Abstract
The cancer epigenome is fundamentally different than that of normal cells. How these differences arise in and contribute to carcinogenesis is not known, and studies using model organisms such as zebrafish provide an opportunity to address these important questions. Modifications of histones and DNA comprise the complex epigenome, and these influence chromatin structure, genome stability and gene expression, all of which are fundamental to the cellular changes that cause cancer. The cancer genome atlas covers the wide spectrum of genetic changes associated with nearly every cancer type, however, this catalog is currently unidimensional. As the pattern of epigenetic marks and chromatin structure in cancer cells is described and overlaid on the mutational landscape, the map of the cancer genome becomes multi-dimensional and highly complex. Two major questions remain in the field: (1) how the epigenome becomes repatterned in cancer and (2) which of these changes are cancer-causing. Zebrafish provide a tractable in vivo system to monitor the epigenome during transformation and to identify epigenetic drivers of cancer. In this chapter, we review principles of cancer epigenetics and discuss recent work using zebrafish whereby epigenetic modifiers were established as cancer driver genes, thus providing novel insights into the mechanisms of epigenetic reprogramming in cancer.
Major Questions Surrounding the Cancer Epigenome: Answers from Zebrafish
The genetic landscape of cancer cells is dramatically different from normal cells. Mutations, chromosomal losses, gains and rearrangements have been described for virtually every type of cancer in humans [1, 2]. Advances in sequencing technology in the past decade have provided an exquisitely detailed view of the cancer genome in humans, and these have been used to identify candidate driver genes in nearly every type of cancer. Documenting the cancer cell epigenome is more complex, as epigenetics is influenced by DNA modifications, namely methylation and hydroxymethylation on cytosines, histone variants and histone modifications. Moreover, epigenetic modifications are clustered, and are influenced by underlying DNA sequence variations. Thus, integrating the mutational and epigenetic landscapes to generate a comprehensive genetic map of cancer to identify key regions that contribute to carcinogenesis remains a major goal of the field. In particular, sorting through these complex catalogs to identify features that cause or sustain malignancy requires tractable in vivo systems to allow functional assessment of candidates. Since epigenetic marks are well conserved in vertebrates, zebrafish represent an excellent system for such studies.
Those studying cancer epigenomics are addressing similar questions as those studying the cancer genome, including: (1) How does the epigenome get restructured in cancer cells? (2) Are there epigenetic signatures that can be used diagnostically or for identifying specific tumor sub-classes? (3) Which epigenetic changes contribute to tumorigenesis and, for those that are carcinogenic, what is the underlying mechanism by which these cause cancer? As with mutational analysis, epigenetic profiling of cancer cells has documented millions of differences compared to their normal counterparts. Importantly, many of these are conserved in zebrafish tumors [3–5]. The functional annotation of epigenetic differences between normal and cancer cells requires in vivo models. As many such epigenetic signatures are conserved in zebrafish, they has proven to be an excellent system to complement the more commonly use models [6].
Epigenetics is implicated in nearly every aspect of embryonic development and, as such, has been heavily investigated by developmental biologists using zebrafish. These studies are now being extended to the study of cancer. Work using zebrafish has recently identified epigenetic modifiers which impact melanoma [7], hepatocellular carcinoma (HCC) [5 ], myelodysplastic syndrome (MDS) [8 ] and rhabdomyosarcoma [9]. Here, the fundamental principles of epigenetic modifications and their relationship to cancer are discussed using zebrafish as a model organism.
The Complexity of the Epigenetic Code
Epigenetics is an umbrella term referring to factors that influence chromatin structure, which in turn regulate chromosome structure, permanent gene silencing and tissue-specific patterns of gene expression. The basic functional unit of chromatin structure is the nucleosome whereby active (euchromatin) and inactive (heterochromatin) domains dictate access of proteins that further modify the epigenome, mediate DNA replication and drive transcription. Many features of the cancer cell phenotype can arise from changes in the epigenome, and indeed, it is speculated that the marked differences in gene expression of malignant cells reflects the massive changes in their epigenome. As cancer cells are typically defined by the suppression of checkpoints that monitor DNA replication, DNA damage and cell cycle progression, it is possible that a monitoring system for epigenome integrity is another one of the checkpoints missing in cancer cells [10, 11].
Epigenetic modifications are added and removed by chromatin-modifying enzymes in a dynamic and tightly regulated fashion without impacting the underlying nucleotide sequence of the DNA (Fig. 1). Epigenetic control of gene activity and overall chromatin structure operates on three levels: DNA, histone proteins, and the nucleosome. Interplay between permissive and repressive domains dictate differential gene expression profiles and maintain a central, functional role during differentiation and development. Most notably, zebrafish research on the developmental functions of epigenetic modifiers has led to discoveries of the importance of epigenetics in fate decisions, gene expression patterning, and zygotic genome activation [12–18]. For instance, a large screen using morpholinos to knock down the expression of 425 chromatin modifiers in zebrafish embryos identified a distinct subset of modifiers that regulate erythroid cell formation and another that was important for hematopoietic stem and progenitor cells [19]. This demonstrates that multiple epigenetic modifiers are required for cell fate decisions and differentiation.
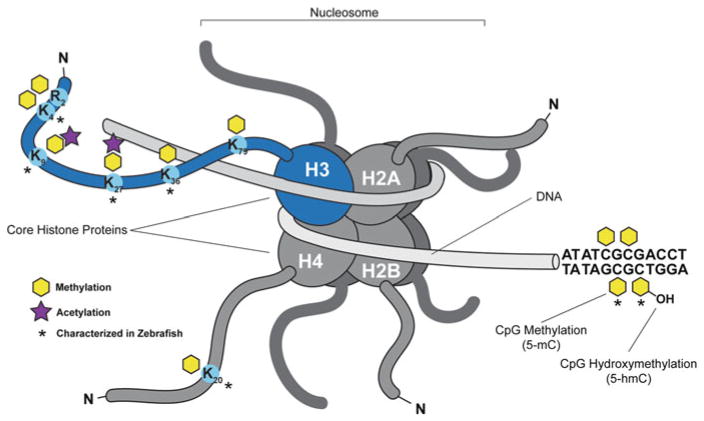
Fig. 1.
Epigenetic modifications are conserved in zebrafish. Representation of the basic structure of the nucleosome composed of 147 base pairs of DNA wrapped around an octamer of histone H3/ H4 and histone H2A/H2B dimmers. The core histone proteins have long N-terminal tails that extend out from the core particle that is rich in basic amino acid residues, lysine (K) and arginine (R), which can be extensively modified in a reversible, covalent manner. Lysine residues that are mono-, di-, or tri- methylated on histone H3 and H4 that are characterized in zebrafish have been indicated and are conserved across species including H3K4, H3K9, H3K27, H3K36, and H4K20. H3 lysine methylation elicits different transcriptional and structural responses depending on chromatin context and the residues that are modified. Histone acetyl marks have also been indicated and are associated with euchromatic regions amenable to gene transcription. Cytosine residues in DNA can be methylated in a CpG dinucleotide context throughout the genome by DNA methyl-transferases (DNMT), which is a conserved process across vertebrate species and plants. DNA methylation is typically associated with irreversibly silenced regions in heterochromatin. Methylation of DNA can be reversed passively or actively through oxidation of the methyl mark. TET family enzymes carry out active demethylation. Extensive DNA methylation profiling of the zebrafish has been performed in a number of studies
Historically, studies in cancer epigenetics have focused on differences of single epigenetic modifications at the gene regulatory region of a gene of interest—such as a tumor suppressor or a key oncogene—and have correlated these marks with gene expression. This has lead to the conclusions that there is a causative relationship between the mark under study and the expression level of the gene. However, as genome-wide techniques advance, it is clear that the relationship can work both ways (i.e. gene expression levels can influence the epigenetic landscape of the gene) and that the influence of single epigenetic marks is balanced by the conformation of the region as a whole. For instance, chromatin is not simply open or closed, but instead there are also intermediate states where epigenetic marks that are cataloged as repressive co-exist with marks that are associated with gene activation. Such “poised” genes are thus held in a repressed, but not completely closed, state that can rapidly be triggered when the signal arrives. Moreover, epigenetic marks have been shown to act at a distance [20]. Thus, the path forward for understanding how epigenetic modifications impact cancer gene expression should incorporate multiple epigenetic marks and a wide-angle view of the genomic region of interest.
Epigenetic readers and writers recognize and target DNA and histone modifications and the erasers remove these marks. The best-studied modifications of DNA and histones are predominantly mediated through writers such as methyltransferase enzymes (both DNA- and histone-methyltransferases; DNMTs and HMTs, respectively), histone demethylases, histone acetyltransferases (HATs), and histone deacetylases (HDACs). Furthermore, incorporating histone variants in place of the canonical histones can impact gene expression, chromatin structure, and can define specialized regions of the chromatin. Readers of the epigenetic code serve to target the writers and erasers and serve as a link between DNA and histone modification to generate a complex and overlapping set of modifications across the genome. DNA methylation is present in all vertebrates, yeast [21] and plants, but not in commonly used invertebrate animal models. Moreover, the factors that modify DNA are well conserved from humans to zebrafish, and make zebrafish an ideal model to study DNA methylation. Histones are over the most highly conserved proteins in vertebrates and conserved from human to zebrafish [22] and the epigenetic marks that regulate them are also well conserved. Thus far, all histone modifications described in humans that have been investigated in zebrafish have been identified, and the readers and writers that mediate these modifications are well conserved in zebrafish (Fig. 1 [23]).
