Abstract
D-amino acids, the enantiomers of naturally abundant L-amino acids, bears unique stereochemistry properties that lead to the resistance towards most of endogenous enzymes. Previous works have demonstrated the applications of D-amino acids in therapeutics development with the aid of mirror-image phage display and retro-inverso peptide synthesis. In this review, we highlight the recent progresses and challenges in the exploration of D-amino acids at the interface of chemistry and life science. First we will introduce some progresses of traditional application of D-amino acids to enhance biostability of peptide therapeutics. Then we discuss some works that explore the relatively underexplored interactions between enzyme and D-amino acids and enzymatic reactions of D-amino acids. To highlight the enzymatic reactions of D-amino acids, we will describe several emerging works on the enzyme-instructed self-assembly (EISA) and their potential application in selective anti-inflammatory or anticancer therapies. At the end, we briefly mention the challenges and possible future directions.
Keywords: Anti-inflammatory and anticancer drugs, biostability, D-amino acids, enzyme-instructed self-assembly, enzymatic reaction
Introduction
As the fundamental build blocks of proteins, natural L-amino acids always serve as a starting point for protein related research. Reflecting the L-amino acids by a mirror is their enantiomers—D-amino acids, which shares identical chemical and physical properties with L-amino acids, except for its ability to rotate plane-polarized light in opposite directions. While L-amino acids serve as the elements of the natural proteins translated in ribosome, D-amino acids are found in some posttranslational modification and peptidoglycan cell walls of bacteria. One notable example of naturally used D-amino acid is D-Ala-D-Ala, which is the stem terminal of the peptidoglycan side-chain pentapeptide found in cell walls of Gram positive bacteria as well as the target for the microbe inhibition by antibiotics like vancomycin.(1) The miraculously use of D-amino acids in nature also stimulates the exploration of D-amino acids for a variety of applications. For example, Kent and co-workers synthesized D-proteins—that is, the proteins consist of solely D-amino acids—through total chemical synthesis via native chemical ligation(2). They have found out that the D-proteins and natural L-proteins display reciprocal chiral specificities on their substrates, from which they proposed that the L- and D-enzyme have entirely mirrored structure of each other(3). In another seminal work, Kim et al. demonstrated the concept of mirror-image phage display, which displays genetically encoded libraries on bacteriophages, for screening D-peptides that would bind to natural protein targets that are made of L-amino acids(4). That elegant approach allows the use of phage display to identify biologically encoded L-peptides specifically binding to the given D-protein that is the enantiomer of the native target (e.g., a protein consisting of L-amino acid). According to the mirror symmetry, the corresponding D-peptide, as the enantiomer of that specific L-peptide, should be able to specifically bind to the natural L-target with high affinity. Despite it is costly to synthesis a D-protein, the D-peptide from such screening process, being resistant to proteolytic degradation, represents a breakthrough in manipulating D-amino acid for developing biostable inhibitors as potential therapeutics(5, 6).
To avoid the time consuming synthesis of D-proteins required for mirror-image phage display, Goodman and Chorev have developed a more effective approach—retro-inverso peptides which consist of D-amino acids in reverse sequence of the naturally occurring L-peptides—for the development of bioactive D-peptides(7). Bearing high degree of topochemical equivalence of the L-peptides, the retro-inverso peptides, presumably, would exhibit the similar bioactivity of the corresponding L-peptides. Regardless of the target, focusing only on the equivalent transformation of small molecules, the retro-inverso peptides show great advantage in the accessibility. Although the inherent differences of L-peptide and its retro-inverso isomer at secondary and tertiary structure prevent the complete recapitulation of the activity of the L-peptides, the retro-inverso peptides, indeed, achieve certain binding activity in a few cases(8–10). Besides the conventional applications of the D-amino acids, Xu and co-workers unexpectedly observed that D-peptides improve the selectivity of non-steroid anti-inflammatory drugs (NSAIDs)(11), as well as the conversion of D-peptides by enzymes (e.g., phosphatases(12, 13) and carboxylesterase(14, 15)).
Recent developments of D-peptides in biomedical research warrant a short review on the development of D-amino acid-based molecules in biomedicine, an emerging area of amino acid research(16). This review briefly describes several representative applications of D-amino acids in biomedicine in the following order. First, we will start from the conventional application of D-amino acids, such as increasing the biostability of the peptides for making peptide hydrogels, drug delivery systems, and protein inhibitors. Then, we discuss the less expected enzyme reactions of D-amino acid, especially highlighting the recent study of D-amino acids for enzyme-instructed self-assembly (EISA), which results in selectivity in potential anti-inflammatory or anticancer drug approaches. We hope that these discussions will help readers to appreciate the study of D-amino acids in achieving biostability(17, 18), investigating interactions between enzymes and D-peptides(19, 20), and improving bioactivities via the introduction of D-amino acids(21, 22).
