Summary
Neuronal differentiation is a multistep process that shapes and re-shapes neurons by progressing through several typical stages, including axon outgrowth, dendritogenesis, and synapse formation. To systematically profile proteome dynamics throughout neuronal differentiation, we took cultured rat hippocampal neurons at different developmental stages and monitored changes in protein abundance using a combination of stable isotope labeling and high-resolution liquid chromatography-tandem mass spectrometry (LC-MS/MS). Almost one third of all 4,500 proteins quantified underwent a more than 2-fold expression change during neuronal differentiation, indicating extensive remodeling of the neuron proteome. To highlight the strength of our resource, we studied the neural-cell-adhesion molecule 1 (NCAM1) and found that it stimulates dendritic arbor development by promoting actin filament growth at the dendritic growth cone. We anticipate that our quantitative map of neuronal proteome dynamics is a rich resource for further analyses of the many identified proteins in various neurodevelopmental processes.
Keywords: neuronal differentiation, neuronal development, dendrite outgrowth, neuroproteomics, NCAM1, adhesion molecule, proteome, quantitative proteomics, stable isotope labeling, mass spectrometry
Graphical Abstract
Highlights
-
•
Systematic profile of proteome dynamics throughout neuronal development in vitro
-
•
Approximately 1,800 proteins show significant expression changes during differentiation
-
•
Six expression profile clusters describe stage-specific patterns of protein dynamics
-
•
This protein database may help to identify neurodevelopment mechanisms
In this study, Frese et al. generate a systematic quantitative map of proteome dynamics during the specific stages of rat hippocampal neuron development in vitro. Six expression profile clusters give a comprehensive overview of protein abundances over time, providing a database of stage-specific protein expression patterns in neurons.
Introduction
The ability to generate in vitro cultures of primary rat or mouse neuronal cells has been fundamental to advancing our understanding of the development and functioning of the nervous system. A major advantage of performing experiments in primary hippocampal culture systems is that neurons resynchronize in culture prior to further differentiation (Dotti et al., 1988). This allows systematic processing and analyses of neuronal populations at the same developmental stage, which is particularly helpful for global system-wide analyses. Pyramidal neurons, the principal neuronal cell type in the hippocampus, account for the vast majority of the total neuronal cell population within these preparations and express many key phenotypic features of various neuronal cell types. The stages of neuron development in culture have been categorized and provide a starting point for studying neurodevelopmental processes, such as neuronal polarity, dendrite development, and synapse formation (Dotti et al., 1988). Directly after plating and attaching to the substrate, the cells develop lamellipodia around the soma (stage 1) that, within a few hours, transform into several short and highly motile neurites (stage 2). Quickly after, the neurons polarize upon which one of the several neurites develops into an axon (stage 3). Around day in vitro (DIV) 4 (DIV4), dendrites start to develop from the minor neurites (stage 4); this process is much slower than axonal outgrowth and specification and lasts for several days. After 1 week in culture, synaptogenesis begins, and neurons form functional synaptic contacts and, in the following weeks, undergo further neuronal maturation (stage 5).
Several studies have been reported monitoring global gene expression changes in developing rodent neurons (Mody et al., 2001, Dabrowski et al., 2003), albeit nearly all at the mRNA transcript level. These studies revealed that large changes in mRNA expression occur across different stages of hippocampal neuronal development (Dabrowski et al., 2003). Several earlier studies have been reporting differential gene expression during the specialization of the axon and dendrites in hippocampal primary culture systems. In more recent studies, also, the expression of non-coding regulatory RNAs has been profiled in developing neurons (van Spronsen et al., 2013). Transcriptome analysis of different brain regions and different cell types, including cortical primary neurons, has been compared with data from large-scale quantitative proteomic screens (Sharma et al., 2015). However, in many cases, the gene expression changes measured with transcriptomics approaches do not completely reflect the changes measured at the protein level. Several studies have shown that the correlation between mRNA and protein expressions can be low due to several factors such as protein turnover, half-lives, and other post-transcription mechanisms (Low et al., 2013). Moreover, a single mRNA can be translated multiple times, introducing another level of complexity in correlating such data. In this context, we argue that applying advanced MS methods and quantitative proteomics approaches to systematically profile protein expression during neuronal differentiation will provide a better proxy for changes in protein expression.
Here, we perform a systematic and in-depth proteome analysis of hippocampal neurons in culture and established a quantitative map of neuron-specific proteome dynamics during developmental stages 2–3, 4, and 5. Our dataset comprises 6,753 protein identifications, of which more than 4,300 were quantified over all time points, covering crucial neuronal developmental processes, including axon outgrowth, dendrite formation, and synaptogenesis. About one third of the proteins reveal substantial changes in protein expression throughout the neuronal differentiation, clearly highlighting the extensive reprogramming of the proteome. Our analysis revealed an unappreciated role for neural-cell-adhesion molecule 1 isoform 180 (NCAM180) in dendritic development. NCAM180 is strongly upregulated during dendrite outgrowth, is highly enriched in dendritic growth cones, and interacts with a large variety of actin-binding proteins.
Results
Global Proteomic Signatures of Developing Hippocampal Neurons
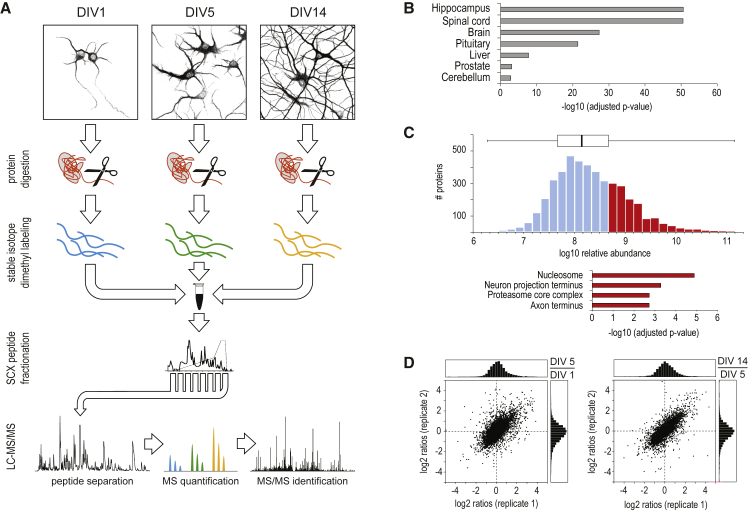
To profile neuronal proteome alterations during differentiation, primary hippocampal neurons were grown in serum-free neurobasal medium, enabling rapid differentiation and maturation (Dotti et al., 1988). Cells were harvested after DIV1, DIV5, and DIV14, corresponding to neuronal developmental stages 2–3, 4, and 5, respectively, as described recently (van Spronsen et al., 2013). These stages are associated with the distinct neuronal differentiation processes axon formation and specification (stages 2–3/DIV1), dendrite outgrowth (stage 4/DIV5), synaptogenesis, and maturation (stage 5/DIV14). For in-depth quantitative proteome analysis, cell lysates of the three time points were subjected to tryptic digestion (Figure 1A), triplex stable-isotope dimethyl labeling (Boersema et al., 2009), strong cation-exchange (SCX)-based fractionation, and nano-ultra-performance liquid chromatography (nano-UPLC) coupled to high-resolution liquid chromatography-tandem mass spectrometry (LC-MS/MS). Quantification of relative protein expression changes was performed based on the MS signal intensities of the stable-isotope-labeled peptide ions (Figure 1A). In total, 46,869 unique peptides from 6,753 unique proteins were identified (median unique peptides per protein = 6.9). Tissue enrichment analysis against the whole Rattus norvegicus proteome as background revealed that the dataset covers large parts of hippocampal neuron-specific proteins (Fisher’s exact test, adjusted p = 2.3E−51; Figure 1B). Overall, 4,354 proteins were quantified across all time points (median ratio count = 14; Table S1). The averaged relative protein abundances span five orders of magnitude (Figure 1C). Within the 25% most abundant proteins, we observed numerous neuron-specific proteins. Gene Ontology (GO) enrichment analysis, with respect to cellular component, highlights proteins located in the axon and nerve terminal (“neuron projection terminus”) as being among the most abundant proteins, besides more commonly detected nucleosomal proteins and the proteasome (Figure 1C). Scatterplots of the log2-transformed protein ratios (DIV5/DIV1 and DIV14/DIV5) and a heatmap of Pearson correlation scores illustrate the good correlation between biological replicates (Figure 1D; Figure S1A). In total, 1,793 proteins show more than 2-fold (log2 scale < −1 and >1) expression changes, indicating extensive remodeling of at least one third of the neuron proteome during differentiation (Figure S1B).
Figure 1.
MS-Based Quantitative Proteomic Analysis of Developing Hippocampal Neurons in Culture
(A) Workflow of quantitative neuroproteomic analysis.
(B) Tissue enrichment analysis of all identified proteins using the Database for Annotation, Visualization, and Integrated Discovery (DAVID).
(C) Distribution of protein abundances, spanning five orders of magnitude.
(D) In total, 4,354 proteins were quantified across all time points, and good correlation between biological replicates is observed.
See also Table S1.
Quantitative Analysis of Proteome Dynamics during Neuron Differentiation
We assessed the global proteome changes of 4,354 proteins quantified across all time points. Close inspection of proteins that were considered not significantly changing (Supplemental Experimental Procedures) revealed general protein metabolism and catabolism as the main processes that are not substantially differentially regulated during neuronal differentiation. This is reflected by several GO terms, such as “translation” (adjusted p value 9.8E-8) and “protein catabolic process” (p = 7.5E−6), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) terms “ribosome” (p = 9.9E−5) and “proteasome” (p = 6.8E−4) that were found overrepresented (Figure S1C). Together, these findings imply that the global protein turnover rates are not substantially affected in the course of proteome remodeling during neuronal differentiation.
