Abstract
Chemical cross-linking and mass spectrometry are now widely used to analyze large-scale protein-protein interactions. The major challenge in cross-linking approaches is the complexity of the mass spectrometric data. New approaches are required that can identify cross-linked peptides with high-confidence and establish a user-friendly analysis protocol for the biomedical scientific community. Here, we introduce a novel cross-linker that can be selectively cleaved in the gas phase using two differential tandem mass-spectrometric fragmentation methods, such as collision-induced or electron transfer dissociation (CID and ETD). This technique produces two signature mass spectra of the same cross-linked peptide, thereby producing high confidence in identifying the sites of interaction. Further tandem mass spectrometry can also give additional confidence on the peptide sequences. We demonstrate a proof-of-concept for this method using standard peptides and proteins. Peptides and proteins were cross-linked and their fragmentation characteristics were analyzed using CID and ETD tandem mass spectrometry. Two sequential cleavages unambiguously identified cross-linked peptides. In addition, the labeling efficiency of the new cross-linker was evaluated in macrophage immune cells after stimulation with the microbial ligand lipopolysaccharide and subsequent pulldown experiments with biotin-avidin affinity chromatography. We believe this strategy will help advance insights into the structural biology and systems biology of cell signaling.
INTRODUCTION
Current biochemical methods are not very efficient at analyzing systems-level or large-scale protein interaction networks. Most studies utilize a technique called “co-immunoprecipitation,” in which a protein is isolated along with its interacting partners (i.e., the protein complex) using an antibody or by incorporating an affinity group in the protein, which can be used as a hook to selectively purify it.1 This method is applicable for very strong and stable interactions, but most protein-protein interactions in cells are likely to be transient and weak. During the purification process these interactions may be lost completely. Moreover, co-immunoprecipitation is qualitative and generally provides little detailed information on the protein-to-protein interaction domains involved.
To solve this very important analytical shortcoming, a chemistry-based fixation method combined with mass spectrometry has come into the limelight, in which a reactive compound, referred to as a cross-linker, is utilized to stabilize a protein with its interaction partners by derivatizing certain side chains of the protein before cell lysis is performed.2 A cross-linker can then fix adjacent proteins or protein complexes using a chemical reaction, holding them tightly so they will not detach during cell lysis or subsequent strict purification conditions. In addition, a cross-linker can only react within a limited distance; hence, protein reactive sites can be measured by calculating the distance between the cross-linked sites. This method has two advantages: 1) it can identify large-scale protein interactions; and 2) it can identify protein structures in their native biological conditions.
The widespread use of this technology is hindered due to several bottlenecks. Traditional cross-linking strategies generate an enormous amount of mass spectrometry data, which is extremely difficult to analyze with routine software tools. Finding these interactions in large datasets is equivalent to finding a needle in a haystack. In this regard, several researchers have contributed by designing new strategies that add either enrichment functionality or cleavable bonds in the cross-linkers.2-12 To deal with the enormous complexity of data-analysis from traditional cross-linking, researchers have developed cleavable cross-linking approaches.
The first efficient cleavable cross-linker, called PIR, used reporter ions and was developed by the James Bruce group at Washington State University and the University of Washington.5,13
Although there were some concerns about the reactive distances, this cross-linking approach has been successfully applied in several large-scale systems and is currently the most efficient cleavable cross-linker reported to date.14-17 Another efficient cleavable cross-linker was developed by Lan Huang's group at UC Irvine. This DSSO cross-linker utilized the labile nature of the sulfoxide group and was also successfully used in several large-scale applications.16,17 Goshe's group at NC State University utilized an Asp-Pro (DP) peptide bond in their cross-linkers, which are efficiently cleaved during low-energy CID-MS/MS.4,18 In addition, a significant number of cross-linkers have been reported by Petrotchenko et al.3,19 The Heck group has also recently developed an integrated workflow for proteome-wide profiling of protein cross-linking using the CID cleavable cross-linker DSSO.20
Although often advantageous, a cleavable cross-linker can also have several disadvantages compared with other traditional cross-linkers. Particularly after cleavage, it requires further tandem mass spectrometry of the cross-linked peptide for sequencing applications. Sometimes these fragmentations are very ambiguous due to the cleavage of the attached cross-linker parts.
All of these cross-linkers have contributed tremendously to the cross-linking field, but confident data analysis is still a major hurdle. Two advances will be critically important to make cleavable cross-linking technology widely amenable for analyzing large-scale protein interactions: 1) the design of effective cleavable chemical cross-linkers with innovative features, which will help reduce the complexity of mass-spectrometry data of large-scale protein interactions; and 2) the development of robust and user-friendly software tools.20-24
To develop a cutting-edge cross-linking technology that will overcome the bottlenecks of existing cross-linking strategies, we have designed a DUal Cleavable Cross-linking Technology (DUCCT), which will improve the confidence in identifying cross-linked peptides by mass spectrometry. At the core of this method is the dual-mass-spectrometry-cleavable cross-linker, which can be fragmented by two differential tandem mass spectrometric techniques. These two differential tandem mass-spectrometric fragmentations will produce different signatures in the mass spectra for the same cross-linked peptide. The two complementary fragmentation signatures can help identify that cross-linked peptide with high confidence. Further MS/MS of the cross-linked peptide, if necessary, will produce additional confidence in identification.
We have synthesized the DUCCT cross-linker with two differential cleavable bonds.4,25 The individual cleavable groups have been tested by several groups, but due to the complexity of synthesis, no cross-linkers have been constructed using both of these bonds.4,25 In this report, we show our primary analysis of the effectiveness of this novel cross-linker. The technical simplicity and the quick screening capabilities of DUCCT will provide high confidence in data analysis. We believe this will significantly enhance confidence in the unambiguous identification of protein interactions in large-scale protein-to-protein cross-linking experiments.