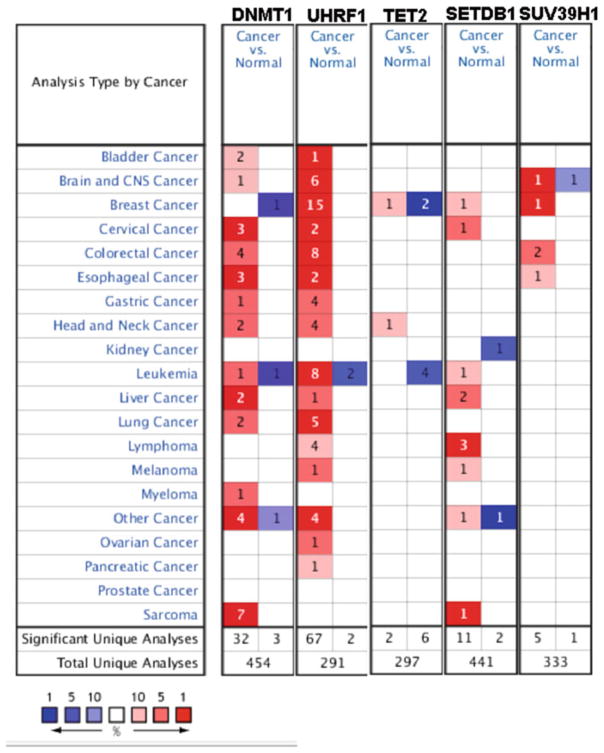
Genome-wide expression studies have documented that the expression pattern of chromatin modifiers is dramatically altered in malignant cells (Fig. 2). Interestingly, while some of genes show similar expression changes across cancer types, others appear more specific. For instance, expression of Dnmt1 and Uhrf1, which are the key components of the DNA methylation machinery, are elevated across cancer types while TET2, a gene involved in cytosine demethylation, is down-regulated primarily in leukemia (Fig. 2). Moreover, as chromatin modifiers are largely regulated by their interacting partners and pre-existing epigenetic modifications can direct whether the writers are able to make changes at each locus, altered expression of a single modifier may be less important than the combinatorial changes for multiple members of a complex.
Fig. 2.
Epigenetic modifiers discovered as cancer genes in zebrafish show unique expression patterns in human cancers. Oncomine expression analysis for each of the key DNA and histone modifying enzymes that have been discovered in zebrafish to play a role in cancer. Expression of each gene was monitored across a series of cancer samples compared to normal counterparts and their expression levels (high = red; low = blue) are indicated. Expression thresholds were set with a fold change of 2 or greater, p value <0.001; gene rank in the top 10 % of deregulated genes and
DNA Methylation
Methylation of cytosine (5-mC) when it is paired with a guanine residue (i.e. CpG) is a critical mechanism of X-chromosome inactivation, imprinting, silencing repeats and transposons and heterochromatin formation [24–28]. Additionally, many studies implicate DNA methylation in transcriptional repression of differentially expressed genes, however whether DNA methylation is sufficient for fine-tuning gene expression is controversial [25]. The most heavily methylated regions of the genome are in intragenic regions, which are largely composed of repetitive sequences, transposable elements and the regions of the chromosome that give it structure (i.e. the centromeres and telomeres). Methylation of these regions is thought to provide an irreversible mechanism of repression and the ability to form higher order chromatin structure. Additionally, gene bodies are heavily methylated and this typically corresponds to actively transcribed genes [24], however, it the function of this region of DNA methylation is not clear [25]. An excellent graphical overview of DNA methylation has recently been published [29].
Methyl groups are transferred from S-adenosyl-L-methionine (SAM) to cytosine by DNMTs. DNMT1 has been extensively studied across species: it preferentially methylates hemimethylated CpG dinucleotide sequences generated during DNA replication, and is thus considered the maintenance methyltransferase. This role has been confirmed in zebrafish through studying dnmt1 mutants [11, 30, 31] and in mutants that cannot generate sufficient SAM [32]. DNMT3A and 3B, however, initiate methylation of those regions that were previously unmethylated (i.e. de novo DNA methylation), which is best studied in imprinted genes, which zebrafish do not appear to have. While DNMT1 can bind DNA [33], it is not very efficient at targeting CpG sites. This is significantly improved accomplished by the Ubiquitin-like with PHD and RING Finger domains 1 (UHRF1) which recognizes hemimethylated CpG sequences during DNA replication and directly recruits DNMT1 to facilitate DNA methylation [34, 35]. 5mC is removed by the Ten-eleven translocation (TET) proteins, which convert 5mC to 5-hydroxymethyl cytosine [36].
Patterns of DNA methylation are dramatically different between cancer cells and their normal counterparts. In non-transformed cells, approximately 2–8 % of the genome is methylated. Certain regions of the genome contain high frequencies of CpG sites, which are termed CpG Islands (CGIs). CGIs occur in approximately 50 % of promoters of human genes and are largely unmethylated, while CpG sites in gene bodies and intergenic regions are mostly methylated. DNA methylation is thought to act as a repressive epigenetic mark and it was first described as a mechanism to assure that some regions of the genome are always maintained in a transcriptionally inactive state such retrotransposable elements [37] repeat sequences [38] imprinted genes [27] and centromeric and pericentromeric regions [39]. Most malignant cells have less DNA methylation than their normal counterparts, and this is largely attributed to loss of methylation at these regions which are typically heavily methylated.
Clusters of CpGs are found in some gene promoters (i.e. CpG islands), but methylation in these gene regulatory regions is largely absent in normal cells. Nevertheless, CpG island methylation patterns has received the most attention in the field. This is largely based on the hypothesis that since DNA methylation is required for silencing those regions that are typically heavily methylated, then DNA methylation in gene regulatory regions must also lead to formation of ‘closed’ chromatin structures rendering the underlying sequence inaccessible to transcription factors and RNA polymerase II. The finding that the promoters of some tumor suppressors are hypermethylated in cancer cells has fueled much of the study on DNA methylation as a mechanism of regulating gene expression in favor of promoting cell proliferation and transformation. However, although there are several clear examples where DNA methylation is inversely correlated with the expression of nearby genes, many studies also show that methylation in promoter regions can also be positively correlated with gene expression [40–43]. Moreover, since DNA methylation and other epigenetic modifications may act at a distance [20], it may be that the focus on differential methylation of CpG islands may be too narrow.
DNA methylation is frequently found to be co-localized with repressive histone modifications and histone variants, and thus it may also perform an instructive role for the recruitment of other chromatin modifiers to promote gene repression and higher order chromatin structure. Thus, in some cases, the correlation of DNA methylation in the regulatory regions of repressed genes may reflect the placement of DNA methylation side by side with other epigenetic marks that play a bigger role in the repression. In such cases, DNA methylation may serve a supportive role. Indeed, a recent study whereby the TET1 hydroxylase was targeted to methylated regulatory regions of specific genes to specifically demethylated these loci. In most cases tested, only modest demethylation was achieved and while demethylation of some sites significantly increased gene expression, this was not a universal finding [44]. While this exciting study is the first to show that reducing DNA methylation in specific promoters can increase gene expression, it is clear that other factors provide important and even dominant regulatory roles.
DNA Methylation in Zebrafish
Zebrafish mutants in the key genes regulating DNA methylation all have DNA hypomethylation, including S-adenosylhomocysteine hydrolase [32], dnmt1 [11, 30, 31] and uhrf1 [11, 45]. Additionally, tet2 mutants show a loss of 5-HmC [8, 46], illustrating that the machinery that mediates the methylome is highly conserved.
Methylation patterning of the zebrafish genome is a highly dynamic process during development of the organism. In a very similar manner to mammals, zebrafish sperm is hypermethylated compared to its oocyte counterpart, which is then rapidly demethylated upon fertilization [47]. Following fertilization, the DNA methylation levels of the 1- to 2- cell stage of the zygote are lower then what is initially observed in the oocyte mediated by a wave of DNA demethylation [47]. De novo DNA methylation resets DNA methylation levels to levels seen in terminally differentiated tissues during the mid-blastula transition and in gastrula [47–49]. Next, a wave of DNA demethylation and remethylation occur during somitogenesis [49] and tissue specific DNA methylation patterns being established and maintained as cells differentiate. However, the evidence in support of DNA methylation as a biochemical system that regulates gene expression during early development through programmed methylation and demethylation patterns are correlative, at best [14, 50]. Therefore, the functional significance of the dynamic changes in DNA methylation remains unclear, and is under intense investigation.
In summary, the DNA methylome can be re-patterned during differentiation and transformation. The global loss of DNA methylation during carcinogenesis is seen across cancer types, but the mechanism leading to the demethylation of the cancer genome has not been established. Since DNA methylation impacts the interaction between DNA and histones within nucleosomes and promotes higher order chromatin formation, it clearly contributes to gene expression however, it does not appear to be the major mechanism that resets the transcriptome to favor cancer-promoting genes and suppress tumor suppressors. Instead, loss of DNA methylation can result in euchromatinization of the genome, which promotes chromosomal translocations, breaks, transposon activation and could thus serve as a major mechanism driving genomic instability.
Histone Modification
Histones can be post-translationally modified in a number of ways, including acetylation, methylation, ubiquitination, and phosphorylation. A large majority of these post-translational modifications occur on the unstructured N-terminal tails of the core histones, which are rich in basic amino acid residues lysine (K) and arginine (R), which confer a positively charged surface that extends out from the nucleosome and interacts with the negatively charged DNA (Fig. 1). The abundance of lysine and arginine residues on the N-terminal tails also make the core histone proteins amenable to post-translational modification by epigenetic enzymes and complexes to modify the local nucleosome structure. In turn, epigenetic modification of the core histone proteins can serve to recruit transcriptional repressors and activators to specific loci or maintain a baseline quiescent state. Here, we review the major types of histone modifications and focus on those that have been specifically shown to play a prominent role in cancer using zebrafish.
Histone Methylation
Histone methylation directly impacts chromatin structure and gene transcription. Lysine (K) and arginine (R) residues along the N-terminal tails of the core histones are methylated by histone methyl transferases (HMTs). The histone methylome is extremely complex, as amino acids can be bi or tri methylated, and the various combinations of the methylated residues ultimately dictate chromatin state. The well- established correlations between some histone methyl marks and gene expression allows the categorization of these marks as “activating” or “repressive”. For instance, trimethylation of H3 on lysine 4 (H3K4Me3) is associated with actively transcribed genes and the di- and tri- methylation of lysine 9 of H3 (H3K9me2/3) is correlated with heterochromatin formation and suppression of gene transcription. Interestingly, a recent study found that SMYD3, a methyltransferase for K3K4Me3, promoted invasion of human tumor cells transplanted into zebrafish embryos. This was attributed to the induction of a metalloproteinase which aids tumor cell mobility [51]. H3K9 methylation is mediated by a number of proteins, including SETDB1 and SUV39H1, which are evolutionarily conserved and expressed in zebrafish [7, 9, 23, 52]. In addition to these, several other epigenetic marks also display a similar distribution across the zebrafish and mammalian genome [17, 45, 53]. Suv39h1 has been shown to cooperate with Dnmt1 to regulate the terminal differentiation of the intestine, exocrine pancreas, and the retina during zebrafish development [52]. Furthermore, Rai et al. demonstrated that H3K9me3 is also positively correlated with levels of DNA methylation in heterochromatic regions of the genome. It is thought that these two marks can collaborate to maintain genes in a repressed state [54]. Histone methyltransferases have been implicated in a wide range of cancers [55] and we discuss how zebrafish have been used to identify functional roles for SETDB1 and SUV39H1 in cancer.