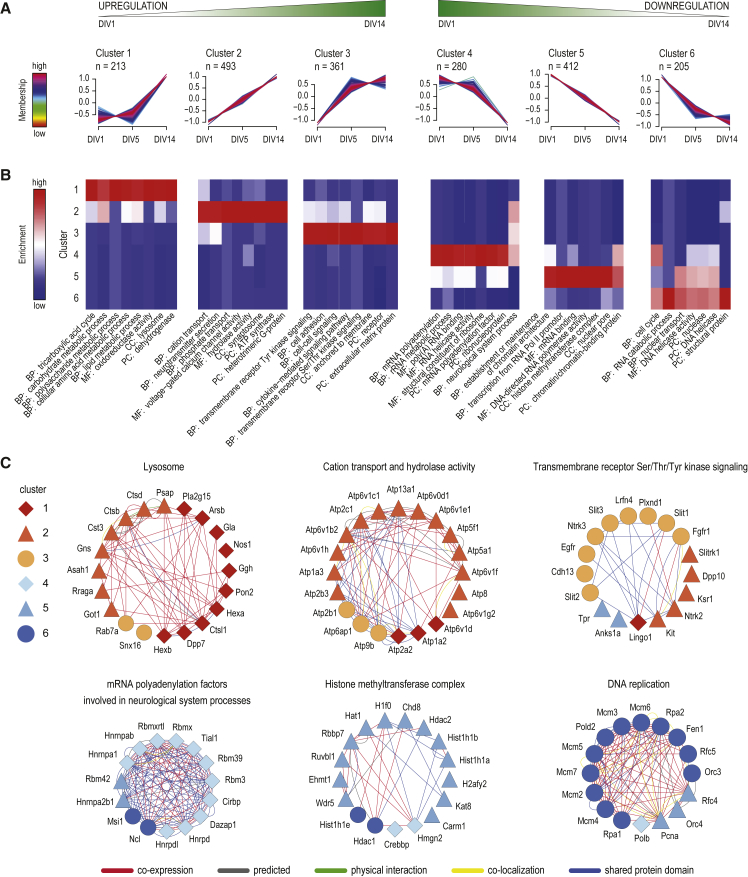
To obtain an unbiased view of the proteome expression dynamics during neuronal differentiation, we performed unsupervised fuzzy clustering of all significantly changing proteins. This analysis resulted in six clusters of distinct expression profiles (Figure 2A), with proteins upregulated during differentiation in clusters 1, 2, and 3 and proteins downregulated present in clusters 4, 5, and 6. We next investigated whether the clusters revealed proteins with developmental stage-specific expression patterns. Indeed, many proteins within specific clusters shared functionalities related to their cellular component, molecular function, or their contribution to distinct biological processes (Figure 2B). Complete GO annotations for each cluster can be found in Figure S2.
Figure 2.
Dynamic Proteome Remodeling during Neuronal Differentiation
(A) Unsupervised clustering of proteome dynamics revealed six clusters with distinct protein expression profiles. n represents the number of proteins per cluster.
(B) Gene ontology enrichment analysis of each cluster was performed using a Fisher’s exact test (cutoff p < 0.002, Benjamini-Hochberg corrected). Representative biological processes (BP), molecular functions (MF), cellular components (CC), and protein classes (PC) that are overrepresented in one of the clusters are visualized.
(C) Protein network analysis showing highly connected proteins that are representative for each cluster.
Cluster 6 contains proteins strongly downregulated from DIV1 to DIV5 and, to a lesser extent, from DIV5 to DIV14, indicating a precisely timed downregulation during early neuronal differentiation. Proteins in this cluster are mainly involved in early differentiation processes of post-mitotic neurons, which do not proliferate and remain arrested in G0 phase, exemplified by multiple proteins involved in the regulation of cell cycle and DNA replication, such as DNA helicases. This includes minichromosome maintenance complex proteins (Mcm2–Mcm7) as well as DNA polymerase delta (Pold2), replication factor 5 (Rfc5), and Flap endonuclease 1 (Fen1) (Figure 2C). Downregulation of proteins related to cell cycle and DNA replication have been similarly observed in other terminally differentiating cells (Kristensen et al., 2013). In line with these findings, we detected slight upregulation of cyclin-dependent kinase 5 (Cdk5) (Figure S3A), which is known to be exclusively expressed in post-mitotic neurons and is involved in suppressing cell-cycle reentry (Zhang and Herrup, 2011).
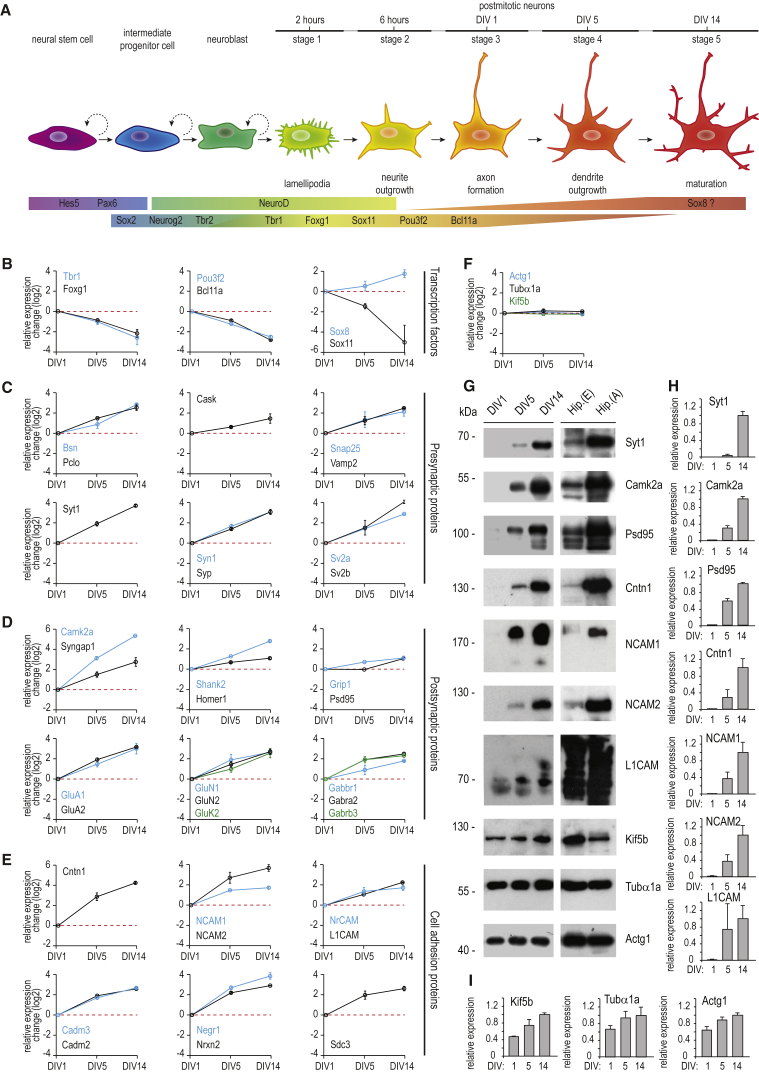
Proteins in cluster 4 exhibit a strong downregulation from DIV5 to DIV14 (Figure 2A). Many proteins in this cluster are associated with RNA metabolism (Figure 2B), exemplified by a highly connected group of mRNA polyadenylation factors (Figure 2C). Interestingly, several transcription factors, which play key roles in neuronal differentiation, are also allocated to this cluster. A prominent example of this subset of proteins is the neuron-specific transcription factor Sox11, which is the most downregulated protein in the total dataset (expression level decreases about 40-fold from DIV1 to DIV14; see also Figures 4A and 4B). The onset of Sox11 expression is known to overlap with decreased expression of Sox2 (not quantified in our data) in neuronal committed proliferating progenitor cells, promoting differentiation into premature neurons (Haslinger et al., 2009). Likewise, transcription factor Bcl11a, which has been identified as a negative regulator of axon branching and dendrite outgrowth (Kuo et al., 2009), follows the expression profile of cluster 4. Other related transcription factors are allocated to cluster 5 (sustained decrease of relative protein levels from DIV1 to DIV14); for instance, Foxg1 and Sox11-interacting protein Pou3f2, both of which play important roles in the early to mid-neurogenesis of postmitotic neurons (Dominguez et al., 2013, Hsieh, 2012). Another transcription factor found in cluster 5 is Tbr1, which is expressed in early postmitotic neurons, promotes neurogenesis, and functions as a repressor of astrocyte formation (Méndez-Gómez et al., 2011). Moreover, Tbr1 has been reported to be involved in regulation of axon pathfinding (Hevner et al., 2006), a process that is part of the major developmental changes from stage 2 to stage 3. Collectively, our data show that neuronal differentiation and maturation from stages 2–3 to stage 5 resonate with a gradual decrease of several transcription factors that are key regulators in the early events of neurogenesis and initial axon formation. Notably, Sox8, another member of the SRY-related HMG (high-mobility group)-box gene family, shows a minor increase from DIV1 to DIV5 and an elevated increase from DIV5 to DIV14 (cluster 1). Sox8 is reportedly involved in the regulation of terminal differentiation of oligodendrocytes (Stolt et al., 2004). Late upregulation from developmental stage 4 to stage 5 suggests that Sox8 might also be involved in the terminal differentiation of neurons. However, its functional role in the later stages of neuronal development remains to be unveiled.
Figure 4.
Protein Expression Changes during Neuronal Differentiation
(A) Schematic overview of neurogenesis, illustrating the distinct developmental stages and representative transcription factors involved.