EXPERIMENTAL SECTION
Materials
Neurotensin (pyr-LYNKPRRPYIL) was purchased from Anaspec, Inc. (San Jose, CA). Bovine serum albumin (BSA), ubiquitin, Tris-HCl, dimethyl sulfoxide (DMSO), ammonium bicarbonate, and formic acid were obtained from Sigma-Aldrich (St. Louis, MO). Liquid chromatography-mass spectrometry (LC-MS) grade methanol and acetonitrile were obtained from VWR (Radnor, PA). A reducing agent, dithiothreitol (Biorad, CA), an alkylating agent, iodoacetamide (Sigma-Aldrich, MO), and a protease, trypsin (Promega, Madison, WI) were utilized to digest the proteins properly. For synthesis of the cross-linker, Fmoc amino acid starting materials were obtained from EMD Millipore (Billerica, MA), formylbenzoic acid from Sigma-Aldrich (St. Luis, MO), Fmoc hydralink 6-Fmoc-HNA from Advanced Automated Peptide Protein Technology (Louisville, KY), and Dicyclohexylcarbodiimide (DCC) from Life Technologies (Pittsburgh, PA). 18 Milli-Q water was used for all the studies and was obtained from a water filtration system purchased from Aries Filterworks (West Berlin, NJ).
Cross-Linking of Neurotensin (Peptide)
The studied cross-linking agent (DUCCT) was prepared in DMSO. Neurotensin was treated with DUCCT in a 1:10 molar ratio in PBS buffer (pH 7.2). The reaction was permitted to continue for 30 mins at ambient temperature, after which the reaction was quenched with 50 mM Tris-HCl buffer (PH 8.0). The samples were desalted by ZipTip (Thermo Scientific, Waltham, MA), dried by speed vacuum, and finally reconstituted in 0.1% formic acid solution in H2O. Cross-linked peptide samples were analyzed with a Linear Ion Trap (LTQ) Velos Pro mass spectrometer (Thermo Scientific, USA).
Cross-Linking of Ubiquitin and BSA
The cross-linking reaction of two proteins, ubiquitin and BSA, with the DUCCT cross-linking agent was carried out in a 1:50 molar ratio in PBS buffer (pH 7.2). The reaction was allowed to proceed for 30 mins and then quenched by 50 mM Tris-HCl buffer. A protein concentrator (3 kDa molecular weight cut-off, Thermo Scientific, USA) was used to remove excess cross-linking agent. The protein concentration was determined using a BCA protein assay. Next, the cross-linked proteins were digested, using both in-gel and in-solution methods with trypsin.
In-Solution Digestion
The cross-linked protein was reduced with 10 mM dithiothreitol (Biorad, CA), alkylated with 55 mM iodoacetamide (Sigma-Aldrich, MO), and then digested by trypsin (Promega, Madison, WI). The protein-trypsin ratio was set at 50:1, and the sample was incubated overnight at 37 °C. The tryptic digestion was quenched using 0.1% Formic Acid (FA) in H2O. The sample was dried using speed vacuum, desalted by ZipTip, and re-suspended in 0.1% FA solution in H2O and finally transferred to an LC vial for mass spectrometric analysis.
In-Gel Digestion
The cross-linked protein was prepared with Laemmli buffer (Bio-Rad, OH), and heated for 5 min at 95 °C for denaturation. Next, the protein sample was loaded on a 10% SDS-PAGE gel. Separation of the proteins was observed according to their molecular weights. The gel bands were excised and digested after reduction and alkylation. After tryptic digestion, the peptides were extracted by 50% acetonitrile, dried by speed vacuum, and reconstituted in a solution of 0.1% FA in H2O.
Instrumental Analysis
For sample analysis, we utilized an LTQ Velos Pro mass spectrometer coupled with a UHPLC (UltiMate 3000, Dionix, USA). Cross-linked peptides were separated by reverse-phase chromatography, using a nano-viper analytical C18 column (Acclaim™ Pep Map™ 100 C18 LC Columns, Thermo Scientific). Separation was performed with a binary gradient system, in which the organic and aqueous mobile phases contained 95% acetonitrile and 98% water, respectively. The nano-column flow rate and injection volume was set at 300 nl/min and 5 μl (partial injection mode), respectively.
For ionization, the nano-electrospray ionization (ESI) source was utilized with a fixed spray voltage and heated capillary temperature of 2.0 V and 275 °C, respectively. Full scan spectra (AGC 3×04) were obtained from 350 to 2000 m/z. Data-dependent MS/MS spectra (AGC 1×104) were collected from the five most abundant precursor ions. The dynamic exclusion time was fixed at 30 ms for separating consecutive ions. Data acquisition was set for 90 mins. XCalibur software was utilized for data processing.
In CID fragmentation mode, the activation energy was set to 45%, along with an isolation width of 1.5 Da, activation Q of 0.25, and activation time of 10 ms. For ETD operation mode, we set the emission current, reagent ion electron energy, reagent ion source CI pressure, and reagent ion source temperature to 50 μA, −70 V, 20 psi, and 110 °C, respectively. The ETD reaction time was fixed at 80 ms, with an isolation width of 2 Da. Direct infusion was also conducted in the LTQ Velos Pro instrument to analyze the cross-linked peptides.