D-amino acids enhancing biostability
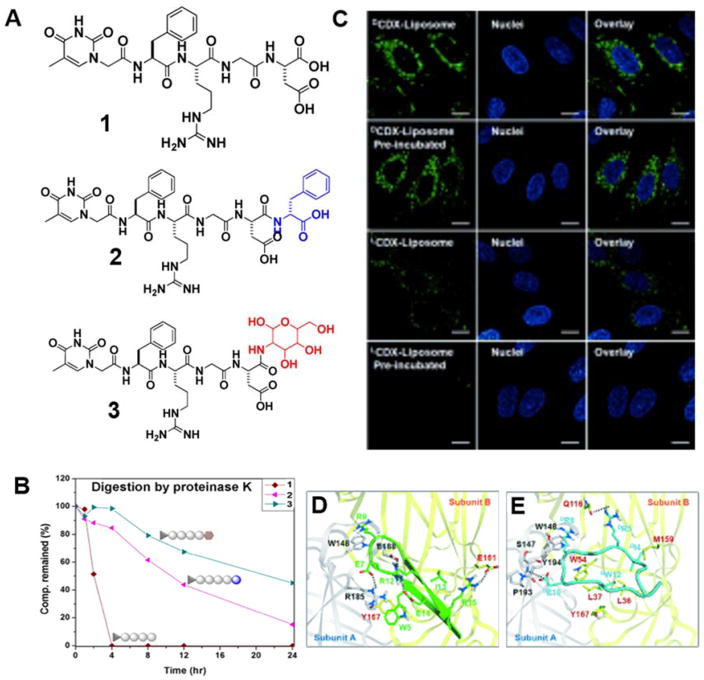
Peptides have become increasingly attractive candidates of drugs, as an alternative of protein therapeutics, for targeting protein-protein interactions because of their low cost and relative easiness of production. However, the susceptibility of L-peptides to proteolysis limits their applications. Unlike L-amino acids, D-amino acids rarely act as the substrates of endogenous proteases, resulting in the resistance of D-peptides towards proteolysis. Taking this stereochemical advantage to improve in vivo proteolytic stability of bioactive molecules, Xu and co-workers have introduced D-amino acid and glycoside (Figure 1A) for enhancing the proteolytic resistance of small peptide hydrogelators after they demonstrated the in vivo stability of small D-peptides using a mice model(23). Using HPLC to monitor the concentration of remaining hydrogelators, the biostability of hydrogelator 1, 2 and 3 were assessed in vitro. As shown in Figure 1B, modifying the C-terminal of short L-peptides with D-amino acids or glycoside has significantly boosted the stabilities of these peptide derivatives towards proteases (e.g., proteinase K)(24). While all of the L-peptides undergo proteolysis catalyzed by proteinase K in 4 hours, the D-amino acid and glycoside modified peptides have 15 % and 45 % remaining after 24 hours, respectively. This result demonstrates that C-terminal conjugation of D-amino acids or glycosides to L-peptides provides a simple way to gain long-term biostability of the L-peptides for the development of biostable hydrogels. Although PEGylation increases the stability of proteins, it may decrease the efficacy of peptides because it reduces effective collision. Thus, D-peptides may be advantageous if they have the same activity as the L-peptides. Moreover, D-peptides may resist PARSylation (25), a key step in the protein degradation machinery.
Figure 1.
A) Molecular structures of the hydrogelators; B) Remaining concentration of hydrogelators during digestions of proteinase K; (Adapted from ref 24, Copyright 2012 American Chemical Society); C) Targeting ability of CDX peptides modified liposomes with brain capillary endothelial cells; D and E) Binding mode of LCDX (D, green) and DCDX (E, aqua) with α7 nicotine acetylcholine receptors (Adapted from ref 9, Copyright 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim).
