fe65 cofactor in neuroblastoma cells decreased mitochondrial membrane potential, ATP levels, and superoxide production. The AICD/Fe65 complex is known to induce genes which disrupt actin dynamics. Overexpression of the actin binding protein, transgelin, recapitulated these results; Transgelin is a downstream target gene of AICD/Fe65 (Ward et al., 2010).
Overall APP and its fragments, and in particular Aβ, interact with, localize to, and alter the function of mitochondria. Studies showing these effects have utilized various in vitro and in vivo model systems. We will next discuss how mitochondria and bioenergetics influence APP processing and sorting pathways.
III. APP Processing and Bioenergetics
In vitro studies
The first studies describing a bioenergetic influence on the fate of APP processing were reported in the 1990s. Initial studies in which COS cells were exposed to a COX inhibitor (sodium azide) and a mitochondrial un-coupler (CCCP) noted the formation of an 11.5 kDa APP CTF that contained the Aβ motif (possibly the CTF99 fragment). Bioenergetic perturbations produced by these mitochondrial toxins also lead to APP accumulation in the Golgi. The production of this CTF was also increased when protein transport in the secretory pathway was inhibited (Gabuzda et al., 1994). Further studies in COS cells showed glucose deprivation, in combination with a glycolysis inhibitor (2-Deoxy-D-glucose), reduced sAPPα levels. Inhibition of COX by sodium azide also decreased sAPPα levels. These effects were reversed by an antioxidant, GSH (Gasparini et al., 1997).
The effects observed by Gasparini et. al. were recapitulated in a separate study using both COS and PC12 cells. It was further found that inhibition of glycolysis by 2-Deoxy-D-glucose resulted in reduced APP glycosylation. The changes observed could be inhibited by phorbol 12-myristate 13-acetate, a protein kinase C (PKC) activator, and increased co-localization was observed between APP and protein phosphatase 1 α (PP1α). This study suggests altered bioenergetics may influence signal transduction pathways which regulate APP processing (Henriques et al., 2005). Other studies have provided evidence that upon glycolysis and COX inhibition (i.e. treatment of cells with 2-Deoxy-D-glucose and sodium azide), APP accumulates in the ER and Golgi, a process also dependent on phosphorylation and signal transduction (Domingues et al., 2007).
The influence of bioenergetics on APP processing has been depicted in various cell types. In PC12 cells, secretion of APP-like peptides was reduced by glycolysis and/or mitochondrial respiration inhibition (either through glucose starvation, 2-Deoxy-D-glucose treatment, or ATP synthase inhibition using oligomycin) (Webster et al., 1998). In fibroblasts derived from control and sporadic AD subjects, glucose deprivation (i.e. reduced glycolysis flux) lowered sAPP production. COX inhibition induced by sodium azide, in addition to glucose deprivation, further reduced sAPP production in sporadic AD fibroblasts. Unlike in the prior studies, antioxidants did not abolish these observations (Gasparini et al., 1999).
In astrocytes, glycolysis flux influences APP processing pathways. In human fetal astrocytes inhibition of the glycolysis regulating enzyme 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase (PFKFB3) through genetic or pharmacological approaches lead to increased Aβ accumulation and toxicity. It was further found that PFKFB3 is upregulated in astrocytes from 12 month old TgCRND8 AD mice, another transgenic AD mouse model (Fu et al., 2015).
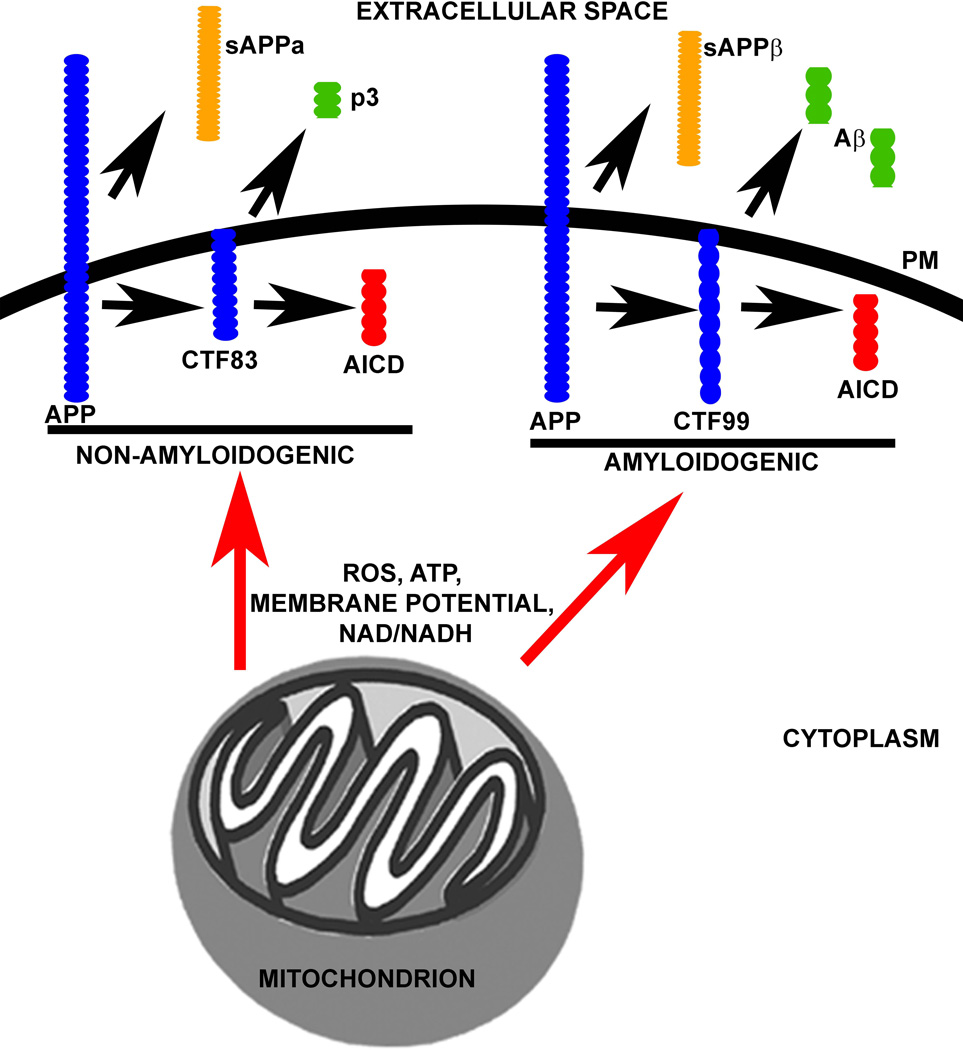
It is clear from these studies that glycolysis flux and mitochondrial function (particularly respiration) influence APP processing pathways. It is important to note, however, that changes in glycolysis flux also affect mitochondrial respiration, and vice versa. For example, in SH-SY5Y neuroblastoma cells, inhibition of glycolysis flux using either 2-Deoxy-D-glucose, glucose deprivation, or iodoacetate increases mitochondrial respiratory flux (Swerdlow et al., 2013). An inverse relationship can thus exist between glycolysis and mitochondrial respiratory flux. Changes in the flux of these bioenergetic pathways will further alter other bioenergetic pathways and intermediates, mitochondrial membrane potential, ROS production rates, ATP/ADP, and ROS scavengers (i.e. NAD+/NADH, NADP+/NADPH, GSH/GSSG) (Graham et al., 2012; Jiang et al., 2013; Korenic et al., 2014; Russell et al., 2002; Shutt et al., 2010; Swerdlow et al., 2013). Such changes influence the fate of APP processing (Figure 2).