Synthesis of the Cross-Linker
The cross-linker was synthesized using Fmoc peptide synthesis reagents with an Applied Biosystems 431 Peptide Synthesizer at the 0.25 mmol scale. Fmoc-Gly was first coupled to the resin, followed by the super-sensitive form of Fmoc-Asp, Fmoc-Asp(O-2-PhiPr) or Fmoc-Gly-Wang resin was purchased (Anaspec Inc), which was followed by the super-sensitive Fmoc-Asp. This was then followed by coupling Pro and then the typical form of Asp using Fmoc Asp-OtBu. The free amine of this Asp residue was coupled to the reagent Hydralink 6-Fmoc-HNA. After Fmoc was released, we added 4-formylbenzoic acid, which formed the hydrazone link and a terminal carboxylic acid. The protection group of the super-sensitive Asp was removed by 2% Trifluroaceticacid (TFA) in dichloromethane to create the second carboxylic acid. These two carboxylic acids were activated by DCC, to which N-hydroxy succinimide (NHS) esters were then formed by adding 2 mmol N-hydroxysuccinimide (Sigma-Aldrich, St. Louis, MO) with the DCC. The final compound was cleaved from the resin by 95% TFA with 5% H2O for 1 h. The product was precipitated with diethyl ether and centrifuged. The pellet was dissolved in 0.1% TFA in H2O and quickly lyophilized.
LPS-Biotin Pulldown Studies
Lipopolysaccharide (LPS) was purchased from Invivogen (San Diego, CA). Dr. Michael B. Fessler at NIEHS, NIH gifted us RAW 264.7 macrophage cells. The cells were grown in Dulbecco's Modified Eagle Medium (DMEM) media (500 mL) with Penicillin-Streptomycin (Pen-Strep) and 10% Fetal Bovine Serum (FBS) at 37° C in a Forma incubator (Thermo Scientific). After 80% confluency, the cells were scraped and collected in a 15 mL centrifuge tube with DMEM media. 25 μg of LPS-biotin was added in the media (10 mL) and the cells were incubated at 37 °C for 15 min with mild stirring. After 30 min, two quick 500 μL PBS washes were done to remove the LPS. 10 mL of PBS buffer was then added in the samples, followed by 1 mg of bis(sulfosuccinimidyl)suberate (BS3 croslinker to this solution. DUCCT cross-linker was added in the same amount (1 mg dissolved in 5 μL of DMSO). The solution was incubated for 30 min with mild stirring at room temp. After 30 minutes, 50 mM of 10 μL Tris-HCl was used to stop the reaction. Next, we washed the cells several times with PBS buffer. The cells were lysed with RIPA buffer (1h) and centrifuged to collect the proteins. After that, the proteins were incubated with 50 μL dynabeads streptavidin (Thermo Scientific). After several washes with PBS in a magnet stand, the proteins were eluted from the beads using 2X Laemmli buffer. SDS-PAGE gels were run and stained with SYPRO® ruby (Thermo Scientific, Pierce). The gel lanes were cut, digested (24 sections for each lane), and analyzed by LC-MS/MS in an Agilent XCT ETD mass spectrometer with CID and ETD-MS/MS fragmentation methods. A heat map using protein spectral counts was generated (Figure S11). All MS/MS samples were analyzed using Spectrum Mill (Agilent, Santa Clara, CA; version unknown). Spectrum Mill was set to search the NCBInr.rodent database (selected for All, unknown version, 14227560 entries) using the digestion enzyme trypsin. Spectrum Mill was searched with a fragment ion mass tolerance of 0.70 Da and a parent ion tolerance of 2.5 Da. Scaffold (version Scaffold_4.3.2, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at a greater than 80.0% probability by the Peptide Prophet algorithm. Protein identifications were accepted if they could be established at a greater than 95.0% probability and contained at least one identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm.26
RESULTS AND DISCUSSION
Cross-Linker Design and Synthesis
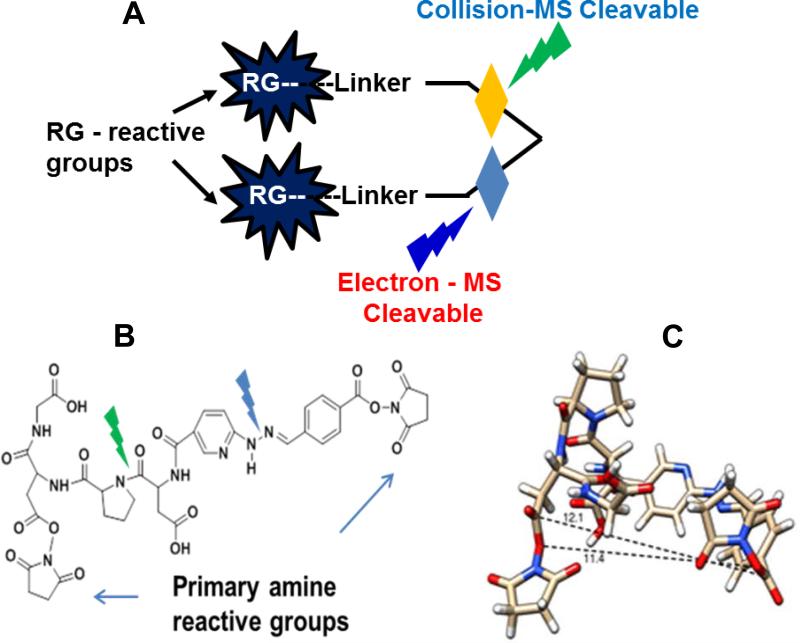
In Figure 1A, we show the design of the novel cross-linker. The cross-linker contains two gas-phase-cleavable bonds, which can be selectively cleaved using collision-based or electron transfer-based dissociations. The cross-linker was synthesized using Fmoc peptide synthesis reagents (details are provided in the Experimental Section). The reactive groups used were NHS esters for conjugation of lysine residues. Two gas-phase-cleavable bonds were added to the design. One is the Asp- Pro (DP) bond, which has been reported to be cleaved by low-energy CID.4 We added another bond (hydrazone), which researchers have reported can be cleaved using ETD.25 Both of these bonds were sandwiched in between two reactive NHS ester groups, which altogether constituted a lysine-reactive cross-linker (Figure 1B).
