Abstract
The intestinal epithelial cells function to gain nutrients, retain water and electrolytes, and form an efficient barrier against foreign microbes and antigens. Researchers employed cell culture lines derived from human or animal cancer cells as experimental models in vitro for understanding of intestinal infections. However, most in vitro models used to investigate interactions between bacteria and intestinal epithelial cells fail to recreate the differentiated tissue components and structure observed in the normal intestine. The in vitro analysis of host-bacterial interactions in the intestine has been hampered by a lack of suitable intestinal epithelium culture systems. Here, we established a new experimental model using an organoid culture system to study bacterial infection.
Keywords: Host-bacterial interactions, inflammation, intestine, infection, organoid, Salmonella, stem cells, tight junctions
1. Introduction
The gastrointestinal (GI) tract is lined by a single layer of epithelial cells that serve to facilitate digestion and absorption of nutrients.[1, 2] Intestinal epithelial cells (IECs) are consistently exposed to pathogenic microorganisms and foreign antigens, which play a key role in normal intestinal development and innate immunity.[3–5] Researchers have employed several in vitro models used to investigate interactions between bacteria and intestinal epithelial cells, including culture lines derived from human or animal cancer cells, suspension culture technology using a rotating wall vessel bioreactor that allows cells to remain in suspension with bubble-free aeration.[6, 7] Studies by Clevers and colleagues established the isolation and culture of primary small intestinal epithelial stem cells.[8–11] Isolated crypts form "organoid structures" contain multiple cell types, including enterocytes, goblet cells, enteroendocrine cells, Paneth cells. Furthermore, some functional and physiological properties of the organoids have been demonstrated by the presence of brush borders on enterocytes, production of mucin by goblet cells. This culture system is particularly useful for studying the regulation of intestinal stem cell self-renewal and differentiation, and of host-pathogen interactions. We sought to establish a bacteria-infected organoid culture system using crypt-derived intestinal organoids.[12] In the infected organoids, we were able to visualize the invasioness of Salmonella and the morphologic changes of the organoids.
2. Materials
2.1 Reagents
-
●
Sterile PBS (Ca2+/Mg2+ free)
-
●
Buffer #1: 2 mM EDTA in PBS
-
●
Buffer #2: 54.9 mM D-sorbitol and 43.4 mM sucrose in PBS
-
●
Growth factor reduced, Phenol red free Matrigel
- Minigut Medium:
-
■Advanced DMEM/F12
-
■Penicillin/Streptomycin (100U/ml)
-
■10 mM Hepes
-
■1:100 N2 supplement
-
■1:50 B27 supplement
-
■
-
●
Mouse Recombinant EGF (50µg/ml) (Note 1)
-
●
Mouse Recombinant Noggin (100µg/ml) (Note 1)
-
●
Rho Kinase Inhibitor Y27632 (10mM)
-
●
R-spondin solution (Note 2)
3. Methods
3.1 Organoids isolation
Murine small intestine (mostly jejunum and ileum) was removed immediately after cervical dislocation, using scissors to remove fat/mesentery. Dissect out 10cm of jejunum and 10 cm of ileum.
The stool was flushed out with ice cold PBS (Penicillin, 100 I.U./ml/Streptomycin, 100µg/ml) in small intestine. Cut intestines longitudinally with special intestine scissors (Made in Germany FST 14080-11) and the small intestines were cut into small (~1 cm) pieces.
Use the forceps to transfer all the pieces to a 15 ml conical tube containing 5 ml of ice cold PBS, rock for 10 minutes in 4°C.
Aspirate PBS from the top of the tissue and replace with cold Buffer #1, rock for 30 minutes in 4°C.
-
Aspirate PBS from the top of the tissue and replace with cold Buffer #2, and then shake for 2 minutes mannually (~80 shakes/minute).
Note: Jejunum will dissociate easier than ileum, which may require extra shaking.
Take 20 µl droplet of Buffer #2 after shaking and check under the microscope. Villi and debris are large chunks. Many crypts with granular paneth cells are at the bottom. Do another 1 minute of shaking in the same buffer if many crypts not observed.
Equilibrate 70 µm sterile cell strainer with Buffer #2. Rinse all filters with 5ml of Buffer #2 to ensure that enough crypts have passed through the filters. Only crypts and small fragments should pass through this filter (ie. large chunks of tissue will be removed in this step.)
Count number of crypts: Mix 10 µl of crypts with 10 µl of trypan blue. Apply all 20 µl to a microscope slide and count all the crypts in the entire drop using a quality inverted scope.
