Abstract
Colorectal cancer (CRC) results from the accumulation of gene mutations and epigenetic alterations in colon epithelial cells, which promotes CRC formation through deregulating signaling pathways. One of the most commonly deregulated signaling pathways in CRC is the transforming growth factor β (TGF-β) pathway. Importantly, the effects of TGF-β signaling inactivation in CRC are modified by concurrent mutations in the tumor cell, and these concurrent mutations determine the ultimate biological effects of impaired TGF-β signaling in the tumor. However, many of the mutations that cooperate with the deregulated TGF-β signaling pathway in CRC remain unknown. Therefore, we sought to identify candidate driver genes that promote the formation of CRC in the setting of TGF-β signaling inactivation. We performed a forward genetic screen in mice carrying conditionally inactivated alleles of the TGF-β receptor, type II (Tgfbr2) using Sleeping Beauty (SB) transposon mediated mutagenesis. We used TAPDANCE and Gene-centric statistical methods to identify common insertion sites (CIS) and, thus, candidate tumor suppressor genes and oncogenes within the tumor genome. CIS analysis of multiple neoplasms from these mice identified many candidate Tgfbr2 cooperating genes and the Wnt/β-catenin, Hippo and MAPK pathways as the most commonly affected pathways. Importantly, the majority of candidate genes were also found to be mutated in human CRC. The SB transposon system provides an unbiased method to identify Tgfbr2 cooperating genes in mouse CRC that are functionally relevant and that may provide further insight into the pathogenesis of human CRC.
Keywords: Tgfbr2, Sleeping Beauty, colorectal cancer, genetic screen, colon
Introduction
Colorectal cancer (CRC) is the third most common cause of cancer related deaths for men and women in the United States.1 CRC typically arises through a polyp-to-cancer progression sequence in which polyps evolve to become CRC over 8–15 years; this process results from the progressive accumulation of genetic and epigenetic alterations that transform normal intestinal epithelial cells into CRC cells.2 These alterations deregulate fundamental behaviors of cells, including those related to growth factors and signaling pathways, to drive the formation of CRC.3, 4
Characterization of human CRC by The Cancer Genome Atlas (TCGA) has identified two different general classes of CRCs, the hypermutated class (16%) and the non-hypermutated (84%) class, based on the frequency of mutations per 106 bases.5 In addition, other CRC molecular subgroups that have been identified include the chromosomal instability (CIN) subgroup, CpG Island Methylator Phenotype (CIMP) subgroup, and microsatellite instability (MSI) subgroup.6–8 Unique sets of genes have been identified as frequently mutated in the hypermutated and non-hypermutated tumors. Some of these genes include ACVR2A, APC, TGFBR2, BRAF, TCF7L2 (hypermutated CRCs) and APC, TP53, KRAS (non-hypermutated CRCs).5 Many of these commonly occurring mutations in CRC affect specific signaling pathways in the majority of CRCs, suggesting that these deregulated pathways may cooperate to influence or “drive” CRC formation. These pathways include the WNT, PI3K, MAPK, p53, and TGF-β signaling pathways. Specifically, mutations in the TGF-β signaling pathway have been reported in 87% of hypermutated CRC and 27% of non-hypermutated tumors.5 The identification of mutations in the TGF-β pathway in the majority of CRCs and functional studies of effects of TGF-β signaling deregulation in CRC cell lines and mouse models suggest that this pathway may have a central role in CRC formation.8
The TGF-β signaling pathway consists of the TGF-β ligand, the TGF-β receptor; a heteromeric complex consisting of type I (TGFBR1) and type II (TGFBR2) receptors; and post-receptor signaling elements including the SMAD (SMAD2, SMAD3, and SMAD4), PI3K, NFκB, and p38MAPK signaling pathways, among others. Mutations in TGFBR2 have been reported in approximately 30% of sporadic CRCs and in >50% of hypermutated tumors.5 Stimulation of the TGF-β pathway regulates epithelial cell growth and death, connective tissue deposition, angiogenesis and genomic stability as well as a variety of other fundamental cell behaviors that can influence tumor formation.9
There is considerable genetic evidence that the TGF-β pathway is predominantly tumor suppressive in colon epithelial cells and that it inhibits the progression of benign adenomas to adenocarcinomas.10, 11 Studies by our lab and others have demonstrated that the effects of TGF-β signaling inactivation on cancer formation are dependent on the concurrent gene mutations present in the tumor cells.12 However, given the thousands of mutations found in the average cancer genome, identification of the functionally important genes that “drive” CRC formation and that cooperate with inactivation of the TGF-β signaling pathway presents a significant challenge. Since the TGF-β pathway is one of the major pathways identified as being altered in CRC, we chose to employ a Sleeping Beauty (SB) transposon mediated mutagenesis screen to identify genes that functionally cooperate with Tgfbr2 inactivation to affect CRC formation.13
Materials and Methods
Mice
The 6070;Rosa26-LsL-SB11;T2/Onc2 [STOCK Gt(ROSA)26 Sortm2(sb11)Njen TgTn(sb-T2/Onc2)6070Njen/Nci] and the 6113;Rosa26-LsL-SB11;T2/Onc2 mice [B6;C3-TgTn(sb-T2/Onc2)6113Njen/Nci] were generous gifts from Drs. Adam J. Dupuy, Neal G. Copeland, and Nancy A. Jenkins.13 These mice were crossed with Villin-Cre mice [B6.Cg-Tg(Vil-cre)997Gum/J] and mice that carry either wild-type Tgfbr2 alleles (Tgfbr2wt/wt) or “floxed” Tgfbr2 alleles (Tgfbr2flx/flx) [B6.129 S6-Tgfbr2tm1Hlm/Nci], to generate mice that were either 6070;Rosa26-LsL-SB11;T2/Onc2;Villin-Cre;Tgfbr2wt/wt and 6113;Rosa26-LsL-SB11;T2/Onc2;Villin-Cre;Tgfbr2wt/wt (SB-Twt/wt; Tgfbr2 intact in intestine) or 6070;Rosa26-LsL-SB11;T2/Onc2;Villin-Cre;Tgfbr2flx/flx and 6113;Rosa26-LsL-SB11;T2/Onc2;Villin-Cre;Tgfbr2flx/flx (SB-TT; Tgfbr2 null in intestine).14 Genotypes were determined by PCR following published protocols.11, 13 All animal procedures were approved and performed according to the Fred Hutchinson Cancer Research Center IACUC #1624.
Histopathology
Mouse intestinal tumors larger than ∼9mm2 were cut in half and either snap-frozen in liquid nitrogen for DNA preparations; or fixed in 10% neutral buffered formalin phosphate (Fisher Scientific), paraffin embedded, and cut into 4 µm sections for hematoxylin and eosin or Alcian blue staining. Sections were examined by a pathologist (SEK) blinded to genotype and classified according to published criteria.15 Whole-slide images were acquired with the Aperio ScanScope AT (Aperio) using the 20X objective.