Figure 2. Mitochondrial function and bioenergetic intermediates influence APP Processing Pathways.
Mitochondrial function and bioenergetic pathways (probably through intermediates, such as ROS, ATP, membrane potential, and NAD/NADH for example) can increase or decrease amyloidogenic and non-amyloidogenic APP processing. PM=plasma membrane.
BACE1 transcription is influenced by the same pathways that regulate mitochondrial biogenesis. Peroxisome proliferator activated receptor-γ (PPARγ) regulates the transcription of BACE1 and peroxisome proliferator activated receptor coactivator 1-alpha (PGC1α). PGC1α modulates mitochondrial biogenesis pathways (Scarpulla, 2011). Overexpression of PGC1α in N2a neuroblastoma cells decreased levels of Aβ secretion, while increasing sAPPα. Overexpression of PGC1β also decreased Aβ secretion. The authors attributed these findings to a decrease in BACE1 mRNA and protein expression, and to a decrease in BACE1 activity. Upon genetic ablation of PGC1α expression, BACE1 expression was elevated (Katsouri et al., 2011).
Mitochondrial DNA from sporadic AD subjects also influences APP processing pathways. In cytoplasmic hybrid (cybrid) cell lines in which mtDNA from sporadic AD subjects is transferred into an identical nuclear background, changes in mitochondrial function were observed. In these cells the Aβ1–40/Aβ1–42 ratio was increased with elevated Congo red reactive Aβ deposits as compared to cybrids prepared from non-demented, age-matched subjects (Khan et al., 2000). In a separate study, cybrid cells generated from sporadic AD donors displayed elevated ROS production, reduced ATP production, and decreased COX activity. Upon exposure of these AD cybrid cells line to exogenous Aβ1–40 mitochondrial function further declined to a greater extent than it did in cybrids generated from non-demented, age-matched subjects. Mitochondrial membrane potential depolarization and cell death pathway activation was also observed (Cardoso et al., 2004b).
In vivo studies
In AD animal models bioenergetic manipulations alter APP processing. For example, in Tg2576 transgenic mice overexpression of a mitochondrial targeted catalase, which contributed to the conversion of hydrogen peroxide to water and oxygen, lead to a reduction in brain APP protein levels and reduced levels of BACE1, CTF99, Aβ1–40, and Aβ1–42. Conversely, this increased CTF83 and sAPPα levels. Beyond apparent reductions in amyloidogenic APP processing, these mice also had reduced Aβ deposits (Mao et al., 2012).
In Tg19959 AD mice overexpression of manganese superoxide dismutase (MnSOD, also called SOD2), a mitochondrial antioxidant enzyme that converts superoxide into hydrogen peroxide, reduced total levels of oxidized proteins (protein carbonyls) and cortical plaque burden. No change was observed in soluble or insoluble Aβ oligomer levels (Dumont et al., 2009). A different effect was found in Tg19959 AD mice with heterozygous knockout of MnSOD, as these mice displayed increased Aβ levels and plaques (Li et al., 2004). Together, these in vivo studies support a role for mitochondrial-derived ROS in driving amyloidogenic APP processing.
While in vitro studies found PGC1α overexpression decreased Aβ secretion, in vivo studies report opposite outcomes. PGC1α overexpression in the Tg19959 AD mouse model increased Aβ and tau accumulation. Changes in Aβ included a larger Aβ plaque area, an increased Aβ plaque number, an increased Aβ1–42/Aβ1–40 ratio, and increased prefibrillar Aβ. As part of a separate yet interesting finding, these mice also displayed increased tau phosphorylation. Finally, these mice also showed decreased citrate synthase, complex I, and complex II activities (Dumont et al., 2014).
In another study mtDNA mutator mice, which harbor a D257A polymerase G mutation, were crossed with APP/Ld mice. D257A mice accumulate mtDNA mutations at an elevated rate, including 3–5 fold more mtDNA point mutations, and increased levels of short-linearized mtDNA fragments (Trifunovic et al., 2004). D257A/APP/Ld mice displayed increased Aβ1–42 plaques and levels, but no change in BACE1, PS1, or CTF99/CTF83 fragments. In this study increased Aβ accumulation was attributed to decreased clearance by insulin degrading enzyme (IDE) (Kukreja et al., 2014).