Figure 1.
The design and chemical structure of the DUCCT cross-linker. (A) The design of the dual CID and ETD cleavable cross-linker. (B) The chemical structure of the novel CID and ETD MS-cleavable cross-linker. The green and blue symbols indicate the CID (DP bond) and ETD (hydrazone) MS-cleavable sites, respectively. (C) The structure of DUCCT is shown with spacer chain length.
The distance of the reactive group is a major concern for MS-cleavable cross-linkers. Most of the cleavable cross-linker reactive distances are very large, and this has been a major concern for non-specific labeling. CLIP, a clickable cross-linker, has reactive distances around 10Å, but has been reported to label across a distance as high as 22 Å.2 However, for the DUCCT cross-linker, the distance between reactive groups was found to be ~12.1 Å (Figure 1C), which we believe even with extended conformation will be very suitable for cross-linking.
Cross-Linked Peptide Identification Strategies
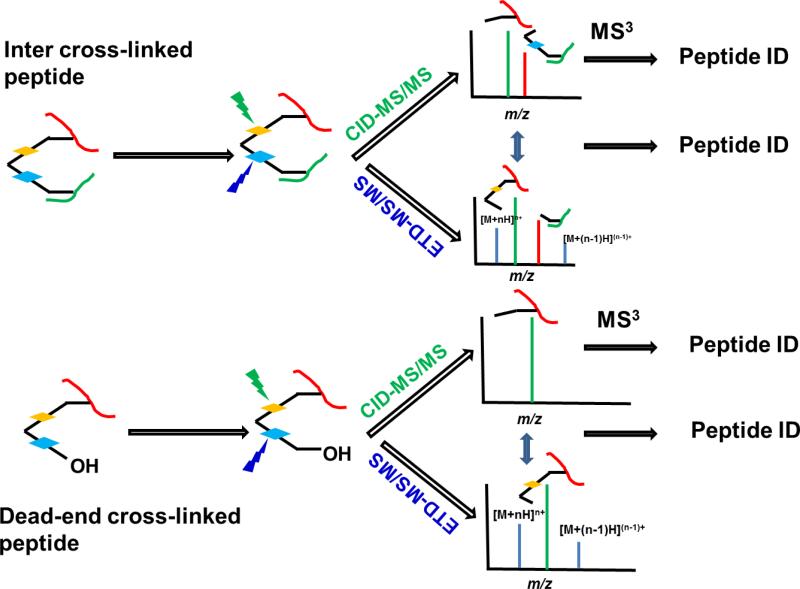
In Figure 2, we illustrate the strategies for the identification of cross-linked peptides using the DUCCT cross-linker. After CID-MS/MS, an inter-cross-linked peptide will produce two peptide peaks attached with cross-linker pieces, whereas ETD will produce two peaks for the same cross-linked peptides with different cross-linker parts (Figure 2, and Figures S1-S2 in the Supporting Information). In addition, ETD will produce charge-reduced ions of the precursor masses. This will also help to identify the charge states of the precursor m/z, even if a low-resolution mass spectrometer is used. These two signature spectra will unambiguously identify cross-linked peptides. Furthermore, dead-end peptides will generate one peptide with residual cross-linker mass due to the hydrolysis of one reactive group in the cross-linker (Figure 2, bottom). Their CID and ETD mass spectra will generate two complementary fragmentation signatures of one cross-linked peptide (Figure S3).
Figure 2.
The scheme to identify inter-cross-linked (top) and dead-end peptides (bottom) by CID and ETD tandem mass spectrometry. MS3 can be used for CID-only identification. Both CID and ETD will identify peptide sequence without further MS/MS.
Proof-of Concept Studies with DUCCT Cross-Linker
Initial characterization of the cross-linker molecular weight (863.2358 Da) and the cleavage of the DP peptide bond is provided in the Supplementary Information (Figure S4). It is important to note that no HPLC purification was conducted for this cross-linker and that high-resolution MS and MS/MS were performed to confirm the structure. Another important issue to note is that the cross-linker will not be cleaved by ETD without conjugation with a peptide, since ETD induces charge-dependent fragmentation.
In validation tests, we next treated a standard peptide, neurotensin, with our cross-linker. Neurotensin has one lysine residue and its N-terminus is blocked with a pyro-Glu modification. The cross-linker labeled the peptide efficiently and produced inter and dead-end cross-linked peptides. Two inter-cross-linked peptides were identified at m/z's of 995.2577 (M+4H+) and 796.4076 (M+5H+), respectively. CID-MS/MS clearly showed cleavage at the DP peptide bond, and ETD-MS/MS showed cleavage at the N-N bonds. In CID, we should observe two peptide ions with added cross-linker residues, and in ETD for inter-cross-linking we should observe the same two peptide ions with different cross-linker residue masses. For neurotensin, we observed two fragment peaks at m/z 971.76 and 1019.25, respectively, which corresponds to the peptide mass and the corresponding CID-cleaved part of the cross-linker (Figure 3A). We also observed 3+ charge states of these peptides (m/z at 679.98 and 648.32). Complete calculations of cross-linked precursors and fragment ions are shown in the Supplementary Information (Table S1 and Figure S5). ETD clearly matched the calculated masses with the cross-linker residues. Charge-reduced precursor ions were also observed with ETD, which clearly provides proof of the charge states of the cross-linked peptides, even though a high-resolution mass spectrometer was not used for this study (Figure 3B). This data clearly shows the CID and ETD signatures for an identically cross-linked peptide. These two mass spectral signatures of the same cross-linked peptide provide a means for the high-confidence identification of this material.
