Abstract
Using a test-negative design, the Canadian Sentinel Practitioner Surveillance Network (SPSN) assessed interim 2016/17 influenza vaccine effectiveness (VE) against dominant influenza A(H3N2) viruses considered antigenically matched to the clade 3C.2a vaccine strain. Sequence analysis revealed substantial heterogeneity in emerging 3C.2a1 variants by province and over time. Adjusted VE was 42% (95% confidence interval: 18–59%) overall, with variation by province. Interim virological and VE findings reported here warrant further investigation to inform potential vaccine reformulation.
Keywords: influenza, influenza virus, influenza-like illness - ILI, vaccine-preventable diseases, vaccines and immunisation, effectiveness
The 2016/17 season in Canada has been characterised by dominant influenza A(H3N2) activity, increasing since late November 2016 but with regional variation in timing and intensity from west to east [1]. We assessed interim 2016/17 vaccine effectiveness (VE) against influenza A(H3N2) viruses collected through the Canadian Sentinel Practitioner Surveillance Network (SPSN). Detailed genetic characterisation of sentinel viruses was undertaken to assess the contribution of emerging clade 3C.2a1 variants and their potential impact on protection conferred by the clade 3C.2a vaccine, specifically the A/Hong Kong/4801/2014(H3N2)-like component.
Virological and vaccine effectiveness evaluation
As previously described [2,3], nasal/nasopharyngeal specimens collected from patients aged 1 year and older presenting within 7 days of influenza-like illness (ILI) onset to community-based sentinel practitioners in four provinces (Alberta, British Columbia, Ontario and Quebec) were included in the interim analysis. Epidemiological information was collected at the time of specimen collection using a standard questionnaire. Ethics review boards in each province approved the study.
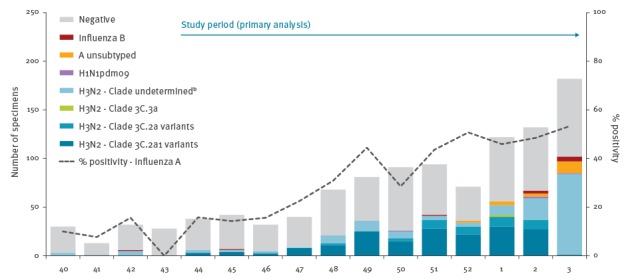
Specimens collected between 1 November 2016 (week 44) and 21 January 2017 (week 3) were included in primary VE analysis, corresponding to the period during which influenza test positivity consistently exceeded 10% (Figure 1).
Figure 1.
Influenza detections by type/subtype/clade and week of specimen collection, Canadian Sentinel Practitioner Surveillance Network, 2 October 2016–21 January 2017 (n = 1,096)a
a Specimens were included if the patient: (i) met the influenza-like illness case definition (requiring fever and cough and at least one or more of sore throat, arthralgia, myalgia or prostration; fever was not required in elderly patients ≥ 65 years-old), (ii) had specimen collection ≤ 7 days after symptom onset, (iii) was ≥ 1-year-old at the time of onset, and (iv) had valid laboratory RT-PCR results. Specimens are displayed in the epidemic curve regardless of the patient’s vaccination status or timing of vaccination. Missing specimen collection dates were imputed as the date the specimen was received and processed at the provincial laboratory minus two days, the average time between specimen collection date and laboratory received date among specimens with complete information for both values.
b Specimens with undetermined clade included those where sequencing was attempted but failed (n = 42) or sequencing was not attempted, e.g. those with insufficient viral load (n = 8), submitted after the start of the mid-season analysis on 21 January 2017 (n = 99), or excluded from primary vaccine effectiveness (VE) analysis (n=23).
Influenza virus testing and influenza A subtyping were conducted using real-time RT-PCR assays validated for use at provincial reference laboratories, including in-house assays in Alberta [4] and British Columbia [5] and commercial assays in Ontario [6] and Quebec [7]. Sequencing of the haemagglutinin (HA) gene was attempted directly on all influenza A(H3N2)-positive patient specimens contributing to VE analysis that had sufficient viral load and that were available up to 21 January 2017 in order to determine clade designation and to identify mutations in established antigenic sites labelled A–E for H3N2 viruses [8,9].