(B) Expression profiles for representative transcription factors including the most downregulated protein in this dataset, Sox11.
(C–F) Expression profiles of selected presynaptic proteins (C), postsynaptic proteins (D), cell-adhesion proteins (E) and proteins with no relative expression change over the developmental stages (F).
(G) Change in the protein expression during neuronal development shown by western blot analysis from cultured hippocampal neurons, embryonic tissue (Hip.(E)), and adult brain tissue (Hip.(A)).
(H and I) Graphs showing the quantifications of relative intensities of the western blot analysis in (G), shown in (H), and control proteins, shown in (I). n = 3 experiments per condition. Error bars indicate mean ± SEM.
In (B)–(F), error bars represent SD.
Cluster 5, representing a constant decrease of relative protein levels over time (Figure 2A), is enriched for proteins associated to chromatin (Figure 2B). Steady downregulation of histone methyltransferases (Ehmt1 and Wdr5), acetyltransferases (Kat8 and Hat1), and histones (Hist1h1b, Hist1h1a, and H2afy2) indicate an ongoing remodeling of the chromatin architecture during neuronal differentiation (Figure 2C). The GO term “mRNA binding” was also found specifically enriched in cluster 5. In fact, 42 out of 103 proteins classified as RNA-binding proteins (RBPs) (Table S2) are allocated to this cluster and undergo, on average, 3-fold downregulation (log2 scale = −1.6) from DIV1 to DIV14. Post-transcriptional regulation by RBPs controls gene expression and diversification and is known to accurately coordinate spatiotemporal differentiation of neurons (DeBoer et al., 2013). A prominent member of this cluster is Polypyrimidine tract-binding protein 2 (Ptbp2). Ptbp2 is predominantly expressed in brain tissue during the early stages of neurogenesis, and its main function is the precisely timed negative regulation of alternative splicing events of a distinct set of target genes that play key roles in neuronal differentiation (Sawicka et al., 2008). Therefore, we mined our data for proteins whose transcripts are targets of Ptbp2 (Licatalosi et al., 2012). We observed that the decrease in Ptbp2 levels coincides with increased expression of several proteins whose transcripts are targets of Ptbp2 (including Camk2b, Camk2g, Psd95, Dnm1, Ppp3cb, and Numb). Interestingly, we observed for three other target proteins, whose transcript levels were found elevated in that study, no significant expression changes (Sorbs2 and Pum2) or only a slight decrease (Sympk, cluster 5) in our dataset. Notably, the neuron-specific RBPs Nova1 and Nova2, which are direct interactors of Ptbp2, show no significant expression change from DIV1 to DIV14. Among the upregulated RBPs are Pura and Purb, which, besides their molecular function in RNA binding, are known transcriptionally active DNA binders. Pura and Purb undergo a highly similar expression increase from DIV5 to DIV14 (cluster 1), indicating a specific role in synapse formation and later maturation processes. Indeed, reduced synapse formation was observed in Pura knockout mice (White et al., 2009), supporting a presumed role during synaptogenesis. Notably, a third member of the Pur family, Purg, shows stronger and earlier upregulation compared to Pura and Purb, suggesting involvement in differentiation events preceding synapse formation. Together, these findings support highly coordinated reorganization of the protein network orchestrating post-transcriptional gene expression regulation during neuronal differentiation (Loya et al., 2010).
The proteins in cluster 1 show a substantial increase in relative expression levels from DIV5 to DIV14 (Figure 2A). This expression profile corresponds to stage-specific upregulation during maturation from stage 4 to stage 5. Distinct GO terms, which were found enriched in cluster 1, cover proteins involved in several metabolic processes, including polysaccharide, amino acid, and lipid metabolism (Figure 2B). Closer inspection of the corresponding proteins revealed that most of them function as catalytic enzymes in lysosomes (e.g., cathepsin L1, arylsulfatase B, and dipeptidyl peptidase 2; Figure 2C). Endosomal trafficking is a key control mechanism in synapse formation and modulation of synaptic plasticity. For example, postsynaptic receptor density is governed by the interplay of endosomal recycling and endosome-lysosome trafficking. Therefore, the observed upregulation of lysosomal proteins in the course of neuronal development could reflect an increase in the cellular capacity for the degradation via the lysosomal pathway.
Cluster 3 comprises proteins that undergo increased expression from DIV1 to DIV5, while the expression changes from DIV5 to DIV14 are marginal (Figure 2A). Many proteins in this cluster are associated with cell adhesion and cell-cell signaling-related GO terms (Figure 2B). This includes several transmembrane receptor kinases involved in cell adhesion, migration, axon guidance, and dendrite formation. For instance, prominent members of this cluster are growth factor receptors Egfr and Fgfr1; neurotrophin receptor Ntrk3; semaphorin receptor Plxnd1; and axon guidance cues Slit1, Slit2, and Slit3 (Figure 2C). Moreover, numerous cell-adhesion proteins also follow the expression pattern of cluster 3, which will be discussed in detail later.
Cluster 2 represents a group of proteins whose expression levels constantly increase from DIV1 to DIV5 and from DIV5 to DIV14 (Figure 2A). GO analysis revealed the enrichment of several terms related to cation transport and neurotransmitter secretion in this cluster (Figure 2B), as exemplified by several highly connected subunits of various ATPases (Figure 2C). Furthermore, the majority of the significantly regulated synaptic proteins belong to this cluster (GO term “synaptosome”), which will be discussed in the following sections.
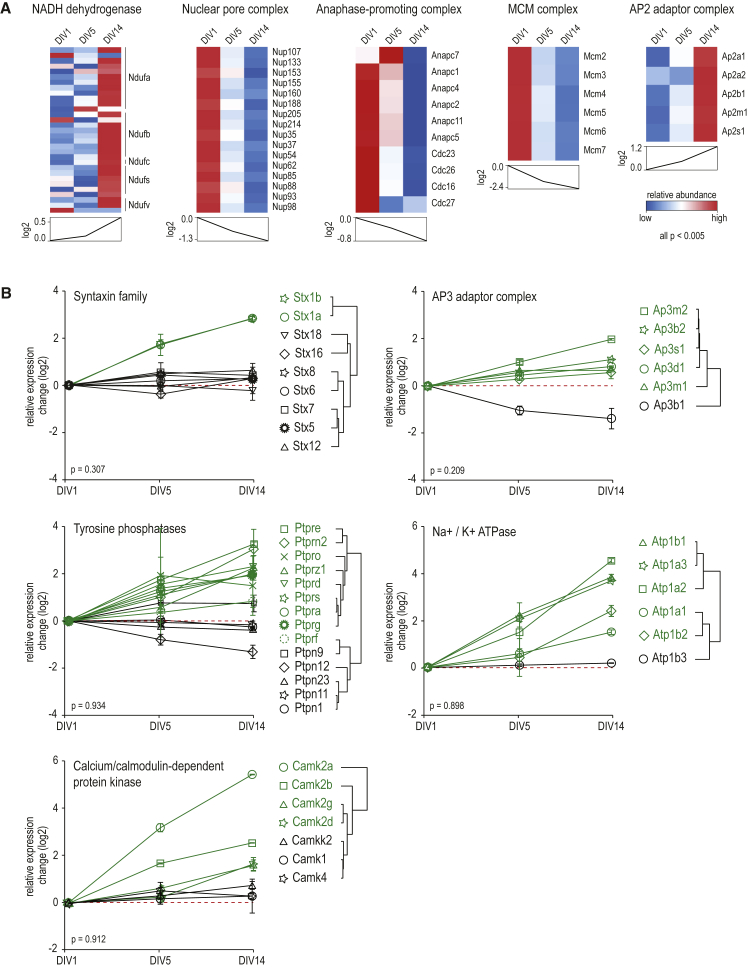
Coordinated Proteome Dynamics of Proteins in Complexes during Differentiation
We next sought to characterize concerted dynamics of proteins within previously annotated protein complexes. Statistical assessment of the significance of co-regulated proteins revealed coordinated expression changes for proteins involved in several cellular processes during neuronal development (Table S3). This includes, for example, nuclear transport (nuclear pore complex), cell-cycle control (anaphase-promoting complex, APC/C), and DNA replication (MCM complex) (all ps < 0.005; Figure 3A), providing further evidence for continuous chromatin remodeling during terminal differentiation of post-mitotic neurons. Conversely, the same statistical analysis can reveal proteins that deviate from the expression profile of the corresponding complex or family, indicating a neuron- or developmental stage-specific function. This is nicely illustrated by expression profile analysis of SNARE proteins from the syntaxin family, revealing neuron-specific expression. Syntaxin proteins are important players in vesicle docking and fusion with target membranes. We observed continuous upregulation of Stx1a and Stx1b from DIV1 to DIV14 (Figure 3B; Figure S3B), both of which are specifically expressed in neuronal and secretory cells, playing a key role in neurotransmitter secretion. All other syntaxins undergo no substantial expression changes (Figure 3B; Figure S3B), indicating that syntaxin-mediated intracellular transport processes, including endosomal (Stx7, Stx8, and Stx12), endoplasmic-reticulum (ER)-to-Golgi- (Stx5 and Stx18), and trans-Golgi (Stx6 and Stx16) trafficking, are not regulated by expression changes of syntaxins in the course of neuronal differentiation.
Figure 3.
Significance Analysis of Protein Expression Profile Similarities for Proteins within Families or Complexes
(A) Examples of tight co-regulation of proteins that are part of larger protein complexes during neuronal development (all ps < 0.001). A complete list is given in Table S3.
(B) The same analysis reveals proteins within complexes or families that undergo divergent expression during differentiation. Hierarchical clustering further differentiates divergent protein expression. Error bars represent SD.