Figure 3.
CID and ETD mass spectra of a cross-linked neurotensin peptide dimer. (A) CID and (B) ETD fragmentation methods show different mass spectra signatures for the same peptide. Please see the Supplementary Information for the added masses from the cross-linker due to differential cleavages.
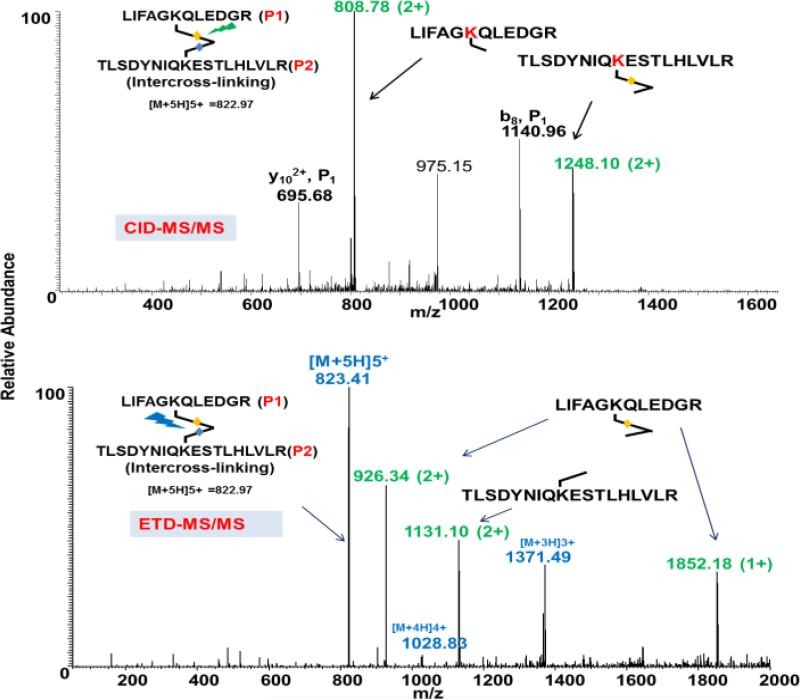
Next, we tested the labeling efficiency of our cross-linker using a small protein, ubiquitin, and a large-protein, BSA. Both studies showed efficient labeling with higher molecular weight dimer bands in the SDS-PAGE gel (Figure S6). After in-solution digestion and subsequent identification, we have clearly identified the proteins’ cross-linked sites in the CID and ETD MS/MS mass spectra. Ubiquitin is a small protein (~8500 Da), and is a very useful model protein for monitoring cross-linking, as lysine 48 of ubiquitin will form a cross-link with lysine 63, as has been reported by several groups.2,27 After cross-linking with our DUCCT compound, we searched the mass spectral data with a newly developed software tool (the development of this software is being described in a separate manuscript in preparation). Even a small protein like ubiquitin can generate ~20,000 spectra in LC-MS/MS. We have identified several cross-linked peptides from ubiquitin, and using careful investigation, have clearly identified the CID and ETD spectra of cross-linked lysine 48 and 63 (Figure 4). Calculation of the fragment mass is provided in Table S2. It is clear that CID produces the expected fragments (peptide mass + cross-linked fragment mass-H) after selective cleavage, and ETD produces similar cleavages in specified sites (Figure S7). Charge-reduced precursor ions were also confirmed by the charge states of the precursor, even though a low-resolution mass spectrometer was used for this study. It is interesting to note that we have found that CID cleavages were efficient in both odd- and even-charge state precursor ions, but ETD-cleavages were observed more efficiently in odd-charge state precursor ions. The DUCCT cross-linker can also be applied using just the CID cleavage parts. Further MS/MS of the fragment masses can be utilized to sequence the cross-linked peptides (Figure S8A). The MS3rd of the fragment peaks also unambiguously confirmed the sequence of the peptides in ubiquitin and neurotensin (Figure S8A-B).
Figure 4.
An example of CID and ETD mass spectra of an inter-cross-linked peptide derived from ubiquitin. The CID and ETD mass spectra show different signatures for the same cross-linked peptide.