Linker-ligation mediated PCR
Following published protocols, T4 DNA Ligase was used to ligate linkers onto BfaI (left side) and NlaIII (right side) digested gDNA isolated from intestinal tumors.16 BamHI digestions were performed to eliminate transposon DNA concatamers. Two rounds of nested PCR were performed using primers specific for the transposon and linker sequences as well as primers containing specific barcodes for each tumor and sequences required for direct sequencing on the Illumina platform.17 PCR products were column purified and pooled (100ng per sample) into either SB-Twt/wt or SB-TT amplicon pools. The two samples were loaded onto two separate lanes of the same flowcell and sequenced using the HiSeq 2000 (Illumina).
Sequence analysis
Sequences were analyzed according to published methods.17, 18 Briefly, sequences were filtered based on the Illumina base-call quality score followed by analysis for the presence of the barcode, transposon sequence, linker sequence and TA integration site. Trimmed sequences were aligned to the mouse genome reference assembly (NCBI37/mm9) using Bowtie.19 Transposon insertion sites that mapped to the same chromosome as the donor transgene were removed to eliminate local hopping bias. Both TAPDANCE and Gene-centric CIS (gCIS) analyses were performed on the data sets and insertion sites were identified as previously published.17, 18 Additionally, each CIS required a minimum of three independent tumors with insertions in the same gene. To calculate the number of genes targeted per tumor as well as the number of shared CIS per tumor set, a tumor matrix was generated in R (version 2.15.1). Statistical analysis was performed using GraphPad Prism version 4.00. The Bonferroni method was used to correct for multiple testing.17 A P value of < 0.05 was considered significant.
Pathway analysis
Genes identified by TAPDANCE were analyzed by QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). Canonical pathway analysis identified the pathways from the IPA library of canonical pathways that were most significant to the data set. Molecules from the data set that were associated with a canonical pathway in the Ingenuity Knowledge Base were considered for the analysis. The significance of the association between the data set and the canonical pathway was measured in 2 ways: 1) A ratio of the number of molecules from the data set that map to the pathway divided by the total number of molecules that map to the canonical pathway is calculated (Overlap). 2) Fisher’s exact test was used to calculate a P-value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone.
Results
Induced deletion of Tgfbr2 in the Rosa26-LsL-SB11;T2/Onc2;Villin-Cre mice results in greater numbers of adenocarcinomas and larger adenocarcinomas
A Sleeping Beauty (SB) transposon-based insertional mutagenesis forward genetic screen was performed to identify genes that cooperate with loss of Tgfbr2 to drive intestinal cancer formation in mice. In order to identify potential cooperating genes across the entire mouse genome and adjust for local hopping events, two different founder strains of T2/Onc2 transgenic mice were used (6113, chromosome 1; 6070, chromosome 4).13 The T2/Onc2 transposon carries a retroviral LTR and splice donor as well as a bidirectional polyA sequence and splice acceptor on both strands of DNA.13 These features render it capable of both activating proto-oncogenes and inactivating tumor suppressor genes. An inducible version of the SB transposase SB11 was used in which expression of Cre recombinase activates the SB11 transposase, which then mobilizes the T2/Onc2 transposon. To induce mobilization of the transgene specifically in the intestinal epithelium, triple transgenic mice expressing Cre under control of the Villin promoter (Rosa26-LsL-SB11;T2/Onc2;Villin-Cre) were generated. The triple transgenic mice were further crossed with mice carrying “floxed” Tgfbr2 alleles in order to obtain quadruple transgenic mice that were Rosa26-LsL-SB11;T2/Onc2;Villin-Cre;Tgfbr2flx/flx (SB-TT, Tgfbr2 null in the intestines).14, 20 The Rosa26-LsL-SB11;T2/Onc2;Villin-Cre;Tgfbr2wt/wt mice (SB-Twt/wt, Tgfbr2 intact in the intestines) were used as controls, since Villin-Cre;Tgfbr2flx/flx mice fail to develop intestinal tumors.11 Mice were monitored for signs of illness and were sacrificed when morbid or at approximately 100 weeks of age.
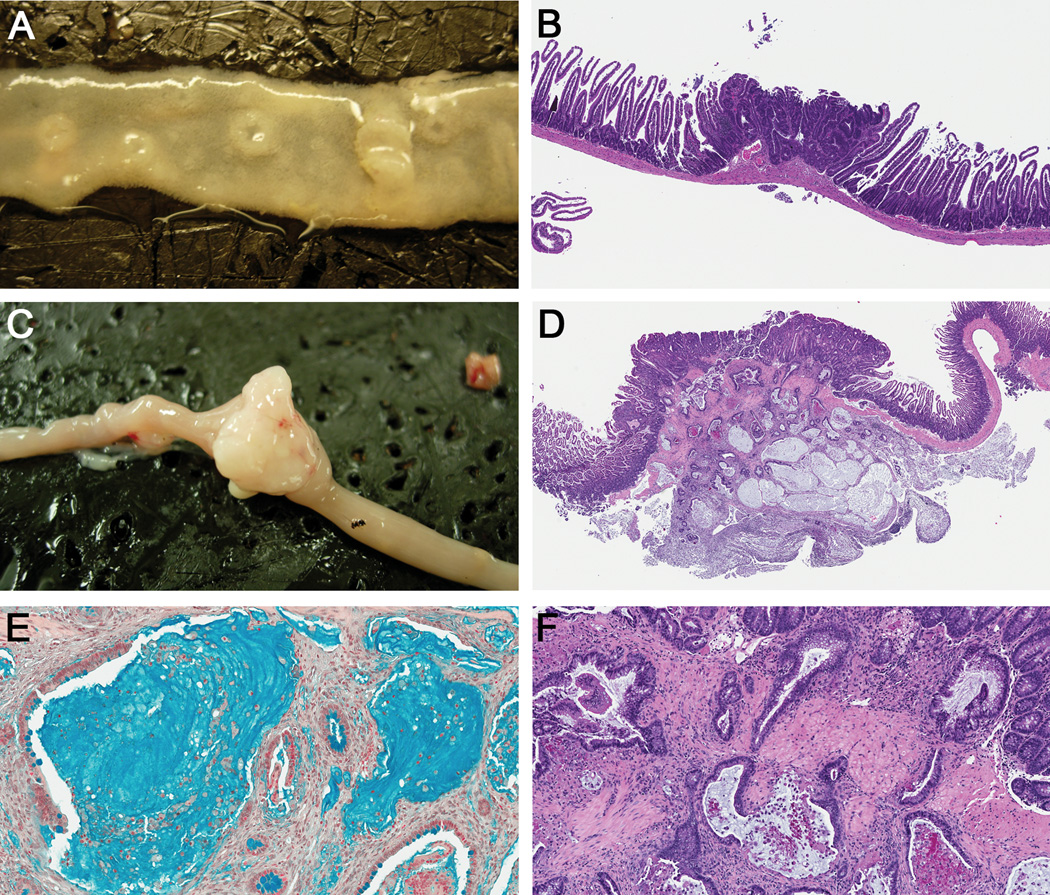
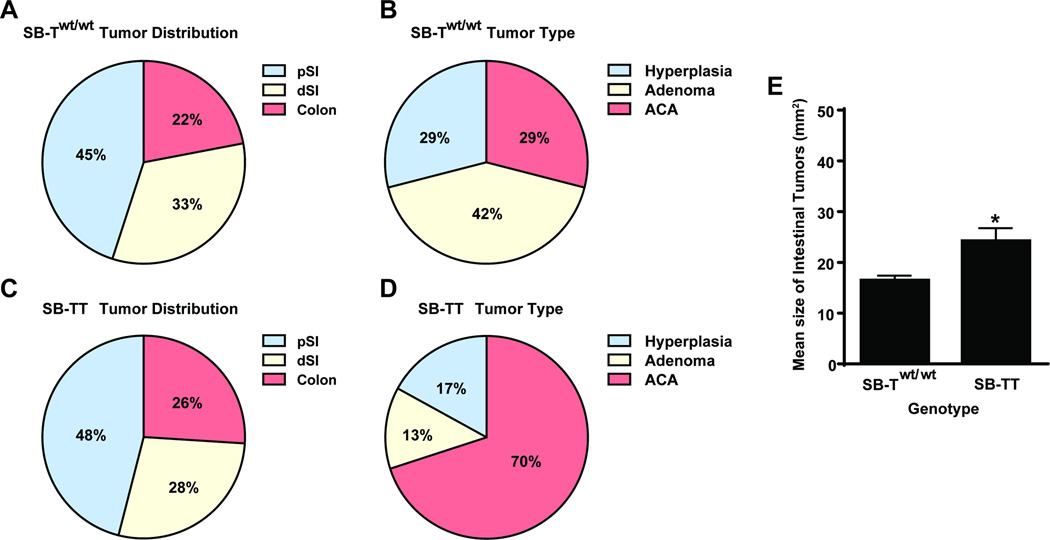