An additional study established mitochondrial congenic mouse lines by crossing females of different strains to male APP transgenic mice. Strains with varying mtDNA on a consistent nuclear background were specifically generated by crossing female FVB/N, AKR/J, or NOD/LtJ mice to male C57BL/6 APP transgenic mice and then back-crossing the female progeny to the male C57Bl/6 mice for a minimum of ten generations. The resulting unique mtDNA strains displayed changes in Aβ burden, including increased plaque numbers and size. The authors also assessed a variety of bioenergetic-related parameters, and concluded high levels of ATP and ATP release increased microglial activation and phagocytosis, which resulted in reduced Aβ plaque numbers and size (Scheffler et al., 2012).
In HEK293 and SH-SY5Y neuroblastoma cells, inhibition of complex I and III by rotenone and antimycin A, respectively, increased the production of Aβ and also ROS. These effects were mitigated by the addition of an antioxidant. Further, mice transgenic for mutant APP (KM670/671NL and V717I) and treated with a complex I inhibitor or crossed with complex I deficient mice (Ndufs4 knockout mice) displayed elevated Aβ levels (Leuner et al., 2012).
In AD transgenic mice reductions in COX activity due to reduced COX holoenzyme limits Aβ production. In a seminal experiment Cox10 knockout mice were crossed with an AD mouse model transgenic for mutant APP and PS1. Cox10 knock-out reduced the ability to generate the COX holoenzyme (Diaz et al., 2006). In the Cox10 knockout/AD mouse model cross, Aβ plaque accumulation, Aβ1–42, and β-secretase activity were reduced (Fukui et al., 2007). Additional studies crossed AD transgenic mice (expressing mutant APP and PS1) with mice expressing a mitochondrial-targeted endonuclease (PstI). This cross resulted in lower mtDNA levels, reduced Aβ plaque burden, and lower Aβ1–42 levels (Pinto et al., 2013).
Under in vivo conditions accumulation of high amounts of mtDNA mutations and alterations in mitochondrial ROS production, as well as reductions in COX activity, complex I activity, or mtDNA levels change the function of APP processing pathways. These alterations may also influence tau phosphorylation and accumulation. Whether the genetic and/or chemical manipulations used in these AD mouse models primarily affect bioenergetics in neurons, glial cells, or both remains undetermined. Further, how bioenergetic interactions between neurons and glial cells influence APP processing pathways has not been addressed. However, based on in vitro data, bioenergetics in both neuronal cells and primary glial cells can alter APP processing pathways.
Conclusions
Mitochondrial function and bioenergetics are perturbed in AD subjects; these changes are not brain-limited. Brain glucose metabolism is reduced in AD subjects (De Santi et al., 2001; Rapoport et al., 1991; Swerdlow et al., 1994). Reduced COX activity in AD subject platelets, brains, and fibroblasts has been reported (Cardoso et al., 2004a; Mancuso et al., 2003; Swerdlow, 2012; Wilkins et al., 2014). Spectral analysis of the COX enzyme from AD subject brains report the enzyme is kinetically altered and therefore structured differently than in control subject brains (Parker and Parks, 1995). Electron microscopy and molecular studies of AD brains also show that despite an overall reduction in the number of intact mitochondria, a heterogeneous picture exists in which amounts of mitochondrial components, such as mtDNA, can vary from markedly reduced (in abnormal appearing neurons) to increased (in relatively healthy-appearing neurons) (Brown et al., 2001; Hirai et al., 2001; Swerdlow, 2012). Moreover, the amount that a component is necessarily increased or decreased can depend on what intracellular compartments are included in the analysis, as autophagosomes that contain the contents of degrading mitochondrial material may contain large amounts of mtDNA and mitochondrial protein (Hirai et al., 2001; Swerdlow, 2012). Overall, these studies support the view that mitochondrial mass varies during the course of AD, with mitochondrial mass increasing in relatively preserved neurons, and decreasing in relatively more damaged neurons.
Consistent with this, brain mtDNA content reportedly increases during aging in cognitively intact individuals (Barrientos et al., 1997; Hirai et al., 2001). This is consistent with the possibility that increased Aβ production may occur in the run-up to clinical AD while neurons are still able to compensate for age-related declines in mitochondrial function, and decline later on in the symptomatic stages as cells can no longer compensate for impaired aerobic function and begin to actually downregulate their aerobic infrastructure.
It is important to note APP localizes to synapses and increased synaptic activity associates with increased processing of APP to Aβ (Brody et al., 2008; Cirrito et al., 2005; Groemer et al., 2011). This may partly arise from a synaptic activity-related retargeting of APP from the plasma membrane to intracellular structures that are more likely to produce Aβ, but based on extensive existing literature it seems likely that another synaptic activity-associated physiologic parameter, aerobic metabolism, also contributes to variations in Aβ production (Cavallucci et al., 2013; Du et al., 2010; Reddy et al., 2012).
In vivo and in vitro studies support a major role of mitochondria and bioenergetics in the fate of APP processing and cellular trafficking. Future studies are warranted to elucidate mechanisms surrounding the relationships between APP (and its fragments), mitochondria, and bioenergetic pathways.
A relationship between bioenergetics, mitochondria, and APP exists.
APP and amyloid beta alter bioenergetic pathways and mitochondrial function.
Bioenergetics and mitochondrial function may determine how APP is processed.
Acknowledgments
This project was supported by the University of Kansas Alzheimer’s Disease Center (P30 AG035982), the Frank and Evangeline Thompson Alzheimer’s Treatment Program fund, the Kansas IDeA Network for Biomedical Research Excellence (KINBRE, P20GM103418), the University of Kansas Medical Center Biomedical Research Training Program, and a Mabel Woodyard Fellowship award.