Methylation of H3K27 is another well-studied epigenetic mark that is commonly incorporated into gene promoters and is mediated by Polycomb group proteins, namely the PRC2 complex [56]. High levels of H3K27Me3 were identified in zebrafish [3] and in humans [57]. On its own, H3K27me3 is typically associated with silenced genes and is thus seen as a repressive mark. However, H3K27 tri- methylation is also associated with ‘bivalent’ transcriptional states. Promoter regions marked with H3K4me3 and H3K27me3 are thought to adopt these ‘bivalent’ states where transcription is primed and RNA polymerase II occupancy is permitted, yet the gene remains inactive.
Histone methylation plays a critical role during embryonic development. Patterning of histone methylation regulates zygotic genome activation in zebrafish [13, 14, 17, 58, 59]. Histone methylation directly influences nucleosome dynamics and stability and serves as a platform for the specific recruitment of transcription factors and remodeling complexes. Mounting evidence supports the crosstalk between DNA methylation and histone methylation. H3K4me3 is negatively correlated with DNA methylation, while H3K9me3 is significantly and positively correlated with DNA methylation. This direct cooperation indicates that a very fine tuned epigenetic signaling network regulates cell function.
Histone Phosphorylation, Acetylation and Histone Variants
Histone core particles can be phosphorylated and acetylated. Phosphorylation of serine, threonine, and tyrosine residues has been found to be critical in regulating chromatin condensation during mitosis, gene expression, and DNA repair. For instance, H3S10 phosphorylation in some contexts, promotes chromatin de-compaction to facilitate transcription, whereas during mitosis H3S10 phosphorylation is critical for chromatin condensation [60, 61]. Phosphorylation of the histone variant H2A.X marks areas of DNA damage and double-stranded breaks to facilitate the DNA repair pathway. This has been shown as a robust marker of DNA damage in mammalian and zebrafish cells [62]. It illustrates the complexity of the histone code, where the context can dictate the impact of the modification.
Lysine acetylation is a conserved, reversible, and highly regulated post- translational modification of core histone proteins that is mediated by HATs. The availability of multiple target sites enables stepwise regulation of acetylation allowing for the fine-tuning of chromatin remodeling mediating gene transcription. Histone deacetylation is mediated by HDACs. A large body of work has demonstrated that, most commonly, acetylated histones lead to an open chromatin configuration and thus HDAC recruitment serves to promote heterochromatin formation.
Histone acetylation is dynamically regulated during zebrafish embryogenesis [63–67 ]. H3K9ac levels were high at 24 h post fertilization (hpf), but rapidly diminished by 48 and 72 hpf [68 ], suggesting that this mark may play a regulatory role during early embryogenesis, further supported by findings of failed development in Hdac deficient embryos [64–67, 69 ]. Histone acetylation has also been linked to maintenance of genomic integrity and DNA repair and the acetylation and de-acetylation of histones has long been a targeted focus of cancer therapies [70] as disrupted histone acetylation dynamics have been associated with a number of cancers.
There are a large number of highly comparable forms of histones that are collectively defined as ‘histone variants’. These variants can be incorporated into the nucleosome, replacing their canonical counterpart, resulting in changes in structural regions of the chromatin, gene expression, response to genotoxic events, genomic stability, and DNA repair. Furthermore, a growing body of evidence has demonstrated that histone variants can play critical roles during cancer development. Particularly in plants, the DNA wrapped in nucleosomes tends to be more methylated, suggesting that the position of the nucleosome influences the methylome pattern [71].
The concept of the ‘histone code’ indicates that the sum of histone modifications at a specific locus during a particular cellular process dictates nucleosome structure. Indeed, both developmental biologists and cancer biologists are now utilizing to a more complicated, but likely more accurate, model whereby the sum of the histone marks, DNA marks, associated co-factors, and transcription factors that creates a chromatin microenvironment which can either repress, induce or poise genes for expression. Studying the overlapping roles of covalent epigenetic marks with histone variant deposition to profile the epigenomic landscape of various cancers is a daunting, but important goal that has recently been tackled by the Roadmap Epigenomics Consortium who have generated a reference epigenome for 111 tumors [72]. The conservation of epigenetic marks and mechanisms of carcinogenesis from humans to zebrafish makes this an accessible and tractable model to study cancer epigenomics.
Common Epigenetic Changes in Cancer: Mechanisms and Questions
As the field of cancer epigenomics evolves, common themes are emerging. In nearly every cancer type, DNA methylation is reduced genome-wide (i.e. global DNA hypomethylation) yet in the same tumors that display global DNA hypomethylation, specific loci can be hypermethylated [73]. The use of ChIPSeq, histone array platforms and nucleosome positioning analysis (i.e. ENCODE) on tumor samples is yielding a rich encyclopedia of the integrated epigenome of over a 100 cancers [72]. Exciting discoveries combining both in silico analysis and in vivo functional studies using model organisms have begun to sift through the differences to uncover mechanisms by which key epigenetic modifiers contribute to cancer formation. In particular, experiments using the power of zebrafish genetics have demonstrated that two key epigenetic marks—DNA methylation and H3K9Me3—are functionally relevant to hepatocellular carcinoma (HCC), myelodysplastic syndrome (MDS), melanoma and rhabdomyosarcoma. These are reviewed below.
The Cancer Methylome: Causes, Consequences and Insight from Zebrafish
It has long been established that the global DNA methylation is significantly reduced in cancer cells [73]. This was first observed using techniques that monitor whole genome 5mC levels [74] which serves as an assay for methylation levels on the most heavily methylated and abundant regions of the genome: introns, gene bodies and intragenic regions containing repeats and transposons. However, those regions that are thought to serve regulate gene expression, such as those for imprinted genes [75] and CpG islands in differentially regulated genes do not influence the signal derived from global assessment of 5mC levels. More sophisticated approaches to monitor the cancer methylome has confirmed that it is the intragenic regions which are less methylated in cancer cells and, in some cases, the regulatory regions of specific genes within the same tumor become hypermethylated [73]. While it is clear that genome-wide approaches that allow locus specific resolution or base pair resolution provide the most comprehensive perspective of the cancer cell methylome, the mechanism by which and the significance of global DNA hypomethylation in cancer are not well understood.
Studies using zebrafish have both confirmed the loss of methylation in tumors [4] and have highlighted that proteins which modify the methylome are cancer genes [5]. Moreover, while it is well established that loss of DNA methylation can promote cancer-causing events, such as genome instability (Fig. 3), the mechanism of this is not yet clear. Three major questions in the field are: (1) How is DNA methylation lost during transformation (2) How does altering the mechanism impact cancer? (3) What is the mechanism of DNA methylation mediated carcinogenesis? Here, we describe how recent studies using zebrafish have provided the answers to these questions.
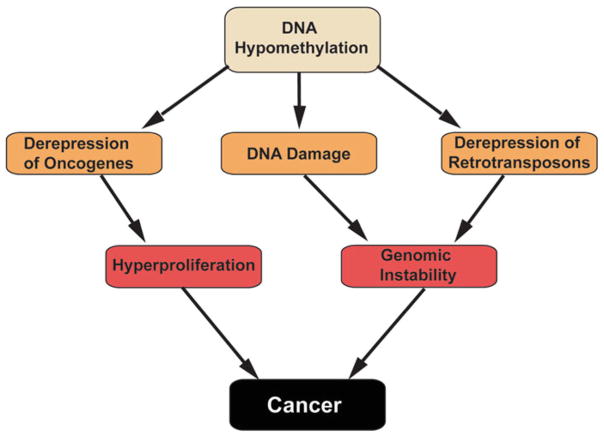
Fig. 3.
Proposed mechanisms by which DNA hypomethylation contributes to cancer. Schematic depicting several pathways by which hypomethylation of the genome can contribute to cancer development. Loss of CpG methylation can lead to general genomic instability and mutation or de-repress typically silenced oncogenes and tumor promoting factors
Loss of DNA methylation can occur via a passive mechanism, whereby methylation of a cytosine is not maintained after DNA replication, or via active demethylation either by the TET proteins which convert 5mC to 5-hydroxymethylcytosine (5hmC), or by spontaneous deamination followed by the repair of the deaminated cytosine [49]. TET2 mutation is found in patients with MDS [76] and it has been proposed that loss of TET2 reduces 5hmC which thereby promotes a more “stem- like” fate by suppressing the expression genes that promote differentiation. One study that used morpholinos to transiently knockdown tet1, tet2 and tet3 in zebrafish embryos found that this caused anemia associated with the loss of genes that promote erythropoiesis, such as scl, gata1 and cmyb. This was associated with a moderate increase in the 5mC levels on a few CpG sites in the promoters of these genes [46], suggesting that the failure to convert 5mC to 5hmC resulted in the suppression of these genes. Another study using gene editing techniques to mutate tet2 in zebrafish demonstrated reduced 5hmC levels in the kidney, the site of hematopoiesis in zebrafish. Similar to the phenotype of tet1, 2 and 3 morphant embryos [46], tet2 mutation was associated with a marked decrease in the number of erythrocytes coupled with an increase in myelomonocytes by 11 months, leading to the development of myelodysplasia in 2 year old fish [8]. While these interesting and clinically relevant studies suggest that blocking Tet activity can reduce the expression of pro- differentiation genes due to retention of DNA methylation marks in their promoters, it is possible that the loss of 5hmC could affect erythrocyte development and MDS by other mechanisms, as 5hmC also can directly affect gene expression [77].