Adenocarcinomas (ACAs), adenomas (Ads), and regions of hyperplasia (Hyp) were observed in the proximal small intestine (pSI), the distal small intestine (dSI) and the colon in both lines of mice (Table 1, Fig. 1A – D). Gross inspection of the intestinal mucosa at necropsy revealed that the overall percent distribution of tumors found in the different anatomical locations of the intestines was similar in the SB-Twt/wt (45% pSI, 33% dSI, 22% colon) and SB-TT (46% pSI, 28% dSI, 26% colon) mice (Fig. 2A and C). However, measurement of the tumor size in two dimensions revealed that the neoplasms in the SB-TT mice were larger than the neoplasms in the SB-Twt/wt mice, (P = 0.0256, Fig. 2E), suggesting that loss of Tgfbr2 promotes the growth of intestinal neoplasms. Interestingly, the SB-Twt/wt mice had more neoplasms and adenomas compared to the SB-TT mice (Wilcoxon rank sum P < 0.05, Table 1), which could indicate that Tgfbr2 mutations inhibit tumor initiation in the SB mouse model and instead have their main effect on tumor progression. Histological analysis determined that the SB-TT mice had significantly more ACAs compared to the SB-Twt/wt mice (P < 0.0001, Table 1; Fig. 2B and D). Additionally, nearly 46% of the SB-TT ACAs were diagnosed as mucinous adenocarcinomas based on >50% of the tumor being composed of extracellular mucin, which is consistent with the histology of intestinal neoplasms seen in other Tgfbr2 null models.11, 15, 21 Mucinous ACAs were almost exclusive to the SB-TT tumors. (Table 1; Fig. 1D – F).
Table 1.
SB intestinal tumor summary.
| Genotype | Number of Mice | Hyperplasia | Adenoma | Adenocarcinomaa (mucinous type)b |
Total |
|---|---|---|---|---|---|
| SB-Twt/wt | 43 | 36 | 53 | 36 (1) | 125 |
| SB-TT | 44 | 14 | 11 | 57 (26) | 82 |
P < 0.0001, Fisher’s Exact Test, Two-Tailed (36/89 vs. 57/25)
P < 0.0001, Fisher’s Exact Test, Two-Tailed (1/35 vs. 26/31)
Figure 1. SB-induced intestinal tumor histology.
Representative intestinal (A) adenomas and (C) adenocarcinoma from SB mice. H&E stained sections of representative intestinal (B) adenoma (Magnification, 4X), and (D) mucinous adenocarcinoma (2X). (E) Alcian blue stained mucinous adenocarcinoma (20X). (F) Higher magnification of panel D, (10X), shows neoplastic glands penetrating through the muscularis mucosa.
Figure 2. Summary of tumor distribution, type and size in SB-Twt/wt and SB-TT mice.
The percentage of lesions found in the proximal small intestine (pSI), the distal small intestine (dSI) and colon of (A) SB-Twt/wt and (C) SB-TT mice. The percentage of hyperplasias, adenomas and adenocarcinomas (ACA) in (B) SB-Twt/wt and (D) SB-TT mice. (E) A significant increase in the mean size of intestinal tumors in SB-TT mice compared to SB-Twt/wt mice was observed, as measured by calipers in two dimensions (P = 0.0256, Mann-Whitney Test).
Of interest, Tgfbr2 deletion in the intestines of the Rosa26-LsL-SB11;T2/Onc2;Villin-Cre mice also associated with changes in survival of the mice compared to those with intact Tgfbr2 (SB-Twt/wt). A total of 16 out of 60 SB-TT mice analyzed died at an average age of 13.75 weeks due to various hematopoietic neoplasias, including T and B-cell lymphomas, and erythroid and myeloid leukemia. In addition, 3 out of 46 SB-Twt/wt mice died at approximately 12.33 weeks of age due to similar neoplasms. Since these early deaths were not the result of intestinal tumors, these mice were not included in the subsequent analyses. Kaplan-Meier survival curve analysis of the SB-Twt/wt and SB-TT cohorts revealed that the SB-TT mice died an average of 2 months sooner than the SB-Twt/wt mice (72.5 weeks vs. 81 weeks) secondary to the development of obstructing intestinal neoplasms, however, there was no statistically significant difference in the overall survival rate (P = 0.2384, Supporting Information, Fig. 1).
Analysis of Common Insertion Sites identifies candidate driver genes that are dependent on induced Tgfbr2 disruption
Transposon insertion sites were analyzed from 130 SB-Twt/wt and 130 SB-TT tumors from 38 and 40 mice, respectively. Based on the available histology of the tumors, 23 ACAs, 53 Ads, and 18 Hyp were sequenced from the SB-Twt/wt mice; and 52 ACAs, 17 Ads, 21 Hyp were sequenced from the SB-TT mice. Given their gross appearance, the additional lesions sequenced (36 from SB-Twt/wt and 40 from SB-TT mice) were most likely adenomas or hyperplasias, however formal histologic examination was not possible given the small size of the tumors, which precluded histologic assessment concurrently with gDNA extraction.