VE was derived using a test-negative design [2,3]. Patients testing positive for influenza A(H3N2) were considered cases; those testing negative were considered controls. Patients who self-reported receiving at least one dose of influenza vaccine at least 2 weeks before ILI onset were considered vaccinated; those vaccinated less than 2 weeks before onset or who had unknown vaccination status or timing were excluded. Patients who did not meet the ILI case definition, those with specimen collection more than 7 days since ILI onset or ILI onset date unknown and those with indeterminate RT-PCR results were also excluded. Odds ratios (OR) were estimated using a logistic regression model, adjusted for age group, province, time from onset to specimen collection and specimen collection date (grouped into 2-week intervals). VE was derived as (1–OR) × 100%, comparing influenza A(H3N2) test positivity between vaccinated and unvaccinated participants.
Virological and vaccine effectiveness findings
A total of 932 specimens met study inclusion criteria. Influenza viruses were detected in 396 (42%) specimens, including 387 (98%) influenza A and nine (2%) influenza B. Of the 374 (97%) influenza A viruses with available subtype information, almost all (n = 370; 99%) were A(H3N2); four A(H1N1)pdm09 viruses were detected. VE analyses are presented for A(H3N2) only, including 370 test-positive cases and 536 test-negative controls (n = 906 overall). Working-age adults 20–64-years-old comprised the majority (57%) of the study sample (Table 1).
Table 1. Participant characteristics, interim vaccine effectiveness evaluation, Canadian Sentinel Practitioner Surveillance Network, 1 November 2016–21 January 2017 (n = 906).
| Characteristic | Overall % (column) |
Distribution by case status % (column) |
Vaccinated % (row) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H3N2 cases | Negative controls |
p valuea | Overall | p valuea | H3N2 cases | Negative controls | ||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | |||
| n % (row) | 906 | 100 | 370 | 41 | 536 | 59 | NA | 246 | 27 | NA | 87 | 24 | 159 | 30 |
| Age group (years) | ||||||||||||||
| 1–8 | 137 | 15 | 51 | 14 | 86 | 16 | 0.19 |
24 | 18 | < 0.01 |
8 | 16 | 16 | 19 |
| 9–19 | 133 | 15 | 66 | 18 | 67 | 13 | 18 | 14 | 8 | 12 | 10 | 15 | ||
| 20–49 | 359 | 40 | 141 | 38 | 218 | 41 | 74 | 21 | 26 | 18 | 48 | 22 | ||
| 50–64 | 155 | 17 | 59 | 16 | 96 | 18 | 54 | 35 | 17 | 29 | 37 | 39 | ||
| ≥ 65 | 122 | 13 | 53 | 14 | 69 | 13 | 76 | 62 | 28 | 53 | 48 | 70 | ||
| Median (range) | 34 (1–97) | 34 (1–91) | 35 (1–97) | 0.99 | 52.5 (1–97) | < 0.01 | 50 (1–90) | 53 (1–97) | ||||||
| Sex | ||||||||||||||
| Female | 524 | 58 | 205 | 56 | 319 | 60 | 0.20 |
154 | 29 | 0.09 |
44 | 21 | 110 | 34 |
| Male | 378 | 42 | 164 | 44 | 214 | 40 | 92 | 24 | 43 | 26 | 49 | 23 | ||
| Unknown | 4 | NA | 1 | NA | 3 | NA | NA | 0 | NA | NA | 0 | NA | 0 | NA |
| Co-morbidityb | ||||||||||||||
| No | 664 | 80 | 270 | 81 | 394 | 79 | 0.52 |
147 | 22 | < 0.01 |
49 | 18 | 98 | 25 |
| Yes | 166 | 20 | 63 | 19 | 103 | 21 | 77 | 46 | 28 | 44 | 49 | 48 | ||
| Unknown | 76 | NA | 37 | NA | 39 | NA | NA | 22 | NA | NA | 10 | NA | 12 | NA |
| Province | ||||||||||||||
| Alberta | 278 | 31 | 110 | 30 | 168 | 31 | 0.03 |
71 | 26 | < 0.01 |
20 | 18 | 51 | 30 |
| British Columbia | 327 | 36 | 134 | 36 | 193 | 36 | 92 | 28 | 37 | 28 | 55 | 29 | ||
| Ontario | 179 | 20 | 87 | 24 | 92 | 17 | 64 | 36 | 25 | 29 | 39 | 42 | ||
| Quebec | 122 | 13 | 39 | 11 | 83 | 15 | 19 | 16 | 5 | 13 | 14 | 17 | ||
| Specimen collection interval from ILI onset (days)c | ||||||||||||||
| ≤ 4 | 687 | 76 | 316 | 85 | 371 | 69 | < 0.01 |
174 | 25 | 0.03 |
70 | 22 | 104 | 28 |
| 5–7 | 219 | 24 | 54 | 15 | 165 | 31 | 72 | 33 | 17 | 31 | 55 | 33 | ||
| Median (range) | 3 (0–7) | 3 (0–7) | 3 (0–7) | < 0.01 | 3 (0–7) | 0.03 | 3 (0–7) | 3 (0–7) | ||||||
| Specimen collection date (2-week interval) | ||||||||||||||
| Weeks 44–45 | 64 | 7 | 10 | 3 | 54 | 10 | < 0.01 |
4 | 6 | < 0.01 |
0 | 0 | 4 | 7 |
| Weeks 46–47 | 61 | 7 | 13 | 4 | 48 | 9 | 12 | 20 | 3 | 23 | 9 | 19 | ||
| Weeks 48–49 | 139 | 15 | 54 | 15 | 85 | 16 | 31 | 22 | 12 | 22 | 19 | 22 | ||
| Weeks 50–51 | 174 | 19 | 65 | 18 | 109 | 20 | 51 | 29 | 11 | 17 | 40 | 37 | ||
| Weeks 52–1 | 184 | 20 | 86 | 23 | 98 | 18 | 58 | 32 | 24 | 28 | 34 | 35 | ||
| Weeks 2–3 | 284 | 31 | 142 | 38 | 142 | 26 | 90 | 32 | 37 | 26 | 53 | 37 | ||
ILI: influenza-like illness; NA: not applicable.