Work in zebrafish has identified a second mechanism by which DNA methylation is actively removed. In an elegant study that capitalized on the power of zebrafish genetics, Rai et al. demonstrated that Aid/Apobec deaminates 5mC, and then Mbp and Gadd45a recognize and excise this aberrant nucleotide [49]. Inappropriate activation of this demethylation program occurs in zebrafish intestinal epithelial cells that lack Apc, a key tumor suppressor in colon cancer. Loss of Apc leads to hypomethylation and upregulation of genes controlling intestinal cell differentiation, including tcf family members, and the downregulation of the pathway that generates retinoic acid, a key driving factor for intestinal cell differentiation. This was shown to be mediated by the failure of Apc mutant cells to activate the Aid/Apobec demethylase program. They conclude that DNA methylation is retained in the regulatory region of genes that keep intestinal cells undifferentiated [78], which could lead to cancer. It is interesting to speculate that demethylation by Aid/Apobec could be one mechanism contributing to DNA hypomethylation in colon and gastric cancer [79].
It has been proposed that DNA hypomethylation gives rise to cancer through a variety of mechanisms (Fig. 3). These include activation of retrotransposons, facilitating chromosome breaks due to loss of heterochromatin, activation of imprinted genes, and deregulation of gene expression. Many of these changes cause genomic instability, a leading cause of transformation. Which of these mechanisms contribute to cancer, and whether they vary by cancer type, remains to be determined.
UHRF1 Overexpression as a Mechanism of Genome Hypomethylation and HCC Formation
Our work on DNA methylation as a mechanism of carcinogenesis suggests another mechanism of DNA hypomethylation which relies on passive removal during cell division. This, we propose, is mediated by overexpression of UHRF1 [5]. UHRF1 overexpression emerged out of the sea of cancer transcriptome analyses as a common feature of many types of cancer (see Fig. 2 and [80–87]). This suggested that UHRF1 might be a conserved mechanism of carcinogenesis across cancer types.
uhrf1 mutation causes a small liver in zebrafish embryos [11, 88] and haploinsufficiency reduces liver regeneration in adult zebrafish [88]. We thus tested the hypothesis that overexpressing human UHRF1 in zebrafish hepatocytes would cause HCC. We found that high UHRF1 levels increased destabilization of Dnmt1, potentially due to its E3 ubiquitin ligase activity [89, 90], and delocalized Dnmt1 away from the chromatin, resulting in global DNA hypomethylation. This phenotype was accompanied by p53 mediated senescence of hepatocytes, a tumor suppressive mechanism that, when overcome, leads to HCC in nearly all fish younger than 20 days old. Analysis of human HCC samples revealed elevated levels of UHRF1 and a general downregulation of the p53 senescence program, as predicted by findings in zebrafish. Moreover, in a classical transformation assay using NIH-3 T3 cells, UHRF1 overexpression was shown to cooperate with another senescence- inducing oncogene, Ras, to mediate transformation. These data indicate that UHRF1 is an epigenetic modifier that acts as an oncogene in HCC and, given its widespread overexpression in a range of cancers types, it may also function as an oncogene in other cancers. Moreover, since high levels of UHRF1 paradoxically caused DNA hypomethylation, it is possible that this is a mechanism by which DNA methylation is lost in cancer cells.
Histone Methyltransferases Discovered as Cancer Genes Using Zebrafish
There are multiple histone methyltransferases which target the same residue for methylation, for instance, both SETDB1 and SUV39H1 mediate H3K9 trimethylation, and it is unclear how these two proteins that perform a similar function achieve specificity. Interestingly, each of these enzymes have a unique expression profile across cancer types (Fig. 2) and have been implicated in distinct cancers. For instance, down regulation of SETDB1 is observed in metastatic lung cancer [91], yet overexpression of SUV39H1 has been reported in HCC [92]. This suggests that the functional similarity between these two HMTs may become more distinct depending on cancer type.
SETDB1
SETDB1 was identified as the first epi-oncogene in zebrafish through a screen for genes that cooperate with the most common mutations in melanoma, activating BRAF and loss of p53 [7]. These mutations are found in nearly 25–60 % of melanomas and a previous study demonstrated that they cooperate to cause melanoma in zebrafish [93]. Since melanomas carry more mutations than virtually any other tumor type likely due to the life-long exposure of melanocytes to UV induced DNA damage and there is a high variability in tumor latency, even after these mutations occur, it has been proposed that cooperating mutations are important for tumor onset. A common hypothesis in cancer genomics is that amplicons which are conserved across tumors harbor oncogenes or other genes required for tumor survival. Chromosome 1q21 is commonly amplified in melanoma [94], hence the authors selected 17 genes in the 1q21 region to screen for their capacity to increase tumor- induced mortality in zebrafish engineered to have melanocytes with deleted p53 and overexpressed activated BRAF. Of these, only SETDB1 overexpression increased the incidence of melanoma from 53 to 94 % and decreased survival by nearly half. Importantly, mutant versions of SETDB1 that lack MT activity were not cancer promoters in this system, implicating H3K9Me3 in this phenotype. Since human melanoma cells transfected with SETDB1 show significant enrichment of SETDB1 on the regulatory regions of genes that are expressed at low levels, but not on those that are upregulated in this system, it is assumed that SETDB1 mediates gene repression through H3K9Me3 deposition. However, they report the unanticipated finding that a subset of the presumed SETDB1 target genes in human cells were not marked by H3K9Me3 in cells overexpressing SETDB1, suggesting that this epigenetic mark alone cannot account for the mechanism by which the genes that are bound by SETDB1 become repressed. Since SETDB1 has recently been reported to exist in a complex with SuV39H1 and other factors [95], there is a possibility that SETDB1 is primarily required for increased targeting of the HMT complex to H3K9. Moreover, catalytically inactive SETDB1 was still able to associate with the HMT complex without any reduction in H3K9me3 and drive the onset of melanoma [7]. This raises the interesting possibility that all of the cancer causing effects of SETDB1 overexpression in melanoma may not be all mediated through its function as an H3K9 methyltransferase.
SUV39H1
Another of the H3K9 methyltransferases, SUV39H1, was found to repress rhabdomyosarcoma formation in the zebrafish [9]. Furthermore, SUV39H1 has been implicated in promoting a number of other cancers including HCC [92], however, the study in zebrafish was the first to demonstrate the oncogenic potential of this important epigenetic regulator. In humans, activating mutation in a ras-pathway gene is common in rhabdomyosarcomas, and overexpressing activated KRAS in zebrafish muscle stem cells causes a high incidence of tumors with an onset prior to 20 days post fertilization (dpf) with over 50 % lethality by 2 months of age [96]. These tumors in zebrafish share many common genetic features with human rhabdomyosarcomas [97], making this a useful model for translational studies on this cancer type. Albacker et al. evaluated human RMS samples to identify chromatin modifiers that were commonly overexpressed, and then used zebrafish to assay the impact of overexpressing 19 of these modifiers on the survival of zebrafish overexpressing KRAS G12D in muscle cells. Only SUV39H1 overexpression imparted a survival advantage compared to controls expressing KRAS G12D plus GFP. Gene expression analysis revealed that cell cycle regulator genes were suppressed in the samples expressing SUV39H1 and KRAS G12D compared to KRAS G12D alone [9]. Interestingly, the effects of SUV39H1 were observed prior to any tumor formation, as early as 7 dpf. It remains unclear whether the suppression of these genes is mediated by enhanced H3K9Me3 deposition in their regulatory regions, and the effect of SUV39H1 overexpression on the epigenetic landscape has yet to be determined.
All of the H3K9 MT proteins—SETDB1 SUV39H1, G9a and GLP—have been identified in a complex [95] raising the possibility that they may work together or co-regulate to control H3K9Me3 domains. However, the mechanism of loci selection for H3K9 methylation by individual methyltransferases has not been identified. Interestingly, overexpression of both SETDB1 and human SUV39H1 was also capable of promoting melanoma in the zebrafish system, and a mutant of SETDB1 lacking methyltransferase activity was not a pro-cancer gene [7]. This indicates that H3K9 methylation is pro-tumorigenic in this cancer type, however, since only SETDB1 is overexpressed in human melanoma (Fig. 2), it suggests that some feature of SETDB1 specifically provides a selective advantage for melanoma cells (Table 1).
Table 1.
Frequent epigenetic marks which are deregulated in cancers
| Deregulated mark | Change in cancer | Modifier | Associated with | References |
|---|---|---|---|---|
| DNA methylation | Regional hypermethylation | DNMT1, DNMT3, UHRF1a | Gene repression | Herman et al. (1995) [98], Ai et al. (2006) [40], Bae et al. (2004) [41], Kornegoo et al. (2012) [99], Roll et al. (2013) [43], Zou et al. (2009) [100] |
| Global hypomethylation | Genomic instability | Bedford and van Helden (1987) [101], Cadieux et al. (2006) [102], Feinberg and Vogelstein (1983) [74], Fraga et al. (2005) [103], Mudbhary et al. (2014) [5], Rauch et al. (2008) [104] | ||
| Activation of retrotransposons | Jackson-Grusby et al. (2001) [105], Howard et al. (2007) [106], Minoguchi and Iba (2008) [107] | |||
| H3K9me3 | Hypermethylation | G9a, SETDB1, SUV39H1 | Increased gene repression | Frigola et al. (2006) [108], Nguyen et al. (2002) [109] |
| H3k27me2–me3 | Hypermethylation | EZH1, EZH2 | DNA hypermethylation | Chinaranagari et al. (2014) [110], Ohm et al. (2007) [111], Popovic et al. (2014) [112], Schlesinger et al. (2007) [113] |
| H3K4me3 | Hypomethylation | MLL4 | Gene repression | Kang et al. (2015) [114], Nguyen et al. (2002) [109], Lee et al. (2013) [115] |
List of common epigenetic marks and their analogous regulators are shown in relation to reported changes in chromatin or gene expression resulting from their misregulation
Although it does not modify the DNA methylome, UHRF1 is an essential necessary component of the DNMT1 complex
With the tools in hand to manipulate the expression of these and other MTs in several zebrafish cancer models, it will be possible to assess their impact on gene expression and on other epigenetic marks. The ability to dissect both their individual and combinatorial effect on chromatin structure will further contribute to understanding the formation of melanoma [7] and rhabdomyosarcoma [9], in addition to other cancers where these genes are overexpressed.
