Abstract
Comparative analyses of the human microbiome have identified both taxonomic and functional shifts that are associated with numerous diseases. To date, however, microbiome taxonomy and function have mostly been studied independently and the taxonomic drivers of functional imbalances have not been systematically identified. Here, we present FishTaco, an analytical and computational framework that integrates taxonomic and functional comparative analyses to accurately quantify taxon-level contributions to disease-associated functional shifts. Applying FishTaco to several large-scale metagenomic cohorts, we show that shifts in the microbiome’s functional capacity can be traced back to specific taxa. Furthermore, the set of taxa driving functional shifts and their contribution levels vary markedly between functions. We additionally find that similar functional imbalances in different diseases are driven by both disease-specific and shared taxa. Such integrated analysis of microbiome ecological and functional dynamics can inform future microbiome-based therapy, pinpointing putative intervention targets for manipulating the microbiome’s functional capacity.
Keywords: Human microbiome, Functional metagenomics, Taxonomic profiling, Compositional shifts, Comparative analysis, Microbial drivers, Multi-omics, Integrative Analysis
eTOC blurb
Comparative human microbiome analyses have identified both taxonomic and functional shifts associated with diseases. Manor and Borenstein present an integrative multi-omic framework that identifies the microbial drivers of functional shifts. Its application reveals that shifts in the microbiome’s functional capacity can be traced back to function-, site-, and disease-specific taxa.
Graphical Abstract
Introduction
The human microbiome – the collection of microorganisms that inhabit the human body – is tightly linked to our health and impacts several crucial host processes (Kinross et al., 2011). Recently, a plethora of comparative studies have identified intriguing associations between the composition of the microbiome and numerous diseases including various metabolic disorders (Greenblum et al., 2012; Karlsson et al., 2014; Qin et al., 2013), malignancies (Schulz et al., 2014), autoimmune diseases (Scher et al., 2013), and neurological developmental disorders (Hsiao et al., 2013). Such studies can take two different approaches to profile the composition of the microbiome (Noecker et al., 2016). The first approach focuses on taxonomy, aiming to profile the abundances of different microbial clades in each sample, either by targeted sequencing of the ribosomal 16S gene and operational taxonomic units clustering (Caporaso et al., 2010; Schloss et al., 2009) or by shotgun metagenomic sequencing and quantification of clade-specific marker genes’ abundances (Segata et al., 2012). The second approach aims to characterize the functional capacity of the community, quantifying the abundances of genes or pathways through shotgun metagenomic sequencing (Abubucker et al., 2012; Carr and Borenstein, 2014). The obtained taxonomic or functional profiles can then be compared across samples to identify shifts in the abundance of specific taxa or functions associated with the host state.
To date, however, disease-associated taxonomic and functional shifts (i.e., significant differences in abundance observed between case and control samples) are often studied independently, and efforts to link these two facets of the microbiome have been mostly anecdotal and often reported only qualitative associations. For example, in a study of the skin microbiome, researchers concluded that changes in the abundance of the NADH dehydrogenase module are driven by shifts in the prevalence of P. acnes, based on a strong correlation observed between the two (Oh et al., 2014). Similarly, Turnbaugh et al. compared microbiome sequences to a custom database of 44 gut microbes’ genomes and concluded that obesity-related functional enrichments in carbohydrate metabolism were a product of elevated abundances of Actinobacteria and Firmicutes (Turnbaugh et al., 2009). Studying the dynamics of the gut microbiome in the first year of life, Bäckhed et al. reported an association between functional enrichment of genes involved in degradation of complex sugars and starch and the abundance of B. thetaiotaomicron (Bäckhed et al., 2015).
The studies above emphasize the importance of identifying taxonomic drivers of disease-associated functional imbalances and highlight the potential for integrating taxonomic and functional comparative analyses, yet do not offer a systematic, rigorous, and comprehensive methodology toward such integration. This gap in our ability to systematically define and quantify taxonomic contributions to functional shifts hinders our understanding of disease-associated functional dynamics and leaves multiple fundamental questions unanswered. It is not clear, for example, whether functional shifts tend to be driven by a change in the abundances of a small set of taxa or by community-wide dysbiosis, and whether the same set of taxa generally drive shifts in multiple functions. It is also not clear how taxon prevalence, inter-sample variation, and co-variation with other taxa come together to induce a functional shift. Most importantly, without linking taxonomic and functional shifts, our ability to ultimately pinpoint species that could be targeted in order to restore desired microbiome-level functional capacity remains limited.
To address this challenge, here we introduce FishTaco, an analytical and computational framework for comprehensively quantifying taxon-level contribution to observed functional shifts and identifying key taxa that drive such shifts. We first apply our framework to a large set of samples from different body sites of healthy individuals, and reveal marked variation in the taxonomic drivers of different functional shifts. Next, we apply our framework to type 2 diabetes (T2D) and inflammatory bowel disease (IBD) cohorts, and show that seemingly similar disease-associated functional shifts are in fact driven by a function-specific and disease-specific set of microbial taxa. Combined, our findings suggest a route towards systematic integration of taxonomic and functional comparative analyses and informed design of precise microbiome-based interventions.
Results
Challenges and opportunities in linking microbiome taxonomic and functional data
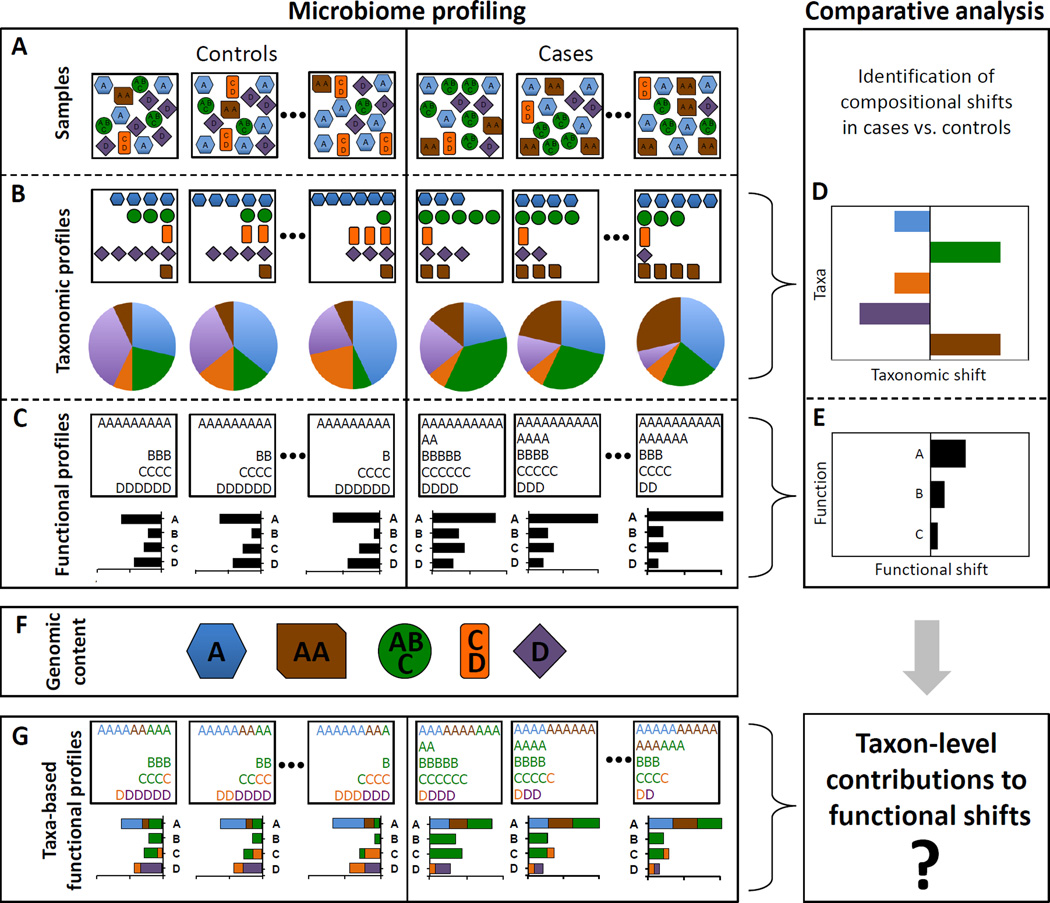
Consider a standard metagenomic comparative analysis, where samples from different habitats or from a disease cohort (Figure 1A) are assayed to characterize the taxonomic (Figure 1B) and functional (Figure 1C) abundance profiles in each sample. Taxonomic and functional profiles can then be compared across samples using some statistical test (e.g., Wilcoxon rank-sum test or fold ratio) to identify compositional shifts and to discover disease-associated taxa (Figure 1D) or functions (Figure 1E).
