Summary
Targeted mutagenesis using programmable DNA endonucleases has broad applications for studying gene function in planta and developing approaches to improve crop yields. Recently, a genetic method that eliminates the need to emasculate the female inbred during hybrid seed production, referred to as Seed Production Technology, has been described. The foundation of this genetic system relied on classical methods to identify genes critical to anther and pollen development. One of these genes is a P450 gene which is expressed in the tapetum of anthers. Homozygous recessive mutants in this gene render maize and rice plants male sterile. While this P450 in maize corresponds to the male fertility gene Ms26, male fertility mutants have not been isolated in other monocots such as sorghum and wheat. In this report, a custom designed homing endonuclease, Ems26+, was used to generate in planta mutations in the rice, sorghum and wheat orthologs of maize Ms26. Similar to maize, homozygous mutations in this P450 gene in rice and sorghum prevent pollen formation resulting in male sterile plants and fertility was restored in sorghum using a transformed copy of maize Ms26. In contrast, allohexaploid wheat plants that carry similar homozygous nuclear mutations in only one, but not all three, of their single genomes were male fertile. Targeted mutagenesis and subsequent characterization of male fertility genes in sorghum and wheat is an important step for capturing heterosis and improving crop yields through hybrid seed.
Keywords: endonuclease, male sterile, rice, sorghum, wheat
Introduction
Crop improvement on a global scale is required to meet growing food demands (Parry and Hawkesford, 2012; Ray et al., 2013). The expansion of yield gains made by developing hybrid crops, rather than varieties, would be an important step to achieve such progress where harnessing the advantages of heterosis is the main objective of hybrid breeding (Duvick, 1999; Melchinger, 1999; Tester and Landrige, 2010). Genetic divergence between parent lines has been demonstrated to correlate with the extent to which heterosis of a hybrid increases. Hybrid seed production requires that a male parent inbred pollinates a genetically distinct female inbred parent that is either male sterile or has been emasculated. This is straightforward in monoecious plants such as maize that have separate male (tassel) and female (ear) flowers on the same plant, where detasseling can emasculate the female parent during hybrid seed production. While detasseling has been used for hybrid maize seed production, physical emasculation is not a scalable solution for crop species with perfect flowers such as rice, sorghum, barley, millet and wheat. Historically, cytoplasmic male sterility (CMS) has been used in these crops for hybrid seed production; however this method is not effective in all germplasm due to the dependence on nuclear restorer genes specific for a given cytoplasm (Dwivedi et al., 2008; Li and Yuan, 1999; Reddy et al., 2005; Whitford et al., 2013). Recently, a novel hybrid seed production system has been described for maize that is not limited by the genetic background (Wu et al., 2015). This Seed Production Technology (SPT) system involves restoring pollen formation while maintaining a homozygous nuclear recessive mutation in the maize male fertility gene Ms45. Although homozygous ms45 maize plants are male sterile, pollen formation can be restored in plants that contain a transformed version of the Ms45 gene. In the SPT system, only the ms45 allele and not the Ms45 restoration cassette is transmitted through pollen. The resultant ms45 progeny are used as females during hybrid production; thus, emasculation by detasseling is not necessary as these females are genetically male sterile. Although this system originally incorporated the maize Ms45 gene, it is applicable to other crops and to other nuclear male sterile mutations, whether dominant or recessive.