To identify genes that cooperate with loss of Tgfbr2 to promote neoplasm progression, we compared transposon common insertion sites (CIS) from the SB-Twt/wt and SB-TT tumors using both the TAPDANCE and Gene-centric (gCIS) statistical methods (Supporting Data).17, 18 CISs are the Common Integration Sites of the T2/Onc2 transposon that occur with a higher frequency than would be expected by chance alone and, thus, indicate a likely functionally important mutation in the tumor. TAPDANCE analysis identified 743 SB-Twt/wt and 673 SB-TT candidate genes, while gCIS analysis identified 520 SB-Twt/wt and 372 SB-TT candidate loci (Supporting Information, Fig. 2). To determine the extent of CIS gene overlap between the SB-TT and SB-Twt/wt mice, we compared the candidate gene lists from each genotype identified by either TAPDANCE or gCIS. Comparison of likely candidate genes from the TAPDANCE analysis revealed that 441 candidate genes were shared between the two cohorts, while 302 genes were only identified in SB-Twt/wt tumors and 232 were only identified in SB-TT tumors. Additionally, comparison of genes identified by gCIS analysis revealed 185 genes shared in common, while 335 were only in SB-Twt/wt tumors and 187 were in SB-TT tumors. The list of CIS genes unique to the Tgfbr2 mutant tumors will be useful in determining pathways that cooperate with loss of Tgfbr2, although not all of these are expected to be specific to tumors that have lost Tgfbr2.
Furthermore, to determine whether or not the location of the tumors influenced the CIS list, we used Fisher’s exact test to check for associations between either colon tumors (n = 65) or between small intestine tumors (n = 195) and every CIS that we had identified as described above. After correcting for multiple testing (Bonferroni corrected P < 4.3E-5) we did not find any significant associations that were only found in colon tumors or in small intestine tumors.
Analysis of mutations that were identified only in Tgfbr2 mutant mice
After identifying CISs in tumors arising in the SB-TT and SB-Twt/wt mice, we carried out an analysis of the candidate driver genes identified by either TAPDANCE or gCIS only identified in the SB-TT tumors. In order to enhance the stringency for identifying genes that were targets of SB mutagenesis in the setting of Tgfbr2 inactivation and thus candidate cooperating driver genes, we first identified those genes that were present in both the TAPDANCE and gCIS gene sets. A total of 17 genes were found to be identified and shared by both analyses (Table 2), which demonstrates the complementary nature of these two methods. Interestingly, all 17 genes had human orthologs that were found to carry mutations in human CRCs in either the TCGA human colon and rectal cancer database or in the COSMIC large intestine database.5, 22, 23 Some of these genes include LRP6, PPP2R1A, TCF7L2 and YAP1, which are involved in the Wnt and Hippo signaling pathways, respectively. Furthermore, PPP2R1A and TCF7L2 were also found in the Cancer Gene Consensus (CGC) catalogue, which is a list of genes for which mutations have been causally implicated in cancer formation.24 IPA analysis confirmed that the top two pathways associated with these 17 commonly disrupted genes are the Wnt/β-catenin signaling pathway (P = 2.32E-04) and the Hippo pathway (P = 1.76E-03).
Table 2.
Novel Tgfbr2 cooperating candidate genes.
| CIS Gene | Chr | TAPDANCE P-value |
gCIS Q-value |
SB-TT Tumors£ |
TCGA# | COSMIC€ | Predicted Effect of Insertion |
Function/Pathway |
|---|---|---|---|---|---|---|---|---|
| Acbd3 | 1 | 2.58E-09 | 9.48E-06 | 13.1 (17) | 2.7 | 2.0 | Disruption | Golgi structure/function |
| Arhgef38 | 3 | 2.56E-07 | 3.65E-06 | 11.5 (15) | 0.4 | 0.8 | Disruption | Rho GEF |
| Cdk13 | 13 | 3.24E-07 | 6.67E-09 | 15.4 (20) | 2.2 | 3.9 | Disruption | Cyclin-dependent kinase |
| Dnajc5 | 2 | 3.07E-03 | 4.98E-05 | 6.9 (9) | 0.0 | 1.0 | Disruption | Regulates heat shock proteins |
| Gna13 | 11 | 5.83E-04 | 4.42E-05 | 10.0 (13) | 1.3 | 2.0 | Disruption | Guanine nucleotide-binding protein |
| Lrp6 | 6 | 1.29E-08 | 7.56E-08 | 15.4 (20) | 7.6 | 5.0 | Disruption | Wnt signaling pathway |
| Metap2 | 10 | 2.78E-05 | 2.32E-14 | 8.5 (11) | 1.3 | 1.1 | Disruption | Methionyl aminopeptidase |
| Nudt3 | 17 | 4.44E-03 | 1.08E-05 | 7.7 (10) | 0.0 | 0.5 | Disruption | Cleaves beta-phosphate |
| Pacsin2 | 15 | 5.60E-07 | 7.52E-05 | 19.2 (25) | 2.2 | 2.5 | Disruption | Vesicle formation/transport |
| Ppp2r1a§ | 17 | 3.67E-04 | 1.42E-06 | 6.9 (9) | 2.7 | 3.3 | Disruption | Wnt signaling pathway |
| Pptc7 | 5 | 2.70E-05 | 1.40E-05 | 13.1 (17) | 1.3 | 1.8 | Disruption | Phosphatase |
| Psma3 | 12 | 2.54E-10 | 2.37E-11 | 13.8 (18) | 1.8 | 1.2 | Disruption | Proteasome subunit |
| Rai1 | 11 | 5.42E-05 | 2.58E-12 | 13.8 (18) | 2.7 | 5.0 | Disruption | Transcriptional regulator |
| Rsbn1l | 5 | 5.12E-11 | 7.36E-05 | 21.5 (28) | 0.4 | 2.1 | Disruption | Unknown |
| Tcf7l2§ | 19 | 9.80E-12 | 3.21E-09 | 22.3 (29) | 9.8 | 9.3 | Disruption | Wnt signaling pathway |
| Yap1 | 9 | 1.23E-06 | 1.94E-07 | 12.3 (16) | 1.3 | 0.7 | Disruption | Hippo signaling pathway |
| Zzz3 | 3 | 3.63E-06 | 1.03E-06 | 13.1 (17) | 3.1 | 2.5 | Disruption | Chromatin binding |
Chr, Mouse Chromosome (NCBI37/mm9); TCGA, The Cancer Genome Atlas; COSMIC, Catalogue Of Somatic Mutations In Cancer.
Ppp2r1a and Tcf7l2 found in the Cancer Gene Census.
Percent mutated samples from TAPDANCE analysis (mouse number in parentheses).
Percent mutated samples out of human tumor samples tested.
Percent mutated samples out of human large intestine cancer samples tested.