Figure 1. An illustrative example of taxonomic and functional comparative metagenomic analyses.
(A) Community composition of case (right) and control (left) samples. (B) Profiling of the samples’ taxonomic composition, showing the species counts (upper boxes) and relative abundances (pie charts). (C) Profiling of the samples’ functional composition, showing the function counts (upper boxes) and abundances (bar charts). (D–E) Comparative analysis aims to explore compositional shifts in the microbiome, identifying case-associated taxa and/or case-enriched functions. (F) The genomic content of the various community members (denoting the copy number of each gene in their genomes) links the community’s taxonomic and functional profiles. (G) Taxa-based functional profiles provide a mapping from taxonomic to functional compositions, detailing the fraction of each gene’s abundance that originated from each species.
Clearly, however, the obtained taxonomic and functional profiles are tightly coupled since the abundance of each gene family (or any other genomic element) in a mixed aggregate of genomes (i.e., the metagenome) is a simple derivative of the prevalence of this gene in each genome and the relative abundance of each genome in the mixture. More formally, given the genomic content of each taxon in the community – denoting the copy number of each gene (or gene-family) in the genome of that taxon (Figure 1F) – the functional composition of the metagenome can be represented as a linear combination, aggregating the genomic content of all taxa weighted by the taxa relative abundances in the community. Such taxa-based functional profiles have been shown to accurately predict the abundance of the various functions in the metagenome, offering a promising route for conducting functional analysis when the gene composition of the metagenome was not assayed directly (Langille et al., 2013). This view of the metagenomic functional content as an aggregate of the member species’ genomic contents provides a mapping from taxonomic abundances to functional abundances, detailing the fraction of each gene’s abundance that originated from each species (Figure 1G).
Importantly, however, even when this mapping is known (e.g., as in the case of taxa-based functional profiles) and the contribution of each taxon to the measured abundance of a given function in a sample can be estimated, the contribution of each taxon to an observed shift in the abundance of this function between two sets of samples cannot be easily quantified. Specifically, a taxon’s contribution to the abundance of some function may not be very indicative of this taxon’s contribution to an observed functional shift since, for example, a taxon could be highly but similarly abundant across cases and controls, contributing significantly to the abundance of a function but not to its disease-associated shift. It is also not clear how to take into account co-variation between taxa that may, for example, allow one taxon to compensate for the functional shifts induced by another taxon. More generally, while the total abundance of a certain gene can be viewed as a linear sum of all taxon-level contributions, an analytical approach that rigorously defines how to partition among the various taxa a measure of the gene’s differential abundance in the context of comparative analysis is lacking.
These challenges call for the development of a framework for defining and calculating taxonomic contributions to functional shifts. Ideally, such a framework should meet several key requirements: First, since comparative studies employ a wide variety of statistical metrics to measure shifts (e.g., a Wilcoxon rank-sum test, fold ratio, or a Student’s t-test), to be widely applicable such a framework should support the use of any shift metric. Moreover, to make the calculated contribution scores intuitive and meaningful, taxon-level contributions should be measured using the same units and the same scale as those used to measure functional shifts. Additionally, to allow contribution scores to be interpreted as a decomposition of the observed shift into its taxon-level components, the sum of calculated contribution scores across all taxa for a given function should equal the shift observed in this specific function. Finally, since, as described above, the contribution of each taxon to observed functional shifts depends on both its abundance relative to other taxa and on its co-variation with other taxa, such a framework should account for community-wide context (rather than considering each taxon in isolation) and for taxa co-variation patterns. The framework described below aims to address these challenges and requirements.
An integrative computational framework for identifying taxonomic drivers of functional shifts in the human microbiome
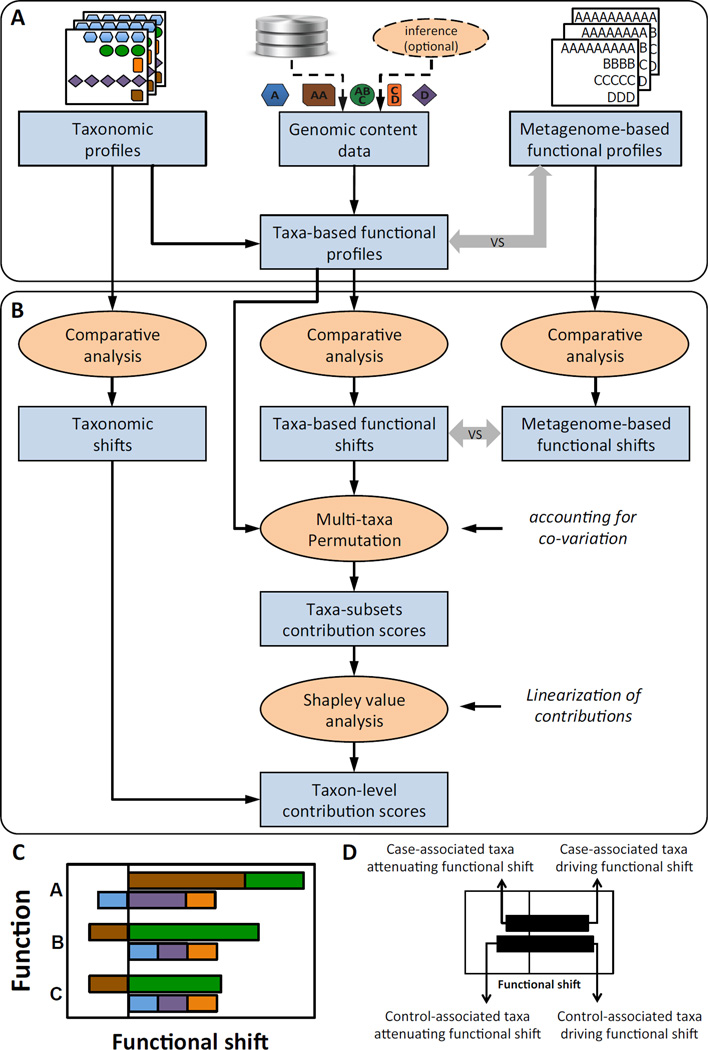
Here we introduce a computational framework termed Functional Shifts’ Taxonomic Contributors (FishTaco, Figure 2) for linking taxonomic and functional comparative analyses. Given two sets of microbiome samples (e.g., cases and controls), FishTaco takes as input both the taxonomic abundance profile and the functional abundance profile of each sample. Taxonomic profiles can be based on either clade-specific markers from metagenomic sequencing (e.g., using MetaPhlAn; Segata et al., 2012) or targeted 16S sequencing followed by OTU picking. Functional profiles can be based on metagenomic shotgun sequencing and annotation (e.g., using HUMAnN; Abubucker et al., 2012), or other similar pipelines, (e.g., Carr and Borenstein, 2014). When available, the input can further include corresponding genomic content data for each taxon. Such genomic content data can be obtained from reference genomes that match identified marker genes (when using marker-based taxonomic profiles) or from closely related reference genomes and phylogenetic-inferred genomes (when using 16S-based taxonomic profiles, e.g., via PICRUSt; Langille et al., 2013). Our framework then integrates these inputs to rigorously define and quantify the contribution of each taxon to the observed shift in the abundance of each function between these two sets.
Figure 2. The FishTaco framework.