a Differences between cases and controls and vaccinated and unvaccinated participants were compared using the chi-squared test or Wilcoxon rank-sum test.
b Includes chronic co-morbidities that place individuals at higher risk of serious complications from influenza as defined by Canada’s National Advisory Committee on Immunization (NACI), including: heart, pulmonary (including asthma), renal, metabolic (such as diabetes), blood, cancer, or immunocompromising conditions, conditions that compromise management of respiratory secretions and increase risk of aspiration, or morbid obesity (body mass index ≥ 40).
c Missing specimen collection dates were imputed as the date the specimen was received and processed at the provincial laboratory minus two days, the average time between specimen collection date and laboratory received date among specimens with complete information for both values. Specimen collection interval was derived based on the number of days between ILI onset and the specified or imputed specimen collection date.
Overall 24% of cases and 30% of controls were considered vaccinated (p=0.04), corresponding to an unadjusted VE of 27% (95% confidence interval (CI): 1–46) against medically attended influenza A(H3N2) illness (Table 2). After adjustment for relevant covariates, VE was 42% (95% CI: 18–59).
Table 2. Interim vaccine effectiveness estimates for influenza A(H3N2), Canadian Sentinel Practitioner Surveillance Network, 1 November 2016–21 January 2017 (n = 906).
| Model | n total | Cases | Controls | VE % (95% CI) |
||
|---|---|---|---|---|---|---|
| n | % vaccinated | n | % vaccinated | |||
| Primary analysisa | ||||||
| Unadjusted | 906 | 370 | 24 | 536 | 30 | 27 (1 to 46) |
| Individual covariate adjustment | ||||||
| Age group (1–8, 9–19, 20–49, 50–64, ≥ 65 years) | 30 (4 to 50) | |||||
| Provinceb | 32 (7 to 50) | |||||
| Specimen collection interval from ILI onset (≤ 4, 5–7 days) | 23 (−5 to 44) | |||||
| Specimen collection date (2-week interval) | 38 (15 to 55) | |||||
| Full covariate adjustment | ||||||
| Adjusted | 42 (18 to 59) | |||||
| Restricted by provincec | ||||||
| Alberta | ||||||
| Unadjusted | 278 | 110 | 18 | 168 | 30 | 49 (8 to 72) |
| Adjusted | 62 (26 to 80) | |||||
| British Columbia | ||||||
| Unadjusted | 327 | 134 | 28 | 193 | 29 | 4 (−56 to 41) |
| Adjusted | 28 (−30 to 60) | |||||
| Ontariod | ||||||
| Unadjusted | 179 | 87 | 29 | 92 | 42 | 45 (−2 to 71) |
| Adjusted | 27 (−60 to 66) | |||||
| Quebec | ||||||
| Unadjusted | 122 | 39 | 13 | 83 | 17 | 28 (−118 to 76) |
| Adjusted | NE | |||||
| All provinces excluding Alberta | ||||||
| Unadjusted | 628 | 260 | 26 | 368 | 29 | 16 (−19 to 42) |
| Adjustede | 34 (−1 to 57) | |||||
CI: confidence interval; ILI: influenza-like illness; NE: not estimated (insufficient sample size); VE: vaccine effectiveness.
a Analysis adjusted for age group, province, specimen collection interval from ILI onset and specimen collection date (2-week interval).
b Alberta, British Columbia, Ontario, Quebec.
c Analysis adjusted for age group, specimen collection interval and specimen collection date (2-week interval).
d Due to logistical issues, specimen collection for the 2016/17 season began late in Ontario. The study period for Ontario-specific VE analysis was defined as 12 December 2016 (week 50) to 21 January 2017 (week 3).
e Analysis adjusted for age group, province (British Columbia, Ontario, Quebec), specimen collection interval and specimen collection date (2-week interval).