Dissecting the Cancer Epigenome: Benefits and Limitations of Zebrafish
The ability to interrogate the epigenome has been transformed over the past decade with the advent of next generation sequencing (NGS) technologies. Additionally, using histone modifications and DNA methylation patterns as biomarkers for diagnosing diseases has generated new opportunities for the use of epigenome data. Typically, increasing resolution requires a trade off in genome coverage. An array of techniques for both genome-wide and locus specific analysis of specific epigenetic marks as well as for the accessibility of the DNA, reflecting open chromatin have been optimized and are used routinely in mammalian systems. Most of these approaches have been established in early embryos [4, 116] however, using samples composed of heterogeneous cells, such as whole embryos at post-cleavage stages of development or of tissues containing both cancer cells and the normal cells that surround the tumors can complicate analysis, because it is difficulty to assign the cellular origin of peaks in the sequencing data. To circumvent this, separating different tissue types requires the ability to label and isolate cells of interest, which is feasible using transgenic zebrafish expressing fluorescent proteins in the cell type of interest (Table 2).
Table 2.
Chromatin modifiers implicated in cancer and their analogous zebrafish models
| Chromatin modifier | Human cancers | Zebrafish cancer model | Mechanism of carcinogenesis in fish | References |
|---|---|---|---|---|
| UHRF1 | Multiple | HCC | Senescence escape, genomic instability | Cui et al. (2014) [117], Mudbhary et al. (2014) [5] |
| DNMT1 | Multiple | No | Unknown | Li et al. (2010) [118], Chen et al. (2006) [119], Xu et al. (2010) [120] |
| DNMT3a/b | AML, melanoma | No | Unknown | Im et al. (2014) [121], Nguyen et al. (2002) [109] |
| TET2 | Leukemia | Myelodysplasia | Unknown | Gjini et al. (2015) [8], Scourzic et al. (2015) [122] |
| EZH2 | Prostate, HCC, myeloma | No | Unknown | Chinaranagari et at. (2014) [110], Hung et al. (2014) [123] |
| SETDB1 | Melanoma | Melanoma | Cooperation with BRAFV 600E | Ceol et al. (2011) [7] |
| SUV39H1 | HCC, gastric colorectal, breast | Rhabdo- myosarcoma | Cyclin B1 repression | Albacker et al. (2013) [9], Cai et al. (2014) [124], Chiba et al. (2013) [92], Khanal et al. (2013) [125] |
| G9a | Squamous cell carcinoma, glioma | No | Unknown | Tao et al. (2014) [126], Zhong et al. (2015) [127] |
Since the structure of epigenetic marks are not species specific, many of the reagents to probe these marks can be used across phyla. Indeed, several histone marks [3, 4, 58, 128] and 5mC [14, 17, 48] have been profiled in zebrafish embryos and in tumors [3, 5]. However, antibody resources in zebrafish are a limitation in the field, finding antibodies that recognize endogenous chromatin modifiers in zebrafish to perform chromatin immunoprecipitation (ChIP) and other studies are a challenge. There are alternative approaches that can be applied to zebrafish samples to yield exciting and informative perspectives on the epigenome. Here, we review some of the common approaches in epigenomic analysis and discuss their applicability to zebrafish samples.
Methylome Analysis
Approaches for analyzing the methylome are numerous, and selection is based on cost, sample volume and downstream application, all of which are important considerations when using zebrafish. Genome-wide base pair resolution is the most informative of all the approaches, but it is also the most costly and often the most difficult to obtain, since mapping reads that are comprised entirely of repeat sequences is an informatic challenge [129], and this is confounded when mapping to the zebrafish genome, which is not as well assembled as the human or mouse. An alternative is the use of arrays, which provide a quick and relatively affordable means to investigate a subset of the genomic regions that are pre-selected, and the read out is as simple as a spreadsheet with relative—but not actual—values in methylation levels for each locus. On the other end of the spectrum is the use of methylation-sensitive restriction enzymes. The range of approaches used to investigate DNA methylation and hydroxymethylation and their applicability to zebrafish samples is presented in Table 3.
Table 3.
Approaches to interrogating the epigenome and their utility in zebrafish samples
| Approach | Application | Level | Resolution | Genome coverage | Enriched | Cost | Zebrafish reference |
|---|---|---|---|---|---|---|---|
| IHC/IF/immunoblot | DNA and Histone | Global | Lowest | High | No | $ | Mudhary et al. (2014) [5], Jacoh et al. (2015) [11] |
| Methylation sensitive restriction digest | DNA methylation | Global | Low | High | No | $ | Mudbhary et al. (2011) [130] |
| MNase assay | Nucleosome position | Global | Low | High | No | $ | |
| Methylation array | DNA methylation | Locus specific | Moderate | Moderate | No | $$ | Mirbahai et al. (2011) [4] |
| MSP | DNA methylation | Locus specific | High | Lowest | No | $$ | |
| BSP | DNA methylation | Locus specific | Highest | Lowest | No | $$ | |
| Chip-seq | Histone modification | Global/specific | Moderate | Moderate | Yes | $$$ | Lindeman et al. (2009) [116] |
| RRBS | DNA methylation | Global/Specific | Moderate | Moderate | Yes | $$$ | Chatterjee et al. (2013) [131] |
| WG-BS | DNA methylation | Global/Specific | Highest | Highest | No | $$$$ | – |
Different techniques used for analyzing the prevalence and distribution of different epigenetic marks are ranked based on genomic coverage, resolution and cost
Briefly, DNA methylation analyses can be subdivided into those that focus on locus-specific vs. genome-wide assessment (with varying resolutions for each) and the ability to obtain base pair resolution compared to global genomic DNA methylation status. For a list of techniques for assessing DNA methylation status please refer to detailed reviews [129, 132, 133] and Table 3.
Identifying whether a cytosine is methylated or unmethylated in the genome relies on the mutagen, bisulfite, which converts any unmethylated cytosine to an uracil, but 5mC is protected and remains a C. Thus, sequencing bisulfite converted DNA will provide information about the methylation status of each cytosine. PCR- based DNA methylation analyses are the most common way of assessing locus specific methylation patterns and are routinely used in research using zebrafish. They fall into two categories; methylation specific PCR (MSP) and bisulfite sequencing PCR (BSP). Methylation status of specific loci can be presented as total methylation at that locus or as percent methylation for a particular CpG dinucleotide at a specific position. The utility and advantage of locus specific assessment of DNA methylation patterns is counterbalanced by the fact that methylation changes are not isolated events but phenomenon that occur across the entire genome.
Genome-wide analysis of specific base pairs or loci is provided by array and sequencing platforms. Affinity enriched microarrays provide a detailed, genome- wide look at certain parts of the genome such as promoters, gene bodies, CpG islands and shores, and have the advantage of being customizable. MeDIP uses an antibody to 5mC to bind the methylated regions of the genome and then precipitate them using the standard approaches for chromatin immunoprecipitation (ChIP). The precipitated DNA can then be used for microarray or sequencing. The advantages of using MeDIP for arrays are cut short by the inherent limitation of the array design. Although high-throughput sequencing can eliminate array based design biases it still faces the shortcomings associated with MeDIP such as mapping difficulty (see the section “Overcoming Limitations of the Zebrafish Model”), low coverage and the fact that ChIP only identifies those regions that are enriched for 5mC, but does not indicate the actual methylation status of CpGs in this region.
The most recent development in DNA methylation analysis, sequence-based profiling, couples bisulfite conversion with NGS. Since no enrichment is required for this method, all regions of the genome are equally represented and single base pair resolution can be achieved. This technology, however, is still costly and often such depth is unnecessary if one wishes to focus primarily on CpG rich regions. RRBS is one method that utilizes NGS with methylation enrichment and bisulfite conversion. Since RRBS features a digestion step with MspI, which recognizes CpGs, it ensures that the sample is enriched for those dinucleotides after size selection and PCR amplification. The amount of required input DNA is also a lot less for RRBS than some of the other methods, which makes this ideal for working with small sized samples such as zebrafish tissues or tumors. However, it is important to note that even though the frequency of CpG dinucleotides is more common in zebrafish genome relative to human or mouse [131], there are fewer MspI restriction sites and thus less enrichment of CpG high regions for these samples.
WGS of bisulfite converted DNA provides the most amount of information, however, this is a challenge, as it requires 1–5 μg of starting genomic material [73]. Amounts such as these can be difficult to obtain from zebrafish tissues or tumors unless samples are pooled. In addition, NGS based approaches often take many weeks to months to carry out, produce many gigabytes of data and require sophisticated analytic ability that is not available to all laboratories. Finally, the relative cost is an important consideration: ranging from a few 100 dollars to analyze hundreds of samples by methylation restriction digestion to several 1000 dollars for NGS of a single WGS sample. Therefore, while these approaches are powerful, it is important to balance the drive to obtain an omics-level perspective of all data with the ability to generate large sample sizes quickly using zebrafish.
Histone Modifications, Variants and Nucleosome Positioning
Detecting the presence and absence of certain histone marks and positions of nucleosomes on chromatin can lend insight into the development and nature of different cancers. Immunofluorescence and Western blotting can give a general overview of histone variant abundance and epigenetic marks since these are well conserved in zebrafish and most commercially developed antibodies for histone and DNA modifications cross react with zebrafish. Thus, analyzing global levels of histone marks or variants are good first approach methods. However, often times it is necessary to know how certain histone marks change across the genome, or how they change at specific loci, where these techniques fall short.
ChIP can be used to analyze changes in histone modifications as they relate to certain regions of the genome, such as promoters or enhancers. ChIP relies on the idea that histones and DNA are close enough in proximity to become covalently linked. Briefly, cells undergo formaldehyde cross-linking and sonication to fragment their DNA. An antibody recognizing the PMT of choice is used to immunoprecipitate the sample followed by cross-link reversal. The freed DNA can then undergo NGS (ChIP-seq) or microarray analysis (ChIP-chip) to determine which genomic regions were enriched for that PMT. ChIP-seq can be easily applied to zebrafish and a methods paper—“Fish and ChIPs”—has been recently published [116].