(A) FishTaco first processes the input data (including taxonomic profiles, metagenome-based functional profiles, and optionally the genomic content of each taxon) to generate taxa-based functional profiles. If genomic content is not provided, a previously introduced inference method can be used. Taxa-based and metagenome-based functional profiles are compared to assess how well they recapitulate the abundances of the various functions in the metagenome. (B) FishTaco then quantifies the contribution of each taxon to observed shifts in each function. Taxa-based functional shifts between cases and controls are compared to metagenome-based shifts to confirm that the taxa-based profiles exhibit similar shifts to those observed in the metagenome. A multi-taxa permutation analysis is then used (accounting for taxa co-variation) to assess the contribution of a large ensemble of taxa subsets to observed shifts, followed by a Shapley value analysis to obtain taxon-level linearized contribution scores (see Methods). (C–D) An illustration of a FishTaco-based taxon-level contribution profile, decomposing functional shifts into taxon-level contribution scores, and of the 4 different contribution modes identified by FishTaco.
Briefly, our framework works as follows: First the taxonomic profiles and the genomic content of the member taxa are used to generate taxa-based functional profiles for all the samples (Figure 2A), detailing how much of the abundance of each function in each sample is accounted for by each taxon. When complete genomic content is not available for all resident taxa, our framework can further employ a previously described machine learning-based method (Carr et al., 2013) to analyze the taxonomic and functional profile of each sample and to infer the genomic content of the different taxa. The obtained taxa-based functional profile for each sample and the functional shifts between the two sets of samples observed in these profiles are compared to those observed in the metagenome to evaluate their accuracy (Figure 2B). Next, to quantify the contribution of taxa to observed functional shifts, our framework employs a permutation-based approach, estimating how these functional shifts would change if the abundances of taxa are shuffled across samples. To this end, our framework utilizes the fact that taxa-based functional profiles allow for translating perturbations in the abundance of species across samples into changes in the corresponding functional profiles, and compares the functional shifts observed in the original taxa-based functional profiles to the shifts observed when the relative abundances of a set of taxa are randomly permuted across samples (Figure 2B). Notably, this approach allows maintaining community context and quantifying contribution scores using the same metric used for measuring functional shifts. Finally, the Shapley value analysis (originally developed for estimating the contribution of individual players in a multi-player game) is applied to determine the individual contribution of each taxon within this multi-taxa setting (Figure S1). The complete details of the FishTaco framework could be found in the Methods and in the Supplemental Experimental Procedures.
Ultimately, this process results in a taxon-level shift contribution profile for each function, decomposing the overall enrichment score of this function into its taxon-level components. Notably, assuming a case vs. control setting, a taxon’s contribution to the enrichment of a specific function in cases can be either positive (i.e., the taxon is driving this enrichment) or negative (i.e., the taxon is attenuating this enrichment). Using these calculated contributions and the shifts observed in the taxonomic profile, FishTaco further classifies each taxon into one of four distinct groups representing different modes of contribution to a given functional shift (Figure 2C–D). The first group includes taxa that drive the enrichment of that function and are case-associated (i.e., have higher abundance in cases). These taxa encode the function in question in their genomes and increase in abundance in cases, hence contributing positively to the function’s enrichment. The second group includes taxa that drive the enrichment of the function but are control-associated taxa (i.e., higher abundance in controls). Such taxa likely lack the function in question in their genomes (or encode it with a relatively low copy number) and their increased relative abundance in control samples therefore decreases the average abundance of that function in controls, ultimately contributing positively to the enrichment of this function in cases. Similarly, taxa that attenuate the enrichment of the function in question in cases (even though that function is ultimately still identified as significantly enriched) can again be classified into two groups, one that includes case-associated taxa (that likely lack the function in question in their genomes), and another that includes control-associated taxa (that likely encode the function in their genomes). These four groups of taxa are then plotted separately (and in the appropriate direction) as stacked bar plots for each function to provide a comprehensive visualization of all taxon-level shift contributions (Figure 2C–D and see also Figure S2 for a numerical example).
To confirm that calculated taxon-level shift contribution profiles correctly reflect the impact that each species may have on observed functional shifts, we implemented a large-scale simulation study, using several perturbation assays to mimic the impact of taxonomic intervention schemes. We found that contribution scores calculated by our framework were highly correlated with these perturbation-based estimates (Figure S3). The complete details of this simulation analysis can be found in the Supplemental Text.
Differences in the functional capacity of the microbiome across body sites can be traced to rich and complex ecological changes
To examine whether differences in the overall functional capacity of distinct microbial communities tend to be driven by changes in the abundance of a small number of taxa or rather by broader and more complex changes in community ecology, we first applied our framework to decompose functional shifts observed between different body sites in healthy individuals. These site-specific communities represent adaptations to distinct niches, yet are similar enough to allow focusing on specific biological pathways that shift in abundance between these sites. We specifically compared samples from the tongue (tongue dorsum) with samples from the inner cheek (buccal mucosa) obtained through the Human Microbiome Project (HMP; Human Microbiome Project Consortium, 2013). These two anatomical sites are among the most highly sampled in this study (106 and 107 samples for tongue and cheek, respectively) and are in close spatial proximity. We first obtained the taxonomic profiles of these samples (at species-level resolution across 76 species, generated by MetaPhlAn; Segata et al., 2012; see also Figure S4) and identified shifts in taxonomic composition between tongue and cheek samples. Next, we used these taxonomic profiles and the genomic content of each taxon (based on reference genomes obtained from the Integrated Microbial Genomes database, IMG; Markowitz et al., 2013) to construct a taxa-based functional profile for each sample (as described above). We finally obtained metagenome-based functional profiles for these samples (as generated by HUMAnN; Abubucker et al., 2012).
We first evaluated the agreement between taxa-based and metagenome-based functional profiles and the corresponding functional shifts observed between tongue and cheek samples. Focusing on the 22 pathways that were significantly enriched in the tongue (Methods), we found a good overall agreement between the taxa-based and metagenome-based functional profiles (Figure 3A, median Pearson’s correlation across functions of R=0.91). Functional shift scores (using Wilcoxon test statistic, W) calculated from these taxa-based functional profiles similarly agreed with those calculated based on the metagenome (Figure 3B and Table S1, Spearman correlation of shift scores ρ=0.65, p<10−5). These findings confirm that our taxa-based functional profiles capture the underlying functional composition of the metagenomes and can be used for downstream analysis of each taxon’s contribution to observed shifts in functional capacity.
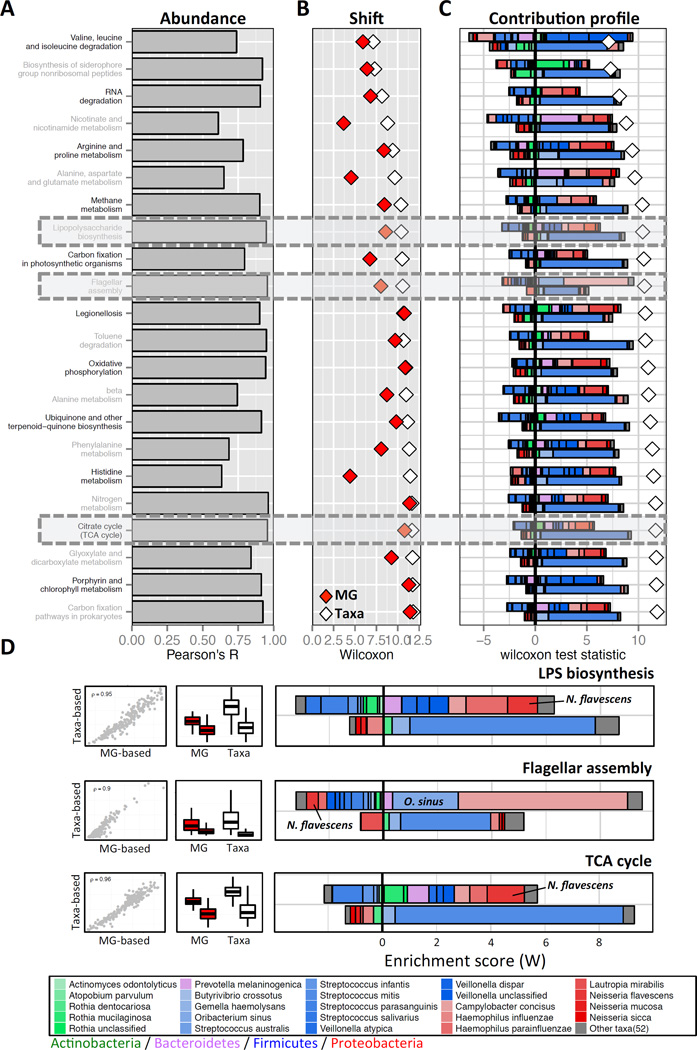
Figure 3. Identifying the taxonomic contributors to differences in the functional capacity of tongue versus cheek microbiome samples.