Of interest, although loss of APC is a hallmark of CRC, our study suggests that loss of Tgfbr2 may decrease the selective pressure for loss of Apc (Supporting Information, Table 1). Only 27% (35/130) of SB-TT tumors had Apc transposon insertions, compared to 45% (58/130) of SB-Twt/wt tumors. This finding was also observed in an SB screen performed using mice with a heterozygous germline loss of Smad4.25 Our findings suggest that in the setting of TGF-β signaling inactivation, the Wnt signaling pathway is important for tumor formation but genes other than Apc mediate deregulation of Wnt signaling in these tumors.
Comparison of tumors that have lost Tgfbr2 to tumors that have lost Smad4
After performing our SB screen for genes that cooperate with loss of Tgfbr2, the Jenkins and Copeland group independently reported their findings from a similar SB screen using a heterozygous Smad4 germline knockout mouse strain.25 Although there are likely to be independent pathways affected by these two models due to Smad-independent signaling via Tgfbr2 26 and the converse, Tgfbr2-independent signaling via Smad4 27, we hypothesized that there would be significant overlap in the candidate genes discovered by these two screens. As mentioned above, in both screens Apc mutations were found at a lower frequency and adenocarcinomas were found at a higher frequency when compared to wild-type mice (Fig. 3, Table 1 and Supporting Information). Comparison of the candidate gene lists indicated that 54% of the 449 genes identified in the study of the Smad4 null mouse model were also identified in our study, indicating a highly significant overlap (Fisher’s exact test P < 2.9E-245).
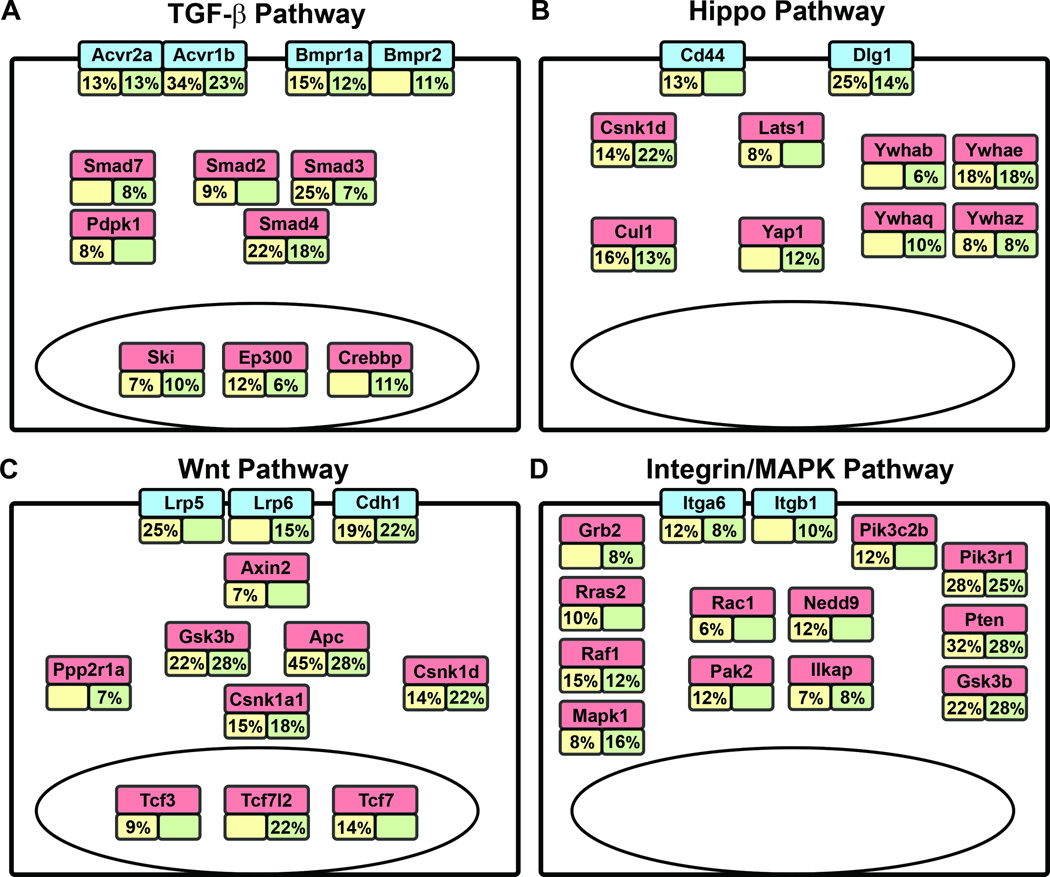
Figure 3. Deregulated signaling pathways in CRC.
The percentages of tumors with transposon insertions in genes for various signaling pathways identified by TAPDANCE analysis are indicated. (A) TGF-β signaling pathway, (B) Hippo signaling pathway, (C) Wnt signaling pathway, and (D) Integrin/MAPK signaling pathways. Yellow boxes (left) indicate SB-Twt/wt tumors and green boxes (right) indicate SB-TT tumors. Empty boxes indicate that < 6% of tumors contained an insertion in that particular gene, the cut-off chosen for our analysis.
To analyze differences between the two gene lists we performed pathway enrichment analysis using the list of CIS genes found only in the Smad4 null screen and the CIS genes found only in our Tgfbr2 null screen. Using the ToppCluster web tool 28, which identifies enriched pathways from multiple pathway databases based on gene lists, Erbb1 downstream signaling and both miRNA and mRNA splicing pathways were enriched in the Tgfbr2 null-only CIS genes, while MAP kinase, cell-cell communication, CDC42 signaling, tight junction, and EGFR1 signaling pathways were enriched in the Smad4 null-only screen (Supporting Data). These differences could reflect signaling by Tgfbr2 or Smad4 outside of the canonical TGF-β pathway.
Pathway analysis of CISs in SB mice reveal differences in the most commonly affected pathways based on the Tgfbr2 status of the tumors
We next performed pathway analysis of the genes identified by TAPDANCE using QIAGEN’s IPA software. The top 5 pathways for the SB-Twt/wt and SB-TT tumor gene sets are shown in Table 3. The integrin signaling pathway was the most commonly deregulated pathway in tumors arising in the SB-Twt/wt mice, while the Hippo pathway was the most commonly affected pathway for the neoplasms arising in the SB-TT mice. Other top pathways identified include the ERK/MAPK pathway, the Wnt/β-catenin pathway and the TGF-β signaling pathway. Based on this pathway analysis we calculated the percent of tumors with an insertion in the gene within those deregulated pathways and found differences in the frequency of alterations in a subset of the pathway members between the SB-TT and SB-Twt/wt tumors (Fig. 3). These results suggest that TGF-β signaling interacts preferentially with certain members of these common pathways and provides further insight into the cooperation of signaling pathways altered in CRC.
Table 3.