The significantly improved biostability of D-peptides usually leads to longer in vivo circulation half-time, making the D-peptide based drug delivery system(23) more attractive and efficient than their L-peptide counterparts. For example, receptor-mediated transcytosis (RMT), which recognizes and facilitates drug delivery by transferring ligands, promises to overcome the major challenge—the blood-brain barrier (BBB)—for treating the central nervous system diseases(26). While the substrates of RMT are normally L-peptides, Lu and co-workers, surprisingly, found that DCDX (a D-stereoisomer), the retro-inverso peptide of L-peptide nicotine acetylcholine receptors (nAChR) ligand LCDX (FKESWREARGTRIERG, the L-stereoisomer) of the neuronal α7 nAChR, is more efficient than LCDX to bind to nAChR.(9) According to the report by the authors, the IC50 value of DCDX is five times lower than LCDX due to a better match with the pocket, as shown in the binding model (Figure 1D and E). Because it resists proteases, DCDX shows considerably higher RMT efficiency (Figure 1C). Together with other promising results of utilizing D-peptides or D-peptides containing nanostructures as drug carriers(23), these results underscore the importance of biostability of the drug carriers in vivo. However, the rather long sequence of this peptide (16 amino acid residues) and the requirement of the assistance of liposomes for the delivery of the drug, unfortunately, diminish the significance of this interesting observation. In addition, the presence of multiple arginine residues in the sequence of CDX suggests that the uptake may be mediated by heparin(27).
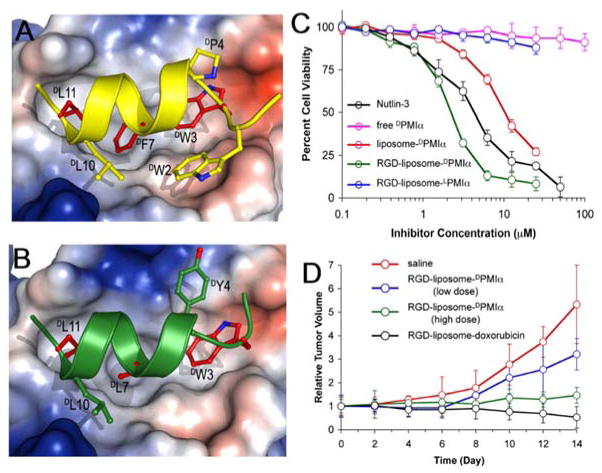
The increased stability of peptide drugs could significantly boost treatment efficacy for treating life threatening diseases, such as cancers. Activation of the tumor suppressor protein p53, which is responsible for genetic stability and tumor suppression, is a promising approach to develop anticancer drugs(28). One indirect strategy is to block the binding between p53 and its negative regulator MDM2/MDMX that is expressed in 50% of cancer cells(29). Being explored as promising inhibitors for protein-protein interaction, peptide inhibitors suffer from the instability against protease digestion, which prevents their success applications as human therapeutics. Lu and co-workers, using mirror phage display(6), developed the new inhibitors DPMI-α (Figure 2A) and DPMI-γ (Figure 2B) for disrupting p53-MDM2/MDMX interactions. With the help of a liposome delivery system, the D-peptide inhibitors have shown significant inhibition to human glioma cell U87 while the L-peptide inhibitor LPMI-α hardly inhibit the cells at the same condition (Figure 2C). In further in vivo experiments, DPMI-α also shows significant inhibition of tumor (Figure 2D). This work suggests that it would be useful and worthwhile to develop a way to enhance the cellular uptake of D-peptides(14), such as DPMIs.
Figure 2.
A and B) Comparison between DPMI-γ (yellow) and DPMI-α (green) in a Cα ribbon diagram bound to the p53-binding pocket of MDM2; C) Growth inhibition of U87 cells by Nutlin-3, free DPMI-α, liposome-DPMI-α, RGD-liposome-DPMI-α, and RGD-liposome-LPMI-α; D) Growth inhibition of U87 s.c. xenografts in nude mice by DPMI-α and doxorubicin (Adapted from ref 6, Copyright 2010 National Academy of Sciences).