(A) Pearson correlation coefficients between the taxa- and metagenome-based functional profiles for each tongue-enriched pathway. (B) Agreement between metagenome-based (MG; red diamonds) and taxa-based (Taxa; white diamonds) functional shift scores (C) Taxon-level shift contribution profiles for tongue-enriched pathways (see Figure 2C–D for more details on the meaning of each bar). For each function, the sum of taxa contribution scores matches the observed taxa-based functional shift score. (D) Taxon-level shift contribution profiles of 3 pathways of interest (highlighted by grey boxes in panels A-C): LPS biosynthesis, flagellar assembly, and TCA cycle. The scatter plots on the left demonstrate the high agreement between metagenome- and taxa-based abundances, where each dot represents a sample, and the Spearman correlation across all samples is reported. The box plots in the middle column illustrate the distribution of metagenome-based (MG; red) and taxa-based (Taxa; white) abundance of this function in the tongue (left boxes) versus the cheek (right boxes).
We next applied FishTaco to calculate a taxon-level shift contribution profile for each tongue-enriched function, quantifying the contribution of each taxon to the observed shift in each function. Our analysis revealed diverse, and non-trivial patterns in these shift contribution profiles (Figure 3C and Table S2). Some species, for example, drove the observed shift in one function while attenuating the shift in another (compare the contribution of Neisseria flavescens to Lipopolysaccharide biosynthesis and Flagellar assembly in Figure 3D). Moreover, different species from the same phylum often had dramatically different impacts on the shift observed in a given function (see, for example, the contribution of Proteobacteria species to the shift in the pathways in Figure 3D).
To demonstrate in detail the complex ways in which ecological changes drive functional differences, we focused on three pathways previously reported as tongue-enriched (Human Microbiome Project Consortium, 2013; Goll et al., 2012): Flagellar assembly, Lipopolysaccharide (LPS) biosynthesis, and the TCA cycle. Notably, as also reported above, taxa-based functional profiles were highly similar to the metagenome-based functional profiles of these pathways (Pearson’s correlation R>0.95), and the corresponding functional shifts were consistent (Figure 3D). Examining the calculated taxon-level shift contribution profiles highlighted several intriguing patterns. First, even though these three pathways exhibited similar overall functional shift scores (Wilcoxon test statistic, W, ranging from ~8 to ~11), the shift contribution profiles were complex and markedly variable (Figure 3D and Table S2). For example, our analysis demonstrated that the shift in the flagellar assembly pathway was driven mostly by tongue-associated taxa, whereas the shift in the TCA cycle pathway was driven largely by cheek-associated taxa. Moreover, some taxa contributed mostly to the shift observed in a single pathway, with relatively little contribution to others. For example, the species Oribacterium sinus was found to contribute substantially to the enrichment of the flagellar assembly pathway, but not to the enrichment of the TCA cycle or LPS biosynthesis pathways, in line with the characteristics of O. sinus, a motile, strictly anaerobic, Gram-positive organism. We further confirmed this specificity in the contribution of O. sinus and of other species to observed functional shifts by analyzing subsets of samples in which species exhibited marked differences in abundance (Supplemental Text). Importantly, such taxon-level characterization and the detection of species that contribute to a restricted set of functional shifts can be used to pinpoint specific driver taxa and ultimately to identify taxonomic intervention targets for manipulating (or restoring) the functional capacity of the microbiome.
Shifts in various type 2 diabetes-associated functions are driven by distinct bacterial clades
Having shown that functional shifts between different anatomical sites of healthy individuals are attributed to a diverse and highly variable set of taxonomic drivers, we set out to examine whether a similar relationship exists between community dysbiosis and disease-associated functional shifts. We specifically applied our framework to a type 2 diabetes (T2D) cohort (Qin et al., 2013), which represents one of the largest studies to date of the association between the composition of the microbiome and a disease state. We used the genus-level taxonomic abundance profiles across 48 genera generated in the study (since species-level abundances were not reported) coupled with corresponding reference genomes for each genus (taken from IMG; Markowitz et al., 2013) to construct taxa-based functional profiles. In addition, we obtained metagenome-based functional profiles generated in the study for these samples. We again confirmed that constructed taxa-based functional profiles were in good agreement with metagenome-based functional profiles (median Pearson’s correlation R=0.76), as were the taxa- and metagenome-based functional shifts (R=0.37, P<0.005, Pearson’s correlation test; Figure S5 and Tables S3–S4).
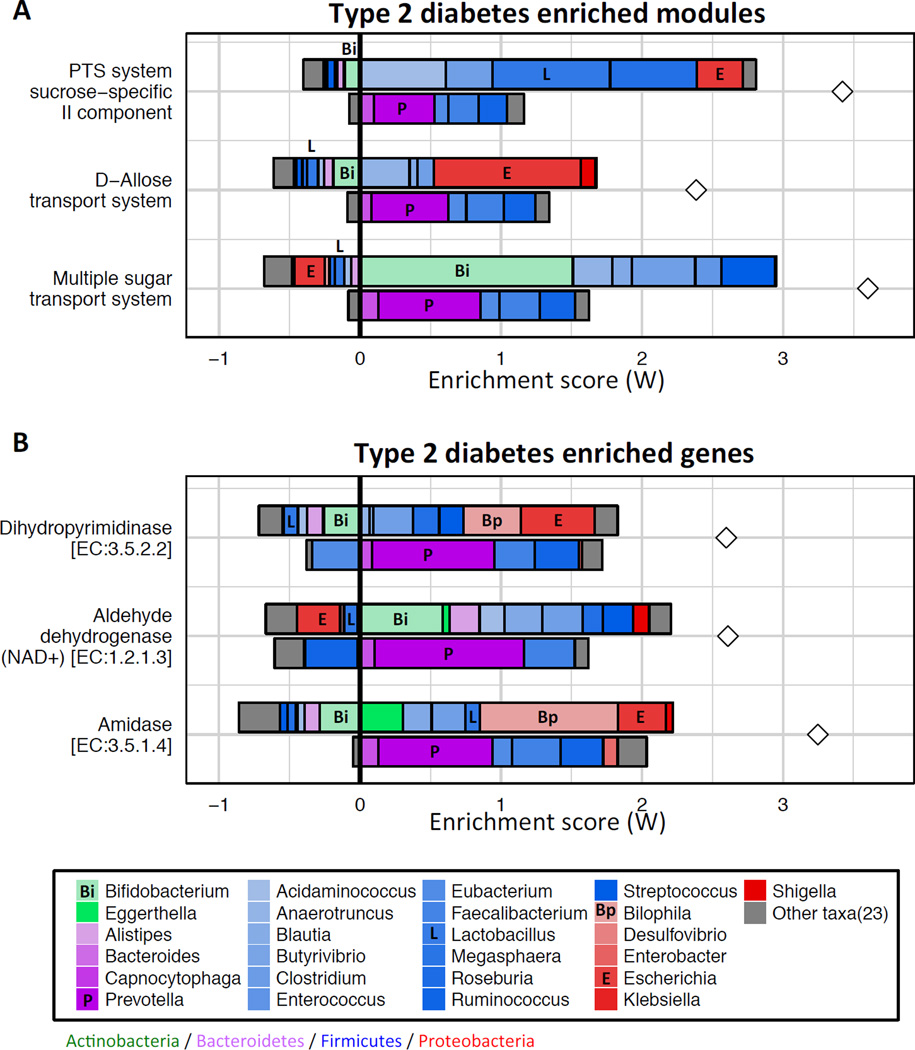