Ingenuity Pathway Analysis of TAPDANCE identified CISs.
| SB-Twt/wt Tumors | ||
|---|---|---|
| Top 5 Canonical Pathways | P-value | Overlap |
| Integrin Signaling | 3.63E-10 | 28/201 (13.9%) |
| Glucocorticoid Receptor Signaling | 5.35E-10 | 33/275 (12.0%) |
| Hippo Signaling | 6.86E-10 | 18/86 (20.9%) |
| Wnt/β-catenin Signaling | 8.90E-10 | 25/169 (14.8%) |
| Rac Signaling | 2.62E-09 | 19/104 (18.3%) |
| SB-TT Tumors | ||
| Top 5 Canonical Pathways | P-value | Overlap |
| Hippo Signaling | 1.42E-11 | 19/86 (22.1%) |
| ERK/MAPK Signaling | 4.48E-09 | 24/187 (12.8%) |
| RAR Activation | 6.17E-09 | 24/190 (12.6%) |
| Ephrin Receptor Signaling | 2.60E-08 | 22/174 (12.6%) |
| TGF-β Signaling | 7.07E-08 | 15/87 (17.2%) |
TGF-β signaling inactivation is subject to strong selective pressure in intestinal cancer formation
In light of prior publications, an unexpected finding of our studies was that the loss of Tgfbr2 did not significantly accelerate the mortality rate or tumor incidence in the SB-TT mice as compared to the SB-Twt/wt mice (Supporting Information, Fig. 1) 11, 21, 29. Analysis of the CISs in the SB-Twt/wt mice revealed that the majority of these tumors had CISs in genes in the TGF-β pathway, suggesting that the reason for the lack of the anticipated effects in the SB-Twt/wt mice was because the tumors in these mice acquired somatic alterations that inactivated TGF-β signaling in the intestinal tumors. This observation suggests that mutations in TGF-β pathway genes and the resulting deregulation of the TGF-β signaling pathway may be a very favorable event for CRC formation. Furthermore, we found it interesting that one of the top 5 pathways identified in SB-TT mice by IPA analysis was the TGF-β pathway itself. Therefore we determined if canonical TGF-β pathway genes were targets of CISs more frequently than would be expected by chance. A list of 19 TGF-β pathway genes was obtained from BioCarta (http://cgap.nci.nih.gov/Pathways/BioCarta/h_tgfbPathway) (Supporting information, Table 2; Supporting Data). A total of 8 TGF-β pathway genes out of 673 TAPDANCE CIS genes were identified in the SB-TT tumors, and 6 out of 743 CIS genes were identified in the SB-Twt/wt tumors. Fisher’s exact test analyses revealed that indeed in both Tgfbr2 mutant and Tgfbr2 wild-type tumors the number of TGF-β pathway genes identified by TAPDANCE were significantly higher than would be expected (P = 2.54E-8 and 1.90E-5, respectively). These findings highlight the importance TGF-β pathway disruption in the development of CRC and also suggest that a mutation in one member of the pathway (i.e. Tgfbr2) does not necessarily prohibit mutations in other members of the same pathway, which is similar to what is observed for the Wnt signaling pathway.30
A subset of candidate driver genes identified by SB mutagenesis are independent of Tgfbr2 status
Our analysis identified a number of candidate driver genes that were commonly altered in both the SB-TT and SB-Twt/wt mice. Apc was the most commonly disrupted gene in both TAPDANCE and gCIS analyses in both the Tgfbr2 intact and Tgfbr2 null mouse lines, although, as noted above, Apc was disrupted less commonly in the SB-TT tumors compared to the SB-Twt/wt tumors. The common disruption of Apc has been found in other intestinal transposon mutagenesis studies as well, which provided validation for our CIS identification methods and provided further support for the importance of Apc disruption in intestinal tumorigenesis.31, 32 In addition to Apc, other candidate driver genes, including Pten, Smad4 and Wac were found to be shared between the SB-Twt/wt tumors, SB-TT tumors, as well as to be targets for CIS’s in other SB intestinal screens.25, 31, 32 The top 123 loci for CIS’s identified by TAPDANCE analysis in SB-Twt/wt tumors (ranked by p-value region) were also sites for CIS’s in SB-TT tumors. Similarly, the top 93 affected loci in SB-TT neoplasms (ranked by p-value region) were also found in the SB-Twt/wt neoplasms. In aggregate, these results show that a number of genes are frequently targeted in SB induced CRC independent of the Tgfbr2 status in the intestines, which raises the possibility that Tgfbr2 inactivation may be playing a role in progression events in the SB-Twt/wt tumors rather than in tumor initiation events or that the genes found in both the SB-TT and SB-Twt/wt tumors affect intestinal neoplasm formation independent of TGF-β signaling.
Genes identified in the screen are commonly mutated in human CRC
In order to determine the relevance of our findings to primary CRC, the gene lists generated by TAPDANCE and gCIS were queried against the TCGA list of somatic mutations in human colon and rectal cancer.5 Over 80% of both SB-Twt/wt and SB-TT mouse CISs identified were found to correlate with human orthologs that have documented somatic mutations in human CRC. These genes include Acvr2a, Apc, Arid1a, Casp8, Fbxw7, Kras, Map7, Ptpn12, Smad2, Smad4, Tcerg1, Tcf7l2 and Trp53, genes that were found to be highly mutated in both hyper and non-hypermutated human CRCs.5 Of note, there were some commonly mutated oncogenes in human CRC that were not identified by our CIS analyses. Some of these genes include the oncogenes Braf, Pik3ca, and Ctnnb1. This result may be due to the observation that the majority of CISs identified in our screen are predicted tumor suppressors and that oncogenes were less commonly identified.
We next queried the top 50 CISs identified by TAPDANCE against the TCGA and COSMIC databases.22, 23 All top 50 CISs had corresponding human orthologs (Supporting Information, Table 3), and 48/50 (96%) of these genes had mutations documented in either the TCGA human colon and rectal cancer database or in the COSMIC large intestine database. Furthermore, 12/50 (24%) genes were found in the CGC catalogue.24 This large level of overlap illustrates the similarity between SB insertions in mouse intestinal cancers and somatic mutations detected in human CRC, and provides evidence that this approach is relevant to the study of human CRC.