Extending this hybrid seed production system to other monoecious crops could afford the ability to maximize yield potentials through heterosis despite important differences such as asynchronous flower development and architecture. This is particularly relevant for rice, sorghum and wheat. However, deployment of a genetically based hybrid seed production system in these crops requires the identification and isolation of male fertility genes. While male fertility genes and mutants have been described and isolated in maize (Cigan et al., 2001; Neuffer et al., 1997) and rice (Guo and Liu, 2012; Yamagata et al., 2007) by classical genetic approaches, there is a paucity of available mutants in crops such as sorghum and wheat. As opposed to the use of chemicals or irradiation to generate random mutations and the associated time‐consuming screening of mutagenized plants, recent progress in DNA double‐strand break (DSB) technologies holds great promise for the systematic generation of male sterility mutants in these agriculturally important crops. A variety of nucleases can be employed for the generation of targeted DSBs; zinc‐finger nucleases (ZFN), I‐CreI‐based homing endonucleases (meganucleases), transcription activator‐like effector nucleases (TALEN) and the Cas9‐guide RNA system (often referred to as ‘CRISPR‐Cas’) have been used to produce mutations in various plant species (Kumar and Jain, 2015; Osakabe and Osakabe, 2015; Svitashev et al., 2015; Voytas and Gao, 2014). DSBs are repaired through endogenous cellular processes of nonhomologous end‐joining (NHEJ) or homology‐directed repair; the NHEJ repair pathway can often be inexact resulting in mutations at the site of the DSB and in this way targeted mutagenesis is achieved. Such a directed mutagenesis strategy utilizing a redesigned I‐CreI homing endonuclease to target the Ms26 male fertility gene in maize has recently been reported (Djukanovic et al., 2013). As demonstrated in rice, the rice Ms26 ortholog encodes for a cytochrome P450 mono‐oxygenase enzyme (OsCYP704B2) and is expressed in the tapetum and microspores in the developing anther (Hobo et al., 2008; Li et al., 2010). The homozygous recessive ms26 mutants in maize and rice are defective in both tapetum and microspore development resulting in male sterility (Li et al., 2010; Loukides et al., 1995). The redesigned endonuclease described by Djukanovic et al. (2013), Ems26+, targets a 22‐nt sequence found in the 5th exon of Ms26. DSB generated by Ems26+ within the predetermined chromosomal recognition site and NHEJ repair resulted in mutations in the Ms26 gene and loss of function. Maize plants containing homozygous recessive ms26 mutant alleles were phenotypically normal, producing tassels that contained developed spikelets, with the exception that anthers did not shed pollen. These mutant ms26 alleles were transmitted to progeny at the expected segregation ratio (Djukanovic et al., 2013). The male sterile phenotype generated by targeted gene disruption using the Ems26+ endonuclease was indistinguishable from ms26 maize mutants generated by transposon‐tagging (Loukides et al., 1995; Fox, T., DuPont Pioneer, unpublished observation).
This report demonstrates that the Ems26+ endonuclease can generate targeted mutations in the orthologs of Ms26 genes of rice (Oryza sativa, Os), sorghum (Sorghum bicolor, Sb) and common wheat (Triticum aestivum, Ta). Orthologous Ms26 mutations in rice and sorghum plants, such as maize (Zea mays L., Zm), confer a recessive male sterile phenotype, and restoration of fertility in these mutant sorghum plants was achieved by a transformed copy of maize Ms26. In addition, using a conditionally regulated Ems26+, mutations in the wheat Ms26 ortholog were recovered in all three genomes of hexaploid spring wheat. These results demonstrate the utility of nuclease‐facilitated gene editing to produce male fertility gene mutants for the purpose of unlocking yield potential through exploiting heterosis in these major food crops.
Results
The maize Ms26 gene is a member of a P450 protein family whose expression is limited to developing anthers (Cigan et al., 2005). Biochemical characterization indicated putative roles of the rice Ms26 ortholog (OsCYP704B2) catalysing ω‐hydroxylated C16 and C18 fatty acids as well as synthesis and transport of sporopollenin precursor components important to pollen wall formation (Li et al., 2010). Maize and rice plants carrying homozygous recessive mutations disrupting the coding‐frame of the Ms26 and OsCYP704B2 genes, respectively, resulted in an inability of these plants to generate fertile pollen grains (Djukanovic et al., 2013; Li et al., 2010). Consistent with the importance of this P450 for pollen development, protein sequences with high homology to the maize Ms26 were identified in sorghum and wheat by searching plant sequence databases. The sorghum and wheat Ms26‐like sequences along with the rice OsCYP704B2 gene were, therefore, referred to herein as SbMs26, TaMs26 and OsMs26, respectively. Although there is significant sequence identity across these crop species (Figure S1 and Text S1), nothing is known regarding the functional conservation of this P450 in sorghum and wheat. As shown in Figure 1, the 22‐nt Ems26+ recognition site is conserved across these monocots and resides in the last exon of each gene, and this exon contains the important haem‐binding loop common to haem‐thiolate cytochrome P450s (Werck‐Reichhart and Feyereisen, 2000). To determine whether the Ms26 gene is required for male fertility, several directed mutagenesis approaches using the Ems26+ endonuclease were developed and tested in rice and then used to determine whether this gene is required for male fertility in sorghum and wheat.