Genetic clade information was available for 221 of 263 (84%) influenza A(H3N2) sentinel specimens for which sequencing was attempted. The majority of viruses (176/221; 80%) clustered with the newly emerging clade 3C.2a1, defined by N171K +/− N121K mutations in site D, with most (165/176; 94%) having between one and three additional antigenic site mutations (Table 3). Other clade 3C.2a variants, each with two or three antigenic site mutations, comprised 43 (19%) sequenced influenza A(H3N2) specimens.
Table 3. Clade distribution and antigenic site mutations for influenza A(H3N2) viruses contributing to interim vaccine effectiveness evaluation, Canadian Sentinel Practitioner Surveillance Network, 1 November 2016–16 January 2017 (n = 221)a.
| Clade | Clade-defining amino acid substitutions (antigenic site)b,c | Distribution by province, % (column) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alberta (n = 81) |
BC (n = 81) |
Ontario (n = 48) |
Quebec (n = 11) |
Total (n = 221) |
|||||||
| n | % | n | % | n | % | n | % | n | % | ||
| Clade 3C.2a | N145S (A) + N144S (A) ( − CHO) + F159Y (B) + K160T (B) ( + CHO) + N225D (RBS) + Q311H (C) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Clade 3C.2a variants | Clade 3C.2a + Q197K (B) + R261Q (E) | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| Clade 3C.2a + T131K (A) + R142K (A) + R261Q (E) | 6 | 7 | 3 | 4 | 21 | 44 | 2 | 18 | 32 | 14 | |
| Clade 3C.2a + N121K (D) + S144K (A) +/ − S219Y (D) | 1 | 1 | 6 | 7 | 1 | 2 | 2 | 18 | 10 | 5 | |
| 3C.2a subtotal | 7 | 9 | 10 | 12 | 22 | 46 | 4 | 36 | 43 | 19 | |
| Clade 3C.2a1 | Clade 3C.2a + N171K (D) | 0 | 0 | 6 | 7 | 0 | 0 | 0 | 0 | 6 | 3 |
| Clade 3C.2a1 variants | Clade 3C.2a + N171K (D) + N121K (D) | 0 | 0 | 5 | 6 | 0 | 0 | 0 | 0 | 5 | 2 |
| Clade 3C.2a + N171K (D) + R142G (A) | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 9 | 2 | 1 | |
| Clade 3C.2a + N171K (D) + N121K (D) + R142G (A) | 9 | 11 | 23 | 28 | 10 | 21 | 0 | 0 | 42 | 19 | |
| Clade 3C.2a + N171K (D) + N121K (D) + R142G (A) + I242V (D) | 63 | 78 | 10 | 12 | 1 | 2 | 0 | 0 | 74 | 33 | |
| Clade 3C.2a + N171K (D) + N121K (D) + T135K (A) ( − CHO) +/ − R142G (A) or T167S (D) or I242M (D) | 2 | 2 | 23 | 28 | 6 | 13 | 2 | 18 | 33 | 15 | |
| Clade 3C.2a + N171K (D) + N121K (D) + K92R (E) + H311Q (C) +/ − Q197R (B) | 0 | 0 | 3 | 4 | 9 | 19 | 2 | 18 | 14 | 6 | |
| 3C.2a1 subtotal | 74 | 91 | 71 | 88 | 26 | 54 | 5 | 45 | 176 | 80 | |
| Clade 3C.3a | T128A (B) ( − CHO) + R142G (A) + N145S (A) + A138S (A) + F159S (B) + N225D (RBS) | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 18 | 2 | 1 |
BC: British Columbia; CHO: carbon-hydrogen-oxygen (glycosylation motif); RBS: receptor binding site.
a Sequencing was attempted on all influenza A(H3N2) sentinel specimens contributing to VE analysis that had sufficient viral load and that were available up to 21 January 2017, with the last included collection date 16 January 2017. Genetic clade information was available for 221 of 263 (84%) viruses for which sequencing was attempted. Sequencing was not attempted on influenza A(H3N2) specimens with insufficient viral load (i.e. high CT value in the RT-PCR assay; n = 8) or those submitted after 21 January 2017 (n = 99).
b Letters A through E refer to established antigenic sites in influenza A(H3N2) viruses [8,9]. RBS refers to the receptor binding site. Substitutions indicated with −CHO refer to mutations resulting in the loss of a potential glycosylation site; those indicated with +CHO refer to mutations resulting in the gain of a potential glycosylation site.
c Additional substitutions in the egg-adapted high-growth reassortant vaccine strain are not considered here.