We first examined FishTaco’s calculated taxon-level contributions to observed shifts in various functional modules, focusing initially on three previously reported T2D-associated sugar transport modules (Karlsson et al., 2014; Qin et al., 2013): D-Allose transport system (M00217), Multiple sugar transport system (M00216), and the PTS system sucrose-specific II component (M00269). We again found marked variation in the identified taxonomic drivers of the enrichments observed in these three modules in T2D samples (Figure 4A and Table S5). For example, the T2D-associated genus Escherichia, as well as several other Proteobacteria genera, were major drivers of the enrichment in the D-Allose module, while the genus Bifidobacterium attenuated that enrichment. In contrast, Bifidobacterium was the main driver of the enrichment of the multiple sugar module in T2D samples, whereas all Proteobacteria species attenuated the enrichment of this module. In support of this functional shift decomposition, we found that metagenome-based abundance of the D-Allose module across samples was correlated with the abundances of Escherichia, as was the abundance of the multiple sugar module with Bifidobacterium (Spearman’s correlation R=0.48 and R=0.25, respectively). Notably, in the subset of samples that had high abundances of Bifidobacterium but low abundances of Escherichia, the D-Allose module did not display significant enrichment in T2D (P=0.55, Wilcoxon rank-sum test) while the multiple sugar module remained significantly enriched (P=0.001, Wilcoxon rank-sum test). In addition, in both modules the genus Prevotella, which is strongly depleted in T2D (fold-ratio of 1.91 in mean abundance between controls and cases), was a major driver of functional shift. Our analysis further revealed that Firmicutes species dominated the driving contributions to the PTS module among T2D-associated taxa, with the genera Clostridium and Lactobacillus accounting for a large fraction of the observed shift. Interestingly, in an independent study that identified this module as part of the most enriched pathway in T2D patients, these two taxa were found to be the most strongly associated with T2D (Karlsson et al., 2014). As our framework can be applied to decompose shifts at various functional levels (ranging from pathways to individual gene families), we further applied it to characterize the taxonomic drivers of several T2D-enriched genes involved in Xenobiotics degradation and amino acid metabolism, revealing similarly diverse, rich, and complex patterns (compare, for example, the contributions of Escherichia and Bifidobacterium; Figure 4B and Table S6).
Figure 4. Identifying the taxonomic contributors in a Type 2 diabetes (T2D) cohort.
Taxon-level shift contribution profiles for several T2D-associated functional modules (A) and gene orthology groups (B). Certain taxa are labeled by representative letters for convenience.
The findings described above highlight the complex nature of disease-associated functional imbalances, and suggest that functional shifts are a convoluted outcome of multiple taxonomic shifts. The calculation of taxon-level contribution profiles is therefore an essential step toward the rational design of targeted interventions aiming to manipulate the functional capacity of the microbiome.
Similar functional imbalances in different body sites or in different diseases have different microbial drivers
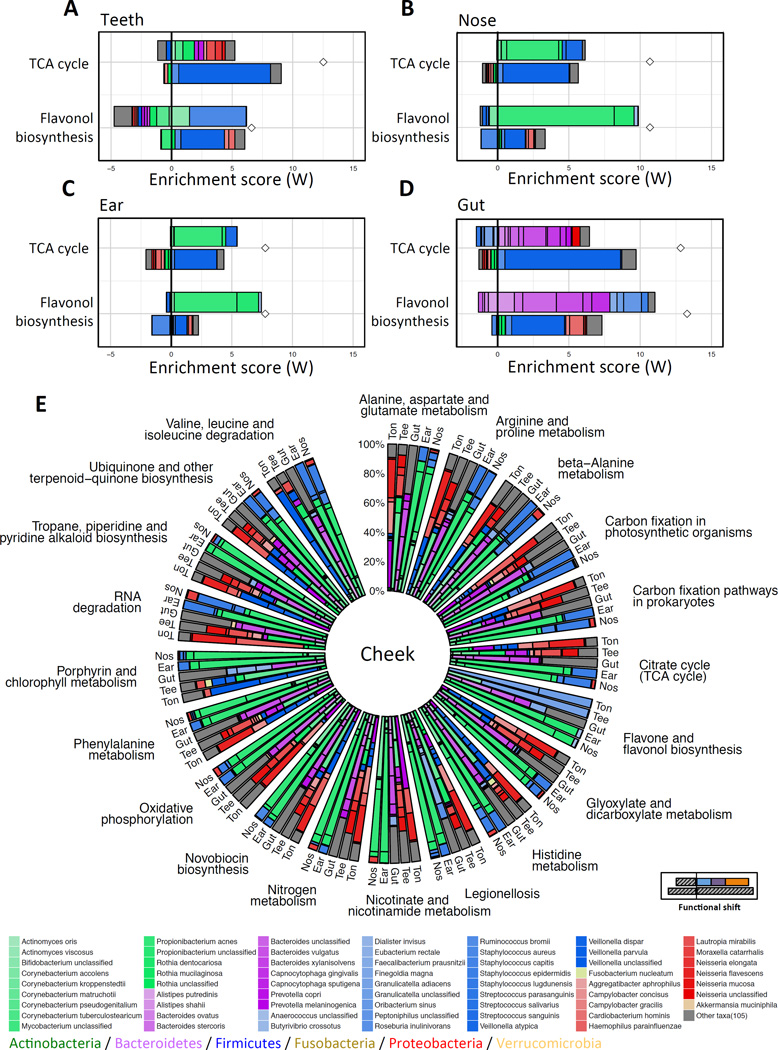
Our analysis of tongue and cheek samples above demonstrated that functional shifts in different functions are often driven by markedly different sets of taxa. To examine whether this trend is universal or limited to these body sites, we identified the taxonomic drivers of observed functional shifts when comparing cheek samples to 4 additional body sites (teeth, gut, ear, and nose). We again found that in each of these body sites, different functions had markedly different shift contribution profiles (Figure 5A–D). Importantly, however, not only did different functions show differences in their taxon-level contribution profiles in each site, but the same function often exhibited substantially different contribution profiles across the different body sites (with less variability between the two skin sites; Figure 5A–D). For example, while the TCA cycle pathway was significantly enriched in each of the 5 body sites when compared to cheek (i.e., including tongue; see Figure 3D), the set of taxa that drove this enrichment in each body site and the level of contribution of each taxon differed greatly, with many Proteobacteria driving the shift in oral sites, Bacteroidetes (e.g., B. ovatus) driving the shift in the gut, and Actinobacteria and Firmicutes (e.g., P. acnes and S. epidermidis) driving the shift in the skin sites. Indeed, such site-specific contribution profiles were also observed when examining all 20 pathways that are significantly enriched in these 5 body sites compared to cheek (Figure 5E).
Figure 5. Identifying taxa driving functional shifts in multiple body sites compared to cheek.
The taxon-level contribution profiles for the enrichment (compared to cheek) of the TCA cycle and the flavone and flavonol biosynthesis pathways in teeth (A), nose (B), ear (C) and gut (D). (E) The shift contribution profiles of all 20 pathways that are enriched in all body sites compared to cheek. Ton: Tongue (tongue dorsum); Tee: Teeth (supragingival plaque); Gut (stool); Ear (retroauricular crease); Nos: Nose (anterior nares). For easier comparison, only the case-associated taxa (case being the body site compared to cheek) that drive the enrichment are presented (see bottom right illustration) and the shift contribution scores are normalized to 1.