Next, we determined whether this approach was able to resolve neuron-specific isoforms that modulate distinct functions within protein complexes. Adaptor protein (AP) complexes are involved in intracellular vesicular transport and cargo selection by binding to sorting signals. The AP2 complex mainly plays a role in endocytosis from the plasma membrane, whereas AP3 is important for trans-Golgi sorting events (Boehm and Bonifacino, 2001). All subunits of the AP2 adaptor complex show a significant (p < 0.005) coordinated upregulation from DIV1 to DIV14 (Figure 3A), following the expression profile of most synaptic proteins (cluster 2; Figure 2A). Strikingly, this does not hold for the AP3 complex (p = 0.209), where one isoform of the Ap3b subunit (Ap3b1) diverges from the common expression profile of the other complex proteins, including the neuron-specific isoform Ap3b2 (Figure 3B), implying functional regulation of the complex activity by Ap3b. It has recently been shown that neuronal AP3 complexes are assembled by one of the two Ap3b isoforms, regulating distinct vesicular sorting processes (Seong et al., 2005). Neuron-specific Ap3b2-containing AP3 mediates cargo sorting preferentially into synaptic vesicles, while ubiquitous Ap3b1-containing AP3 more likely promotes cargo delivery toward the lysosomes (Newell-Litwa et al., 2009). Moreover, neuronal AP3 was shown to be 4-fold more abundant in axons and dendrites than ubiquitous AP3 (Seong et al., 2005). Together, this substantiates our data across developmental stages 2–3 to 5, which are accompanied with dendrite formation, synaptogenesis, and maturation.
The richness of the neuron-specific proteome allowed us to further explore expression profiles of several other protein families. We observed distinct regulation of members of the tyrosine-protein phosphatase family, where receptor-type tyrosine phosphatases (Ptpr) undergo upregulation during neuronal differentiation, whereas most non-receptor-type tyrosine phosphatases (Ptpn) showed no expression changes (Figure 3B; Figure S3B). Hierarchical clustering further supports this observation by clearly separating the two subclasses. Ptps are important in signal transduction via a highly coordinated interplay with kinases (Tonks, 2013), and both classes are involved in cell-adhesion processes. Upregulation of the receptor-type Ptps implies an increased demand for cell-adhesion molecules in the course of dendrite outgrowth and synapse formation.
Similar observations were made for members of proteins from the Atp1- and Ca2+/calmodulin-dependent protein kinase families. The main function of the Na/K-ATPase complex in neurons is to maintain the resting potential by using large amounts of the total cellular ATP supply. Interestingly, we observed that expression levels of Atp1b3 remain unchanged during neuronal differentiation, whereas all other subunits undergo upregulation, indicating a potential functional regulation through a stoichiometric change of the complex (Figure 3B; Figure S3B) (Ori et al., 2016). The ratio between the α and β subunit is 1:1 during assembly in the ER; however, the degradation rate of β subunits is known to be significantly higher. Moreover, each β isoform confers a different ATPase activity; thus, a change in stoichiometry indicates a potential cellular mechanism to regulate pump activity according to changes in Na+ or K+ concentrations, allowing maintenance of the resting potential or adjustment for changes in cell volume during neuronal differentiation. For the Ca2+/calmodulin-dependent protein kinase family, we observed that Camk2 proteins, overall, upregulated during neuronal differentiation, whereas expression levels of the members of the Camk cascade (Camkk, Camk1, and Camk4) remained unchanged (Figure 3B; Figure S3B). The various Cam kinases differ in their subcellular localization. Camk2 predominantly localizes in dendrites, whereas Camkk and Camk1 are ubiquitous cytoplasmic proteins, and Camk4 is nuclear (Wayman et al., 2008). Camkk phosphorylates and activates Camk1 and Camk4, which act on further downstream targets in the MEK/Erk (mitogen-activated protein kinase/extracellular-signal-related kinase) pathway (Camk1) or modulate CREB (cAMP response element-binding protein)-mediated transcription (Camk4) (Wayman et al., 2008). In contrast, Camk2 is one of the major constituents of the post-synaptic density (PSD) and is a key player in mediating Ca2+-influx-triggered signal transduction, modulation of synaptic plasticity, and induction of long-term potentiation. Notably, Camk2a is the most upregulated protein in our dataset. Our data clearly show an increased demand for PSD-localized Camk2 in growing and developing neurons.
Synaptic and Cell-Adhesion Protein Profiling during Neuronal Development
Next, we assessed the proteome dynamics during synaptogenesis, a key event in neuronal development that requires the coordinated assembly of a highly connected protein network that includes scaffolding proteins, receptors, and their downstream targets, signaling molecules, and cell-adhesion proteins. In our hippocampal neuron cultures, dividing non-neuronal cells are mostly represented by astrocytes. They expand after DIV5 (Figure S4A), thus promoting synapse formation and transmission. Indeed, synapse formation during the neuronal development of hippocampal neurons in culture occurs from stage 4 (DIV5) to stage 5 (DIV14) (Figure 4A). Interestingly, the majority of all differentially expressed synaptic proteins are allocated to cluster 2 (Table S4), which corresponds to a steady increase in relative protein levels from DIV1 to DIV14 (Figure 2A). This cluster includes important presynaptic proteins involved in vesicle trafficking (Syp and Syt1), cytoskeleton organization and protein anchoring (Cask, Pclo, Bsn), and neurotransmitter release (Syn1, Vamp2, Snap25, Sv2a, and Sv2b) (Figure 4C). Prominent members of the upregulated postsynaptic proteins are scaffold and anchoring proteins (Shank2, Psd95, Grip1, Homer1), and several glutamate and GABA receptors (Figure 4D; Figure S4B). To validate our quantitative proteomics data, we additionally plotted the relative expression of several proteins where we do not expect large changes (Figure 4F) and confirmed expression of several depicted proteins by western blot analysis (Figures 4G–4I) of lysates from the corresponding time points. These results were paralleled by immunoblotting of embryonic and adult rat hippocampal tissue, Hip.(E) and Hip.(A), respectively (Figure 4G), substantiating the observation of higher expression levels at later developmental stages. Presynaptic markers such as Bassoon (Bsn) and Synaptophysin (Syp) (Figure 4C) are known to accumulate at new synapses before postsynaptic proteins such as Psd95 (Friedman et al., 2000, Okabe et al., 2001). This is supported by our data, which show a later expression increase of Psd95 (Figure 4D). However, for the majority of postsynaptic proteins, we do not observe a timely delayed expression increase, which is also evident from hierarchical clustering analysis of all synaptic proteins (Figure S4D). This is likely because the temporal resolution of this study is limited by the experimental design. Nevertheless, our dataset demonstrates the substantial upregulation of many synaptic proteins already at early developmental stages, long before synaptogenesis takes place. Similar expression profiles are observed for subunits of various neurotransmitter receptors and voltage-gated ion channels (Figures S4B and S4C).
Cell-adhesion molecules are another important class of proteins with a key role in neuronal development. They mediate axon pathfinding and axon-dendrite contact formation, and they further modulate dendritic spine morphology and synaptic plasticity. In total, 82 cell-adhesion proteins were quantified in our dataset (Table S5), including Cntn1, NrCAM, Nrxn2, NCAM1, and NCAM2 (Figure 4E). Interestingly, 24 of these are allocated to cluster 3 (Figure 2A), highlighting that their major expression increase proceeds during the transition from stage 2–3 to stage 4, indicating an important role in dendrite formation. Neural cell-adhesion molecule 1 (NCAM1) is a prominent member of the cell-adhesion proteins that share the expression profile of cluster 3 (Figure 2A).
NCAM1 Is Highly Enriched in Dendritic Growth Cones
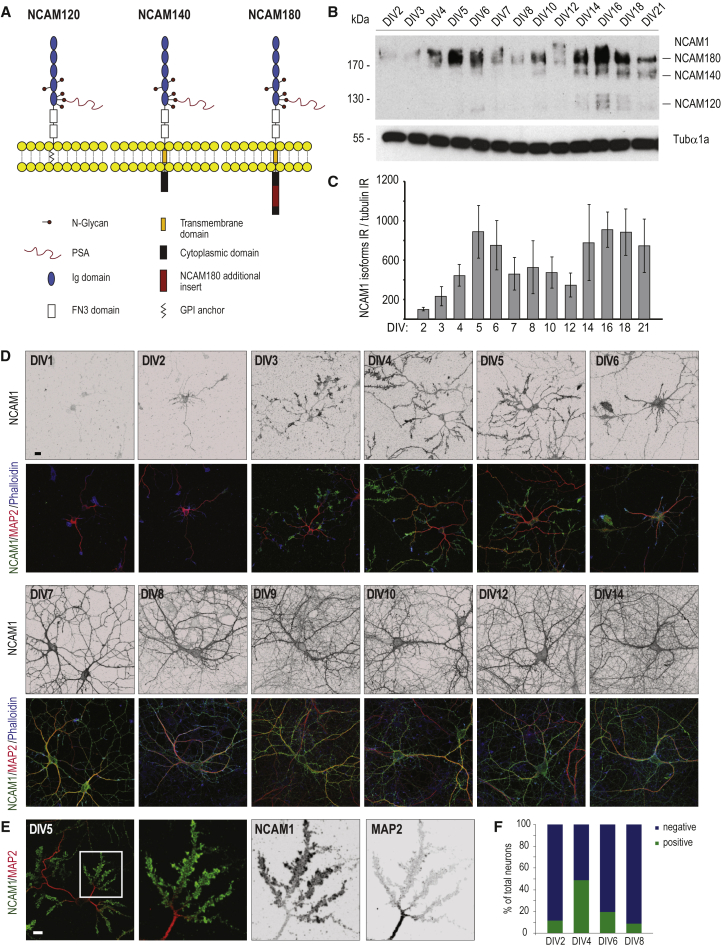
NCAM1 belongs to the immunoglobulin-like family of cell-adhesion molecules and is one of the most abundant neuronal adhesion proteins (Sharma et al., 2015). NCAM1 expresses in three main isoforms produced by alternative splicing of the NCAM1 gene. Two of the isoforms (NCAM140 and NCAM180) are transmembrane proteins, while NCAM120 is glycosylphosphatidylinositol (GPI) anchored (Figure 5A). We first analyzed by western blot isotype-specific protein expression levels in cultured rat hippocampal neurons extracts from DIV2 to DIV21 (Figure 5B). In agreement with previous observations, NCAM180 is the main isoform expressed in neurons (Noble et al., 1985). Quantification revealed that NCAM180 shows two main peaks of protein expression at DIV5 and DIV16 (Figure 5C), confirming our quantitative MS data and indicating that, in addition to its well-known role in synaptic plasticity (Doherty et al., 1995), NCAM1 may be important at earlier stages of neuronal development (DIV5).