Abbreviations
- ABAD
Aβ binding alcohol dehydrogenase
- AD
Alzheimer’s disease
- ADAM
A Disintegrin and metalloproteinase
- AICD
APP intracellular domain
- APH1
anterior pharynxdefective 1
- APP
amyloid precursor protein
- ATP
adenosine triphosphate
- Aβ
amyloid beta
- BACE1
β-secretase 1
- BACE2
β secretase 2
- CCCP
Carbonyl cyanide m-chlorophenyl hydrazine
- COX
cytochrome oxidase or complex IV
- CTF
C-terminal fragment
- CTF83
C-terminal fragment 83
- CTF99
C-terminal fragment 99
- cybrid
cytoplasmic hybrid
- DRP1
dynamin related protein 1
- EGF
epidermal growth factor
- ER
endoplasmic reticulum
- GSH
glutathione
- IDE
insulin degrading enzyme
- IL6
interleukin 6
- MnSOD/SOD2
manganese superoxide dismutase
- mtDNA
mitochondrial DNA
- PEN2
Presenilin enhancer 2
- PET
positron emission tomography
- PFKFB3
6-phosphofructo-2-kinase-fructose-2,6-biphosphatase 3
- PGC1α
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- PGC1β
Peroxisome proliferator-activated receptor gamma coactivator 1-beta
- PKC
protein kinase C
- PPARγ
Peroxisome proliferator-activated receptor gamma
- PP1α
protein phosphatase 1 α
- PS1
Presenilin 1
- PS2
Presenilin 2
- ROS
reactive oxygen species
- sAPPα
soluble APP α
- sAPPβ
soluble APP β
- TACE
tumor necrosis factor-alpha converting enzyme
- TIM23
translocase of the inner mitochondrial membrane 23
- TNFα
tumor necrosis factor α
- TOMM40
translocase of the outer mitochondrial membrane 40
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
No conflicts of interest, financial or otherwise, are declared by the authors.
Bibliography
- Aho L, et al. Immunohistochemical visualization of amyloid-beta protein precursor and amyloid-beta in extra- and intracellular compartments in the human brain. J Alzheimers Dis. 2010;20:1015–1028. doi: 10.3233/JAD-2010-091681. [DOI] [PubMed] [Google Scholar]
- Allinson TM, et al. ADAMs family members as amyloid precursor protein alpha-secretases. J Neurosci Res. 2003;74:342–352. doi: 10.1002/jnr.10737. [DOI] [PubMed] [Google Scholar]
- Anandatheerthavarada HK, et al. Mitochondrial targeting and a novel transmembrane arrest of Alzheimer's amyloid precursor protein impairs mitochondrial function in neuronal cells. J Cell Biol. 2003;161:41–54. doi: 10.1083/jcb.200207030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai M, et al. Putative function of ADAM9, ADAM10, and ADAM17 as APP alpha-secretase. Biochem Biophys Res Commun. 2003;301:231–235. doi: 10.1016/s0006-291x(02)02999-6. [DOI] [PubMed] [Google Scholar]
- Barrientos A, et al. Reduced steady-state levels of mitochondrial RNA and increased mitochondrial DNA amount in human brain with aging. Brain Res Mol Brain Res. 1997;52:284–289. doi: 10.1016/s0169-328x(97)00278-7. [DOI] [PubMed] [Google Scholar]
- Bennett DA, et al. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology. 2006;66:1837–1844. doi: 10.1212/01.wnl.0000219668.47116.e6. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya R, Barren C, Kovacs DM. Palmitoylation of amyloid precursor protein regulates amyloidogenic processing in lipid rafts. J Neurosci. 2013;33:11169–11183. doi: 10.1523/JNEUROSCI.4704-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc DM, et al. Nicastrin functions to sterically hinder gamma-secretase-substrate interactions driven by substrate transmembrane domain. Proc Natl Acad Sci U S A. 2016;113:E509–E518. doi: 10.1073/pnas.1512952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody DL, et al. Amyloid-beta dynamics correlate with neurological status in the injured human brain. Science. 2008;321:1221–1224. doi: 10.1126/science.1161591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AM, et al. Correlation of the clinical severity of Alzheimer's disease with an aberration in mitochondrial DNA (mtDNA) J Mol Neurosci. 2001;16:41–48. doi: 10.1385/JMN:16:1:41. [DOI] [PubMed] [Google Scholar]
- Burns JM, Swerdlow RH. Backwaters and rapids on the amyloid river. Neurology. 2013;80:878–879. doi: 10.1212/WNL.0b013e3182840d14. [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, et al. Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the Alzheimer amyloid protein precursor. J Biol Chem. 1998;273:27765–27767. doi: 10.1074/jbc.273.43.27765. [DOI] [PubMed] [Google Scholar]
- Cai H, et al. BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat Neurosci. 2001;4:233–234. doi: 10.1038/85064. [DOI] [PubMed] [Google Scholar]
- Caille I, et al. Soluble form of amyloid precursor protein regulates proliferation of progenitors in the adult subventricular zone. Development. 2004;131:2173–2181. doi: 10.1242/dev.01103. [DOI] [PubMed] [Google Scholar]
- Calkins MJ, Reddy PH. Amyloid beta impairs mitochondrial anterograde transport and degenerates synapses in Alzheimer's disease neurons. Biochim Biophys Acta. 2011;1812:507–513. doi: 10.1016/j.bbadis.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canevari L, Clark JB, Bates TE. beta-Amyloid fragment 25–35 selectively decreases complex IV activity in isolated mitochondria. FEBS Lett. 1999;457:131–134. doi: 10.1016/s0014-5793(99)01028-5. [DOI] [PubMed] [Google Scholar]
- Cardoso S, Swerdlow RH, Oliveira CR. Induction of cytochrome c-mediated apoptosis by amyloid beta 25–35 requires functional mitochondria. Brain Res. 2002;931:117–125. doi: 10.1016/s0006-8993(02)02256-4. [DOI] [PubMed] [Google Scholar]
- Cardoso SM, et al. Cytochrome c oxidase is decreased in Alzheimer's disease platelets. Neurobiol Aging. 2004a;25:105–110. doi: 10.1016/s0197-4580(03)00033-2. [DOI] [PubMed] [Google Scholar]
- Cardoso SM, et al. Mitochondria dysfunction of Alzheimer's disease cybrids enhances Abeta toxicity. J Neurochem. 2004b;89:1417–1426. doi: 10.1111/j.1471-4159.2004.02438.x. [DOI] [PubMed] [Google Scholar]
- Casley CS, et al. Beta-amyloid inhibits integrated mitochondrial respiration and key enzyme activities. J Neurochem. 2002a;80:91–100. doi: 10.1046/j.0022-3042.2001.00681.x. [DOI] [PubMed] [Google Scholar]