Considerable genetic heterogeneity was also observed among dominant but emerging clade 3C.2a1 variants by province and time (Figure 2).
Figure 2.
Distribution of clade 3C.2a1 variants by provincea and week of specimen collection, Canadian Sentinel Practitioner Surveillance Network (SPSN), 1 November 2016−16 January 2017 (n = 176)
a Alberta and British Columbia are adjacent provinces located in western Canada; Ontario and Quebec are adjacent provinces located in central Canada, > 2,500 km away from Alberta and British Columbia.
b Due to logistical issues, specimen collection did not begin until week 48 in Ontario.
c +/− R142G or T167S or I242M mutations.
d +/− Q197R mutation.
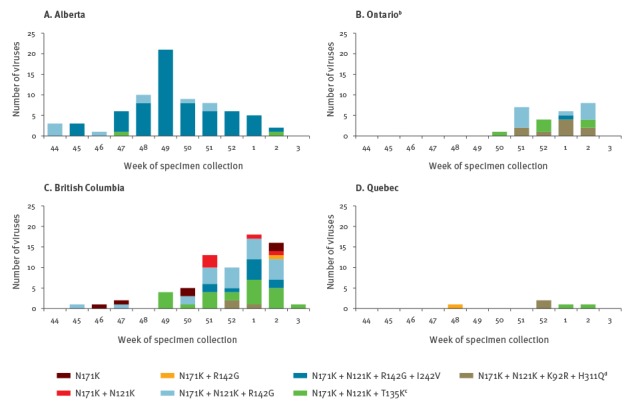
In exploratory analyses, VE was highest and significantly protective in Alberta where an earlier epidemic start included a more limited range of clade 3C.2a1 variants dominated by N121K + R142G + I242V mutations (Figure 2, Table 2). Conversely, in the adjacent westernmost province of British Columbia and also further east in the provinces of Ontario and Quebec in central Canada, delayed epidemic activity was associated with lower VE and greater diversity in circulating clade 3C.2a1 variants, although confidence intervals overlapped for all four provinces.
Discussion
Whereas the 2015/16 season was mild overall with late-season circulation of influenza A(H1N1)pdm09 viruses, the current 2016/17 season has been characterised to date by dominant influenza A(H3N2) activity, more comparable to the 2014/15 or 2012/13 seasons [1,10-12]. In the 2016/17 interim VE analysis reported here, we found overall vaccine protection of 42% (95% CI: 18–59) against medically-attended A(H3N2) illness, with variation by province that may reflect genetic heterogeneity in circulating A(H3N2) variants. This overall estimate is consistent with a recent meta-analysis of global studies based on the test-negative design that reported a pooled VE, including both interim and end-of-season estimates, of 33% (95% CI: 26–39) against seasonal A(H3N2) viruses [13]. Early VE estimates for the 2016/17 season available from Finland and Sweden found significant protection of 20–30% against laboratory-confirmed influenza in adults 65 years and older [14]; however, methodological details and influenza virus characterisations are not available for these estimates, limiting their interpretation.
Although still suboptimal given the substantial disease burden associated with influenza A(H3N2) seasons [15,16], our mid-season VE estimate for 2016/17 is considerably higher than the last A(H3N2)-dominated season in 2014/15 during which no vaccine protection was found [2,3]. In 2014/15, with unchanged vaccine components from the prior 2013/14 season and substantial antigenic drift in circulating viruses, negative interference from the prior season’s vaccination may have contributed to the historically low VE observed [3,17]. While more than 80% of vaccinated participants in 2016/17 were also vaccinated in the prior 2015/16 season (data not shown), higher VE than in 2014/15 was anticipated. This expectancy was in part based on the change in vaccine component from the prior 2015/16 season’s A/Switzerland/9715293/2013(H3N2)-like (clade 3C.3a) virus to the A/Hong Kong/4801/2014(H3N2)-like (clade 3C.2a) vaccine strain [18]. The latter is also considered a better antigenic match to circulating viruses than was the case in 2014/15 [18,19]. Specific evaluation of this hypothesis related to less pronounced effects of repeat vaccination for 2016/17 awaits end-of-season analyses.