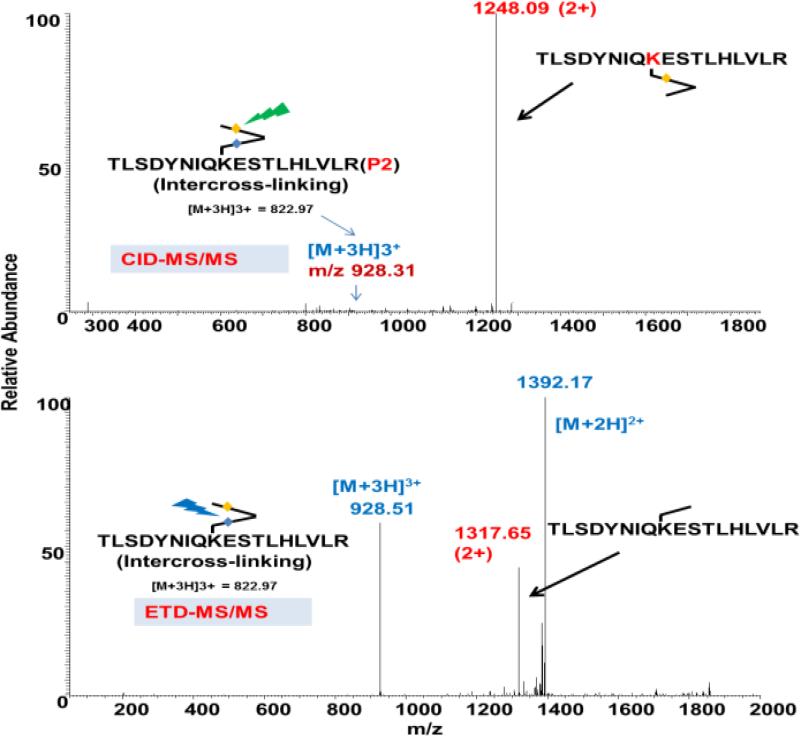
Dead-end cross-linking was identified very efficiently in the neurotensin cross-linked peptides. Dead-end peptides are very good indicators of the surface and reactive lysine residues. Identifying dead-ends with the cleavable cross-linker requires further MS/MS of the cross-linked peptides, due to the generation of a single peptide ion in the mass spectrum. For an asymmetrical cleavable cross-linker, dead-end peptides can react in two different ways depending on the reactive ends. The majority of the mass spectra will be one peptide, but some of them can be a mixture of two peptides. Fortunately, the cross-linked precursor mass database of dead-end peptides will clearly filter these out during the first stage of searching the database.
Since dead-end cross-linked peptides can produce a peptide with added cross-linker masses, sometimes it can produce both peptides due to uneven fragmentation on both cleavage sites as a result of the labeling positions (Figure S3). This problem can easily be solved by the dual-cleavable properties of the DUCCT cross-linker. Both CID and ETD MS/MS will unambiguously identify dead-end peptides. A dead-end precursor peptide was calculated and found at m/z 775 (3+) for neurotensin. After CID-MS/MS, it was cleaved at the DP bond and generated a peptide with the added cross-linker masses at m/z 1019.01 (Figure S9). However, it produced one peptide with ETD cleavage masses (m/z = 1088.81, calculation on page S5 of the Supporting Information), as well as charge-reduced ions. In the ubiquitin cross-linking experiments, we observed several dead-end peptides. One of the dead-end peptides is shown in Figure 5A single peptide was identified in the mass spectrum after CID cleavage. This peptide requires further MS/MS/MS for confident identification. Due to the dual-cleavable properties, further ETD MS/MS produced the same peptides with added cross-linker parts. Charge-reduced precursor ions were also observed and provided the charge states of this precursor. It is clear that CID and ETD pinpointed the same cross-linked peptide with high confidence. However, the cleavable cross-linker is not suitable to analyze intra-peptide cross-links. Hence, we omitted identification of these peptides in our studies.
Figure 5.
Examples of CID and ETD mass spectra of a dead-end cross-linked peptide, derived from ubiquitin. The CID and ETD mass spectra show different signatures for the same cross-linked peptide.
Confidence in Cross-Linked Peptide Identifications
We developed an automated software tool to analyze this data. Even small proteins in a LC-MS/MS experiment generate ~20,000 mass spectra. To test how confidently we can identify cross-linked peptides by two differential cleavages, we created a theoretical database of cross-linked peptides using precursor and fragment masses. We then created an mgf file of an LC-MS/MS experiment of cross-linked ubiquitin, which contained an experimental list of precursors and fragment ion masses. We found approximately 30,500 scan ions in the mgf file. At first the software tool searched the precursor mass spectra database (2.5 Da), then matched spectra were further searched for fragment peaks within a defined mass threshold (0.6 Da). We chose these mass thresholds due to the use of a low-resolution mass spectrometer in this study. Cross-searching our experimental (mgf) file with the theoretical cross-linked database, we found a number of hits in the CID search. It will require further MS/MS of both cross-linked peptides to establish full confidence in the identification of these materials (see the excel files in the Supporting Information). We also searched the mgf file for ETD-MS/MS of the cross-linked ubiquitin using the same process. The search found several hits from the theoretical ETD dataset. After comparing the common sequences identified in both datasets with precursor m/z restricted to ±0.5 Da, we confidently identified several peptide sequences from both datasets (Table S3, and Supporting Information excel files). We manually verified each result and found they efficiently matched our theoretical and experimental calculations. We are currently working on large datasets and developing scoring tools for analyzing the data with proper statistics. The software tool will be published in an appropriate journal soon. It is very clear that CID and ETD results pinpointed with confidence the cross-linked peptides in ubiquitin (see the crystal structure in Figure S10 for identified sites with distance) from these CID- and ETD-MS/MS experiments. We have also demonstrated MS/MS of these cleavage parts and confirmed some of the peptide sequences with high confidence. These three steps: 1) CID-MS/MS of the cross-linked peptides; 2) ETD-MS/MS of the cross-linked peptides; and an 3) optional MS3rd of the cleaved cross-linked peptide parts, can simultaneously identify the cross-linked peptides without ambiguity. It is also important to note that a high-resolution mass spectrometer will significantly reduce the number of hits due to the search threshold of precursor and fragment masses.