- Casley CS, et al. Beta-amyloid fragment 25–35 causes mitochondrial dysfunction in primary cortical neurons. Neurobiol Dis. 2002b;10:258–267. doi: 10.1006/nbdi.2002.0516. [DOI] [PubMed] [Google Scholar]
- Cavallucci V, Ferraina C, D'Amelio M. Key role of mitochondria in Alzheimer's disease synaptic dysfunction. Curr Pharm Des. 2013;19:6440–6450. doi: 10.2174/1381612811319360005. [DOI] [PubMed] [Google Scholar]
- Chasseigneaux S, et al. Secreted amyloid precursor protein beta and secreted amyloid precursor protein alpha induce axon outgrowth in vitro through Egr1 signaling pathway. PLoS One. 2011;6:e16301. doi: 10.1371/journal.pone.0016301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasseigneaux S, Allinquant B. Functions of Abeta, sAPPalpha and sAPPbeta : similarities and differences. J Neurochem. 2012;120(Suppl 1):99–108. doi: 10.1111/j.1471-4159.2011.07584.x. [DOI] [PubMed] [Google Scholar]
- Choy RW, Cheng Z, Schekman R. Amyloid precursor protein (APP) traffics from the cell surface via endosomes for amyloid beta (Abeta) production in the trans-Golgi network. Proc Natl Acad Sci U S A. 2012;109:E2077–E2082. doi: 10.1073/pnas.1208635109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirrito JR, et al. Synaptic activity regulates interstitial fluid amyloid-beta levels in vivo. Neuron. 2005;48:913–922. doi: 10.1016/j.neuron.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Cole SL, Vassar R. The Alzheimer's disease beta-secretase enzyme, BACE1. Mol Neurodegener. 2007;2:22. doi: 10.1186/1750-1326-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins E, Small DH. Insights into the physiological function of the beta-amyloid precursor protein: beyond Alzheimer's disease. J Neurochem. 2014;129:756–769. doi: 10.1111/jnc.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santi S, et al. Hippocampal formation glucose metabolism and volume losses in MCI and AD. Neurobiol Aging. 2001;22:529–539. doi: 10.1016/s0197-4580(01)00230-5. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Iwatsubo T, Wolfe MS. Presenilins and gamma-secretase: structure, function, and role in Alzheimer Disease. Cold Spring Harb Perspect Med. 2012;2:a006304. doi: 10.1101/cshperspect.a006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi L, et al. Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer's disease brain is associated with mitochondrial dysfunction. J Neurosci. 2006;26:9057–9068. doi: 10.1523/JNEUROSCI.1469-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi L, Ohno M. Mitochondrial dysfunction and accumulation of the beta-secretase-cleaved C-terminal fragment of APP in Alzheimer's disease transgenic mice. Neurobiol Dis. 2012;45:417–424. doi: 10.1016/j.nbd.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, Kim TW. Linking lipids to Alzheimer's disease: cholesterol and beyond. Nat Rev Neurosci. 2011;12:284–296. doi: 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F, et al. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol Cell Biol. 2006;26:4872–4881. doi: 10.1128/MCB.01767-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson DW, et al. Identification of normal and pathological aging in prospectively studied nondemented elderly humans. Neurobiol Aging. 1992;13:179–189. doi: 10.1016/0197-4580(92)90027-u. [DOI] [PubMed] [Google Scholar]
- Domingues SC, et al. Altered subcellular distribution of the Alzheimer's amyloid precursor protein under stress conditions. Ann N Y Acad Sci. 2007;1096:184–195. doi: 10.1196/annals.1397.085. [DOI] [PubMed] [Google Scholar]
- Dries DR, Yu G. Assembly, maturation, and trafficking of the gamma-secretase complex in Alzheimer's disease. Curr Alzheimer Res. 2008;5:132–146. doi: 10.2174/156720508783954695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, et al. Cyclophilin D deficiency attenuates mitochondrial and neuronal perturbation and ameliorates learning and memory in Alzheimer's disease. Nat Med. 2008;14:1097–1105. doi: 10.1038/nm.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, et al. Early deficits in synaptic mitochondria in an Alzheimer's disease mouse model. Proc Natl Acad Sci U S A. 2010;107:18670–18675. doi: 10.1073/pnas.1006586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M, et al. Reduction of oxidative stress, amyloid deposition, and memory deficit by manganese superoxide dismutase overexpression in a transgenic mouse model of Alzheimer's disease. FASEB J. 2009;23:2459–2466. doi: 10.1096/fj.09-132928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M, et al. PGC-1alpha overexpression exacerbates beta-amyloid and tau deposition in a transgenic mouse model of Alzheimer's disease. FASEB J. 2014;28:1745–1755. doi: 10.1096/fj.13-236331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar JR, et al. ESCRTs regulate amyloid precursor protein sorting in multivesicular bodies and intracellular amyloid-beta accumulation. J Cell Sci. 2015;128:2520–2528. doi: 10.1242/jcs.170233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehehalt R, et al. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, et al. aph-1 and pen-2 are required for Notch pathway signaling, gamma-secretase cleavage of betaAPP, and presenilin protein accumulation. Dev Cell. 2002;3:85–97. doi: 10.1016/s1534-5807(02)00189-2. [DOI] [PubMed] [Google Scholar]
- Fu W, et al. Bioenergetic mechanisms in astrocytes may contribute to amyloid plaque deposition and toxicity. J Biol Chem. 2015;290:12504–12513. doi: 10.1074/jbc.M114.618157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui H, et al. Cytochrome c oxidase deficiency in neurons decreases both oxidative stress and amyloid formation in a mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2007;104:14163–14168. doi: 10.1073/pnas.0705738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabuzda D, et al. Inhibition of energy metabolism alters the processing of amyloid precursor protein and induces a potentially amyloidogenic derivative. J Biol Chem. 1994;269:13623–13628. [PubMed] [Google Scholar]