Circulating influenza A(H3N2) viruses in Canada and elsewhere this season have continued to evolve, with an increasing proportion since June 2015 clustering with the newly emerging clade 3C.2a1 that is distinguished by the HA1 substitution N171K, often combined with N121K, both in antigenic site D [20,21]. These clade 3C.2a1 variants are considered antigenically similar to the egg-adapted clade 3C.2a vaccine strain based on haemagglutination inhibition (HI) assay [1,19]. However, recent A(H3N2) viruses continue to be difficult to characterise antigenically by HI assay [20]. A potential glycosylation motif present at positions 158–160 in all clade 3C.2a and descendant viruses has resulted in variable agglutination of erythrocytes; loss or partial loss of this glycosylation motif during cell-culture passage may enable HI characterisation of a subset of clade 3C.2a viruses but also limit the generalisability of antigenicity findings on that basis [20,22].
In sequencing analysis, we identified considerable diversity among circulating influenza A(H3N2) strains, including a mix of genetic variants that differed geographically and with time. The majority (80%) of A(H3N2) viruses included in our VE analysis belonged to the newly emerging clade 3C.2a1, but with continuing genetic evolution compared with the vaccine strain. Almost all (95%) 3C.2a1 viruses had both the N171K and N121K mutations in site D that distinguish this clade. About two-thirds had acquired an additional R142G (site A) mutation, also present in all clade 3C.3 viruses and the majority of clade 3C.2a variants detected in this study, with or without an I242V mutation (site D). The clinical implications of accumulated antigenic site D mutations, representing a shift away from the heavily glycosylated but immunodominant sites A and B, requires further investigation [8,23]. Another 3C.2a1 variant, detected more frequently in the later study period but comprising only 15% of study viruses overall, had an additional T135K mutation in site A. T135K is associated with loss of a potential glycosylation site at positions 133–135 that has otherwise been present in all descendant A(H3N2) viruses since A/Sydney/5/1997 [24]. Changes in glycosylation motifs may be relevant to antigenicity, viral fitness and/or pathogenicity [24-26]. The ecological correlation between greater genetic diversity and lower VE by geographic region warrants further investigation in other countries, as well as end-of-season analyses.
Limitations of this analysis include the observational study design for which residual bias and confounding cannot be ruled out, and the small sample size resulting in wide confidence intervals, particularly in subgroup analyses. Although interim estimates are generally considered a reliable predictor of final estimates, this reliability depends in part upon the stage of the epidemic and virus evolution, and contributing virological and participant profiles, at the time of the mid- and end-of-season analyses [27]. Of particular note, Alberta had an earlier start to the influenza season and findings may not reflect the full diversity or distribution of evolved variants or VE estimates for the remainder of the season. Given the high specificity of RT-PCR assays for influenza viruses, differences in diagnostic test characteristics between provinces are unlikely to have influenced VE findings [28]. VE estimates are subject to change and are provided here only for influenza A(H3N2); if feasible, VE against other types/subtypes, as well as clade-specific VE, will be explored and compared with findings from other settings in end-of-season analyses.
Conclusion
We report interim VE of ca 40% for the 2016/17 influenza A(H3N2) epidemic in Canada, which is higher than in 2014/15 and consistent with expected but suboptimal VE estimates for influenza A(H3N2) more generally. Given that a substantial proportion of vaccinated people may remain unprotected against influenza A(H3N2) illness, other adjunct measures should be considered to minimise associated morbidity and mortality, particularly among high-risk individuals. Continued evolution in circulating 3C.2a variants and their derivatives, and the impact on vaccine protection, warrants ongoing monitoring to inform potential vaccine reformulation.
Acknowledgements
The authors gratefully acknowledge the contribution of sentinel sites whose regular submission of specimens and data provide the basis of our analyses. We wish to acknowledge the coordination and technical support provided by epidemiological and laboratory staff in all participating provinces. We wish to thank the following for network coordination and data entry activities in each province including: Lisan Kwindt for the British Columbia Centre for Disease Control; Elaine Douglas, Virginia Goetz and Dylan Kendrick for TARRANT in Alberta; Romy Olsha for Public Health Ontario; Sophie Auger and Isabelle Petillot for the Institut national de santé publique du Québec; and Joel Ménard at the Laboratoire de santé publique du Québec for data compilation. We thank those who provided laboratory support in each of the British Columbia Centre for Disease Control Public Health Laboratory, the Alberta Provincial Laboratory for Public Health (ProvLab), the Public Health Ontario Laboratory, and the Laboratoire de santé publique du Québec.
Funding was provided by the British Columbia Centre for Disease Control, Alberta Health and Wellness, Public Health Ontario, Ministère de la santé et des services sociaux du Québec, and l’Institut national de santé publique du Québec.