Enzymatic transformation of unnatural substrates
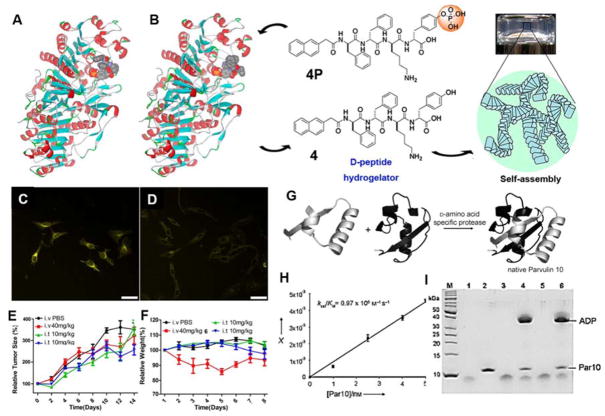
Based on that phosphorylation/dephosphorylation of proteins is a fundament process for regulating the functions of proteins for signal transduction, Xu and co-workers demonstrated the use of phosphatases to dephosphorylate a precursor to form a supramolecular hydrogel, which has been applied to D-peptidic substrate as well. Being segments of natural proteins, L-peptides tend to be good substrates for most enzymes; however, their D-counterparts, due to chirality mismatch with the binding pockets of the protein, usually are poor substrates of endogenous enzymes, particularly proteases. Therefore, few reports focus on the interactions between natural enzymes and unnatural D-amino acids. Xu and co-workers recently explored the interactions between alkaline phosphatase and its substrates—a N-terminal capped L-tetrapeptide and its D-counterpart(12). According to the binding model (Figure 3A and B), it is clear that the phosphate group of the D-peptide is able to bind to the active site of alkaline phosphatase with little hindrance. The NMR data shows that the chirality of the substrate hardly has any effect on the rate of enzymatic dephosphorylation in water, which agrees with the model. Conjugating the tetrapeptide with an environment sensitive dye, NBD, makes a substrate ideal for imaging intracellular self-assembly catalyzed by the phosphatases (Figure 3C and D). Compared to its L-counterpart, the D-peptide conjugate exhibits faster emergence of fluorescence in HeLa cells, agreeing with the higher stability of the D-peptide towards proteolytic degradation. Furthermore, after connection to an anticancer drug, paclitaxel (i.e., taxol), the D-peptide derivative exhibits similar in vivo anti-cancer effect as that of L-isoform (Figure 3E and F). This result demonstrates the feasibility of applying D-peptides in the development of drug candidates for chemotherapy.
Figure 3.
A and B) Binding model of the L- and D-peptides (presented as CPK model: yellow, phosphorus; red, oxygen) to the active site of ALP (presented as solid ribbons) and the chemical structures of precursors and hydrogelators; 500 μM of NapFFK(NBD)Y without (C) or with (D) the PTP1B inhibitor (25 μM) (scale bar is 50 μm); E) Relative tumor sizes and (F) relative weights of mice treated with Taxol (red line), L-NapFFK(Taxol)Yp (green line), and D-NapFFK(Taxol)Y (blue line) for in vivo tests; (Adapted from ref 12, Copyright 2013 American Chemical Society). G) Scheme of using D-amino acid specific protease in the chemoenzymatic synthesis of a native L-protein; H) Specificity constants kcat/KM of synthetic Par10; I) SDS-Page of ligated par10 product with/without ADP (Adapted from ref 31, Copyright 2008 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim).
The increasing knowledge of the interactions between the substrates and enzymes facilitates the use of various enzymes for the synthesis of a wide range of molecules. Proteases represent an important class of established reagent in stereospecific reactions in organic synthesis, especially in the total synthesis of peptides and proteins(30). However, risks associated with unexpected proteolytic cleavage during ligation reactions limit their use. Bordusa et al. developed an innovative method to use D-amino acid specific protease for L-peptide ligation (Figure 3G), which prevents unexpected cleavage by the protease(31). Using the substrate-mimetic strategy, the D-amino acid specific protease is able to catalyze the ligation of nonspecific L-peptides. After identifying a suitable enzyme, alkaline D-peptidase (ADP), from Bacillus cereus and their mimetic substrates, the authors synthesized protein peptidyl prolyl cis–trans isomerase parvulin 10 (Par10) from E. coli as a model protein for evaluating the above method. The activity and SDS-PAGE results (Figure 3H and I) show that this method is capable of synthesizing active native proteins in 61% yield without proteolytic side reactions.