Discussion
Remarkable progress has been made over the last decade with regards to our understanding of the molecular genetics of CRC. The results of genome-wide sequencing using next-generation high throughput methods have identified a large number of mutations and alterations that occur in colorectal cancers.5, 33 These studies have demonstrated that the average CRC genome has 100’s-1000’s of mutations and that there are a few dozen commonly occurring mutations in the average colorectal cancer genome. These genes include those classically associated with colorectal cancer, like APC, KRAS, and TP53, and a large number of other genes mutated at lower frequencies. Currently, the biological relevance of many of these mutations is not clear and determining their relevance in CRC is a major challenge that will require extensive studies. Another challenge that the data from the cancer genome studies has revealed is that it is likely that the mutant genes cooperate with each other in affecting the biological responses of CRC cells. The need to determine the functionally relevant genetic alterations and to identify the “driver gene” mutations in CRC prompted us to develop a forward genetic screen using the Sleeping Beauty transposon system.34, 35 We carried out studies that identified novel genes that occurred in the context of mice with an intact intestinal TGF-β signaling pathway as well as in the context of an initially inactivated TGF-β signaling pathway. We chose this model because the TGF-β signaling pathway is one of the most commonly deregulated signaling pathways in CRC and because the effects of TGF-β signaling deregulation depend on the context of concurrent mutations.36, 37
The value of the SB model system is that it identifies functionally relevant genes that affect intestinal cancer in an unbiased fashion. Initial analysis of the genes disrupted in our SB mouse models revealed enrichment for genes in pathways commonly altered in cancer, including the Wnt/β-catenin, ERK/MAPK and PI3K/AKT pathways. We specifically found that Apc, Pten, and Gsk3b are commonly disrupted in SB-TT tumors, which suggests these genes cooperate with inactivated TGF-β signaling to promote intestinal cancer formation. The biological significance of these results is supported by prior studies that demonstrate that mice carrying mutant Apc or mutant Pten in combination with inactivated Tgfbr2 develop intestinal cancer.11, 29.
In addition to demonstrating that genes mutated in CRC cooperate with inactivated TGF-β signaling to promote CRC, we have also identified many novel genes that likely play a role in CRC formation. Many of these newly identified genes disrupt signaling pathways believed to play a role in CRC, but there are others that appear to have potentially novel roles in CRC. For instance, Tcf7l2 is down regulated in many human CRCs and appears to function as a tumor suppressor in the colon in cooperation with mutant APC.38 Other genes, like Csnk1d, identified in the SB-TT screen have not been previously shown to have a role in CRC, but are plausible tumor suppressor genes or oncogenes based on their known functions. In the case of Csnk1d, it has been shown to regulate Mdm2 protein levels, which could affect p53 activity.39, 40
In the Smad4 null SB screen performed by Takeda, et al., the authors highlighted two CIS genes, Rspo1 and Rspo2. They noted that these CISs were identified in the Smad4 null screen, but not in previous screens with mice that have an Apc mutation. They reported that Rspo1 and Rspo2 mutant tumors were mutually exclusive of each other. In contrast, our screen also identified Rspo1 and Rspo2 as CIS genes, but of the 28 tumors with Rspo insertions, three tumors had insertions in both Rspo1 and Rspo2, indicating a lack of mutual exclusivity (Fisher’s Exact Test, P = 0.91). Takeda et al., also hypothesized that Rspo1/2 mutations may be an alternative to APC mutations based on data from human patients with RSPO fusion genes 41, however both the Tgfbr2 null screen and the Smad4 null screen had a large number of tumors with concomitant insertions in both Apc and Rspo1/2. In our study, 30% of Tgfbr2 mutant tumors with Apc insertions also had insertions in Rspo1 or Rspo2. It is worth noting however, that the hypothesis of mutual exclusivity of Rspo and Apc mutations could still hold true for the 70% of Tgfbr2/Rspo mutant tumors lacking insertions in Apc. In addition, it is possible that Rspo1, Rspo2 and Apc mutations could occur in different subclones of a cancer and not be identified as mutually exclusive by our method of analysis.
A particularly interesting observation is that disruption of genes in the Hippo pathway are found commonly in the SB-TT mice. The Hippo pathway is known to regulate tissue homeostasis, including proliferation and differentiation, in the intestines and modulates a number of signaling pathways in the intestinal epithelium, including the WNT, Notch, and TGF-β pathways. Yap1, a key member of the Hippo pathway, promotes tumorigenesis and is associated with poor prognosis in CRC when it is unphosphorylated, the state in which it can enter the nucleus and act as a transcriptional activator.42–44 When Yap1 is phosphorylated it is sequestered in the cytoplasm by 14-3-3 and is rendered inactive. Several Hippo pathway members were identified as CISs in the SB-TT tumors including Yap1, Dlg1, Csnk1d, Cul1, and the 14-3-3 genes Ywhab, Ywhae, Ywhaq, and Ywhaz (Figure 3). Analysis of the SB insertion patterns in these genes indicates that SB mutagenesis is creating loss-of-function mutations in these genes (Supporting Information, Fig. 3). For the genes that negatively regulate Yap1 signaling, including Dlg1, Csnk1d, Cul1 and the 14-3-3 genes, this pattern is consistent with the hypothesis that SB insertions are resulting in activation of Yap1.45 However, the mutation pattern in Yap1 does not appear to be consistent with Yap1 activation because it predicts a loss-of-function outcome in these tumors. One possible explanation for this observation is that the transposon insertions in the second intron, which account for > 50% of the insertions mapped to Yap1, are creating a truncated protein product that is acting in a dominant active fashion. Further analysis of individual Yap1 variants will be required to test this possibility. An additional confounding factor is the presence of multiple Yap1 insertions within a tumor. It is possible that one of the insertions could be activating, while the others are the result of local hopping events in minor subclones of the tumor. Our PCR amplification based methods do not allow us to be able to accurately assess the allele frequencies and, thus, this possibility.
In summary, through the use of functional genomics, we have identified a number of genes that cooperate with mutant Tgfbr2 to drive intestinal neoplasm progression and that disruption of the TGF-β signaling pathway is strongly selected for in intestinal cancer formation. The significance of the genes identified in the SB-TT model can be assessed when considered in the context of our understanding of the molecular genetics of CRC formation as well as in the context of the investigation for novel treatment approaches for CRC. The analysis of these candidate genes carries the promise to reveal further insights into the relevant molecular and cell biology of human CRC.
Supplementary Material
Acknowledgments
The authors wish to acknowledge support from Jeff Delrow and Andy Marty in the Genomics Shared Resource, the staff of the Comparative Medicine Shared Resource, and the Experimental Histopathology Core at the Fred Hutchinson Cancer Research Center. We would also like to thank Neal Copeland, Nancy Jenkins, and Martin McIntosh for their support.
It was funded by NIH grants R01CA194663, P30CA15704, U01CA152756, U54CA143862, 5R00CA151672, P30-CA77598, T32 CA080416, and 5P50CA150964
Abbreviations
- ACA
adenocarcinoma
- Ad
adenoma
- APC
adenomatous polyposis coli
- CGC
Wellcome Trust Sanger Instituted Cancer Gene Census
- CIMP
CpG Island Methylator Phenotype
- CIN
chromosomal instability
- CIS
common insertion site
- COSMIC
Catalog of Somatic Mutations in Cancer
- CRC
colorectal cancer
- dSI
distal small intestine
- gCIS
Gene-centric CIS
- Hyp
hyperplasia
- IPA
Ingenuity Pathway Analysis
- MSI
microsatellite instability
- pSI
proximal small intestine
- SB
Sleeping Beauty
- TCGA
The Cancer Genome Atlas
- TGF-β
transforming growth factor-beta
- Tgfbr1
transforming growth factor-beta receptor, type I
- Tgfbr2
transforming growth factor-beta receptor, type II
Footnotes
Article Category
Cancer Genetics and Epigenetics
Novelty and Impact
Many of the genes and pathways that functionally cooperate with deregulated TGF-β signaling to drive intestinal cancer are unknown. Using the Sleeping Beauty mouse model we discovered functionally relevant candidate pathways and driver genes that mediate intestinal cancer formation in vivo in the setting of inactivation of the TGF-β receptor. Most of these are novel genes and the majority were also found to be mutated in human colorectal cancers, demonstrating the relevance of our findings to human primary cancer. The identification of these factors advances our understanding of the molecular pathogenesis of colorectal cancer and provides insight into the possible treatment options for colorectal cancer patients.