GenBank Accession Numbers
Conflict of interest: Within 36 months of manuscript submission, GDS has received grants unrelated to influenza from GSK and Pfizer and travel reimbursement to attend an ad hoc advisory board meeting of GSK also unrelated to influenza; he has provided paid expert testimony in a grievance against a vaccinate-or-mask healthcare worker influenza vaccination policy for the Ontario Nurse Association. JBG has received research grants from GlaxoSmithKline Inc. and Hoffman-La Roche Ltd to study antiviral resistance in influenza, and from Pfizer Inc. to conduct microbiological surveillance of Streptococcus pneumoniae. MK has received research grants from Roche, Merck, Siemens, Hologic, and Boerhinger Ingelheim for unrelated studies. Other authors have no conflicts of interest to declare.
Authors’ contributions: Principal investigators (epidemiological): DMS (National and British Columbia); JAD (Alberta); ALW (Ontario); and GDS (Québec). Principal investigator (laboratory): AJ and MK (British Columbia); SD (Alberta); JBG (Ontario); HC (Québec); and NB and YL (National Microbiology Laboratory). Virus sequencing: SS. Data analysis: CC and DMS (epidemiological); RB (statistical support); SS (molecular). Preparation of first draft: CC and DMS. Draft revision and approval: all.
References
- 1.Public Health Agency of Canada (PHAC). FluWatch report: January 8 to January 14, 2017 (week 2). Ottawa: PHAC; 2017. Available from: http://www.healthycanadians.gc.ca/publications/diseases-conditions-maladies-affections/fluwatch-2016-2017-02-surveillance-influenza/index-eng.php?_ga=1.147496691.591173311.1486470400
- 2. Skowronski DM, Chambers C, Sabaiduc S, De Serres G, Dickinson JA, Winter AL, et al. Interim estimates of 2014/15 vaccine effectiveness against influenza A(H3N2) from Canada’s Sentinel Physician Surveillance Network, January 2015. Euro Surveill. 2015;20(4):21022. 10.2807/1560-7917.ES2015.20.4.21022 [DOI] [PubMed] [Google Scholar]
- 3. Skowronski DM, Chambers C, Sabaiduc S, De Serres G, Winter AL, Dickinson JA, et al. A perfect storm: Impact of genomic variation and serial vaccination on low influenza vaccine effectiveness during the 2014-2015 season. Clin Infect Dis. 2016;63(1):21-32. 10.1093/cid/ciw176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360(25):2605-15. 10.1056/NEJMoa0903810 [DOI] [PubMed] [Google Scholar]
- 5. Chen Y, Cui D, Zheng S, Yang S, Tong J, Yang D, et al. Simultaneous detection of influenza A, influenza B, and respiratory syncytial viruses and subtyping of influenza A H3N2 virus and H1N1 (2009) virus by multiplex real-time PCR. J Clin Microbiol. 2011;49(4):1653-6. 10.1128/JCM.02184-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allplex Respiratory Panel Assay. Seoul: Seegene. Available from: http://seegene.com/neo/en/products/respiratory/allplex_Rp_fp.php
- 7.NxTAG Respiratory Pathogen Panel (RUO). Luminex. Available from: https://www.luminexcorp.com/clinical/ruo-products/nxtag-respiratory-pathogen-panel/ [DOI] [PMC free article] [PubMed]
- 8. Ndifon W, Wingreen NS, Levin SA. Differential neutralization efficiency of hemagglutinin epitopes, antibody interference, and the design of influenza vaccines. Proc Natl Acad Sci USA. 2009;106(21):8701-6. 10.1073/pnas.0903427106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342(6161):976-9. 10.1126/science.1244730 [DOI] [PubMed] [Google Scholar]
- 10.Public Health Agency of Canada (PHAC). FluWatch report: August 14 to August 27, 2016 (weeks 33-34). Ottawa: PHAC; 2016. Available from: https://www.canada.ca/en/public-health/services/publications/diseases-conditions/fluwatch-report-august-14-27-2016-weeks-33-34.html
- 11.Public Health Agency of Canada (PHAC). FluWatch report: August 16 to August 29, 2015 (weeks 33 & 34). Ottawa: PHAC; 2015. Available from: https://www.canada.ca/en/public-health/services/publications/diseases-conditions/fluwatch-report-august-16-29-2015-weeks-33-34.html
- 12.Public Health Agency of Canada (PHAC). FluWatch report: August 11 to August 24, 2013 (weeks 33 & 34). Ottawa: PHAC; 2013. Available from: http://publications.gc.ca/collections/collection_2013/aspc-phac/HP58-1-2013-34-eng.pdf