Selectivity arising from D-amino acid (or the aggregates of D-amino acids)
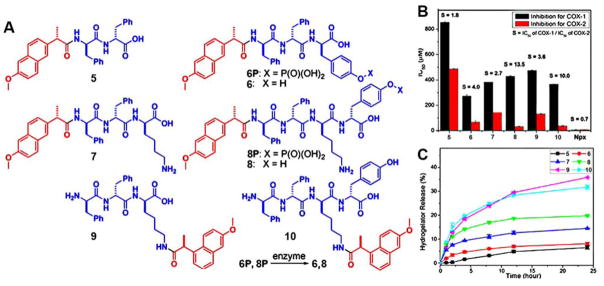
Severe adverse effects, including inhibition of un-targeted proteins and toxicity to normal tissues, restrict applications of many therapeutic reagents(32). Molecular hydrogels could trap therapeutics around the lesion and keep them away from normal tissues, which is ideal for localized drug delivery(33, 34). Recently, Xu and co-workers developed a unconventional strategy to minimize the adverse effects of nonsteroidal anti-inflammatory drug (NSAID) using multifunctional molecular hydrogels(11). Based on the crystal structure of cyclooxygenase-2 (COX-2), the C-terminal modification of naproxen (Npx) hardly hinders the drug-protein interaction. A series of D-amino acids-Npx conjugates (Figure 4A), which afford homogenous hydrogels, exhibit highly selective COX-2 inhibition. In vitro inhibition assays show that (Figure 4B) the attachment of small D-peptides to Npx greatly increases its inhibition selectivity towards COX-2, as the ratio of IC50 for COX-1 to IC50 for COX-2 increased from 0.7 to 13.5. The selectivity gained by using D-peptide in Npx-containing hydrogelators promises effectively decreasing the adverse gastrointestinal and renal effects associated with COX-1 binding. In addition, the steady release of Npx-containing hydrogelator from the hydrogels (Figure 4C) indicates that these hydrogels may serve as topical gels for sustained drug delivery of pain medicine.
Figure 4.
A) The structures of hydrogelators 5–10 consisting of D-Amino acids and Npx; B) IC50 values of Npx-based hydrogelators for inhibiting COX enzyme (selectivity, defined as the ratio of the IC50 values towards COX-1 and COX-2, is labeled on top of bars); C) Release profiles of Npx-based hydrogelators from hydrogels 5–10. (Adapted from ref 11, Copyright 2013 American Chemical Society)
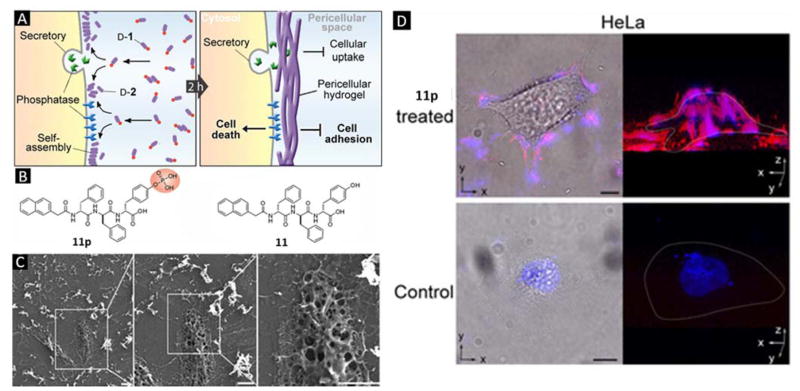
In comparison with normal cells, certain cancer cells overexpress certain enzymes such as phosphatase and esterase, which are commonly used as cancer biomarkers(35). Taking advantage of the overexpression of esterases in cancer cells, Xu et al. developed a series of enzyme generated hydrogels that form inside the cells for controlling the cell fate(36). Besides the intracellular hydrogelation, Xu and co-workers recently found that a small D-peptide derivative (Figure 5B) forms fibrous nanonets on cancer cells, but not around normal cells (Figure 5A)(37). The formed pericellular hydrogel further disturb cell communication, block cellular mass exchange, and lead to cancer cell apoptosis. A visible layer of hydrogel-like material, which has fibrous nanostructure under scanning electron microscopy (Figure 5C), builds up on HeLa cells after incubating the cells with 11p, the pre-cursor of 11 and the substrate of alkaline phosphatases. However, the compound 11 is unable to form directly such pericellular structure at the same concentration of 11p. This result reveals the importance of enzymatic dephosphorylation in the formation of the hydrogel. The confocal images (Figure 5D) shows that the HeLa cells incubated with 11p have intense red fluorescence after Congo red staining. DAPI, a nucleus dye, co-localizes with Congo red, which validates the existence of nanofibers and their function to block mass exchange. This work demonstrates that molecular nanofibers of D-peptides, formed by localized enzyme-instructed self-assembly of innocuous monomers on cell surface, can selectively inhibit cancer cells.
Figure 5.
A) Enzyme-catalyzed formation of pericellular hydrogel/nanonets to induce cell death; B) Molecular structures of the precursor (11p) and the hydrogelator (11); C) SEM images of freeze-dried HeLa cells treated with 11p at 560 μM for 2 h; D) Overlaid images and 3D stacked z-scan images of Congo red and DAPI stained HeLa cell treated by 11p or culture medium as control for 12 h (Adapted from ref 37, Copyright 2014 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim).