Figure 1.
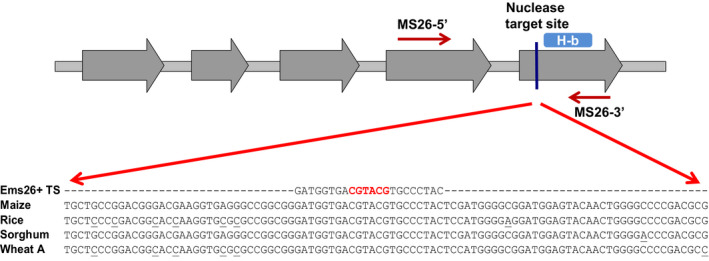
Ems26+ endonuclease target site is conserved across maize, rice, sorghum and wheat. Gene structure illustration of maize Ms26; exons (grey arrows) and the haem‐binding (H‐b) domain are shown. The haem‐binding domain resides in the 5th exon of the maize Ms26 (Gene ID: 100191749) gene and sorghum (Gene ID: 8082128) ortholog. This domain is also found in the last exon of the rice (Gene ID: 4331756) and the wheat (Fielder A‐genome) Ms26 orthologs, which each consists of four exons. The Ems26+ target site is depicted as a vertical line upstream of the H‐b domain. Sequence differences in rice, sorghum and wheat A‐genome as compared to the maize Ms26 gene are indicated as underlined nucleotides. The UNIMS26 primer pair (arrows labelled MS26‐5′ and MS26‐3′) was used to amplify the region of interest from isolated genomic DNA to screen for Ms26 mutations. A Bsi WI restriction enzyme site (red font) present in the 22‐nt Ems26+ target site allowed for Ms26 mutation screening by determining resistance to Bsi WI digestion.
Ems26+‐generated mutations in rice
Young rice callus [(Oryza sativa L. ssp. Japonica (cv. Kitaake)] initiated from germinating seed was used as transformation targets for biolistic‐ or Agrobacterium‐mediated delivery of vectors that contained the custom endonuclease, Ems26+. For biolistic transformation, two vectors were constructed: (i) a vector containing a codon‐optimized Ems26+ under the transcriptional regulation of the maize ubiquitin promoter (UBIpro:Ems26+) enabling constitutive expression, and (ii) a vector containing a herbicide resistance selectable marker phosphinothricin acetyltransferase (PAT) fused to the red fluorescence protein (RFP) gene and placed under the regulation of the maize END2 promoter. This vector allowed transformed rice cells to grow in the presence of the herbicide glufosinate while monitoring red fluorescence, when codelivered into 2‐week‐old, seed‐derived rice callus. Ems26+ contains a nuclear localization signal at the N‐terminus and an intron to eliminate expression in bacterial cells used for vector propagation (Djukanovic et al., 2013). Approximately 6 weeks after bombardment, callus sectors that grew on media containing glufosinate were screened for mutations at the 22‐nt Ms26 target site. Due to the presence of a BsiWI restriction site (5′‐CGTACG‐3′) across the centre of the Ems26+ recognition sequence and at the site of Ems26+‐mediated DNA cleavage (Figure 1), PCR amplification and digestion by BsiWI was used as an initial screening tool to detect Ms26 mutations. PCR amplicons resistant to BsiWI digestion would be indicative of sequence changes at this recognition site due to cutting by Ems26+ and imprecise repair by NHEJ (Djukanovic et al., 2013). Genomic DNA isolated from these glufosinate‐resistant callus sectors was amplified using the primer pair UNIMS26 5′‐2 and UNIMS26 3′‐1 to generate a 653‐bp amplicon that was then subjected to digestion with BsiWI. Products of these reactions were electrophoresed on 1% agarose gels and screened; amplicons containing wild‐type sequence across the Ems26+ recognition site would produce 376‐ and 277‐bp DNA digestion products (Figure S2, lanes 2, 4). Screening of 292 glufosinate‐resistant events generated by cobombardment with the Ems26+ vector identified 22 callus sectors that contained PCR products resistant to BsiWI digestion. Subcloning and DNA sequence analysis of these PCR products revealed a variety of mutations across the Ms26 target site, which included single‐ and multiple‐nucleotide insertions and deletions (Figure 2).