Nucleosome positioning is another useful epigenetic marker when looking at the cancer genome. Nucleosome occupancy is a critical factor in determining chromatin structure and density and is thus strongly linked to the balance between hetero- and euchromatin and DNA methylation [71]. Methods for studying nucleosome positioning, such as MNAse digestion or DNAse hypersensitivity assay, take advantage of enzymes that are able to cut DNA on nucleosome barren regions thus generating short fragments that can be subjected to sequencing and mapped to specific loci.
Additional insights can be gained by combining epigenomics data with previously reported mRNA expression profiles. Databases, such as ENCODE, allow researchers to overlay genome-wide epigenetic interrogations with expression profiles to gain a better understanding of gene regulation. However, such large-scale, genomic collections are still in their early phases, with limited sample data.
Overcoming Limitations of the Zebrafish model
Although many epigenetic techniques have been successfully coupled with the zebrafish model, there are still several limitations that must be overcome before the fish system can rival the current mammalian standards. The small size of zebrafish, which enables high-throughput genetic screens, becomes problematic when trying to collect a sufficient amount of sample to carry out genome-wide epigenome interrogations. Obtaining sufficient genomic DNA from a larval liver is impossible and thus calls for pooling of samples. This can limit research applications—for example, pooling samples is acceptable for assessing liver development, where it is presumed that there is low animal-to-animal variability in hepatoblasts, but becomes unacceptable for assessing tumors, which are inherently heterogeneous even within a single animal. Since it is unlikely that the size of the zebrafish will increase in the near future, resolution to this problem depend on enhancing the sensitivity of techniques used to generate epigenetic data.
A second problem stems from the limited annotation of the zebrafish genome. This, coupled with the generation of short DNA fragments from NGS makes it difficult to map repetitive elements to correct positions in the genome. Only ~30 % of the zebrafish library generated from RRBS maps to unique sites in the genome—the rest maps to multiple regions [131]. Incomplete annotation results in “orphaned” data that could contain potentially useful information but cannot be accessed without knowing its genomic address. It is anticipated that the continual efforts to improve the annotation and assembly of the zebrafish genome—the tenth version of the assembled genome was just released (GRCz10; http://www.sanger.ac.uk/resources/zebrafish/genomeproject.html_)—and advances in bioinformatics approaches and tools for mapping will overcome this limitation.
Clinical Impacts and Future Perspectives
The cancer genomic landscape is complex, and dissecting the functional relevance of complex genetic and epigenetic changes in cancer cells requires in vivo systems. Zebrafish provide an excellent system to carry these out, as all epigenetic marks investigated to date are conserved in zebrafish, and the mechanism for genome editing and tissue specific over-expression is straightforward, as demonstrated by many of the studies reviewed here. Moreover, the ability to carry out epigenetic analysis is improving in this system, which provides a unique opportunity to sort the genome- wide data obtained from human tumors into a relevant framework. As the ability to screen drugs for efficacy in zebrafish tumor models advances [6, 134], this provides a tractable system to not only identify important cancer genes and pathways, but also to screen for drugs that will halt cancer.
Contributor Information
Yelena Chernyavskaya, Department of Developmental and Regenerative Biology, Icahn School of Medicine at Mount Sinai, Box 1020, 1 Gustave L. Levy Place, New York, NY 10029, USA. Division of Liver Diseases, Department of Medicine, Icahn School of Medicine at Mount Sinai, Box 1020, 1 Gustave L. Levy Place, New York, NY 10029, USA.
Brandon Kent, Department of Developmental and Regenerative Biology, Icahn School of Medicine at Mount Sinai, Box 1020, 1 Gustave L. Levy Place, New York, NY 10029, USA. Division of Liver Diseases, Department of Medicine, Icahn School of Medicine at Mount Sinai, Box 1020, 1 Gustave L. Levy Place, New York, NY 10029, USA. School of Biomedical Science, Icahn School of Medicine at Mount Sinai, Box 1020, 1 Gustave L. Levy Place, New York, NY 10029, USA.
Kirsten C. Sadler, Biology Program, New York University Abu Dhabi, Saadiyat Campus, P.O. Box 129188, Abu Dhabi, United Arab Emirates. Department of Developmental and Regenerative Biology, Icahn School of Medicine at Mount Sinai, Box 1020, 1 Gustave L. Levy Place, New York, NY 10029, USA. Division of Liver Diseases, Department of Medicine, Icahn School of Medicine at Mount Sinai, Box 1020, 1 Gustave L. Levy Place, New York, NY 10029, USA. School of Biomedical Science, Icahn School of Medicine at Mount Sinai, Box 1020, 1 Gustave L. Levy Place, New York, NY 10029, USA
References
- 1.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anelli V, Santoriello C, Distel M, et al. Global repression of cancer gene expression in a zebrafish model of melanoma is linked to epigenetic regulation. Zebrafish. 2009;6(4):417–424. doi: 10.1089/zeb.2009.0612. [DOI] [PubMed] [Google Scholar]
- 4.Mirbahai L, Williams TD, Zhan H, et al. Comprehensive profiling of zebrafish hepatic proximal promoter CpG island methylation and its modification during chemical carcinogenesis. BMC Genomics. 2011;12:3. doi: 10.1186/1471-2164-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mudbhary R, Hoshida Y, Chernyavskaya Y, et al. Overexpression of UHRF1 drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell. 2014;25:1–14. doi: 10.1016/j.ccr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White R, Rose K, Zon L. Zebrafish cancer: the state of the art and the path forward. Nat Rev Cancer. 2013;13(9):624–636. doi: 10.1038/nrc3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceol CJ, Houvras Y, Jane-Valbuena J, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471(7339):513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gjini E, Mansour MR, Sander JD, et al. A zebrafish model of myelodysplastic syndrome produced through tet2 genomic editing. Mol Cell Biol. 2015;35(5):789–804. doi: 10.1128/MCB.00971-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albacker CE, Storer NY, Langdon EM, et al. The histone methyltransferase SUV39H1 suppresses embryonal rhabdomyosarcoma formation in zebrafish. PLoS One. 2013;8(5):e64969. doi: 10.1371/journal.pone.0064969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milutinovic S, Zhuang Q, Niveleau A, et al. Epigenomic stress response. Knockdown of DNA methyltransferase 1 triggers an intra-S-phase arrest of DNA replication and induction of stress response genes. J Biol Chem. 2003;278(17):14985–14995. doi: 10.1074/jbc.M213219200. [DOI] [PubMed] [Google Scholar]
- 11.Jacob V, Chernyavskaya Y, Chen X, et al. DNA hypomethylation induces a DNA replication-associated cell cycle arrest to block hepatic outgrowth in uhrf1 mutant zebrafish embryos. Development. 2015;142(3):510–521. doi: 10.1242/dev.115980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Almeida RD, Loose M, Sottile V, et al. 5-hydroxymethyl-cytosine enrichment of non- committed cells is not a universal feature of vertebrate development. Epigenetics. 2012;7(4):383–389. doi: 10.4161/epi.19375. [DOI] [PubMed] [Google Scholar]
- 13.Andersen IS, Ostrup O, Lindeman LC, et al. Epigenetic complexity during the zebrafish mid-blastula transition. Biochem Biophys Res Commun. 2012;417(4):1139–1144. doi: 10.1016/j.bbrc.2011.12.077. [DOI] [PubMed] [Google Scholar]
- 14.Andersen IS, Lindeman LC, Reiner AH, et al. Epigenetic marking of the zebrafish developmental program. Curr Top Dev Biol. 2013;104:85–112. doi: 10.1016/B978-0-12-416027-9.00003-6. [DOI] [PubMed] [Google Scholar]
- 15.Bogdanovic O, Fernandez-Minan A, Tena JJ, et al. The developmental epigenomics toolbox: ChIP-seq and MethylCap-seq profiling of early zebrafish embryos. Methods. 2013;62(3):207–215. doi: 10.1016/j.ymeth.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 16.Hirose K, Shimoda N, Kikuchi Y. Transient reduction of 5-methylcytosine and 5-hydroxymethylcytosine is associated with active DNA demethylation during regeneration of zebrafish fin. Epigenetics. 2013;8(9):899–906. doi: 10.4161/epi.25653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindeman LC, Winata CL, Aanes H, et al. Chromatin states of developmentally- regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos. Int J Dev Biol. 2010;54(5):803–813. doi: 10.1387/ijdb.103081ll. [DOI] [PubMed] [Google Scholar]
- 18.Long HK, Sims D, Heger A, et al. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife. 2013;2:e00348. doi: 10.7554/eLife.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang HT, Kathrein KL, Barton A, et al. A network of epigenetic regulators guides developmental haematopoiesis in vivo. Nat Cell Biol. 2013;15(12):1516–1525. doi: 10.1038/ncb2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bert SA, Robinson MD, Strbenac D, et al. Regional activation of the cancer genome by long-range epigenetic remodeling. Cancer Cell. 2013;23(1):9–22. doi: 10.1016/j.ccr.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Tang Y, Gao XD, Wang Y, et al. Widespread existence of cytosine methylation in yeast DNA measured by gas chromatography/mass spectrometry. Anal Chem. 2012;84(16):7249–7255. doi: 10.1021/ac301727c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGhee JD, Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- 23.Sun XJ, Xu PF, Zhou T, et al. Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLoS One. 2008;3(1):e1499. doi: 10.1371/journal.pone.0001499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 25.Bestor TH, Edwards JR, Boulard M. Notes on the role of dynamic DNA methylation in mammalian development. Proc Natl Acad Sci U S A. 2014;112:6796–6799. doi: 10.1073/pnas.1415301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohandas T, Sparkes RS, Shapiro LJ. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981;211(4480):393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- 27.