- Gasparini L, et al. Effect of energy shortage and oxidative stress on amyloid precursor protein metabolism in COS cells. Neurosci Lett. 1997;231:113–117. doi: 10.1016/s0304-3940(97)00536-3. [DOI] [PubMed] [Google Scholar]
- Gasparini L, et al. Energy metabolism inhibition impairs amyloid precursor protein secretion from Alzheimer's fibroblasts. Neurosci Lett. 1999;263:197–200. doi: 10.1016/s0304-3940(99)00155-x. [DOI] [PubMed] [Google Scholar]
- Glabe C. Intracellular mechanisms of amyloid accumulation and pathogenesis in Alzheimer's disease. J Mol Neurosci. 2001;17:137–145. doi: 10.1385/JMN:17:2:137. [DOI] [PubMed] [Google Scholar]
- Graham NA, et al. Glucose deprivation activates a metabolic and signaling amplification loop leading to cell death. Mol Syst Biol. 2012;8:589. doi: 10.1038/msb.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groemer TW, et al. Amyloid precursor protein is trafficked and secreted via synaptic vesicles. PLoS One. 2011;6:e18754. doi: 10.1371/journal.pone.0018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyure KA, et al. Intraneuronal abeta-amyloid precedes development of amyloid plaques in Down syndrome. Arch Pathol Lab Med. 2001;125:489–492. doi: 10.5858/2001-125-0489-IAAPDO. [DOI] [PubMed] [Google Scholar]
- Haapasalo A, Kovacs DM. The many substrates of presenilin/gamma-secretase. J Alzheimers Dis. 2011;25:3–28. doi: 10.3233/JAD-2011-101065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, et al. Normal cellular processing of the beta-amyloid precursor protein results in the secretion of the amyloid beta peptide and related molecules. Ann N Y Acad Sci. 1993a;695:109–116. doi: 10.1111/j.1749-6632.1993.tb23037.x. [DOI] [PubMed] [Google Scholar]
- Haass C, et al. beta-Amyloid peptide and a 3-kDa fragment are derived by distinct cellular mechanisms. J Biol Chem. 1993b;268:3021–3024. [PubMed] [Google Scholar]
- Haass C, et al. Trafficking and proteolytic processing of APP. Cold Spring Harb Perspect Med. 2012;2:a006270. doi: 10.1101/cshperspect.a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson Petersen CA, et al. The amyloid beta-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proc Natl Acad Sci U S A. 2008;105:13145–13150. doi: 10.1073/pnas.0806192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann T, et al. Distinct sites of intracellular production for Alzheimer's disease A beta40/42 amyloid peptides. Nat Med. 1997;3:1016–1020. doi: 10.1038/nm0997-1016. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, et al. Analysis of microdissected human neurons by a sensitive ELISA reveals a correlation between elevated intracellular concentrations of Abeta42 and Alzheimer's disease neuropathology. Acta Neuropathol. 2010;119:543–554. doi: 10.1007/s00401-010-0661-6. [DOI] [PubMed] [Google Scholar]
- Henriques AG, et al. Sodium azide and 2-deoxy-D-glucose-induced cellular stress affects phosphorylation-dependent AbetaPP processing. J Alzheimers Dis. 2005;7:201–212. doi: 10.3233/jad-2005-7302. discussion 255–62. [DOI] [PubMed] [Google Scholar]
- Hicks DA, Nalivaeva NN, Turner AJ. Lipid rafts and Alzheimer's disease: protein-lipid interactions and perturbation of signaling. Front Physiol. 2012;3:189. doi: 10.3389/fphys.2012.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai K, et al. Mitochondrial abnormalities in Alzheimer's disease. J Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes O, et al. Pen-2 is essential for gamma-secretase complex stability and trafficking but partially dispensable for endoproteolysis. Biochemistry. 2014;53:4393–4406. doi: 10.1021/bi500489j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, et al. Brain beta-amyloid load approaches a plateau. Neurology. 2013;80:890–896. doi: 10.1212/WNL.0b013e3182840bbe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, et al. Monosodium iodoacetate induces apoptosis via the mitochondrial pathway involving ROS production and caspase activation in rat chondrocytes in vitro. J Orthop Res. 2013;31:364–369. doi: 10.1002/jor.22250. [DOI] [PubMed] [Google Scholar]
- Katsouri L, et al. PPARgamma co-activator-1alpha (PGC-1alpha) reduces amyloid-beta generation through a PPARgamma-dependent mechanism. J Alzheimers Dis. 2011;25:151–162. doi: 10.3233/JAD-2011-101356. [DOI] [PubMed] [Google Scholar]
- Khan SM, et al. Alzheimer's disease cybrids replicate beta-amyloid abnormalities through cell death pathways. Ann Neurol. 2000;48:148–155. [PubMed] [Google Scholar]
- Koo EH, Squazzo SL. Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J Biol Chem. 1994;269:17386–17389. [PubMed] [Google Scholar]
- Korenic A, et al. Astrocytic mitochondrial membrane hyperpolarization following extended oxygen and glucose deprivation. PLoS One. 2014;9:e90697. doi: 10.1371/journal.pone.0090697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn PH, et al. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010;29:3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukreja L, et al. Increased mtDNA mutations with aging promotes amyloid accumulation and brain atrophy in the APP/Ld transgenic mouse model of Alzheimer's disease. Mol Neurodegener. 2014;9:16. doi: 10.1186/1750-1326-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar DK, et al. Amyloid-beta peptide protects against microbial infection in mouse and worm models of Alzheimer's disease. Sci Transl Med. 2016;8:340ra72. doi: 10.1126/scitranslmed.aaf1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa LR, et al. Y682G Mutation of Amyloid Precursor Protein Promotes Endo-Lysosomal Dysfunction by Disrupting APP-SorLA Interaction. Front Cell Neurosci. 2015;9:109. doi: 10.3389/fncel.2015.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A, Sisodia SS, Trowbridge IS. Characterization of sorting signals in the beta-amyloid precursor protein cytoplasmic domain. J Biol Chem. 1995;270:3565–3573. [PubMed] [Google Scholar]