Transfer enough medium/crypts into one tube so that there are 1,500 crypts in a round bottom 5ml popypropylene tube, centrifuge tubes for 2 min at 150 g at 4°C.
-
While centrifuging mixture, prepare Matrigel™ on ice (MatrigelTM will polymerize at room temperature).
50µl Matrigel/well:-
●0.25µl of 100µg/mL EGF (final concentration 50ng/ml)
-
●1µl of 50µg/ml Noggin (final concentration 100 ng/ml)
-
●1µl of Y27632 (ROCK inhibitor, final concentration 10µM)
-
●5µl of R-spondin
-
●
After the spin, remove supernatant with a pasteur pipet attached to a vacuum, being careful not to touch the pellet. Leave ~50 µl of medium in the tube.
-
Resuspend crypt pellet in the 50 µl that remained in the tube. Add an additional 100 µl of Matrigel and mix well with the pipette, pipette up and down being careful to avoid bubbles.
Note: Remember to place the tube with the matrigel back on ice immediately after using it.
-
Draw up the entire 50 µl volume with Matrigel/crypt into a pipette, being careful to avoid any bubbles. Put 50 µl droplet of mix in center of a well of a 12 well plate. Incubate 24-well plate at 37°C for 30 minutes to polymerize Matrigel. After 30 minutes of polymerization, 500 ml of minigut medium was overlain [10].
Note: Not supplemented with growth factors at this point.
Change medium every 3–4 days with minigut medium containing growth factors (Noggin: 100 ng/ml; EGF: 50ng/ml; Y27632:10 µM; R-spondin: 1/10).
3.2 Observe organoids under microscope
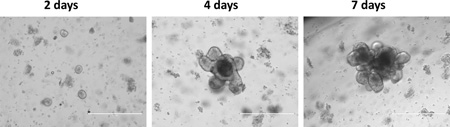
At 2 days post-prep, the organoid begins to take shape. At 3–4 days post-prep, the organoid begins to bud. At 5–7 days post-prep, the organoid has many buds with lots of luminal debris, which is a point for passage.
3.3 Passage of organoids
The passage was performed every 7–10 days with a 1:4 split ratio.
Leave 12-wells cell culture plate on ice for 30 minutes. Aspirate medium and add 1ml of cold PBS to each well.
Mix twice through a 1ml syringe with a 27G 1/2 needle and transfer into a 5 ml tube. Spin down for 5 minutes at 200 g at 4°C.
Remove supernatant, and leave ~50 µl of medium in the tube for resuspension.
Add an additional 150 µl of Matrigel and mix well with the pipette avoiding bubbles.
Add 50 µl droplet of mix to the center of a well of a 12 well plate. Incubate cell plate at 37°C for 30 minutes. After 30 minutes of polymerization, 500 µl of minigut medium containing growth factors was overlain.
Change medium every 3–4 days with minigut medium containing growth factors.
3.4 Bacterial colonization of organoids
-
Organoids cells colonized with bacteria: Organoid cells (6 days after passage) were colonized with the indicated bacterial strain for 30 minutes, washed with HBSS, and incubate in minigut medium containing gentamicin (500 mg/ml) for 1 hours.
Note: After extensive HBSS washing, the extracellular bacteria were washed away. Incubation with gentamicin inhibited the growth of extracellular bacteria [13].
Organoid cells Immunoblotting: The organoid cells were rinsed three times in ice-cold HBSS and then suspended in ice cold HBSS. The organoid cells were then spun down at 200 g for 10 minutes at 4°C. Next, using a pipette to aspirate the PBS at the top, the organoid cells were lysed in lysis buffer (1% Triton X-100, 150 mM NaCl, 10 mM Tris pH 7.4, 1 mM EDTA, 1 mM EGTA pH 8.0, 0.2 mM sodium orthovanadate, protease inhibitor cocktail) and then sonicated. The protein concentration was then measured. Nexzt, equal amounts of protein (20 µg/well) were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and immunoblotted with primary antibodies. Following the primary antibody step, the nitrocellulose membranes were incubated with secondary antibodies and visualized by ECL.