Outlooks
The examples described above underscore several opportunities and challenges for the applications of D-amino acids in biomedicine. While the replacement of L-amino acids with D-amino acids greatly increases the stability towards proteolysis, this property alone is unable to compensate the decrease of the affinity in the case of retro-inverso peptides. Despite the impressive achievement in the use of D-proteins to develop high affinity D-peptides, the low cellular uptake of D-peptides remain an unmet challenge. The recent use of taurine to promote the uptake of D-peptides(14) has offered a potential approach to address this problem, but the parameters of that approach remain to be explored and defined. One place to draw inspiration for the application of D-amino acids is Nature. Some pharmaceutical important peptide antibiotics (e.g., gramicidins, actinomycins, bacitracins, and lantibiotics) incorporate D-amino acids, which may serve as a useful blue print. The assemblies of D-peptides (38–42), in fact, represent a useful but less explored type of biological entities, which deserve further exploration and may lead to more surprises. Overall, D-amino acids belongs to a less explored territory for scientists. The studies of D-amino acids likely will receive increased attention, generate more useful results(43), and lead to previously unexplored applications.
Acknowledgments
This work was partially supported by NIH (CA142746) and W. M. Keck Foundation. ZF thanks the Dean’s fellowship from Brandies University.
List of non-standard abbreviations
- EISA
enzyme-instructed self-assembly
- COX
cyclooxygenase
- HPLC
high performance liquid chromatography
- RMT
receptor-mediated transcytosis
- nAChR
nicotine acetylcholine receptors
- BBB
blood-brain barrier
- p53
cellular tumor antigen p53
- MDM2
Mouse double minute 2 homolog
- MDMX
Protein Mdm4
- NMR
Nuclear magnetic resonance
- NBD
7-nitro-1,2,3-benzoxadiazole
- ADP
alkaline D-peptidase
- Npx
naproxen
- NSAIDs
non-steroid anti-inflammatory drugs
Contributor Information
Zhaoqianqi Feng, Department of Chemistry, Brandeis University, 415 South Street, Waltham, Massachusetts 02454, United States.
Bing Xu, Department of Chemistry, Brandeis University, 415 South Street, Waltham, Massachusetts 02454, United States.
References
- 1.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriological reviews. 1972;36(4):407–77. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson PE, Muir TW, Clark-Lewis I, Kent S. Synthesis of proteins by native chemical ligation. Science. 1994;266(5186):776–9. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 3.Milton Rd, Milton S, Kent S. Total chemical synthesis of a D-enzyme: the enantiomers of HIV-1 protease show reciprocal chiral substrate specificity [corrected] Science. 1992;256(5062):1445–8. doi: 10.1126/science.1604320. [DOI] [PubMed] [Google Scholar]
- 4.Schumacher TN, Mayr LM, Minor DL, Milhollen MA, Burgess MW, Kim PS. Identification of D-peptide ligands through mirror-image phage display. Science. 1996;271(5257):1854–7. doi: 10.1126/science.271.5257.1854. [DOI] [PubMed] [Google Scholar]
- 5.Eckert DM, Malashkevich VN, Hong LH, Carr PA, Kim PS. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell. 1999;99(1):103–15. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 6.Liu M, Li C, Pazgier M, Li C, Mao Y, Lv Y, Gu B, Wei G, Yuan W, Zhan C, Lu WY. D-peptide inhibitors of the p53–MDM2 interaction for targeted molecular therapy of malignant neoplasms. Proc Natl Acad Sci USA. 2010;107(32):14321–6. doi: 10.1073/pnas.1008930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodman M, Chorev M. On the concept of linear modified retropeptide structures. Acc Chem Res. 1979;12(1):1–7. [Google Scholar]