Labeling Studies in Cell Lysate
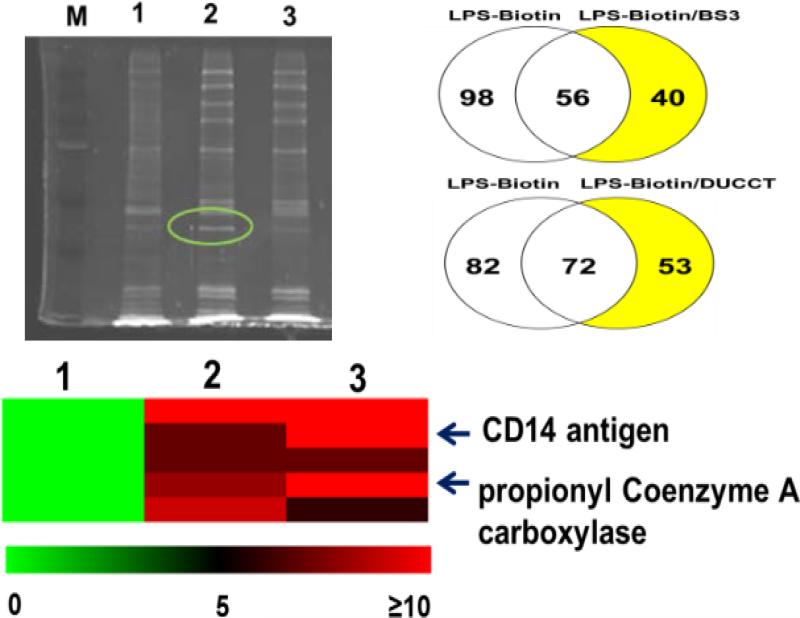
In order to test the labeling efficiencies of DUCCT in in vivo pulldown samples, we tested our cross-linker in macrophages after stimulation with the Toll-Like Receptor 4 (TLR4) ligand LPS. RAW 264.7 macrophage cells were grown and treated for 1 h with LPS-biotin, followed by cross-linking with DUCCT or BS3, a commercial cross-linker. After SDS-PAGE separation of the samples, in-gel digestion was carried out for each gel lane (24 pieces), then we performed the LC-MS/MS experiments (see the Supporting Information for total protein identification). The DUCCT cross-linker-treated sample yielded more protein identifications compared to BS3 (Figure 6).
Figure 6.
(Upper left) The SDS-PAGE gel of biotin-avidin pulldown experiments (M = marker, 1 = LPS biotin, 2 = LPS-biotin/DUCCT, and 3 = LPS-biotin/BS3). (Upper right) A Venn diagram of the number of proteins identified. (Bottom) A partial heat map of selected proteins identified exclusively in cross-linked samples after pulldown studies with avidin (see the full heat map in Figures S11, S12). The scale denotes the spectral counts. For the heat map, 1 = LPS biotin, 2 = LPS-biotin/BS3, and 3 = LPS-biotin/DUCCT.
We also confirmed some known protein interactions in cross-linked samples that had been treated with LPS-biotin, providing feasibilities of our method in large-scale cell signaling studies. CD14, a known LPS-binding protein that associates with the TLR4 receptor, was identified with high spectral counts using DUCCT compared to BS3.28 No CD14 peptides were identified in the control LPS-biotin treated sample (Figure 6, and Figures S11-S12). Our studies clearly showed that labeling efficiencies of our cross-linker are comparable to the widely used commercial cross-linker BS3. We are currently working on software that will allow us to search large-scale samples (> 500 proteins using our CID and ETD cleavage technique with appropriate statistics) with high confidence.
CONCLUSIONS
We show here a novel cross-linker, DUCCT, which has two differential tandem mass spectrometry cleavage properties. Differential cleavage can pinpoint the same cross-linked peptides with high confidence. This cross-linker can also work as a CID-cleavable cross-linker and subsequent MS/MS/MS can confirm the peptide sequence. We have also developed a software tool and are currently improving it for large-scale data analysis. Together with software capability and the DUCCT cross-linker's dual-gas-phase-cleavable properties, we demonstrate that unambiguous identification of protein interactions is feasible. We believe this cross-linking-based proteomics technology will tremendously advance the system-level identification of cell-signaling cascades. DUCCT cross-linking should allow investigators to quickly and efficiently screen interacting partners in discovery proteomics, providing lists of interacting proteins for further biochemical and/or molecular biological validation.
Supplementary Material
ACKNOWLEDGMENT
We acknowledge funding from 1UA5GM113216-01, NIGMS, NIH, and also funding from the UT Proteomics Core Facility Network for a mass spectrometer. The authors also acknowledge the Shimadzu Center for Advance Analytical Chemistry (SCAAC) at the University of Texas at Arlington. Dr. Michael Fessler is funded by the Intramural Research Program of the National Institute of Environmental Health Sciences, NIH (Z01 ES102005). We also thank Dr. Lalith Perera at NIEHS, NIH for molecular modeling support.