- 13. Belongia EA, Simpson MD, King JP, Sundaram ME, Kelley NS, Osterholm MT, et al. Variable influenza vaccine effectiveness by subtype: a systematic review and meta-analysis of test-negative design studies. Lancet Infect Dis. 2016;16(8):942-51. 10.1016/S1473-3099(16)00129-8 [DOI] [PubMed] [Google Scholar]
- 14.European Centre for Disease Prevention and Control (ECDC). Risk assessment of seasonal influenza, EU/EEA, 2016-2017: Update, 25 January 2017. Stockholm: ECDC; 2017. Available from: http://ecdc.europa.eu/en/publications/Publications/Risk-assessment-seasonal-influenza-2016-2017-update.pdf
- 15. Schanzer DL, Sevenhuysen C, Winchester B, Mersereau T. Estimating influenza deaths in Canada, 1992-2009. PLoS One. 2013;8(11):e80481. 10.1371/journal.pone.0080481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289(2):179-86. 10.1001/jama.289.2.179 [DOI] [PubMed] [Google Scholar]
- 17. Smith DJ, Forrest S, Ackley DH, Perelson AS. Variable efficacy of repeated annual influenza vaccination. Proc Natl Acad Sci USA. 1999;96(24):14001-6. 10.1073/pnas.96.24.14001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization (WHO). Recommended composition of influenza virus vaccines for use in the 2016-2017 northern hemisphere influenza season. Geneva: WHO; 2016. Available from: http://www.who.int/influenza/vaccines/virus/recommendations/2016_17_north/en/
- 19.The Francis Crick Institute. September 2016 interim report. London: The Crick Worldwide Influenza Centre (WIC); 2016. Available from: https://www.crick.ac.uk/media/326439/september_2016_interim_report.pdf
- 20.European Centre for Disease Prevention and Control (ECDC). Influenza virus characterisation, summary Europe, September 2016. Stockholm: ECDC; 2016. Available from: http://ecdc.europa.eu/en/publications/Publications/influenza-virus-characterisation-september-2016.pdf
- 21. Neher RA, Bedford T. nextflu: real-time tracking of seasonal influenza virus evolution in humans. Bioinformatics. 2015;31(21):3546-8. . Available from: http://nextflu.org/h3n2/3y/ 10.1093/bioinformatics/btv381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Skowronski DM, Sabaiduc S, Chambers C, Eshaghi A, Gubbay JB, Krajden M, et al. Mutations acquired during cell culture isolation may affect antigenic characterisation of influenza A(H3N2) clade 3C.2a viruses. Euro Surveill. 2016;21(3):30112. 10.2807/1560-7917.ES.2016.21.3.30112 [DOI] [PubMed] [Google Scholar]
- 23. Popova L, Smith K, West AH, Wilson PC, James JA, Thompson LF, et al. Immunodominance of antigenic site B over site A of hemagglutinin of recent H3N2 influenza viruses. PLoS One. 2012;7(7):e41895. 10.1371/journal.pone.0041895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abe Y, Takashita E, Sugawara K, Matsuzaki Y, Muraki Y, Hongo S. Effect of the addition of oligosaccharides on the biological activities and antigenicity of influenza A/H3N2 virus hemagglutinin. J Virol. 2004;78(18):9605-11. 10.1128/JVI.78.18.9605-9611.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tate MD, Job ER, Deng Y-M, Gunalan V, Maurer-Stroh S, Reading PC. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses. 2014;6(3):1294-316. 10.3390/v6031294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. An Y, McCullers JA, Alymova I, Parsons LM, Cipollo JF. Glycosylation analysis of engineered H3N2 influenza A virus hemagglutinins with sequentially added historically relevant glycosylation sites. J Proteome Res. 2015;14(9):3957-69. 10.1021/acs.jproteome.5b00416 [DOI] [PubMed] [Google Scholar]
- 27. Leung VK, Cowling BJ, Feng S, Sullivan SG. Concordance of interim and final estimates of influenza vaccine effectiveness: a systematic review. Euro Surveill. 2016;21(16):30202. 10.2807/1560-7917.ES.2016.21.16.30202 [DOI] [PubMed] [Google Scholar]
- 28. Orenstein EW, De Serres G, Haber MJ, Shay DK, Bridges CB, Gargiullo P, et al. Methodologic issues regarding the use of three observational study designs to assess influenza vaccine effectiveness. Int J Epidemiol. 2007;36(3):623-31. 10.1093/ije/dym021 [DOI] [PubMed] [Google Scholar]