Figure 2.
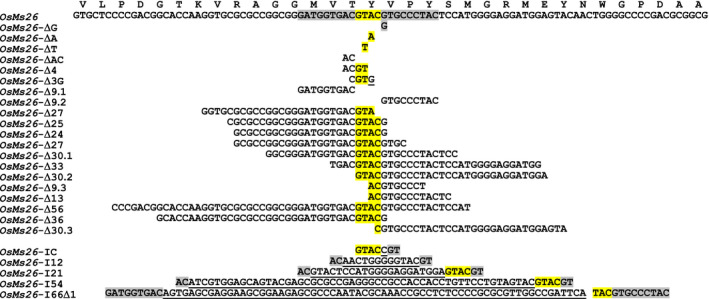
Ems26+‐generated OsMs26 mutations in rice. Shown in the WT reference sequence, the 22‐nt Ems26+ recognition sequence is highlighted in grey and yellow. The latter indicates the 4 bp overhang, 5′‐GTAC‐3′, produced by nuclease cutting and is highlighted throughout for orientation purposes. For each mutant, deleted nucleotides are shown and inserted nucleotides are shown as underlined. Amino acid sequence corresponding to the OsMs26 gene is provided as single letters above the WT nucleotide reference sequence.
To examine the male fertility phenotype of rice plants containing OsMs26 mutations generated by Ems26+, plants were regenerated from glufosinate‐resistant callus sectors, analysed for OsMs26 mutations and allowed to set self‐pollinated seed (T1 seed). T1 seed from three plants containing nonidentical OsMs26 mutations, but lacking the randomly integrated transformation vectors, were advanced for progeny testing. Using the BsiWI digestion assay, genomic DNA collected from progeny plantlets was amplified with the UNIMS26 primer pair and screened for mutations at the Ms26 target site. Heterozygous OsMs26/Osms26 (Figure S2, lane 4) and homozygous Osms26/Osms26 (Figure S2, lane 6) mutant plants were advanced and scored for their ability to form pollen by analysing seed set upon self‐fertilization. Throughout vegetative growth, these plants appeared phenotypically normal and flowered at the same time as wild‐type plants with the exception that dehisced anthers were not observed in the homozygous recessive Osms26 plants (Figure 3a). Microscopic examination of anthers from OsMs26/Osms26 plants staged at late uninucleate microspore development revealed normal appearing microspores, which continued to develop into mature, fertilization‐competent pollen. However, examination of anthers from Osms26/Osms26 plants revealed that microspores arrested development immediately after tetrad release, whereupon these plants contained anthers devoid of mature pollen (Figure 3b). These observations were similar to those reported by Loukides et al. (1995) and Li et al. (2010). Surprisingly, as shown in Figure 3b, the in‐frame deletion of nine nucleotides in Osms26‐Δ9.2 conferred a male sterile phenotype. In summary, all OsMs26/Osms26 plants were male fertile; and all Osms26/Osms26 plants were male sterile (Table 1). In addition, homozygous Osms26/Osms26 plants in this study were female fertile as demonstrated by their ability to set seed when fertilized with wild‐type rice pollen.
Figure 3.
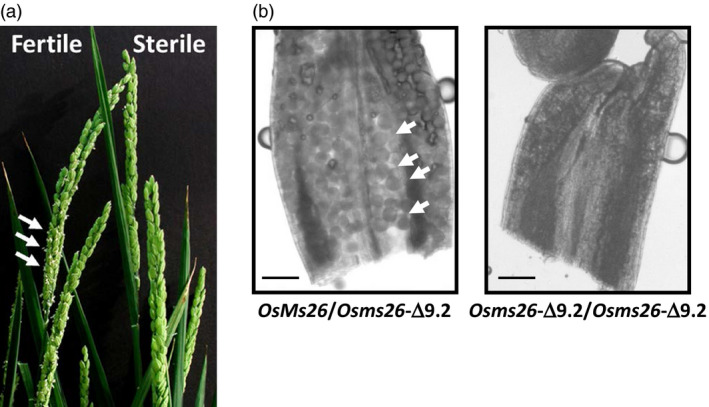
Male sterile rice generated from targeted mutagenesis of the OsMs26 gene via bombardment delivery of Ems26+. (a) Rice panicles from male fertile (left; OsMs26/Osms26‐Δ36) and male sterile (right; Osms26‐Δ36/Osms26‐Δ36) plants. Dehisced anthers are shown with white arrows. (b) Longitudinal anther squash from heterozygous OsMs26/Osms26‐Δ9.2 male fertile plants (left) containing microspores (white arrows) and homozygous Osms26‐Δ9.2/Osms26‐Δ9.2 male sterile plants (right). Magnification bar = 100 μM.