Footnotes
SUPPORTING INFORMATION
Characterization of the molecular weight of the DUCCT cross-linker, chemical drawings of the theoretical fragments including their masses, detailed calculation of the masses of the cross-linked peptides (precursors and fragments) observed for neurotensin and ubiquitin, SDS-PAGE gel images of the ubiquitin and the BSA, MS/MS/MS of cleaved cross-linked fragments for the ubiquitin and the neurotensin, image of the color-coded crystal structure of ubiquitin with identified cross-linked sites, and complete heat map of LPS stimulated pulldown samples were provided in the supplementary data. These Supporting Information are available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.Gingras AC, Gstaiger M, Raught B, Aebersold R. Nat. Rev. Mol. Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 2.Chowdhury SM, Du X, Tolic N, Wu S, Moore RJ, Mayer MU, Smith RD, Adkins JN. Anal. Chem. 2009;81:5524–5532. doi: 10.1021/ac900853k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrotchenko EV, Serpa JJ, Borchers CH. Mol. Cell Proteomics. 2011;10:M110.001420. doi: 10.1074/mcp.M110.001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soderblom EJ, Goshe MB. Anal. Chem. 2006;78:8059–8068. doi: 10.1021/ac0613840. [DOI] [PubMed] [Google Scholar]
- 5.Tang X, Munske GR, Siems WF, Bruce JE. Anal. Chem. 2005;77:311–318. doi: 10.1021/ac0488762. [DOI] [PubMed] [Google Scholar]
- 6.Greber BJ, Boehringer D, Leibundgut M, Bieri P, Leitner A, Schmitz N, Aebersold R, Ban N. Nature. 2014;515:283–286. doi: 10.1038/nature13895. [DOI] [PubMed] [Google Scholar]
- 7.Tan D, Li Q, Zhang MJ, Liu C, Ma C, Zhang P, Ding YH, Fan SB, Tao L, Yang B, Li X, Ma S, Liu J, Feng B, Liu X, Wang HW, He SM, Gao N, Ye K, Dong MQ, Lei X. eLife. 2016;5:e12509. doi: 10.7554/eLife.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu C, Yang Y, Wang X, Guan S, Fang L, Liu F, Walters KJ, Kaiser P, Huang L. Mol. Cell Proteomics. 2016;15:2279–2292. doi: 10.1074/mcp.M116.058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greber BJ, Bieri P, Leibundgut M, Leitner A, Aebersold R, Boehringer D, Ban N. Science. 2015;348:303–308. doi: 10.1126/science.aaa3872. [DOI] [PubMed] [Google Scholar]
- 10.Lauber MA, Rappsilber J, Reilly JP. Mol. Cell Proteomics. 2012;11:1965–1976. doi: 10.1074/mcp.M112.019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arlt C, Goetze M, Ihling CH, Hage C, Schaefer M, Sinz A. Anal. Chem. 2016;88:7930–7937. doi: 10.1021/acs.analchem.5b04853. [DOI] [PubMed] [Google Scholar]
- 12.Gutierrez CB, Yu C, Novitsky EJ, Huszagh AS, Rychnovsky SD, Huang L. Anal. Chem. 2016;88:8315–8322. doi: 10.1021/acs.analchem.6b02240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhury SM, Munske GR, Tang X, Bruce JE. Anal. Chem. 2006;78:8183–8193. doi: 10.1021/ac060789h. [DOI] [PubMed] [Google Scholar]
- 14.Chavez JD, Cilia M, Weisbrod CR, Ju HJ, Eng JK, Gray SM, Bruce JE. J. Proteome Res. 2012;11:2968–2981. doi: 10.1021/pr300041t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeBlasio SL, Chavez JD, Alexander MM, Ramsey J, Eng JK, Mahoney J, Gray SM, Bruce JE, Cilia M. J. Virol. 2016;90:1973–1987. doi: 10.1128/JVI.01706-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu C, Yang Y, Wang X, Guan S, Fang L, Liu F, Walters KJ, Kaiser P, Huang L. Mol. Cell Proteomics. 2016;15:2279–2292. doi: 10.1074/mcp.M116.058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaake RM, Wang X, Burke A, Yu C, Kandur W, Yang Y, Novtisky EJ, Second T, Duan J, Kao A, Guan S, Vellucci D, Rychnovsky SD, Huang L. Mol. Cell Proteomics. 2014;13:3533–3543. doi: 10.1074/mcp.M114.042630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu F, Goshe MB. Anal. Chem. 2010;82:6215–6223. doi: 10.1021/ac101030w. [DOI] [PubMed] [Google Scholar]
- 19.Makepeace KA, Serpa JJ, Petrotchenko EV, Borchers CH. Methods. 2015;89:74–78. doi: 10.1016/j.ymeth.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 20.Liu F, Rijkers DT, Post H, Heck AJ. Nat. Methods. 2015;12:1179–1184. doi: 10.1038/nmeth.3603. [DOI] [PubMed] [Google Scholar]
- 21.Du X, Chowdhury SM, Manes N, Wu S, Mayer-Cumblidge U, Adkins J, Anderson GA, Smith RD. J. Proteome Res. 2010;10:923–931. doi: 10.1021/pr100848a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinner O, Seebacher J, Walzthoeni T, Mueller LN, Beck M, Schmidt A, Mueller M, Aebersold R. Nat. Methods. 2008;5:315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, Luo SK, Ding YH, Wang LH, Hao Z, Xiu LY, Chen S, Ye K, He SM, Dong MQ. Nat. Methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 24.Liu F, Wu C, Sweedler JV, Goshe MB. Proteomics. 2012;12:401–405. doi: 10.1002/pmic.201100352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardner MW, Brodbelt JS. Anal. Chem. 2010;82:5751–5759. doi: 10.1021/ac100788a. [DOI] [PubMed] [Google Scholar]
- 26.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 27.Gardner MW, Vasicek LA, Shabbir S, Anslyn EV, Brodbelt JS. Anal. Chem. 2008;80:4807–4819. doi: 10.1021/ac800625x. [DOI] [PubMed] [Google Scholar]
- 28.Rajaiah R, Perkins DJ, Ireland DD, Vogel SN. Proc. Natl. Acad. Sci. U. S. A. 2015;112:8391–8396. doi: 10.1073/pnas.1424980112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.