- 8.Guichard G, Benkirane N, Zeder-Lutz G, Van Regenmortel M, Briand J-P, Muller S. Antigenic mimicry of natural L-peptides with retro-inverso-peptidomimetics. Proc Natl Acad Sci USA. 1994;91(21):9765–9. doi: 10.1073/pnas.91.21.9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei XL, Zhan CY, Shen Q, Fu W, Xie C, Gao J, Peng CM, Zheng P, Lu WY. A D-peptide ligand of nicotine acetylcholine receptors for brain-targeted drug delivery. Angew Chem Int Ed. 2015;54(10):3023–7. doi: 10.1002/anie.201411226. [DOI] [PubMed] [Google Scholar]
- 10.Li C, Pazgier M, Li J, Li C, Liu M, Zou G, Li Z, Chen J, Tarasov SG, Lu W-Y. Limitations of peptide retro-inverso isomerization in molecular mimicry. J Biol Chem. 2010;285(25):19572–81. doi: 10.1074/jbc.M110.116814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Kuang Y, Gao Y, Du X, Shi J, Xu B. D-amino acids boost the selectivity and confer supramolecular hydrogels of a nonsteroidal anti-inflammatory drug (NSAID) J Am Chem Soc. 2013;135(2):542–5. doi: 10.1021/ja310019x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JY, Gao Y, Kuang Y, Shi JF, Du XW, Zhou J, Wang HM, Yang ZM, Xu B. Dephosphorylation of D-peptide derivatives to form biofunctional, supramolecular nanofibers/hydrogels and their potential applications for intracellular imaging and intratumoral chemotherapy. J Am Chem Soc. 2013;135(26):9907–14. doi: 10.1021/ja404215g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, Liang G, Xu B. Enzymatic hydrogelation of small molecules. Acc Chem Res. 2008;41(2):315–26. doi: 10.1021/ar7001914. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, Du XW, Li J, Yamagata N, Xu B. Taurine boosts cellular uptake of small D-peptides for enzyme-instructed intracellular molecular self-assembly. J Am Chem Soc. 2015;137(32):10040–3. doi: 10.1021/jacs.5b06181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao F, Gao Y, Shi J, Browdy HM, Xu B. Novel anisotropic supramolecular hydrogel with high stability over a wide pH range. Langmuir. 2010;27(4):1510–2. doi: 10.1021/la103982e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brückner H, Fujii N. D-amino Acids: In chemistry, life sciences, and biotechnology. Verlag Helvetica Chimica Acta; 2011. [Google Scholar]
- 17.Tugyi R, Uray K, Iván D, Fellinger E, Perkins A, Hudecz F. Partial D-amino acid substitution: Improved enzymatic stability and preserved Ab recognition of a MUC2 epitope peptide. Proc Natl Acad Sci USA. 2005;102(2):413–8. doi: 10.1073/pnas.0407677102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wade D, Boman A, Wåhlin B, Drain C, Andreu D, Boman HG, Merrifield RB. All-D amino acid-containing channel-forming antibiotic peptides. Proc Natl Acad Sci USA. 1990;87(12):4761–5. doi: 10.1073/pnas.87.12.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eller M, Jarv J, Toomik R, Ragnarsson U, Ekman P, Engstrom L. Substrate-specificity of protein-kinase-c studied with peptides containing D-aminoacid residues. J Biochem. 1993;114(2):177–80. doi: 10.1093/oxfordjournals.jbchem.a124151. [DOI] [PubMed] [Google Scholar]
- 20.Merrifield EL, Mitchell SA, Ubach J, Boman HG, Andreu D, Merrifield RB. D-enantiomers of 15-residue cecropin a-melittin hybrids. Int J Pept Protein Res. 1995;46(3–4):214–20. doi: 10.1111/j.1399-3011.1995.tb00592.x. [DOI] [PubMed] [Google Scholar]
- 21.Shi J, Du X, Yuan D, Zhou J, Zhou N, Huang Y, Xu B. D-amino acids modulate the cellular response of enzymatic-instructed supramolecular nanofibers of small peptides. Biomacromolecules. 2014;15(10):3559–68. doi: 10.1021/bm5010355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Wang Y, Han A, Cai Y, Xiao N, Wang L, Ding D, Yang Z. Cellular membrane enrichment of self-assembling D-peptides for cell surface engineering. ACS Appl Mater Interfaces. 2014;6(12):9815–21. doi: 10.1021/am502250r. [DOI] [PubMed] [Google Scholar]
- 23.Liang G, Yang Z, Zhang R, Li L, Fan Y, Kuang Y, Gao Y, Wang T, Lu WW, Xu B. Supramolecular hydrogel of a D-amino acid dipeptide for controlled drug release in vivo. Langmuir. 2009;25(15):8419–22. doi: 10.1021/la804271d. [DOI] [PubMed] [Google Scholar]