Organoid cells embedded in a paraffin block: The organoid cells were rinsed three times in ice-cold HBSS and then suspended in cold HBSS. The organoid cells were spun down at 200 g for 10 minutes at 4°C. To fix organoid cells, the following steps are used: 10% formalin 30 minutes; 75% alcohol 5 minutes; 100% alcohol 10 minutes; xylene 5 minutes; xylene 10 minutes; and paraffin (65°C) 60 minutes. The paraffin sections were processed with standard techniques.[14, 15]
Immunofluorescent staining: The immunofluorescence measurements were performed on paraffin-embedded sections (4 µm) of organoid cells. After preparation of the slides, as described previously[16], the slides were permeabilized for 20 minutes with 0.2% Triton X-100, followed by three rinses with HBSS, and incubation for 1 hour in 3% BSA + 1% goat serum in HBSS to reduce nonspecific background. The permeabilized organoid cell samples were incubated with primary antibodies overnight in 37°C. The samples were then incubated with goat anti-rabbit Alexa Fluor 488 or goat anti-mouse Alexa Fluor 488 (Molecular Probes, CA; 1:200) and DAPI (Molecular Probes 1:10,000) for 1 hour at room temperature. The organoid cells were mounted with SlowFade (SlowFade® AntiFade Kit, Molecular Probes) followed by a coverslip, and the edges were sealed to prevent drying. The specimens were examined with a Zeiss 710 Laser Scanning confocal microscope.
Acknowledgments
We would like to acknowledge the NIDDK grant R01 DK105118 (JS)
Footnotes
To make stock solution of mouse recombinant EGF, mouse recombinant noggin, reconsitute powder in sterile PBS then followed by sterilization through 0.22 µm filter. Aliquot in 100 µl volumes and store at −20°C.
To make solution of R-spondin Day1: check the confluence of HEK293-RSPO2 (~90%), aspirate normal medium, wash the cells gently with several mls of OPTI-MEM, then replace them with ~30ml of OPTI for every T150 flask. Day4: collect the first wave conditional medium, add fresh OPTI again. Day7: collect the second wave conditional medium. Mix the two waves medium together and aliquot them. Add 1ml of cond.-medium to 9ml of Minigut medium which is good for organoid growth.
References
- 1.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9(11):799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 2.Robinson A, Keely S, Karhausen J, Gerich ME, Furuta GT, Colgan SP. Mucosal protection by hypoxia-inducible factor prolyl hydroxylase inhibition. Gastroenterology. 2008;134(1):145–155. doi: 10.1053/j.gastro.2007.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooper LV. Do symbiotic bacteria subvert host immunity? Nat Rev Microbiol. 2009;7(5):367–374. doi: 10.1038/nrmicro2114. [DOI] [PubMed] [Google Scholar]
- 4.Camp JG, Kanther M, Semova I, Rawls JF. Patterns and scales in gastrointestinal microbial ecology. Gastroenterology. 2009;136(6):1989–2002. doi: 10.1053/j.gastro.2009.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136(1):65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nickerson CA, Ott CM. A New Dimension in Modeling Infectious Disease. ASM News. 2004;70(4):169–175. [Google Scholar]
- 7.Unsworth BR, Lelkes PI. Growing tissues in microgravity. Nat Med. 1998;4(8):901–907. doi: 10.1038/nm0898-901. [DOI] [PubMed] [Google Scholar]
- 8.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 9.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469(7330):415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang N, Zhang H, Zhang BQ, Liu W, Zhang Z, Qiao M, et al. Adenovirus-mediated efficient gene transfer into cultured three-dimensional organoids. PLoS One. 2014;9(4):e93608. doi: 10.1371/journal.pone.0093608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato T, Clevers H. Primary mouse small intestinal epithelial cell cultures. Methods in molecular biology. 2013;945:319–328. doi: 10.1007/978-1-62703-125-7_19. [DOI] [PubMed] [Google Scholar]
- 12.Zhang YG, Wu S, Xia Y, Sun J. Salmonella-infected crypt-derived intestinal organoid culture system for host-bacterial interactions. Physiol Rep. 2014;2(9) doi: 10.14814/phy2.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun J, Hobert ME, Rao AS, Neish AS, Madara JL. Bacterial activation of beta-catenin signaling in human epithelia. Am J Physiol Gastrointest Liver Physiol. 2004;287(1):G220–G227. doi: 10.1152/ajpgi.00498.2003. [DOI] [PubMed] [Google Scholar]
- 14.Wu S, Ye Z, Liu X, Zhao Y, Xia Y, Steiner A, et al. Salmonella typhimurium infection increases p53 acetylation in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2010;298(5):G784–G794. doi: 10.1152/ajpgi.00526.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye Z, Petrof EO, Boone D, Claud EC, Sun J. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am J Pathol. 2007;171(3):882–892. doi: 10.2353/ajpath.2007.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S, Liao AP, Xia Y, Li YC, Li JD, Sartor RB, et al. Vitamin D receptor negatively regulates bacterial-stimulated NF-kappaB activity in intestine. Am J Pathol. 2010;177(2):686–697. doi: 10.2353/ajpath.2010.090998. [DOI] [PMC free article] [PubMed] [Google Scholar]