Figure 5.
The Cell-Adhesion Molecule NCAM1 and Its Expression throughout Neuronal Development
(A) Schematic overview of the three main NCAM1 isoforms. Ig, immunoglobulin.
(B and C) Western blot analysis (B) and quantification (C) of developmental expression of NCAM1 in DIV2–DIV21 rat hippocampal neurons. Error bars indicate mean ± SEM.
(D) Representative images of rat hippocampal neurons from DIV1 to DIV14, stained for NCAM1 (green), MAP2 (red), and phalloidin (blue). Scale bar, 20 μm.
(E) Representative rat hippocampal neuron at DIV5 stained for NCAM1 (green) and MAP2 (red). Scale bar, 20 μm.
(F) Quantification of the percentage of neurons characterized by the NCAM1 dendritic staining shown in (D) and (E). n = 100 cells per DIV.
To further investigate NCAM1 distribution in developing neurons (DIV1–DIV14), we performed triple-labeling immunofluorescence experiments using a NCAM1-specific antibody, the dendritic microtubule marker MAP2, and the F-actin marker phalloidin (Figure 5D). As reported previously, NCAM1 showed some staining in stage 2–3 cells (DIV1), which was mainly present in the axonal growth cones. Interestingly, at DIV4–DIV5, many neurons displayed strong accumulation of NCAM1 in dendritic growth cones. Here, NCAM1-rich dendritic growth cones are positive for F-actin but not for the microtubule marker MAP2, which is more restricted to the dendritic shaft (Figure 5E). Dendritic NCAM1 accumulations are present in approximately 40% of the neurons counted at DIV4, and during later stages (DIV6–DIV8), it gradually disappears (Figure 5F). At later developmental stages, NCAM1 was found diffusely distributed over the dendritic plasma membrane. Similar results were obtained with the mouse primary hippocampal culture, although here, NCAM1 was already localized in dendrites at DIV3 (Figure S5).
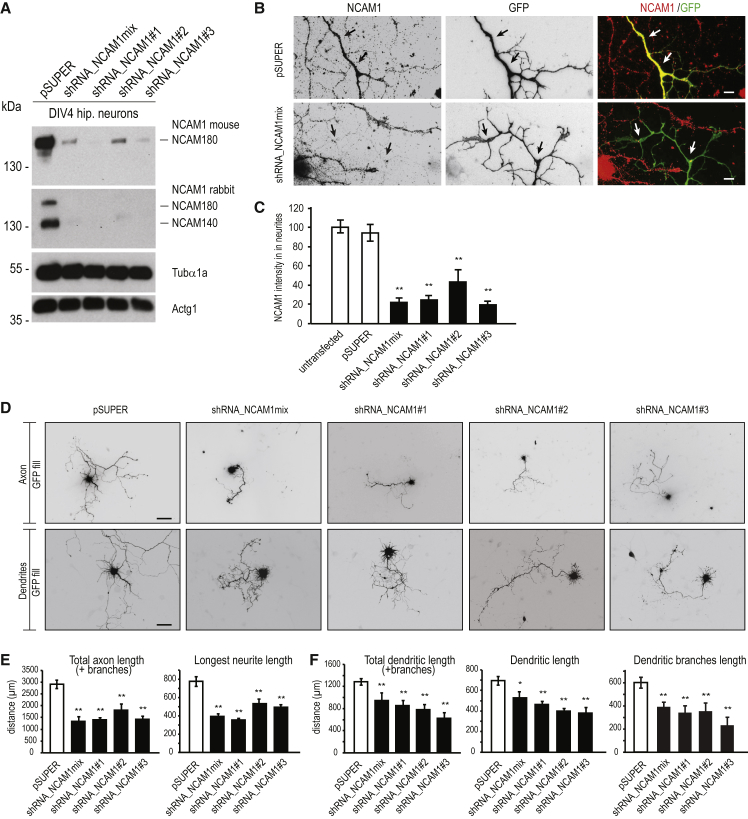
NCAM1 Is Required for Both Axonal and Dendritic Outgrowth and Branching
Since NCAM1 accumulates in dendritic growth cones, we further investigated the role of NCAM1 during dendritic development. Therefore, three specific short hairpin RNA (shRNA) sequences were designed and generated, based on the NCAM180 mRNA rat sequence, to perform knockdown experiments. All three shRNAs (shRNA_NCAM1#1, shRNA_NCAM1#2, and shRNA_NCAM1#3) reduced protein levels by ∼80%, as revealed from both western blot analysis and immunostaining (Figures 6A–6C; Figure S6). We next examined the effect of NCAM1 knockdown on outgrowth of axon and dendrites (Figure 6D). Quantification revealed that both the length of the axon (Figure 6E) and length of the dendrites (Figure 6F) were significantly reduced compared to that of control neurons. The observed reduction of axon length in NCAM1-depleted neurons is consistent with that reported in previous works (Pollerberg et al., 2013). Apart from the axonal phenotype, NCAM1 depletion in young neurons also caused a marked reduction in dendrite development. Quantification indicated that knockdown of NCAM1 using three different shRNAs reduces the length of primary dendrites, total dendrites, and dendritic branches by 40% compared to control neurons (Figure 6F).
Figure 6.
shRNA-Mediated Knockdown of NCAM1 in Neurons
(A) Knockdown of NCAM1 by indicated shRNA constructs and analyzed by western blot in DIV4 cortical neurons.
(B) Representative images of hippocampal neurons at DIV4 co-transfected with indicated constructs and stained with NCAM1 monoclonal antibody. Arrows indicate transfected neurites.
(C) Quantification of NCAM1 fluorescent-staining intensities in neurites of hippocampal neurons co-transfected at DIV4 with indicated constructs.
(D) Representative images of primary hippocampal neurons co-transfected with indicated constructs (DIV1–DIV7) to visualize axon (top) and dendrite (bottom) morphology.
(E and F) Quantification of axon (E) and dendrite (F) morphological parameters.
Error bars indicate mean ± SEM. ∗p < 0.5, t test; ∗∗p < 0.01, t test. Scale bars, 10 μm in (B), 100 μm for the top of (D), and 50 μm for the bottom of (D).
Actin Stabilization Rescues NCAM1 Knockdown Phenotype
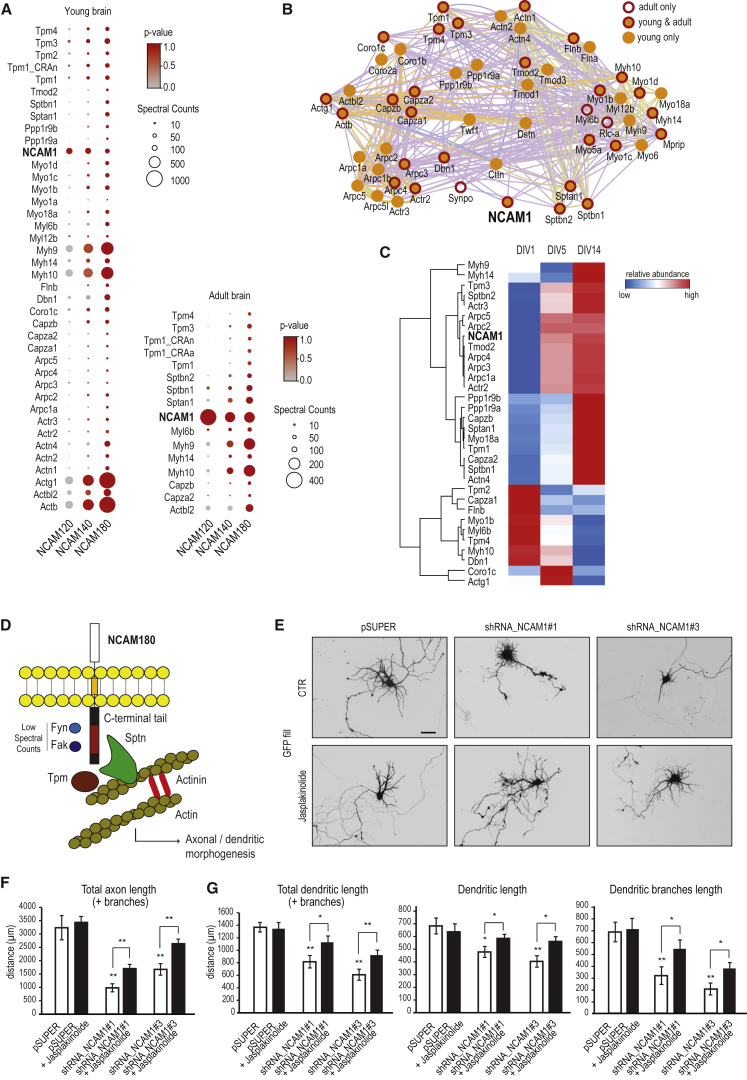
To investigate the mechanism by which NCAM1 influences dendritic development, we searched for NCAM180-binding partners in neurons and used the other two isoforms as controls. The GFP-tagged NCAM1 isoforms were expressed in neurons to check their proper localization and their correct insertion in the cell membranes (Figures S7A and S7B). Subsequently, constructs of all three GFP-NCAM1 isoforms and GFP (as control) were transiently expressed in HEK293 cells; GFP proteins were isolated with GFP beads and incubated with either young (postnatal day 5; P5) or adult rat brain extracts, and then co-isolated proteins were analyzed in an affinity purification (AP)-MS experiment. To identify true NCAM1-binding partners (in adult or young brains), we used the “Significance Analysis of INTeractome (SAINT)” for probabilistic scoring of our AP-MS data (Choi et al., 2011). Among the entire list of putative interacting proteins identified (Table S6), only those with a SAINT probability > 0.75 (false discovery rate [FDR] = 0.1) were considered as true interactors and were then represented as spheres of different size and color according to their relative abundance and obtained p value, respectively (Figure 7A). Interestingly, several members of actin-binding, -stabilizing and/or -polymerizing proteins such as spectrin (Spt) and myosin (Myo/Myh); actin-related proteins (Arps), tropomyosins (Tpms); F-actin-capping proteins (Capzas); and actinins (Actn) are the main neuronal interactors of NCAM1, ubiquitously enriched in young brains and, to a lesser extent, in adult brains. The interaction between the NCAM140 and NCAM180 intracellular tails and actin-cytoskeleton-related proteins (Spt, α-Actn, and Tpm) has been already reported and characterized in previous studies (Pollerberg et al., 2013). In our AP-MS experiments, it is interesting to notice that the length of each NCAM1 isoform used as bait directly correlates with the relative abundance of each specific actin-binding protein identified (as indicated by the amount of peptides and peptide-spectrum matches [PSMs] measured), thus corroborating the hypothesis that NCAM180 (with its longer C terminus) might be the main player involved in the regulation of actin dynamics in dendrites (Figure 7A; Table S6). By drawing a protein interaction network (Cytoscape, Genemania plugin), using as input the actin-binding proteins identified in the NCAM180 pull-down, it becomes evident how these proteins are tightly connected (Figure 7B). Similarly to NCAM1, most actin-interacting proteins are grouped in the earlier defined cluster 3 (Figure 2A) and upregulated at DIV5 (Figure 7C), suggesting that they may strongly cooperate in NCAM1 signaling.