- 24.Li XM, Du XW, Li JY, Gao Y, Pan Y, Shi JF, Zhou N, Xu B. Introducing D-amino acid or simple glycoside into small peptides to enable supramolecular hydrogelators to resist proteolysis. Langmuir. 2012;28(37):13512–7. doi: 10.1021/la302583a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.d’AMOURS D, DESNOYERS S, d’SILVA I, Poirier GG. Poly (ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 26.Pardridge WM. Receptor-mediated peptide transport through the blood-brain barrier. Endocr Rev. 1986;7(3):314–30. doi: 10.1210/edrv-7-3-314. [DOI] [PubMed] [Google Scholar]
- 27.Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J Biol Chem. 2001;276(8):5836–40. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- 28.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351(6326):453–6. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 29.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 30.Wong C-H, Whitesides GM. Enzymes in synthetic organic chemistry. Academic Press; 1994. [Google Scholar]
- 31.Wehofsky N, Pech A, Liebscher S, Schmidt S, Komeda H, Asano Y, Bordusa F. D-Amino acid specific proteases and native all-L-proteins: A convenient combination for semisynthesis. Angew Chem Int Ed. 2008;47(29):5456–60. doi: 10.1002/anie.200800340. [DOI] [PubMed] [Google Scholar]
- 32.Naghavi M, Wang H, Lozano R, Davis A, Liang X, Zhou M, Vollset SE, Ozgoren AA, Abdalla S, Abd-Allah F. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385(9963):117–71. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao F, Ma ML, Xu B. Molecular hydrogels of therapeutic agents. Chem Soc Rev. 2009;38(4):883–91. doi: 10.1039/b806410p. [DOI] [PubMed] [Google Scholar]
- 34.Gao Y, Kuang Y, Guo Z-F, Guo Z, Krauss IJ, Xu B. Enzyme-instructed molecular self-assembly confers nanofibers and a supramolecular hydrogel of taxol derivative. J Am Chem Soc. 2009;131(38):13576–7. doi: 10.1021/ja904411z. [DOI] [PubMed] [Google Scholar]
- 35.Burtis CA, Ashwood ER, Burns DE. Tietz texbook of clinical and molecular diagnosis. Elsevier; Saunders: 2006. [Google Scholar]
- 36.Yang Z, Xu K, Guo Z, Guo Z, Xu B. Intracellular enzymatic formation of nanofibers results in hydrogelation and regulated cell death. Adv Mater. 2007;19(20):3152–6. [Google Scholar]
- 37.Kuang Y, Shi J, Li J, Yuan D, Alberti KA, Xu Q, Xu B. Pericellular hydrogel/nanonets inhibit cancer cells. Angew Chem Int Ed. 2014;53(31):8104–7. doi: 10.1002/anie.201402216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Kuang Y, Shi J, Zhou J, Medina JE, Zhou R, Yuan D, Yang C, Wang H, Yang Z. Enzyme-instructed intracellular molecular self-assembly to boost activity of cisplatin against drug-resistant ovarian cancer cells. Angew Chem Int Ed. 2015;127(45):13505–13509. doi: 10.1002/anie.201507157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang C, Chu L, Zhang Y, Shi Y, Liu J, Liu Q, Fan S, Yang Z, Ding D, Kong D. Dynamic biostability, biodistribution, and toxicity of L/D-peptide-based supramolecular nanofibers. ACS Appl Mater Interfaces. 2015;7(4):2735–44. doi: 10.1021/am507800e. [DOI] [PubMed] [Google Scholar]
- 40.Ou C, Zhang J, Shi Y, Wang Z, Wang L, Yang Z, Chen M. D-amino acid doping peptide hydrogel for the production of a cell colony. RSC Adv. 2014;4(18):9229–32. [Google Scholar]
- 41.Marchesan S, Easton C, Styan K, Waddington L, Kushkaki F, Goodall L, McLean K, Forsythe J, Hartley P. Chirality effects at each amino acid position on tripeptide self-assembly into hydrogel biomaterials. Nanoscale. 2014;6(10):5172–80. doi: 10.1039/c3nr06752a. [DOI] [PubMed] [Google Scholar]
- 42.Marchesan S, Easton CD, Kushkaki F, Waddington L, Hartley PG. Tripeptide self-assembled hydrogels: unexpected twists of chirality. Chem Commun. 2012;48(16):2195–7. doi: 10.1039/c2cc16609g. [DOI] [PubMed] [Google Scholar]
- 43.Rabideau AE, Pentelute BL. A D-amino acid at the N-terminus of a protein abrogates its degradation by the N-end rule pathway. ACS Cent Sci. 2015;1(8):423–430. doi: 10.1021/acscentsci.5b00308. [DOI] [PMC free article] [PubMed] [Google Scholar]