- Lammich S, et al. Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci U S A. 1999;96:3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, et al. APP processing is regulated by cytoplasmic phosphorylation. J Cell Biol. 2003;163:83–95. doi: 10.1083/jcb.200301115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuner K, et al. Mitochondrion-derived reactive oxygen species lead to enhanced amyloid beta formation. Antioxid Redox Signal. 2012;16:1421–1433. doi: 10.1089/ars.2011.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, et al. Increased plaque burden in brains of APP mutant MnSOD heterozygous knockout mice. J Neurochem. 2004;89:1308–1312. doi: 10.1111/j.1471-4159.2004.02455.x. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler SF. alpha-secretase in Alzheimer's disease: molecular identity, regulation and therapeutic potential. J Neurochem. 2011;116:10–21. doi: 10.1111/j.1471-4159.2010.07081.x. [DOI] [PubMed] [Google Scholar]
- Lustbader JW, et al. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science. 2004;304:448–452. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- Mancuso M, et al. Decreased platelet cytochrome c oxidase activity is accompanied by increased blood lactate concentration during exercise in patients with Alzheimer disease. Exp Neurol. 2003;182:421–426. doi: 10.1016/s0014-4886(03)00092-x. [DOI] [PubMed] [Google Scholar]
- Manczak M, Calkins MJ, Reddy PH. Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer's disease: implications for neuronal damage. Hum Mol Genet. 2011;20:2495–2509. doi: 10.1093/hmg/ddr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, et al. Mitochondria-targeted catalase reduces abnormal APP processing, amyloid beta production and BACE1 in a mouse model of Alzheimer's disease: implications for neuroprotection and lifespan extension. Hum Mol Genet. 2012;21:2973–2990. doi: 10.1093/hmg/dds128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, et al. Expression of APP pathway mRNAs and proteins in Alzheimer's disease. Brain Res. 2007;1161:116–123. doi: 10.1016/j.brainres.2007.05.050. [DOI] [PubMed] [Google Scholar]
- Morel E, et al. Phosphatidylinositol-3-phosphate regulates sorting and processing of amyloid precursor protein through the endosomal system. Nat Commun. 2013;4:2250. doi: 10.1038/ncomms3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller T, et al. The amyloid precursor protein intracellular domain (AICD) as modulator of gene expression, apoptosis, and cytoskeletal dynamics-relevance for Alzheimer's disease. Prog Neurobiol. 2008;85:393–406. doi: 10.1016/j.pneurobio.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Oakley H, et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer's disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006;26:10129–10140. doi: 10.1523/JNEUROSCI.1202-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyango IG, et al. Nerve growth factor attenuates oxidant-induced beta-amyloid neurotoxicity in sporadic Alzheimer's disease cybrids. J Neurochem. 2010;114:1605–1618. doi: 10.1111/j.1471-4159.2010.06871.x. [DOI] [PubMed] [Google Scholar]
- Pahlsson P, Shakin-Eshleman SH, Spitalnik SL. N-linked glycosylation of beta-amyloid precursor protein. Biochem Biophys Res Commun. 1992;189:1667–1673. doi: 10.1016/0006-291x(92)90269-q. [DOI] [PubMed] [Google Scholar]
- Park HJ, et al. Beta-amyloid precursor protein is a direct cleavage target of HtrA2 serine protease. Implications for the physiological function of HtrA2 in the mitochondria. J Biol Chem. 2006;281:34277–34287. doi: 10.1074/jbc.M603443200. [DOI] [PubMed] [Google Scholar]
- Parker WD, Jr, Parks JK. Cytochrome c oxidase in Alzheimer's disease brain: purification and characterization. Neurology. 1995;45:482–486. doi: 10.1212/wnl.45.3.482. [DOI] [PubMed] [Google Scholar]
- Pavlov PF, et al. Mitochondrial gamma-secretase participates in the metabolism of mitochondria-associated amyloid precursor protein. FASEB J. 2011;25:78–88. doi: 10.1096/fj.10-157230. [DOI] [PubMed] [Google Scholar]
- Pearson HA, Peers C. Physiological roles for amyloid beta peptides. J Physiol. 2006;575:5–10. doi: 10.1113/jphysiol.2006.111203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C, Santos MS, Oliveira C. Mitochondrial function impairment induced by amyloid beta-peptide on PC12 cells. Neuroreport. 1998;9:1749–1755. doi: 10.1097/00001756-199806010-00015. [DOI] [PubMed] [Google Scholar]
- Perez RG, et al. Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J Biol Chem. 1999;274:18851–18856. doi: 10.1074/jbc.274.27.18851. [DOI] [PubMed] [Google Scholar]
- Pinto M, et al. Mitochondrial DNA damage in a mouse model of Alzheimer's disease decreases amyloid beta plaque formation. Neurobiol Aging. 2013;34:2399–2407. doi: 10.1016/j.neurobiolaging.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placido AI, et al. The role of endoplasmic reticulum in amyloid precursor protein processing and trafficking: implications for Alzheimer's disease. Biochim Biophys Acta. 2014;1842:1444–1453. doi: 10.1016/j.bbadis.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Plant LD, et al. The production of amyloid beta peptide is a critical requirement for the viability of central neurons. J Neurosci. 2003;23:5531–5535. doi: 10.1523/JNEUROSCI.23-13-05531.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Morris JC. Tangles and plaques in nondemented aging and "preclinical" Alzheimer's disease. Ann Neurol. 1999;45:358–368. doi: 10.1002/1531-8249(199903)45:3<358::aid-ana12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Rapoport SI, et al. Abnormal brain glucose metabolism in Alzheimer's disease, as measured by position emission tomography. Adv Exp Med Biol. 1991;291:231–248. doi: 10.1007/978-1-4684-5931-9_18. [DOI] [PubMed] [Google Scholar]