Table 1.
Male fertility phenotypes of T1 progeny plants with heterozygous or homozygous OsMs26 mutations
| Mutation | Plants | OsMs26/Osms26 | Osms26/Osms26 | ||
|---|---|---|---|---|---|
| Male fertile | Male sterile | Male fertile | Male sterile | ||
| Osms26‐Δ9.2 | 18 | 10 | 0 | 0 | 8 |
| Osms26‐Δ36 | 9 | 5 | 0 | 0 | 4 |
| Osms26‐I66Δ1 | 15 | 11 | 0 | 0 | 4 |
Inducible Ems26+ for producing double‐strand breaks
Given the ability to successfully direct mutations to the rice Ms26 ortholog that confer a male sterile phenotype, the requirement of Ms26 for pollen development in sorghum and wheat was investigated. Relative to maize and rice, transformation of sorghum and wheat is challenging (Casas et al., 1997; Zhao et al., 2000; Zhu et al., 1998). Moreover, gene editing in wheat is complicated due to the size and polyploid nature of the genome. Thus, as an alternative to biolistic delivery of Ems26+, an in planta method to stimulate targeted mutagenesis in sorghum and wheat was devised. This strategy depended on the generation of plants containing a stably integrated, temperature‐regulated Ems26+ expression cassette delivered on a T‐DNA vector by Agrobacterium‐mediated transformation. Ems26+ was placed under the transcriptional control of a maize temperature‐regulated promoter, MDH (ZmMDHpro:Ems26+) (Svitashev et al., 2015), which allowed for temperature‐inducible expression of the nuclease in plant tissues. In this strategy, immature embryos from plants containing conditionally expressed Ems26+ would be heated at 37 °C for 24 h to generate mutations. This T‐DNA also contained a blue fluorescence gene (Cyan) regulated by an embryo‐preferred END2 promoter (ZmEnd2pro:Cyan). This reporter served as a visible marker for selection of transformed callus, fertilized immature embryos and mature seed. A disrupted copy of the DsRED gene transcriptionally regulated by a maize Histone 2B promoter was also present in this T‐DNA to monitor endonuclease activity. This nonfunctional colour marker consisted of duplicated DsRED gene fragments (RF‐FP) with a 369‐bp overlap separated by a 136‐bp spacer containing the Ems26+ recognition site (Text S2). DSBs in RF‐FP would either be repaired by NHEJ or intramolecular recombination within the repeated RF‐FP fragments (Figure 4a). Duplicated gene coding regions interrupted by a DSB target site(s) have been demonstrated to be useful visual reporters to detect nuclease activity (Puchta and Hohn, 2012). As such, one outcome of a DSB at the target site would be to stimulate intramolecular recombination within the repeated RF‐FP restoring DsRED gene function as revealed by the appearance of red‐fluorescing callus cells against a background of blue fluorescence. Thus in contrast to control callus where no red fluorescence would be expected, the appearance of red‐fluorescing cells in treated samples would demonstrate activity of Ems26+ endonuclease and suggest potential cutting and, therefore, mutagenesis within the endogenous Ms26 gene.
Figure 4.