Disclosure
There are no conflicts of interest to disclose for any of the authors.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Jones S, Chen WD, Parmigiani G, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogaert J, Prenen H. Molecular genetics of colorectal cancer. Ann Gastroenterol. 2014;27:9–14. [PMC free article] [PubMed] [Google Scholar]
- 7.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grady WM, Markowitz SD. TGF-B signaling pathway in tumor suppression. In: Derynck R, Miyazano K, editors. The TGF-B family. ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2008. pp. 889–938. [Google Scholar]
- 9.Horbelt D, Denkis A, Knaus P. A portrait of Transforming Growth Factor β superfamily signalling: Background matters. Int J Biochem Cell Biol. 2012;44:469–474. doi: 10.1016/j.biocel.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 10.Grady WM, Rajput A, Myeroff L, et al. Mutation of the type II transforming growth factor-beta receptor is coincident with the transformation of human colon adenomas to malignant carcinomas. Cancer Res. 1998;58:3101–3104. [PubMed] [Google Scholar]
- 11.Muñoz NM, Upton M, Rojas A, et al. Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res. 2006;66:9837–9844. doi: 10.1158/0008-5472.CAN-06-0890. [DOI] [PubMed] [Google Scholar]
- 12.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 13.Dupuy AJ, Akagi K, Largaespada DA, et al. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 14.Chytil A, Magnuson MA, Wright CV, et al. Conditional inactivation of the TGF-beta type II receptor using Cre:Lox. Genesis. 2002;32:73–75. doi: 10.1002/gene.10046. [DOI] [PubMed] [Google Scholar]
- 15.Boivin GP, Washington K, Yang K, et al. Pathology of mouse models of intestinal cancer: consensus report and recommendations. Gastroenterology. 2003;124:762–777. doi: 10.1053/gast.2003.50094. [DOI] [PubMed] [Google Scholar]
- 16.Largaespada DA, Collier LS. Transposon-mediated mutagenesis in somatic cells: identification of transposon-genomic DNA junctions. Methods Mol Biol. 2008;435:95–108. doi: 10.1007/978-1-59745-232-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brett BT, Berquam-Vrieze KE, Nannapaneni K, et al. Novel molecular and computational methods improve the accuracy of insertion site analysis in Sleeping Beauty-induced tumors. PLoS One. 2011;6:e24668. doi: 10.1371/journal.pone.0024668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarver AL, Erdman J, Starr T, et al. TAPDANCE: an automated tool to identify and annotate transposon insertion CISs and associations between CISs from next generation sequence data. BMC Bioinformatics. 2012;13:154. doi: 10.1186/1471-2105-13-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biswas S, Chytil A, Washington K, et al. Transforming growth factor beta receptor type II inactivation promotes the establishment and progression of colon cancer. Cancer Res. 2004;64:4687–4692. doi: 10.1158/0008-5472.CAN-03-3255. [DOI] [PubMed] [Google Scholar]
- 21.Trobridge P, Knoblaugh S, Washington MK, et al. TGF-beta receptor inactivation and mutant Kras induce intestinal neoplasms in mice via a beta-catenin-independent pathway. Gastroenterology. 2009;136:1680–1688. doi: 10.1053/j.gastro.2009.01.066. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forbes SA, Bhamra G, Bamford S, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Chapter 10:Unit 10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–1783. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeda H, Wei Z, Koso H, et al. Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nat Genet. 2015;47:142–150. doi: 10.1038/ng.3175. [DOI] [PubMed] [Google Scholar]
- 26.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu AD, Zhang S, Wang Y, et al. A critical role for transcription factor Smad4 in T cell function that is independent of transforming growth factor beta receptor signaling. Immunity. 2015;42:68–79. doi: 10.1016/j.immuni.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaimal V, Bardes EE, Tabar SC, et al. ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res. 2010;38:W96–W102. doi: 10.1093/nar/gkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu M, Trobridge P, Wang Y, et al. Inactivation of TGF-β signaling and loss of PTEN cooperate to induce colon cancer in vivo. Oncogene. 2014;33:1538–1547. doi: 10.1038/onc.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 31.Starr TK, Allaei R, Silverstein KA, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.March HN, Rust AG, Wright NA, et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat Genet. 2011;43:1202–1209. doi: 10.1038/ng.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sjöblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 34.Collier LS, Carlson CM, Ravimohan S, et al. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 35.Dupuy AJ, Jenkins NA, Copeland NG. Sleeping beauty: a novel cancer gene discovery tool. Hum Mol Genet. 2006;15:R75–R79. doi: 10.1093/hmg/ddl061. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 36.Chittenden TW, Howe EA, Culhane AC, et al. Functional classification analysis of somatically mutated genes in human breast and colorectal cancers. Genomics. 2008;91:508–511. doi: 10.1016/j.ygeno.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massagué J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angus-Hill ML, Elbert KM, Hidalgo J, et al. T-cell factor 4 functions as a tumor suppressor whose disruption modulates colon cell proliferation and tumorigenesis. Proc Natl Acad Sci U S A. 2011;108:4914–4919. doi: 10.1073/pnas.1102300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inuzuka H, Tseng A, Gao D, et al. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell. 2010;18:147–159. doi: 10.1016/j.ccr.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knippschild U, Kruger M, Richter J, et al. The CK1 Family: Contribution to Cellular Stress Response and Its Role in Carcinogenesis. Front Oncol. 2014;4:96. doi: 10.3389/fonc.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seshagiri S, Stawiski EW, Durinck S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee KW, Lee SS, Kim SB, et al. Significant association of oncogene YAP1 with poor prognosis and cetuximab resistance in colorectal cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:357–364. doi: 10.1158/1078-0432.CCR-14-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gregorieff A, Liu Y, Inanlou MR, et al. Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature. 2015;526:715–718. doi: 10.1038/nature15382. [DOI] [PubMed] [Google Scholar]
- 44.Yu FX, Meng Z, Plouffe SW, et al. Hippo pathway regulation of gastrointestinal tissues. Annu Rev Physiol. 2015;77:201–227. doi: 10.1146/annurev-physiol-021014-071733. [DOI] [PubMed] [Google Scholar]
- 45.Zhao B, Li L, Tumaneng K, et al. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.