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 28.Onodera Y, Haag JR, Ream T, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120(5):613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Ooi SK, O’Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J Cell Sci. 2009;122(Pt 16):2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson RM, Bosch JA, Goll MG, et al. Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Dev Biol. 2009;334:213–223. doi: 10.1016/j.ydbio.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goll MG, Anderson R, Stainier DY, et al. Transcriptional silencing and reactivation in transgenic zebrafish. Genetics. 2009;182(3):747–755. doi: 10.1534/genetics.109.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthews RP, Lorent K, Manoral-Mobias R, et al. TNFα-dependent hepatic steatosis and liver degeneration caused by mutation of zebrafish s-adenosylhomocysteine hydrolase. Development. 2009;136(5):865–875. doi: 10.1242/dev.027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song J, Rechkoblit O, Bestor TH, et al. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science. 2011;331(6020):1036–1040. doi: 10.1126/science.1195380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bostick M, Kim JK, Esteve PO, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317(5845):1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 35.Sharif J, Muto M, Takebayashi S, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450(7171):908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 36.Hill PW, Amouroux R, Hajkova P. DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: an emerging complex story. Genomics. 2014;104(5):324–333. doi: 10.1016/j.ygeno.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Swindle CS, Kim HG, Klug CA. Mutation of CpGs in the murine stem cell virus retroviral vector long terminal repeat represses silencing in embryonic stem cells. J Biol Chem. 2004;279(1):34–41. doi: 10.1074/jbc.M309128200. [DOI] [PubMed] [Google Scholar]
- 38.Yu W, McIntosh C, Lister R, et al. Genome-wide DNA methylation patterns in LSH mutant reveals de-repression of repeat elements and redundant epigenetic silencing pathways. Genome Res. 2014;24(10):1613–1623. doi: 10.1101/gr.172015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.John RM, Lefebvre L. Developmental regulation of somatic imprints. Differentiation. 2011;81(5):270–280. doi: 10.1016/j.diff.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Ai L, Kim WJ, Kim TY, et al. Epigenetic silencing of the tumor suppressor cystatin M occurs during breast cancer progression. Cancer Res. 2006;66(16):7899–7909. doi: 10.1158/0008-5472.CAN-06-0576. [DOI] [PubMed] [Google Scholar]
- 41.Bae YK, Brown A, Garrett E, et al. Hypermethylation in histologically distinct classes of breast cancer. Clin Cancer Res. 2004;10(18 Pt 1):5998–6005. doi: 10.1158/1078-0432.CCR-04-0667. [DOI] [PubMed] [Google Scholar]
- 42.Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2:e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roll JD, Rivenbark AG, Sandhu R, et al. Dysregulation of the epigenome in triple- negative breast cancers: basal-like and claudin-low breast cancers express aberrant DNA hypermethylation. Exp Mol Pathol. 2013;95(3):276–287. doi: 10.1016/j.yexmp.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 44.Maeder ML, Angstman JF, Richardson ME, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat Biotechnol. 2013;31(12):1137–1142. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng S, Cokus SJ, Zhang X, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 2010;107(19):8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ge L, Zhang RP, Wan F, et al. TET2 plays an essential role in erythropoiesis by regulating lineage-specific genes via DNA oxidative demethylation in a zebrafish model. Mol Cell Biol. 2014;34(6):989–1002. doi: 10.1128/MCB.01061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mhanni AA, McGowan RA. Global changes in genomic methylation levels during early development of the zebrafish embryo. Dev Genes Evol. 2004;214(8):412–417. doi: 10.1007/s00427-004-0418-0. [DOI] [PubMed] [Google Scholar]
- 48.Andersen IS, Reiner AH, Aanes H, et al. Developmental features of DNA methylation during activation of the embryonic zebrafish genome. Genome Biol. 2012;13(7):R65. doi: 10.1186/gb-2012-13-7-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rai K, Huggins IJ, James SR, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135(7):1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGaughey DM, Abaan HO, Miller RM, et al. Genomics of CpG methylation in developing and developed zebrafish. G3 (Bethesda) 2014;4(5):861–869. doi: 10.1534/g3.113.009514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cock-Rada AM, Medjkane S, Janski N, et al. SMYD3 promotes cancer invasion by epigenetic upregulation of the metalloproteinase MMP-9. Cancer Res. 2012;72(3):810–820. doi: 10.1158/0008-5472.CAN-11-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rai K, Nadauld LD, Chidester S, et al. Zebra fish Dnmt1 and Suv39h1 regulate organ- specific terminal differentiation during development. Mol Cell Biol. 2006;26(19):7077–7085. doi: 10.1128/MCB.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tena JJ, Gonzalez-Aguilera C, Fernandez-Minan A, et al. Comparative epigenomics in distantly related teleost species identifies conserved cis-regulatory nodes active during the vertebrate phylotypic period. Genome Res. 2014;24(7):1075–1085. doi: 10.1101/gr.163915.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2(5):657–669. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albert M, Helin K. Histone methyltransferases in cancer. Semin Cell Dev Biol. 2010;21(2):209–220. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 56.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kampilafkos P, Melachrinou M, Kefalopoulou Z, et al. Epigenetic modifications in cutaneous malignant melanoma: EZH2, H3K4me2, and H3K27me3 immunohistochemical expression is enhanced at the invasion front of the tumor. Am J Dermatopathol. 2015;37(2):138–144. doi: 10.1097/DAD.0b013e31828a2d54. [DOI] [PubMed] [Google Scholar]
- 58.Vastenhouw NL, Zhang Y, Woods IG, et al. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464(7290):922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindeman LC, Andersen IS, Reiner AH, et al. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev Cell. 2011;21(6):993–1004. doi: 10.1016/j.devcel.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 60.de la Barre AE, Gerson V, Gout S, et al. Core histone N-termini play an essential role in mitotic chromosome condensation. EMBO J. 2000;19(3):379–391. doi: 10.1093/emboj/19.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Hooser A, Goodrich DW, Allis CD, et al. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. J Cell Sci. 1998;111(Pt 23):3497–3506. doi: 10.1242/jcs.111.23.3497. [DOI] [PubMed] [Google Scholar]
- 62.Pereira S, Bourrachot S, Cavalie I, et al. Genotoxicity of acute and chronic gamma- irradiation on zebrafish cells and consequences for embryo development. Environ Toxicol Chem. 2011;30(12):2831–2837. doi: 10.1002/etc.695. [DOI] [PubMed] [Google Scholar]
- 63.Karmodiya K, Anamika K, Muley V, et al. Camello, a novel family of Histone Acetyltransferases that acetylate histone H4 and is essential for zebrafish development. Sci Rep. 2014;4:6076. doi: 10.1038/srep06076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stadler JA, Shkumatava A, Norton WH, et al. Histone deacetylase 1 is required for cell cycle exit and differentiation in the zebrafish retina. Dev Dyn. 2005;233(3):883–889. doi: 10.1002/dvdy.20427. [DOI] [PubMed] [Google Scholar]
- 65.Cunliffe VT, Casaccia-Bonnefil P. Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mech Dev. 2006;123(1):24–30. doi: 10.1016/j.mod.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 66.Farooq M, Sulochana KN, Pan X, et al. Histone deacetylase 3 (hdac3) is specifically required for liver development in zebrafish. Dev Biol. 2008;317:336–353. doi: 10.1016/j.ydbio.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 67.Noel ES, Casal-Sueiro A, Busch-Nentwich E, et al. Organ-specific requirements for Hdac1 in liver and pancreas formation. Dev Biol. 2008;322(2):237–250. doi: 10.1016/j.ydbio.2008.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y, Wang J, Xie Y, et al. Pattern of change in histone 3 lysine 9 acetylation and histone deacetylases in development of zebrafish embryo. J Genet. 2014;93(2):539–544. doi: 10.1007/s12041-014-0403-y. [DOI] [PubMed] [Google Scholar]
- 69.Yamaguchi M, Tonou-Fujimori N, Komori A, et al. Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and Notch signaling pathways. Development. 2005;132(13):3027–3043. doi: 10.1242/dev.01881. [DOI] [PubMed] [Google Scholar]
- 70.Shabason JE, Tofilon PJ, Camphausen K. HDAC inhibitors in cancer care. Oncology (Williston Park) 2010;24(2):180–185. [PMC free article] [PubMed] [Google Scholar]
- 71.Chodavarapu RK, Feng S, Bernatavichute YV, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466(7304):388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roadmap Epigenomics C. Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stirzaker C, Taberlay PC, Statham AL, et al. Mining cancer methylomes: prospects and challenges. Trends Genet. 2014;30(2):75–84. doi: 10.1016/j.tig.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 74.Feinberg AP, Vogelstein B. Alterations in DNA methylation in human colon neoplasia. Semin Surg Oncol. 1987;3(3):149–151. doi: 10.1002/ssu.2980030304. [DOI] [PubMed] [Google Scholar]
- 75.Rainier S, Johnson LA, Dobry CJ, et al. Relaxation of imprinted genes in human cancer. Nature. 1993;362(6422):747–749. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 76.Jadersten M, Hellstrom-Lindberg E. New clues to the molecular pathogenesis of myelodysplastic syndromes. Exp Cell Res. 2010;316(8):1390–1396. doi: 10.1016/j.yexcr.2010.02.043. [DOI] [PubMed] [Google Scholar]
- 77.Colquitt BM, Allen WE, Barnea G, et al. Alteration of genic 5-hydroxymethylcytosine patterning in olfactory neurons correlates with changes in gene expression and cell identity. Proc Natl Acad Sci U S A. 2013;110(36):14682–14687. doi: 10.1073/pnas.1302759110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rai K, Sarkar S, Broadbent TJ, et al. DNA demethylase activity maintains intestinal cells in an undifferentiated state following loss of APC. Cell. 2010;142(6):930–942. doi: 10.1016/j.cell.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8(12):686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou L, Zhao X, Han Y, et al. Regulation of UHRF1 by miR-146a/b modulates gastric cancer invasion and metastasis. FASEB J. 2013;27(12):4929–4939. doi: 10.1096/fj.13-233387. [DOI] [PubMed] [Google Scholar]