Figure 7.
NCAM1 and Its Role in Actin Cytoskeleton Stabilization
(A) Selected candidates of putative NCAM1 interactors identified in young and adult rat brains by AP-MS experiments (see Table S6 for the complete list of proteins). SAINT probability cutoff > 0.75 corresponds to an average FDR of 0.1. The p values and spectral counts are graphically represented by colors and spheres, respectively.
(B) Network analysis on selected NCAM180-binding proteins linked to actin cytoskeleton. Edge color coding: blue, co-localization; orange, predicted interaction; green, shared protein domain; purple, co-expression.
(C) Heatmap and hierarchical clustering of the expression profiles of selected NCAM1-interacting proteins.
(D) Schematic representation of NCAM180 associated with actin-stabilizing proteins.
(E) Representative images of primary hippocampal neurons co-transfected with indicated constructs (DIV1–DIV7) and treated 24 hr later with DMSO or 10 nM jasplakinolide. Scale bar, 50 μm.
(F and G) Quantification of axon (F) and dendrite (G) morphological parameters.
Error bars indicate mean ± SEM. ∗p < 0.5, t test; ∗∗p < 0.01, t test.
See also Table S6.
To investigate the potential relationship between NCAM1 and the actin cytoskeleton (Figure 7D), we tested whether the dendritic phenotype caused by NCAM1 depletion can be reversed by jasplakinolide-forced F-actin stabilization. Jasplakinolide is a drug known to stabilize actin fibers and to facilitate actin polymerization. Thus, neurons were first transfected with the two most efficient NCAM1 shRNAs (ShRNA_NCAM1#1 and ShRNA_NCAM1#3), and 24 hr later, a low dose of jasplakinolide (10nM) was added to the growth medium for 6 days. The addition of jasplakinolide partially rescues the neuronal developmental defects caused by NCAM1 depletion, as demonstrated by the quantification of the axonal and dendritic parameters (Figures 7E–7G). Total dendritic length, primary dendrite length, and dendritic branch length were significantly increased in jasplakinolide-treated neurons compared to control neurons (Figure 7G). Together, the presented data indicate actin stabilization as a mechanism for NCAM1-mediated dendritic growth.
Discussion
The in-depth analysis of neuronal proteins that are abundantly and differentially expressed during the well-defined stages of differentiation is a promising strategy for understanding neurodevelopment-regulated processes. We provide proof of concept by applying this strategy to identify and characterize the role of NCAM1 in dendritic outgrowth.
A Quantitative Map of Proteome Dynamics during Neuronal Differentiation
In neurons, like in all other cells, proteins are the main functional components, but how the proteomes differ in developing neurons is not known. To resolve the global neuronal proteome and to determine the basis for cellular differentiation, we performed a quantitative analysis of protein levels by combining triplex stable-isotope dimethyl labeling coupled to high-resolution LC-MS/MS of hippocampal neurons in culture. Here, we found that 1,793 proteins show more than 2-fold expression changes during the different developmental steps, indicating extensive remodeling of the neuron proteome during early differentiation. This analysis revealed six clusters of distinct expression profiles, with proteins upregulated during differentiation in clusters 1, 2, and 3 and proteins downregulated present in clusters 4, 5, and 6. We mainly focused our attention on cluster 3, which contains proteins whose expression levels are highly upregulated between stages 2–3 and stage 4 of in vitro neuronal development. According to GO classification, most of the proteins present in this cluster are transmembrane proteins that could play a role by modulating dendritic outgrowth and branching. It is interesting to notice that, during these stages of neuronal development, a very specific group of NCAM-adhesion molecules (NCAM1, NCAM2, NRCAM, and L1CAM) is characterized by exactly the same pattern of protein-level regulation (Figure 4E), suggesting that these proteins may act synergistically. On the other hand, pre- and postsynaptic proteins are highly represented in cluster 2, indicating that their expression levels increase throughout neuronal development. These data strongly support a model in which specific synaptic scaffolding proteins such as Bassoon, Piccolo (pre-synaptic proteins), Shank, and Camk2A (post-synaptic proteins) are expressed prior to synapse formation. The full datasets are available in Tables S1, S2, S3, S4, and S5.
NCAM1 Plays an Important Role in Dendritic Outgrowth
To demonstrate the strength of our resource, we focused on NCAM1 as a regulator for dendritic outgrowth. NCAM1 is best known for its role in axonal wiring through interactions with various ligands via its extracellular domain (Pollerberg et al., 2013). NCAM1 is unique among the adhesion molecules because it carries a polysialic acid (Togashi et al., 2009), which can be reversibly attached to extracellular domains, reducing its homophilic interactions and thereby controlling clustering and downstream signaling of NCAM1 (Pollerberg et al., 2013). The cytosolic part of the longer NCAM1 isoform (NCAM180) interacts with several cytoskeletal elements and signaling molecules. NCAM1 is reportedly involved in neurite outgrowth and axon pathfinding (Walsh and Doherty, 1997). For example, in mice lacking NCAM1, hippocampal axons show abnormal pathfinding (Cremer et al., 1997). Our data suggest that NCAM1 not only plays a role in axon outgrowth and guidance but is also required for proper dendritic outgrowth during first days of neuronal development. We speculate that NCAM180 is the specific isoform playing a role in dendrites based on the developmental expression pattern. Consistently, NCAM180 is strongly upregulated during dendrite outgrowth and highly enriched in dendritic growth cones. Interestingly, other NCAM family proteins have been reported to be involved in dendritic branching and morphology in C. elegans (Dong et al., 2013). It is likely that, similar to its role in axonal growth cones, NCAM1 at the tips of dendrites may sense various types of extrinsic signals, such as other CAMs or various diffusible factors that determine their general orientation and outgrowth (Pollerberg et al., 2013). The extracellular signals may come from crossing axons, dendrites from the same cell, and neighboring arbors and together regulate the immense complexity of dendrite morphology.
Actin Stabilization as a Mechanism for NCAM1-Mediated Dendritic Growth
Our AP-MS experiments revealed that NCAM180 interacts with several members of actin-binding, -stabilizing and/or -polymerizing proteins in young and adult brains. These data are consistent with the previously reported interactions between the NCAM1 intracellular tails and actin-related proteins (Büttner et al., 2003, Leshchyns’ka and Sytnyk, 2016). These observations suggest that NCAM1 can act as a scaffold for multiple cytoskeleton components and that multiple actin-binding proteins can amplify the interaction between NCAM1 with the actin cytoskeleton. However, it is also possible that these interactions do not occur simultaneously but are rather highly regulated in response to the extracellular signals (Leshchyns’ka and Sytnyk, 2016). Interestingly, inducing actin polymerization by jasplakinolide treatment partially rescues the dendritic phenotype of NCAM1 depletion. These results indicate that forced actin stabilization can compensate for the absence of NCAM1 and is one of the driving forces that act during dendrite morphogenesis. These data suggest a model in which NCAM1 stimulates dendritic arbor development by promoting actin filament stabilization at the dendritic growth cone.