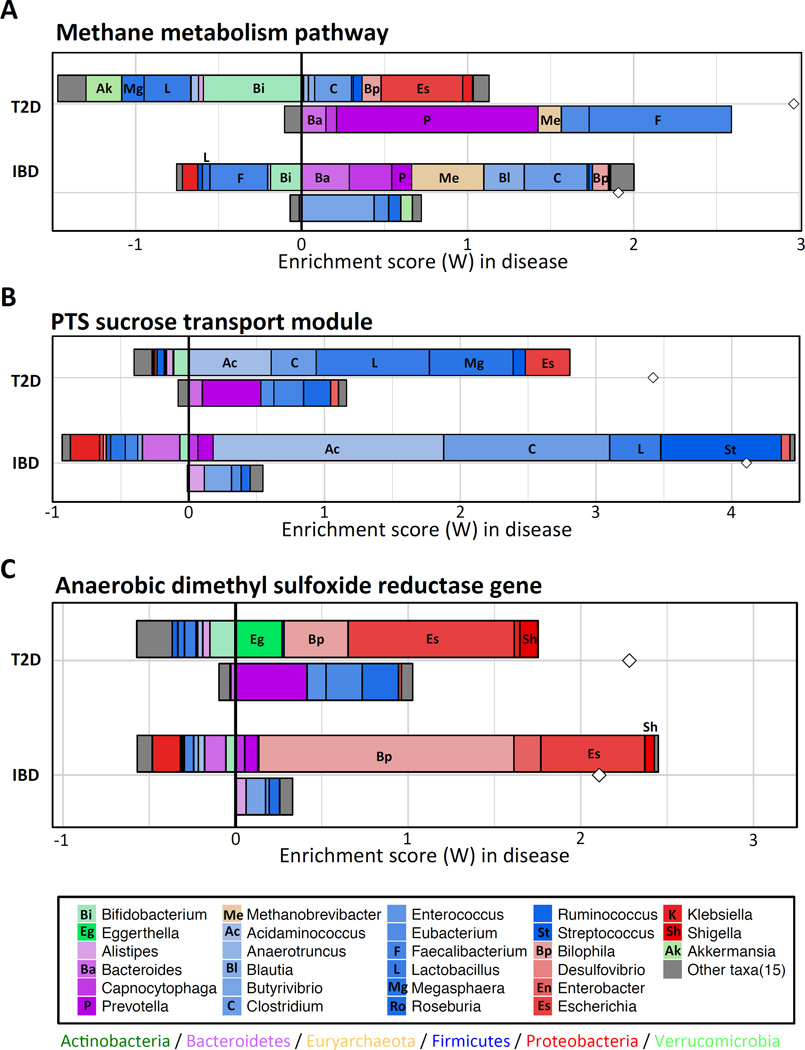
Next, we sought to examine whether contribution profiles are not only site-specific, but also disease-specific (put differently, are similar functional imbalances in different diseases driven by similar taxonomic dynamics or not?). To this end, we applied our framework to a previously obtained inflammatory bowel disease (IBD) cohort (Qin et al., 2010), and identified genera that are driving shifts in functions that were found to be both T2D- and IBD-enriched (Greenblum et al., 2012; Morgan et al., 2012; Qin et al., 2013). We explored such functions at three levels of functional organization, pathways, modules, and gene orthology groups, and below highlight specific examples from each of these levels. We first focused on the Methane metabolism pathway, which was found to be enriched in both T2D and IBD. We found a large set of shared taxa that contributed to the enrichment of this pathway in both diseases, such as Clostridium and Methanobrevibacter (a genus that includes the key gut methanogen M. smithii), although the mode and level of contribution of each taxon differed across diseases (Figure 6A). Importantly, however, the calculated contribution profiles also highlighted a set of disease-specific drivers, such as Escherichia in T2D and Blautia in IBD, suggesting that similar functional imbalances in different diseases may still be attributed, at least in part, to different ecological changes. Indeed, the role of Escherichia in driving enrichment of this pathway in T2D is in agreement with previous observations of an enrichment of the E. coli hxlB gene (a part of the methane metabolism pathway) in T2D (Qin et al., 2013).
Figure 6. Comparing taxon-level contribution profiles of functional shifts in type 2 diabetes (T2D) and inflammatory bowel disease (IBD).
Taxon-level shift contribution profiles in T2D and IBD of the methane metabolism pathway (A), the PTS system sucrose-specific II component module (B), and the Anaerobic dimethyl sulfoxide reductase gene orthology group (C).
We next focused on the PTS sucrose transporter module (M00269; Figure 6B), as an example of the many oligosaccharide transporters found to be significantly enriched in both T2D and IBD patients (Morgan et al., 2012; Qin et al., 2013). In contrast to the diverse set of phyla contributing to the shift in the methane metabolism pathway, the shift in this module was primarily driven by Firmicutes. Nonetheless, as above, the contribution profiles included both shared and disease-specific taxa, with several Firmicutes (such as Acidaminococcus, Clostridium, Lactobacillus) driving the shift in both diseases and other Firmicutes driving the shift in only one (e.g., Megasphaera in T2D and Streptococcus in IBD). These taxonomic contribution profiles are again supported by previous observations. For example, an IBD study demonstrated that patients treated with mesalamine exhibit significant decrease in Clostridium coupled with a significant decrease in the PTS pathway (Morgan et al., 2012), and a T2D study demonstrated enrichment of 2 Clostridium genes (of the 3 genes in the PTS sucrose transporter module) in T2D (Qin et al., 2013). Similarly, Megasphaera was found to be more highly abundant in pre-diabetic individuals (Lambeth et al., 2015), while Streptococcus was found to be associated with IBD (Kojima et al., 2012) but to decrease in abundance in T2D (Zhang et al., 2013). Similar biologically-relevant combinations of disease-specific and shared contributors were also observed at the gene level (Figure 6C and Supplemental Text). Combined, these findings highlight the extremely complex ecological dynamics that underlie shifts in the functional capacity of the microbiome and emphasize the importance of characterizing the taxonomic drivers of these shifts.
Genomic content inference improves agreement between metagenome- and taxa-based functional profiles and uncovers previously unknown drivers of functional shifts
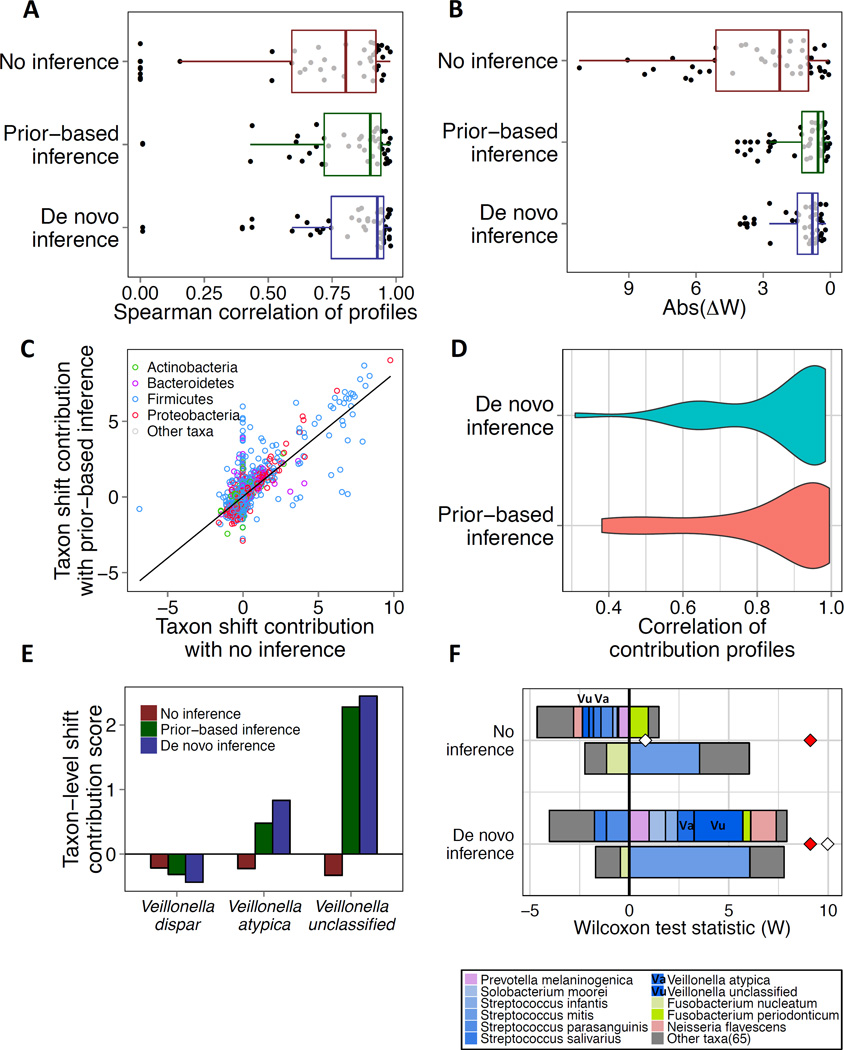
As noted above, when reference genomes are not available for all community members, our framework can employ a previously introduced method (Carr et al., 2013) to infer the genomic content of each taxon based on coupling of taxonomic and functional abundance profiles (see Methods and Supplemental Experimental Procedures). To examine the applicability of this approach and its impact on the calculated FishTaco contribution profiles, we again applied FishTaco to the HMP and T2D datasets, but this time using genomic content inference (rather than available reference genomes). We first confirmed that genomic content inference successfully recovered the genomic content of the member species (R=0.83 median Pearson correlation across pathways in HMP species). Importantly, however, we found that genomic content inference markedly improved the agreement between taxa-based and metagenome-based functional profiles and shifts (Figure 7A–B; and see also Figure S6).