- Reddy PH, et al. Abnormal mitochondrial dynamics and synaptic degeneration as early events in Alzheimer's disease: implications to mitochondria-targeted antioxidant therapeutics. Biochim Biophys Acta. 2012;1822:639–649. doi: 10.1016/j.bbadis.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhein V, et al. Amyloid-beta leads to impaired cellular respiration, energy production and mitochondrial electron chain complex activities in human neuroblastoma cells. Cell Mol Neurobiol. 2009;29:1063–1071. doi: 10.1007/s10571-009-9398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JW, et al. High glucose-induced oxidative stress and mitochondrial dysfunction in neurons. FASEB J. 2002;16:1738–1748. doi: 10.1096/fj.01-1027com. [DOI] [PubMed] [Google Scholar]
- Saitoh T, et al. Secreted form of amyloid beta protein precursor is involved in the growth regulation of fibroblasts. Cell. 1989;58:615–622. doi: 10.1016/0092-8674(89)90096-2. [DOI] [PubMed] [Google Scholar]
- Sandbrink R, Masters CL, Beyreuther K. APP gene family. Alternative splicing generates functionally related isoforms. Ann N Y Acad Sci. 1996;777:281–287. doi: 10.1111/j.1749-6632.1996.tb34433.x. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffler K, et al. Mitochondrial DNA polymorphisms specifically modify cerebral beta-amyloid proteostasis. Acta Neuropathol. 2012;124:199–208. doi: 10.1007/s00401-012-0980-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ, et al. The role of APP processing and trafficking pathways in the formation of amyloid beta-protein. Ann N Y Acad Sci. 1996;777:57–64. doi: 10.1111/j.1749-6632.1996.tb34401.x. [DOI] [PubMed] [Google Scholar]
- Shutt DC, et al. 2-deoxy-D-glucose induces oxidative stress and cell killing in human neuroblastoma cells. Cancer Biol Ther. 2010;9:853–861. doi: 10.4161/cbt.9.11.11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisodia SS. Beta-amyloid precursor protein cleavage by a membrane-bound protease. Proc Natl Acad Sci U S A. 1992;89:6075–6079. doi: 10.1073/pnas.89.13.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soscia SJ, et al. The Alzheimer's disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS One. 2010;5:e9505. doi: 10.1371/journal.pone.0009505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow R, et al. Brain glucose metabolism in Alzheimer's disease. Am J Med Sci. 1994;308:141–144. doi: 10.1097/00000441-199409000-00003. [DOI] [PubMed] [Google Scholar]
- Swerdlow RH. Mitochondria and cell bioenergetics: increasingly recognized components and a possible etiologic cause of Alzheimer's disease. Antioxid Redox Signal. 2012;16:1434–1455. doi: 10.1089/ars.2011.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow RH, et al. Glycolysis-respiration relationships in a neuroblastoma cell line. Biochim Biophys Acta. 2013;1830:2891–2898. doi: 10.1016/j.bbagen.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi RH, et al. Intraneuronal Alzheimer abeta42 accumulates in multivesicular bodies and is associated with synaptic pathology. Am J Pathol. 2002;161:1869–1879. doi: 10.1016/s0002-9440(10)64463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuma K, et al. ABAD enhances Abeta-induced cell stress via mitochondrial dysfunction. FASEB J. 2005;19:597–598. doi: 10.1096/fj.04-2582fje. [DOI] [PubMed] [Google Scholar]
- Thinakaran G, Koo EH. Amyloid precursor protein trafficking, processing, and function. J Biol Chem. 2008;283:29615–29619. doi: 10.1074/jbc.R800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Urano Y, et al. Association of active gamma-secretase complex with lipid rafts. J Lipid Res. 2005;46:904–912. doi: 10.1194/jlr.M400333-JLR200. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. Lost region in amyloid precursor protein (APP) through TALEN-mediated genome editing alters mitochondrial morphology. Sci Rep. 2016;6:22244. doi: 10.1038/srep22244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MW, et al. The amyloid precursor protein intracellular domain(AICD) disrupts actin dynamics and mitochondrial bioenergetics. J Neurochem. 2010;113:275–284. doi: 10.1111/j.1471-4159.2010.06615.x. [DOI] [PubMed] [Google Scholar]
- Webster MT, et al. The effects of perturbed energy metabolism on the processing of amyloid precursor protein in PC12 cells. J Neural Transm. 1998;105:839–853. doi: 10.1007/s007020050098. [DOI] [PubMed] [Google Scholar]
- Wilkins HM, Carl SM, Swerdlow RH. Cytoplasmic hybrid (cybrid) cell lines as a practical model for mitochondriopathies. Redox Biol. 2014;2:619–631. doi: 10.1016/j.redox.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirths O, Dins A, Bayer TA. AbetaPP accumulation and/or intraneuronal amyloid-beta accumulation? The 3xTg-AD mouse model revisited. J Alzheimers Dis. 2012;28:897–904. doi: 10.3233/JAD-2011-111529. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Koo EH, Selkoe DJ. Cell surface amyloid beta-protein precursor colocalizes with beta 1 integrins at substrate contact sites in neural cells. J Neurosci. 1997;17:1004–1010. doi: 10.1523/JNEUROSCI.17-03-01004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, et al. Inhibition of amyloid-beta (Abeta) peptide-binding alcohol dehydrogenase-Abeta interaction reduces Abeta accumulation and improves mitochondrial function in a mouse model of Alzheimer's disease. J Neurosci. 2011;31:2313–2320. doi: 10.1523/JNEUROSCI.4717-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youmans KL, et al. Intraneuronal Abeta detection in 5xFAD mice by a new Abeta-specific antibody. Mol Neurodegener. 2012;7:8. doi: 10.1186/1750-1326-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young-Pearse TL, et al. Secreted APP regulates the function of full-length APP in neurite outgrowth through interaction with integrin beta1. Neural Dev. 2008;3:15. doi: 10.1186/1749-8104-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, et al. APP processing in Alzheimer's disease. Mol Brain. 2011;4:3. doi: 10.1186/1756-6606-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]