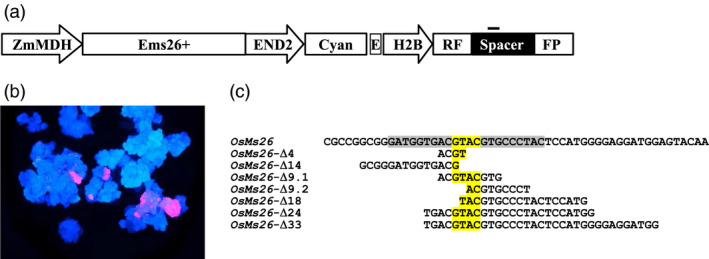
OsMs26 mutations in rice generated using a stably integrated cassette containing Ems26+ under transcriptional control of a maize temperature‐regulated promoter. (a) Agrobacterium vector for stable integration of the ZmMDHpro:Ems26+. The vector also contained a blue fluorescence gene (Cyan) as a visible marker regulated by the maize END2 promoter (END2). The disrupted DsRED gene (H2Bpro:RF‐FP) contained duplicated DsRED gene fragments (RF, FP) separated by a 136‐bp spacer containing the Ems26+ recognition site (position identified by the black bar above spacer sequence). (b) Blue and red fluorescence overlay image of rice calli containing stably integrated ZmMDHpro:Ems26+ after treatment at 37 °C for 48 h. Images captured 7 days after treatment. (c) Deletions in OsMs26 achieved via targeted mutagenesis. Shown in the reference sequence, the 22‐nt Ems26+ recognition sequence is highlighted in grey and yellow. The latter indicates the 4 bp overhang, 5′‐GTAC‐3′, produced by nuclease cutting and is highlighted throughout for orientation purposes. Nucleotides shown are deleted sequence in seven of eight identified mutations, one larger deletion not shown. ZmMDH, Maize Mannitol dehydrogenase promoter; Ems26+, homing endonuclease targeting Ms26 target site; E, CaMV 35S promoter; H2B, maize Histone 2B promoter.
To test this scheme, rice plants were generated to contain a stable, randomly integrated ZmMDHpro:Ems26+ expression cassette as described above in the rice genome. Four independently transformed plants were allowed to self‐pollinate, and blue‐fluorescing seed (ZmEnd2pro:Cyan) were selected to initiate rice calli (Hervé and Kayano, 2006; Toki, 1997). Calli from these four events were allowed to grow at 26 °C (control) for 30 days, whereupon a portion of callus from each event was then incubated at 37 °C (treated) for 48 h. The temperature‐treated calli were returned to 26 °C, upon which control and treated samples were examined for the appearance of red‐fluorescing sectors. Over the course of 2 weeks, red sectors were readily visible in three of the four independent callus isolates treated at 37 °C (Figure 4b) and none in the control calli. Detection of red fluorescence indicated DSBs as a result of active Ems26+; therefore, genomic DNA was isolated from several red‐sectoring callus events and screened for mutations at the endogenous OsMs26 gene. A DNA fragment spanning a portion of the OsMs26 gene was amplified from control and temperature‐treated calli and screened using the BsiWI digestion assay. Analysis of amplified products from the three callus events demonstrated RF‐FP repair and revealed BsiWI‐resistant fragments present only in genomic DNA derived from these callus sectors incubated at 37 °C. Uncut amplified PCR products from control and temperature‐treated callus DNA were sequenced. Analysis of PCR products amplified from callus sectors maintained at 26 °C revealed only wild‐type OsMs26 sequences. In contrast, only four of the 16 PCR products derived from callus exposed to 37 °C contained wild‐type OsMs26 sequences. The remaining 12 products revealed mutations across the Ems26+ recognition site, which consisted of several independent deletions (Figure 4c). These observations demonstrated that a stably integrated and conditionally regulated endonuclease could be used in rice to generate site‐specific mutations and, thus, it has the potential to perform similarly in other monocots such as sorghum and wheat.