Figure 7. Using genomic content inference provides more accurate taxa-based functional profiles in HMP samples.
(A) Spearman correlation between metagenome- and taxa-based functional abundance profiles for HMP samples, without and with inference (each dot represents a single pathway). Median Spearman correlations were ρ=0.8, ρ=0.9 and ρ=0.93 for no inference, prior-based inference, and de novo inference, respectively. (B) Absolute difference between the metagenome- or taxa-based functional shift scores, with no inference, prior-based inference, and de novo inference (median absolute differences were W=2.2, W=0.55 and W=0.8, respectively. (C) Scatter plot comparing taxon-level shift contribution values computed by FishTaco using genomic content from reference genomes to those computed with prior-based genomic content inference. Each circle represents the contribution of a single species (colored by phyla) to a single pathway. Regression line is also depicted. (D) Distributions of Pearson correlation values between the resulting taxon-level shift contribution profiles when using prior-based or de novo inferences of genomic content and the taxon-level shift contribution profiles when genomic content is based on reference genomes (i.e., no inference), for the pathways in the HMP dataset. (E) Taxon-level shift contribution values for the flavonoid biosynthesis pathway for species in the genus Veillonella, with no inference, prior-based inference, and de novo inference. (F) Taxon-level shift contribution profiles for the flavonoid biosynthesis pathway without (top) and with (bottom) de novo inference of the genomic content. See Supplemental Text for additional details.
Moreover, even though the resulting taxon-level shift contribution profiles obtained with these inferred genomes were overall similar to those obtained based on reference genomes (Figures 7C–D, S6), a few taxa were found to have markedly different contributions to specific functional shifts when using genomic content inference vs. reference genomes (Figures 7E–F, S7). These newly uncovered drivers of functional shifts revealed by genome content inference were often supported by experimental observations (see Supplemental Text). These findings suggest that our ability to infer missing genomic information not only supports the application of our framework for studying communities harboring poorly characterized microbial clades, but could also promote discoveries of microbial drivers that may be masked by incomplete genomic annotation.
Discussion
The ultimate goal of comparative metagenomics is to identify potentially meaningful changes in the microbiome’s taxonomic and functional composition that are associated with health. However, to date, the relationship between detected taxonomic and functional shifts remains largely unexplored, hindering our ability to characterize the impact of individual taxa on disease-associate functional imbalances and ultimately to pinpoint critical intervention targets. Our study represents an important first step towards addressing this challenge and provides a comprehensive computational framework for integrating taxonomic and functional comparative analyses.
Importantly, decomposing each functional shift to its taxon-level contributors offers unique insights into both basic and translational research. Specifically, it provides a window to the ecological dynamics at play and their relationship to the host environment, highlighting the complex and non-trivial ways by which changes in taxonomic composition translate into observed shifts in the community-level functional profile. Translationally, the identification of function- or disease-specific taxonomic drivers can inform future targeted microbiome-based therapies that aim to modulate the abundance of these taxa in order to restore desired capacity of specific functions.
Notably, our framework’s analysis involves several simplifying assumptions. First, it constructs taxa-based functional profiles using fixed genomic content (e.g., based on reference genomes) assuming that each species is associated with the same genomic content across all samples and ignoring strain-level variation in the presence and copy number of genes (Greenblum et al., 2015). Higher resolution taxonomic profiling, extended strain-level genomic annotation, and more sophisticated inference techniques can improve the accuracy of our taxa-based functional profiles but are ultimately limited and cannot eliminate the problem altogether. Second, using our framework’s identified driver taxa as putative intervention targets further assumes that removing a single species from the community would leave the composition of other species intact. However, since the various species in the microbiome likely cross-feed and interact, such interventions could impact the relative abundances of other species in the community and induce an unexpected shift in the community’s functional capacity. Addressing this challenge clearly requires a better understanding of microbial interactions and ecological dynamics, which could potentially be incorporated into a predictive model of the expected impact of taxonomic perturbations.
Future research could also integrate other data types or additional comparative analysis settings. For example, integrating community-wide transcriptomic, proteomic, and metabolomics data could allow us to examine whether genomic, transcriptional, and ultimately metabolic shifts are driven by the same taxa, and help distinguish between ‘passenger’ taxa that merely increase the abundance of various gene classes in the metagenome to ‘driver’ taxa that actively play a role in disease etiology. Similarly, extending our framework to decompose functional shifts in longitudinally (rather than only cross-sectional) obtained samples, while correctly accounting for induced autocorrelations in such datasets, would allow us to follow disease dynamics and elucidate how the link between taxonomic and functional shifts evolves over time. Such analyses could ultimately pinpoint preferred intervention windows in the path leading to disease-associated functional imbalances or suggest time-dependent intervention strategies. Finally, going beyond a binary classification of samples (e.g., cases vs. controls) and extending our analysis approach to support multiple categories of functional imbalance, or preferably identify such categories based on both observed functional profiles and the resulting taxonomic contribution profiles, could offer more informed, accurate, and personalized microbiome-based intervention targets.
Clearly, the link between compositional shifts in taxonomy and function in a dysbiotic microbiome is complex and intertwined and distinguishing cause and effect is extremely challenging. Frameworks such as FishTaco that aim to characterize and quantify the relationship between taxonomic and functional dynamics obviously cannot fully resolve these complexities, but are a promising step in gaining a principled understanding of the underlying forces shaping microbiome dysbiosis in disease. We accordingly hope that the framework and analyses presented above will help to elucidate the role that the microbiome plays in human diseases, facilitate the development of microbiome-based therapeutic routes, and inspire future studies of the fascinating linkage between composition and function.
Methods
Software implementation and distribution
FishTaco was implemented in Python and is available for download at http://elbo.gs.washington.edu/software.html. In addition, FishTaco is available as a pip-installable python package (i.e., pip install fishtaco), and the source code is available on GitHub (http://github.com/borenstein-lab/fishtaco). Visualization of FishTaco-based decompositions can be done via a web-based application (https://elbo-spice.gs.washington.edu/shiny/FishTacoPlot/) or by using a dedicated R package (http://github.com/borenstein-lab/fishtaco-plot).
Human Microbiome Project (HMP) data
Data were downloaded from the Data Analysis and Coordination Center website (http://www.hmpdacc.org/), including KEGG Orthology group (KO) abundances (Kanehisa et al., 2012) and sample metadata and species abundances as calculated by MetaPhlAn (Segata et al., 2012). A list of the samples used for each body site is provided in Table S7.