In summary, the analysis of proteins differentially expressed at each specific stage of hippocampal neuron development constitutes a fundamental research tool that may be used in the future for a better understanding of any specific molecular mechanism involved in neurodevelopment. Our quantitative proteomics results give a comprehensive overview of protein abundances in time, thereby providing a unique database regarding protein expression patterns in cultured neurons. Furthermore, thanks to the recent technological advances in MS-based proteomics (tandem mass tags [TMTs] or iTRAQ isobaric mass tags would allow relative quantification of up to ten different time points), this analysis could also constitute an excellent starting point for other studies that aim to further complement our understanding of neuronal protein dynamics.
Experimental Procedures
Animals
All experiments with animals were performed in compliance with the guidelines for the welfare of experimental animals issued by the Government of the Netherlands and were approved by the Animal Ethical Review Committee (DEC) of Utrecht University.
Expression Vectors, shRNA Constructs, and Antibodies
The NCAM120, NCAM140, and NCAM180 constructs were kindly provided by Dr. Landmesser. The following NCAM1 shRNAs were designed and used in this study: NCAM1#1 (5′-GGATCTCATCTGGACTTTG), NCAM1#2 (5′-GATCTTCCAGAAGCTCATG), and NCAM1#3 (CGTTGGAGAGTCCAAATTC) targeting rat NCAM1 mRNA (NM_031521.1). NCAM1 mouse (Millipore) and NCAM1 rabbit (Proteintech) antibodies were used for western blot and immunocytochemistry experiments. For further details, see Supplemental Experimental Procedures.
Primary Hippocampal Neuron Cultures
Primary hippocampal and cortical cultures were isolated from embryonic day 18 (E18) rat brains. Cells were plated on coverslips coated with poly-L-lysine (30 μg/mL) and laminin (2 μg/mL) at a density of 100,000 per well. Hippocampal neurons were transfected at DIV1 using Lipofectamine 2000 (Invitrogen). Cortical neurons were transfected using the Amaxa Rat Neuron Nucleofector Kit (Lonza). For further details, see Supplemental Experimental Procedures.
Sample Preparation, Peptide Fractionation, MS, and Data Analysis
The MS methods and any associated references are available in the online version of this paper.
Statistical Methods
Analysis of significance of proteins changing between developmental stages was carried out using the significance analysis of microarrays (SAM). Clustering was performed using unsupervised fuzzy clustering. Gene Ontology enrichment analyses were performed using a Fisher’s exact test. Co-regulation of proteins within complexes or families was assessed using the R package protein profiles. coIP data were analyzed using the SAINT. For morphometric analyses of hippocampal neurons, statistical significance was determined using Student’s t test assuming a two-tailed variation. The graphs represent mean ± SEM. For further details, see Supplemental Experimental Procedures.
Author Contributions
C.K.F. designed and performed the quantitative proteomics experiments and analyzed the data; Q.L. conducted proteomics sample preparation; M.M. and R.S. designed and performed immunohistochemistry and biochemical experiments and analyzed the data. R.S. and V.G. performed AP/MS experiments, and C.K.F. and R.S. analyzed the results. S.M. and A.J.R.H. supervised the quantitative proteomics experimental setup and the mass spectrometry data. The figures were designed and assembled by C.K.F., M.M., R.S., A.F.M.A., and C.C.H. The manuscript was written by C.K.F., M.M., R.S., A.F.M.A., and C.C.H., with input from A.J.R.H.; A.F.M.A. and C.C.H. supervised the project and coordinated the study.
Acknowledgments
We thank Dr. L.T. Landmesser for sharing the NCAM120, NCAM140, and NCAM180 constructs. This work was supported by the Netherlands Organization for Scientific Research (NWO) (NWO-ALW-VICI 865.10.010 to C.C.H. and NWO-CW-VIDI 723.012.102 to A.F.M.A.) and Proteins@Work, a program of the NWO, as part of the National Roadmap Large-Scale Research Facilities of the Netherlands (project number 184.032.201); the Foundation for Fundamental Research on Matter (FOM programme number 137; to C.C.H.), which is part of the NWO; the Netherlands Organization for Health Research and Development (ZonMW-TOP 91213017 and 91215084; to C.C.H.); the European Research Council (ERC) (ERC Consolidator grant 617050 to C.C.H.); Marie-Curie Actions (FP7-MC-IEF 300814; to M.M.), and the DFG Emmy-Noether Programm (Ml 1923/1-1 to M.M.).
Published: February 7, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2017.01.025.
Contributor Information
A.F. Maarten Altelaar, Email: m.altelaar@uu.nl.
Casper C. Hoogenraad, Email: c.hoogenraad@uu.nl.
Accession Numbers
The accession number for the mass spectrometry proteomics data reported in this paper is PRIDE: PXD005031 (Vizcaíno et al., 2016).
Supplemental Information
Contains the complete proteomics data set containing all identified proteins. ID = unique identifier for each protein; Uniprot = Uniprot accession code; gene name = corresponding gene name; Description = Protein name/description derived from ∗.fasta database; Σ# Unique Peptides = sum of unique peptides per protein; repl.1 DIV5/DIV1 = log2 fold change between DIV5 and DIV1 from replicate 1; repl.2 DIV5/DIV1 = log2 fold change between DIV5 and DIV1 from replicate 2; repl.1 DIV14/DIV5 = log2 fold change between DIV14 and DIV5 from replicate 1; repl.2 DIV14/DIV5 = log2 fold change between DIV14 and DIV5 from replicate 2; quantified = if a protein was quantified at all time points and in both replicates it is flagged “all”; average log2 DIV5/DIV1 = the average log2 fold-change between DIV5 and DIV1; average log2 DIV14/DIV5 = the average log2 fold-change between DIV14 and DIV5; SAM q-value [%] = q-value derived from statistical analysis of global proteome changes (lower values correspond to higher statistical significance); GProX cluster = cluster derived from fuzzy clustering analysis, related to Figure 2; Protein family = Protein family; Panther protein class = protein class retrieved from pantherdb.org; GO molecular function = Gene ontology molecular function; GO biological process = Gene ontology biological process; GO cellular component = Gene ontology cellular component.
Contains quantitative information about all RNA binding proteins (for column key see description of Table S1).
Contains the information on coordinated proteome dynamics of protein families or proteins in complexes. Category = name of protein complex, protein family or gene ontology term; p.value = computed p value for assessment of significance of co-regulation of the corresponding proteins. P-values were computed using the ‘proteinProfile’ package within R/bioconductor; ID = unique identified matching to table S1; Uniprot = Uniprot accession code; Gene.name = gene name; Protein.name = corresponding protein name; average log2 DIV5/DIV1 = the average log2 fold-change between DIV5 and DIV1; average log2 DIV14/DIV5 = the average log2 fold-change between DIV14 and DIV5.
Contains quantitative information about synaptic proteins (for column key see description of Table S1).
Contains quantitative information about cell adhesion proteins (for column key see description of Table S1).
Contains data from affinity purification mass spectrometry analysis of NCAM1 IPs in adult and young rat brain extracts. Uniprot = Uniprot accession code; gene name = gene name; Description = description of the protein; Σ# PSMs = sum of all peptide-spectrum-matches (PSMs) across all IPs; GFP_control = number of PSM from GFP-only control IP; NCAM120/140/180 = number of PSM from NCAM120/NCAM140/NCAM180 IPs; p value NCAM120/140/180 = probability score from SAINT (Significance Analysis of INTeractions, version 2.3.2, http://dx.doi.org/10.1038%2Fnmeth.1541) analysis. 0≤p≤1, the higher the score the higher the probability for a given interaction with NCAM1; SAINT probability >0.75 = proteins with a SAINT probability score >0.75 are flagged as ‘TRUE’.