- 81.Wang F, Yang YZ, Shi CZ, et al. UHRF1 promotes cell growth and metastasis through repression of p16 ink4a in colorectal cancer. Ann Surg Oncol. 2012;19:2753–2762. doi: 10.1245/s10434-011-2194-1. [DOI] [PubMed] [Google Scholar]
- 82.Babbio F, Pistore C, Curti L, et al. The SRA protein UHRF1 promotes epigenetic crosstalks and is involved in prostate cancer progression. Oncogene. 2012;31:4878–4887. doi: 10.1038/onc.2011.641. [DOI] [PubMed] [Google Scholar]
- 83.Daskalos A, Oleksiewicz U, Filia A, et al. UHRF1-mediated tumor suppressor gene inactivation in nonsmall cell lung cancer. Cancer. 2011;117(5):1027–1037. doi: 10.1002/cncr.25531. [DOI] [PubMed] [Google Scholar]
- 84.Unoki M, Daigo Y, Koinuma J, et al. UHRF1 is a novel diagnostic marker of lung cancer. Br J Cancer. 2010;103(2):217–222. doi: 10.1038/sj.bjc.6605717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Unoki M, Kelly JD, Neal DE, et al. UHRF1 is a novel molecular marker for diagnosis and the prognosis of bladder cancer. Br J Cancer. 2009;101(1):98–105. doi: 10.1038/sj.bjc.6605123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jin W, Chen L, Chen Y, et al. UHRF1 is associated with epigenetic silencing of BRCA1 in sporadic breast cancer. Breast Cancer Res Treat. 2009;123:359–373. doi: 10.1007/s10549-009-0652-2. [DOI] [PubMed] [Google Scholar]
- 87.Jenkins Y, Markovtsov V, Lang W, et al. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol Biol Cell. 2005;16(12):5621–5629. doi: 10.1091/mbc.E05-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sadler KC, Krahn KN, Gaur NA, et al. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc Natl Acad Sci U S A. 2007;104(5):1570–1575. doi: 10.1073/pnas.0610774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Du Z, Song J, Wang Y, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3(146):ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qin W, Leonhardt H, Spada F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J Cell Biochem. 2011;112(2):439–444. doi: 10.1002/jcb.22998. [DOI] [PubMed] [Google Scholar]
- 91.Wu PC, Lu JW, Yang JY, et al. H3K9 histone methyltransferase, KMT1E/SETDB1, cooperates with the SMAD2/3 pathway to suppress lung cancer metastasis. Cancer Res. 2014;74(24):7333–7343. doi: 10.1158/0008-5472.CAN-13-3572. [DOI] [PubMed] [Google Scholar]
- 92.Chiba T, Saito T, Yuki K, et al. Histone lysine methyltransferase SUV39H1 is a potent target for epigenetic therapy of hepatocellular carcinoma. Int J Cancer. 2015;136(2):289–298. doi: 10.1002/ijc.28985. [DOI] [PubMed] [Google Scholar]
- 93.Patton EE, Widlund HR, Kutok JL, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15(3):249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 94.Macgregor S, Montgomery GW, Liu JZ, et al. Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nat Genet. 2011;43(11):1114–1118. doi: 10.1038/ng.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fritsch L, Robin P, Mathieu JR, et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell. 2010;37(1):46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 96.Langenau DM, Keefe MD, Storer NY, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21(11):1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen EY, Dobrinski KP, Brown KH, et al. Cross-species array comparative genomic hybridization identifies novel oncogenic events in zebrafish and human embryonal rhabdomyosarcoma. PLoS Genet. 2013;9(8):e1003727. doi: 10.1371/journal.pgen.1003727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 99.Kornegoor R, Moelans CB, Verschuur-Maes AH, Hogenes MC, de Bruin PC, Oudejans JJ, van Diest PJ. Promoter hypermethylation in male breast cancer: analysis by multiplex ligation-dependent probe amplification. Breast Cancer Res. 2012;14:1–9. doi: 10.1186/bcr3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zou X-P, Zhang B, Zhang X-Q, Chen M, Cao J, Liu W-J. Promoter hypermethylation of multiple genes in early gastric adenocarcinoma and precancerous lesions. Hum Pathol. 2009;40:1534–1542. doi: 10.1016/j.humpath.2009.01.029. [DOI] [PubMed] [Google Scholar]
- 101.Bedford MT, van Helden PD. Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res. 1987;47:5274–5276. [PubMed] [Google Scholar]
- 102.Cadieux B, Ching T-T, VandenBerg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 103.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 104.Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, Riggs AD, Pfeifer GP. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 106.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2007;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 107.Minoguchi S, Iba H. Instability of retroviral DNA methylation in embryonic stem cells. Stem Cells. 2008;26:1166–1173. doi: 10.1634/stemcells.2007-1106. [DOI] [PubMed] [Google Scholar]
- 108.Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 109.Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JCY, Liang G, Jones PA. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-Aza-2′-deoxycytidine. Cancer Res. 2002;62:6456–6461. [PubMed] [Google Scholar]
- 110.Chinaranagari S, Sharma P, Chaudhary J. EZH2 dependent H3K27me3 is involved in epigenetic silencing of ID4 in prostate cancer. Oncotarget. 2014;5:7172–7182. doi: 10.18632/oncotarget.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell–like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Popovic R, Martinez-Garcia E, Giannopoulou EG, Zhang Q, Zhang Q, Ezponda T, Shah MY, Zheng Y, Will CM, Small EC, et al. Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS Genet. 2014;10:e1004566. doi: 10.1371/journal.pgen.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 114.Kang J-Y, Song S-H, Yun J, Jeon M-S, Cha Y, Lee S-H, Kim H-P, Jeong E-G, Han S-W, Cho N-Y, et al. Identification of long-range epigenetic silencing on chromosome 15q25 and its clinical implication in gastric cancer. Am J Pathol. 2015;185(3):666–678. doi: 10.1016/j.ajpath.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 115.Lee J-E, Wang C, Xu S, Cho Y-W, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife. 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lindeman LC, Vogt-Kielland LT, Alestrom P, et al. Fish’n ChIPs: chromatin immunoprecipitation in the zebrafish embryo. Methods Mol Biol. 2009;567:75–86. doi: 10.1007/978-1-60327-414-2_5. [DOI] [PubMed] [Google Scholar]
- 117.Cui L, Chen J, Zhang Q, Wang X, Qu J, Zhang J, Dang S. Up-regulation of UHRF1 by oncogenic Ras promoted the growth, migration, and metastasis of pancreatic cancer cells. Mol Cell Biochem. 2015;400:223–232. doi: 10.1007/s11010-014-2279-9. [DOI] [PubMed] [Google Scholar]
- 118.Li A, Omura N, Hong S-M, Goggins M. Pancreatic cancer DNMT1 expression and sensitivity to DNMT1 inhibitors. Cancer Biol Ther. 2010;9:321–329. doi: 10.4161/cbt.9.4.10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen D, Wang P, Huang A, Zhang B. Effects of pshRNA-DNMT1 on the proliferation and apoptosis of gastric cancer: experiments in vitro and in vivo. Zhonghua Yi Xue Za Zhi. 2006;86:1534–1539. [PubMed] [Google Scholar]
- 120.Xu H, Wang Z, Liu D, Li Q, Dai S, Wang E. High expression of DNMT1 was correlated with beta-catenin accumulation and malignant phynotype of lung squamous cell carcinoma and adenocarcinoma. Chin J Lung Cancer. 2010;13:856–860. doi: 10.3779/j.issn.1009-3419.2010.09.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Im AP, Sehgal AR, Carroll MP, Smith BD, Tefferi A, Johnson DE, Boyiadzis M. DNMT3A and IDH mutations in acute myeloid leukemia and other myeloid malignancies: associations with prognosis and potential treatment strategies. Leukemia. 2014;28:1774–1783. doi: 10.1038/leu.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Scourzic L, Mouly E, Bernard OA. TET proteins and the control of cytosine demethylation in cancer. Genome Med. 2015;7:1–16. doi: 10.1186/s13073-015-0134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hung S-Y, Lin H-H, Yeh K-T, Chang J-G. Histone-modifying genes as biomarkers in hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- 124.Cai L, Ma X, Huang Y, Zou Y, Chen X. Aberrant histone methylation and the effect of Suv39H1 siRNA on gastric carcinoma. Oncol Rep. 2014;31:2593–2600. doi: 10.3892/or.2014.3135. [DOI] [PubMed] [Google Scholar]
- 125.Khanal P, Kim G, Lim S-C, Yun H-J, Lee KY, Choi H-K, Choi HS. Prolyl isomerase Pin1 negatively regulates the stability of SUV39H1 to promote tumorigenesis in breast cancer. FASEB J. 2013;27:4606–4618. doi: 10.1096/fj.13-236851. [DOI] [PubMed] [Google Scholar]
- 126.Tao H, Li H, Su Y, Feng D, Wang X, Zhang C, Ma H, Hu Q. Histone methyltransferase G9a and H3K9 dimethylation inhibit the self-renewal of glioma cancer stem cells. Mol Cell Biochem. 2014;394:23–30. doi: 10.1007/s11010-014-2077-4. [DOI] [PubMed] [Google Scholar]
- 127.Zhong X, Chen X, Guan X, Zhang H, Ma Y, Zhang S, Wang E, Zhang L, Han Y. Overexpression of G9a and MCM7 in oesophageal squamous cell carcinoma is associated with poor prognosis. Histopathology. 2015;66:192–200. doi: 10.1111/his.12456. [DOI] [PubMed] [Google Scholar]
- 128.Aday AW, Zhu LJ, Lakshmanan A, et al. Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites. Dev Biol. 2011;357(2):450–462. doi: 10.1016/j.ydbio.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bock C, Tomazou EM, Brinkman AB, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28(10):1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mudbhary R, Sadler KC. Epigenetics, development, and cancer: zebrafish make their mark. Birth Defects Res Part C Embryo Today Rev. 2011;93:194–203. doi: 10.1002/bdrc.20207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chatterjee A, Ozaki Y, Stockwell PA, et al. Mapping the zebrafish brain methylome using reduced representation bisulfite sequencing. Epigenetics. 2013;8(9):979–989. doi: 10.4161/epi.25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 2012;13(10):705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 133.Gupta R, Nagarajan A, Wajapeyee N. Advances in genome-wide DNA methylation analysis. Biotechniques. 2010;49(4):3–11. doi: 10.2144/000113493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rennekamp AJ, Peterson RT. 15 years of zebrafish chemical screening. Curr Opin Chem Biol. 2014;24C:58–70. doi: 10.1016/j.cbpa.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]