Type 2 diabetes (T2D) and Inflammatory bowel disease (IBD) data
Data were downloaded from the human gut microbiome Integrated Reference Catalogue (IGC; Li et al., 2014) website (http://meta.genomics.cn/meta/dataTools), including KO abundances, sample metadata, and genera abundances.
Genomic content data
For each species in the HMP data, we obtained all reference genomes corresponding to this species from the IMG database (Markowitz et al., 2013), and defined the copy number of each KO as the mean copy number across reference genomes. For the T2D and IBD data, we used a list curated by the original T2D study (Li et al., 2014) of 511 prokaryotic gut reference genomes in IMG and defined the copy number of each KO in each genus as the mean copy number across reference genomes corresponding to this genus.
Taxonomic abundance profile filtering and normalization
Taxonomic abundances were normalized to represent relative abundances. For MetaPhlAn data, only species level abundances were used, and samples where the total species level abundance was <95% were removed from further analysis.
Functional profile normalization
Functional profiles were re-normalized using MUSiCC (Manor and Borenstein, 2015), converting relative KO abundances into KO average genomic copy numbers across samples’ community members.
Calculating pathway and module abundance profiles
MUSiCC-normalized abundances of all KOs associated with a specific pathway/module in the KEGG database were summed for each sample. Pathways with <20 KOs, or that are present in <1% of bacterial genomes in KEGG, or that had <5% of their genes present in these genomes on average were removed from downstream analysis. Similarly, modules present in <1% of bacterial genomes in KEGG or that had <20% of their genes present in these genomes on average were removed from downstream analysis. This resulted in a total of 80 pathways and 409 modules.
Calculating taxonomic and functional shifts
As noted above, our framework supports the use of any metric of choice for measuring functional shifts. In this study, we specifically used the Wilcoxon rank-sum test to compare the abundance values of a given taxon (species or genus) or function (KO, module, or pathway) between case and control samples (or different body sites) and reported the Wilcoxon test statistic, W, as a measure of observed shifts. Multiple hypothesis correction was done using Bonferroni (<5%) for HMP samples and false discovery rate (FDR; <10%) for T2D and IBD samples.
Generating taxa-based functional profiles
Let T denote the matrix (size N×M) describing the relative abundances of M taxa across N samples, and let G denote the matrix (size M×K) describing the genomic content of each taxon (wherein each entry Gi,j denotes the genomic copy number of gene (or any other genomic element) j (j = 1 … K) in taxon i (i = 1 … M). F=TxG then denotes the taxa-based functional profile matrix (size N×K), in which every entry Fs,j is the inferred taxa-based abundance of gene j in sample s.
Functional shift decomposition
To assess the contribution of each taxon to functional shifts while maintaining global community composition properties, a permutation-based approach coupled with a game-theory method was used (see also Figure S1). Briefly, the matrix of taxonomic profiles, T, was permuted multiple times, each time permuting the relative abundance of a different subset of taxa across samples and preserving the relative abundance of others. Each permuted matrix was used to generate the corresponding taxa-based functional profiles and to calculate the induced functional shift of each function. Comparing observed shifts when using the permuted vs. original taxonomic profiles provided an estimate for the contribution of the permuted taxa subset to functional shifts. Using a large ensemble of such taxa subset contribution estimates, the average marginal contribution of each taxon to each functional shift was calculated to obtain the final contribution score. This approach was inspired by the Shapley value analysis – a game-theory technique for obtaining the optimal linear estimate of individual contributions in a multi-player game (Shapley, 1953). Since it is not feasible in this setting to analyze all 2M possible multi-taxa subsets, we followed the Shapley value estimation method presented in Keinan et al. (Keinan et al., 2004), using a partial collection of taxa subsets of all sizes from 1 to M. For a formal description of this our framework and the parameters used see Supplemental Experimental Procedures.
Genomic content inference
Both de novo and prior-based (i.e., using available reference genomes as a starting point) machine learning schemes were implemented to computationally infer the genomic content of taxa (e.g., in cases where reference genomes are not available or not complete), extending our previously published method (Carr et al., 2013). Briefly, our method is based on representing the expected relationship between taxonomic profiles, functional profiles, and genomic content as a set of linear equations, and using non-negative elastic-net regularized linear regression to infer the genomic content of each taxa. See Supplemental Experimental Procedures for a detailed description of this method.
Supplementary Material
Highlights.
A framework for identifying taxonomic drivers of functional shifts in the microbiome
Taxon-level contributions to functional shifts are complex and function-specific
Similar functional imbalances in different diseases are driven by different taxa
Inference of microbial genomes uncovers previously unknown drivers of functional shifts
Acknowledgments
We are grateful to all members of the Borenstein lab for helpful discussions. This work was supported in part by New Innovator Award DP2 AT007802-01 to EB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
OM and EB conceived and designed the study, the computational framework, and the analyses. OM implemented the framework and performed the analyses. OM and EB wrote the paper.
References
- Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr R, Borenstein E. Comparative analysis of functional metagenomic annotation and the mappability of short reads. PLoS ONE. 2014;9:e105776. doi: 10.1371/journal.pone.0105776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr R, Shen-Orr SS, Borenstein E. Reconstructing the genomic content of microbiome taxa through shotgun metagenomic deconvolution. PLoS Comput Biol. 2013;9:e1003292. doi: 10.1371/journal.pcbi.1003292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2013;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll J, Thiagarajan M, Abubucker S, Huttenhower C, Yooseph S, Methé BA. A case study for large-scale human microbiome analysis using JCVI’s metagenomics reports (METAREP) PLoS ONE. 2012;7:e29044. doi: 10.1371/journal.pone.0029044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblum S, Carr R, Borenstein E. Extensive strain-level copy-number variation across human gut microbiome species. Cell. 2015;160:583–594. doi: 10.1016/j.cell.2014.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci USA. 2012;109:594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2014;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- Keinan A, Sandbank B, Hilgetag CC, Meilijson I, Ruppin E. Fair attribution of functional contribution in artificial and biological networks. Neural Comput. 2004;16:1887–1915. doi: 10.1162/0899766041336387. [DOI] [PubMed] [Google Scholar]
- Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3:14. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima A, Nakano K, Wada K, Takahashi H. Infection of specific strains of Streptococcus mutans, oral bacteria, confers a risk of ulcerative colitis. Sci. Rep. 2012 doi: 10.1038/srep00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth SM, Carson T, Lowe J, Ramaraj T, Leff JW, Luo L, Bell CJ, Shah VO. Composition, Diversity and Abundance of Gut Microbiome in Prediabetes and Type 2 Diabetes. J Diabetes Obes. 2015;2:1–7. doi: 10.15436/2376-0949.15.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- Manor O, Borenstein E. MUSiCC: a marker genes based framework for metagenomic normalization and accurate profiling of gene abundances in the microbiome. Genome Biology. 2015;16:53. doi: 10.1186/s13059-015-0610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Pillay M, Ratner A, Huang J, Woyke T, Huntemann M, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Research. 2013;42:D560–D567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biology. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noecker C, McNally CP, Eng A, Borenstein E. High-resolution characterization of the human microbiome. Transl Res. 2016;0 doi: 10.1016/j.trsl.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Byrd AL, Deming C, Conlan S, Program NCS, Kong HH, Segre JA. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Jeroen Raes, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2013;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife Sciences. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz MD, Atay Ç, Heringer J, Romrig FK, Schwitalla S, Aydin B, Ziegler PK, Varga J, Reindl W, Pommerenke C, et al. High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature. 2014;514:508–512. doi: 10.1038/nature13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Meth. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapley LS. A value for n-person games. In: Kuhn HW, Tucker aW, editors. Contributions to the Theory of Games. Vol. 2. Princeton, NJ: Princeton University Press; 1953. pp. 307–317. [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang C, Chen Y, Ji L. Human Gut Microbiota Changes Reveal the Progression of Glucose Intolerance. PLoS ONE. 2013;8:e71108. doi: 10.1371/journal.pone.0071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.