References
- Boehm M., Bonifacino J.S. Adaptins: the final recount. Mol. Biol. Cell. 2001;12:2907–2920. doi: 10.1091/mbc.12.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersema P.J., Raijmakers R., Lemeer S., Mohammed S., Heck A.J.R. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- Büttner B., Kannicht C., Reutter W., Horstkorte R. The neural cell adhesion molecule is associated with major components of the cytoskeleton. Biochem. Biophys. Res. Commun. 2003;310:967–971. doi: 10.1016/j.bbrc.2003.09.105. [DOI] [PubMed] [Google Scholar]
- Choi H., Larsen B., Lin Z.Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z.S., Tyers M., Gingras A.C., Nesvizhskii A.I. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer H., Chazal G., Goridis C., Represa A. NCAM is essential for axonal growth and fasciculation in the hippocampus. Mol. Cell. Neurosci. 1997;8:323–335. doi: 10.1006/mcne.1996.0588. [DOI] [PubMed] [Google Scholar]
- Dabrowski M., Aerts S., Van Hummelen P., Craessaerts K., De Moor B., Annaert W., Moreau Y., De Strooper B. Gene profiling of hippocampal neuronal culture. J. Neurochem. 2003;85:1279–1288. doi: 10.1046/j.1471-4159.2003.01753.x. [DOI] [PubMed] [Google Scholar]
- DeBoer E.M., Kraushar M.L., Hart R.P., Rasin M.R. Post-transcriptional regulatory elements and spatiotemporal specification of neocortical stem cells and projection neurons. Neuroscience. 2013;248:499–528. doi: 10.1016/j.neuroscience.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty P., Fazeli M.S., Walsh F.S. The neural cell adhesion molecule and synaptic plasticity. J. Neurobiol. 1995;26:437–446. doi: 10.1002/neu.480260315. [DOI] [PubMed] [Google Scholar]
- Dominguez M.H., Ayoub A.E., Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb. Cortex. 2013;23:2632–2643. doi: 10.1093/cercor/bhs252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Liu O.W., Howell A.S., Shen K. An extracellular adhesion molecule complex patterns dendritic branching and morphogenesis. Cell. 2013;155:296–307. doi: 10.1016/j.cell.2013.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti C.G., Sullivan C.A., Banker G.A. The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman H.V., Bresler T., Garner C.C., Ziv N.E. Assembly of new individual excitatory synapses: time course and temporal order of synaptic molecule recruitment. Neuron. 2000;27:57–69. doi: 10.1016/s0896-6273(00)00009-x. [DOI] [PubMed] [Google Scholar]
- Haslinger A., Schwarz T.J., Covic M., Lie D.C. Expression of Sox11 in adult neurogenic niches suggests a stage-specific role in adult neurogenesis. Eur. J. Neurosci. 2009;29:2103–2114. doi: 10.1111/j.1460-9568.2009.06768.x. [DOI] [PubMed] [Google Scholar]
- Hevner R.F., Hodge R.D., Daza R.A.M., Englund C. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci. Res. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Hsieh J. Orchestrating transcriptional control of adult neurogenesis. Genes Dev. 2012;26:1010–1021. doi: 10.1101/gad.187336.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen A.R., Gsponer J., Foster L.J. Protein synthesis rate is the predominant regulator of protein expression during differentiation. Mol. Syst. Biol. 2013;9:689. doi: 10.1038/msb.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T.Y., Hong C.J., Hsueh Y.P. Bcl11A/CTIP1 regulates expression of DCC and MAP1b in control of axon branching and dendrite outgrowth. Mol. Cell. Neurosci. 2009;42:195–207. doi: 10.1016/j.mcn.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Leshchyns’ka I., Sytnyk V. Reciprocal interactions between cell adhesion molecules of the immunoglobulin superfamily and the cytoskeleton in neurons. Front. Cell Dev. Biol. 2016;4:9. doi: 10.3389/fcell.2016.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D.D., Yano M., Fak J.J., Mele A., Grabinski S.E., Zhang C., Darnell R.B. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low T.Y., van Heesch S., van den Toorn H., Giansanti P., Cristobal A., Toonen P., Schafer S., Hübner N., van Breukelen B., Mohammed S. Quantitative and qualitative proteome characteristics extracted from in-depth integrated genomics and proteomics analysis. Cell Rep. 2013;5:1469–1478. doi: 10.1016/j.celrep.2013.10.041. [DOI] [PubMed] [Google Scholar]
- Loya C.M., Van Vactor D., Fulga T.A. Understanding neuronal connectivity through the post-transcriptional toolkit. Genes Dev. 2010;24:625–635. doi: 10.1101/gad.1907710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Gómez H.R., Vergaño-Vera E., Abad J.L., Bulfone A., Moratalla R., de Pablo F., Vicario-Abejón C. The T-box brain 1 (Tbr1) transcription factor inhibits astrocyte formation in the olfactory bulb and regulates neural stem cell fate. Mol. Cell. Neurosci. 2011;46:108–121. doi: 10.1016/j.mcn.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Mody M., Cao Y., Cui Z., Tay K.Y., Shyong A., Shimizu E., Pham K., Schultz P., Welsh D., Tsien J.Z. Genome-wide gene expression profiles of the developing mouse hippocampus. Proc. Natl. Acad. Sci. USA. 2001;98:8862–8867. doi: 10.1073/pnas.141244998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Litwa K., Salazar G., Smith Y., Faundez V. Roles of BLOC-1 and adaptor protein-3 complexes in cargo sorting to synaptic vesicles. Mol. Biol. Cell. 2009;20:1441–1453. doi: 10.1091/mbc.E08-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble M., Albrechtsen M., Møller C., Lyles J., Bock E., Goridis C., Watanabe M., Rutishauser U. Glial cells express N-CAM/D2-CAM-like polypeptides in vitro. Nature. 1985;316:725–728. doi: 10.1038/316725a0. [DOI] [PubMed] [Google Scholar]
- Okabe S., Miwa A., Okado H. Spine formation and correlated assembly of presynaptic and postsynaptic molecules. J. Neurosci. 2001;21:6105–6114. doi: 10.1523/JNEUROSCI.21-16-06105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori A., Iskar M., Buczak K., Kastritis P., Parca L., Andrés-Pons A., Singer S., Bork P., Beck M. Spatiotemporal variation of mammalian protein complex stoichiometries. Genome Biol. 2016;17:47. doi: 10.1186/s13059-016-0912-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollerberg G.E., Thelen K., Theiss M.O., Hochlehnert B.C. The role of cell adhesion molecules for navigating axons: density matters. Mech. Dev. 2013;130:359–372. doi: 10.1016/j.mod.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Sawicka K., Bushell M., Spriggs K.A., Willis A.E. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem. Soc. Trans. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- Seong E., Wainer B.H., Hughes E.D., Saunders T.L., Burmeister M., Faundez V. Genetic analysis of the neuronal and ubiquitous AP-3 adaptor complexes reveals divergent functions in brain. Mol. Biol. Cell. 2005;16:128–140. doi: 10.1091/mbc.E04-10-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Schmitt S., Bergner C.G., Tyanova S., Kannaiyan N., Manrique-Hoyos N., Kongi K., Cantuti L., Hanisch U.K., Philips M.A. Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci. 2015;18:1819–1831. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt C.C., Lommes P., Friedrich R.P., Wegner M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development. 2004;131:2349–2358. doi: 10.1242/dev.01114. [DOI] [PubMed] [Google Scholar]
- Togashi H., Sakisaka T., Takai Y. Cell adhesion molecules in the central nervous system. Cell Adhes. Migr. 2009;3:29–35. doi: 10.4161/cam.3.1.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks N.K. Protein tyrosine phosphatases--from housekeeping enzymes to master regulators of signal transduction. FEBS J. 2013;280:346–378. doi: 10.1111/febs.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spronsen M., van Battum E.Y., Kuijpers M., Vangoor V.R., Rietman M.L., Pothof J., Gumy L.F., van Ijcken W.F.J., Akhmanova A., Pasterkamp R.J., Hoogenraad C.C. Developmental and activity-dependent miRNA expression profiling in primary hippocampal neuron cultures. PLoS ONE. 2013;8:e74907. doi: 10.1371/journal.pone.0074907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh F.S., Doherty P. Neural cell adhesion molecules of the immunoglobulin superfamily: role in axon growth and guidance. Annu. Rev. Cell Dev. Biol. 1997;13:425–456. doi: 10.1146/annurev.cellbio.13.1.425. [DOI] [PubMed] [Google Scholar]
- Wayman G.A., Lee Y.S., Tokumitsu H., Silva A.J., Soderling T.R. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.K., Johnson E.M., Khalili K. Multiple roles for Puralpha in cellular and viral regulation. Cell Cycle. 2009;8:1–7. doi: 10.4161/cc.8.3.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Herrup K. Nucleocytoplasmic Cdk5 is involved in neuronal cell cycle and death in post-mitotic neurons. Cell Cycle. 2011;10:1208–1214. doi: 10.4161/cc.10.8.15328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contains the complete proteomics data set containing all identified proteins. ID = unique identifier for each protein; Uniprot = Uniprot accession code; gene name = corresponding gene name; Description = Protein name/description derived from ∗.fasta database; Σ# Unique Peptides = sum of unique peptides per protein; repl.1 DIV5/DIV1 = log2 fold change between DIV5 and DIV1 from replicate 1; repl.2 DIV5/DIV1 = log2 fold change between DIV5 and DIV1 from replicate 2; repl.1 DIV14/DIV5 = log2 fold change between DIV14 and DIV5 from replicate 1; repl.2 DIV14/DIV5 = log2 fold change between DIV14 and DIV5 from replicate 2; quantified = if a protein was quantified at all time points and in both replicates it is flagged “all”; average log2 DIV5/DIV1 = the average log2 fold-change between DIV5 and DIV1; average log2 DIV14/DIV5 = the average log2 fold-change between DIV14 and DIV5; SAM q-value [%] = q-value derived from statistical analysis of global proteome changes (lower values correspond to higher statistical significance); GProX cluster = cluster derived from fuzzy clustering analysis, related to Figure 2; Protein family = Protein family; Panther protein class = protein class retrieved from pantherdb.org; GO molecular function = Gene ontology molecular function; GO biological process = Gene ontology biological process; GO cellular component = Gene ontology cellular component.
Contains quantitative information about all RNA binding proteins (for column key see description of Table S1).
Contains the information on coordinated proteome dynamics of protein families or proteins in complexes. Category = name of protein complex, protein family or gene ontology term; p.value = computed p value for assessment of significance of co-regulation of the corresponding proteins. P-values were computed using the ‘proteinProfile’ package within R/bioconductor; ID = unique identified matching to table S1; Uniprot = Uniprot accession code; Gene.name = gene name; Protein.name = corresponding protein name; average log2 DIV5/DIV1 = the average log2 fold-change between DIV5 and DIV1; average log2 DIV14/DIV5 = the average log2 fold-change between DIV14 and DIV5.
Contains quantitative information about synaptic proteins (for column key see description of Table S1).
Contains quantitative information about cell adhesion proteins (for column key see description of Table S1).
Contains data from affinity purification mass spectrometry analysis of NCAM1 IPs in adult and young rat brain extracts. Uniprot = Uniprot accession code; gene name = gene name; Description = description of the protein; Σ# PSMs = sum of all peptide-spectrum-matches (PSMs) across all IPs; GFP_control = number of PSM from GFP-only control IP; NCAM120/140/180 = number of PSM from NCAM120/NCAM140/NCAM180 IPs; p value NCAM120/140/180 = probability score from SAINT (Significance Analysis of INTeractions, version 2.3.2, http://dx.doi.org/10.1038%2Fnmeth.1541) analysis. 0≤p≤1, the higher the score the higher the probability for a given interaction with NCAM1; SAINT probability >0.75 = proteins with a SAINT probability score >0.75 are flagged as ‘TRUE’.