Mutations in the sorghum Ms26 ortholog confer male sterility
To determine whether the ortholog of maize Ms26 in S. bicolor was required for pollen formation, Ems26+ was used to direct mutations to the conserved recognition site in the 5th exon of this single‐copy gene (SbMs26) present on Chromosome 1 (Sb01g045960). Immature embryos from S. bicolor [Tx430 (Miller, 1984)] were transformed with the ZmMDHpro:Ems26+ T‐DNA using Agrobacterium‐mediated delivery. Six independent blue‐fluorescing callus sectors, indicative of the presence of a stably integrated copy of ZmMDHpro:Ems26+ T‐DNA, were used to generate sorghum plants (approximately 2–12 plants per callus sector). Intactness and copy number of the T‐DNA insertion were determined in these plants by DNA hybridization analysis (Southern, 1975) while amplification and sequencing of genomic DNA confirmed the presence of only wild‐type SbMs26 alleles in these plants. In addition, no red fluorescence was detected when progeny seed were screened visually. The presence of only wild‐type SbMs26 alleles and the absence of red‐fluorescing seed suggested that the Ems26+ was not activated in these T0 plants. Several plants containing single‐copy ZmMDHpro:Ems26+ T‐DNA insertions were allowed to self‐pollinate, and blue‐fluorescing immature embryos (10 plants, 16 embryos per plant, representing four independent T‐DNA insertions) were harvested 10–15 days postfertilization to be used for SbMs26 mutational analysis. Visual inspection of these harvested embryos revealed that there were no red‐fluorescing sectors or red‐fluorescing embryos present. These blue‐only‐fluorescing embryos were incubated at 26 °C or 37 °C for 48 h and examined for RF‐FP repair. In contrast to control embryos maintained at 26 °C, red‐fluorescing sectors were readily detected to varying degrees in all embryos incubated at 37 °C indicating the presence of active Ems26+. Calli from embryos treated at 37 °C were grown for 4 weeks and used to regenerate plants that were subsequently screened for mutations at the SbMs26 target site on Chromosome 1. In total, approximately 20% of the plants (n = 440) regenerated from these four independent T‐DNA insertion events revealed heterozygous mutations within the SbMs26 gene when screened by PCR amplification and digestion with BsiWI. Although expected at a low frequency, biallelic mutations were not detected in these sorghum experiments. Similar to the experiments in rice, DNA sequence analysis of these PCR products (n = 50 independent BsiWI‐resistant PCR fragments) revealed a variety of mutations across the SbMs26 region that consisted of deletions and insertions. In total, 16 nonidentical mutations were identified in these regenerated sorghum plants (Figure S3 and Text S3).
Sorghum plants containing heterozygous mutations at SbMs26 were allowed to flower, and seed from self‐pollinated plants was advanced to examine male fertility phenotype in progeny plants. T1 progeny containing heterozygous SbMs26 mutations [36‐bp deletion (Sbms26‐Δ36), 59‐bp deletion with 11‐bp insertion (Sbms26‐Δ59 + I11) and 78‐bp deletion (Sbms26‐Δ78)] were phenotypically identical to wild‐type sorghum Tx430 plants and were male fertile (Table 2). As shown in Figure 5a, panicles from SbMs26/Sbms26‐Δ78 plants extruded anthers that contained pollen and had begun to set seed as demonstrated by the appearance of collapsed stigmas after anthesis. Homozygous Sbms26 plants were also normal in stature and development with the exception of being completely male sterile: Sbms26‐Δ78 plants extruded small shrivelled anthers, did not shed pollen and did not set seed upon self‐pollination (Figure 5b and c). Upon closer examination, pollen was easily detected in anthers from SbMs26/Sbms26‐Δ78 plants (Figure 5d and g), while pollen was not observed in anthers from Sbms26‐Δ78/Sbms26‐Δ78 plants (Figure 5e and h). In summary, all SbMs26/Sbms26 plants were male fertile, while all Sbms26/Sbms26 plants were male sterile (Table 2). These male sterile plants were female fertile as demonstrated by their ability to set seed when pollinated with wild‐type sorghum pollen.
Table 2.
Male fertility phenotypes associated with sorghum mutants and complementation of male sterility using maize Ms26 gene
| SbMs26/Sbms26 | Sbms26/Sbms26 | ||||
|---|---|---|---|---|---|
| Mutation | Plants | Male fertile | Male sterile | Male fertile | Male sterile |
| Sbms26‐Δ36 | 6 | 3 | 0 | 0 | 3 |
| Sbms26‐Δ78 | 6 | 3 | 0 | 0 | 3 |
| Sbms26‐Δ59 + I11 | 6 | 3 | 0 | 0 | 3 |
| Complementation data | |||||
|---|---|---|---|---|---|
| Mutation | Plants | Sbms26/Sbms26 | Sbms26/Sbms26 + ZmMs26 | ||
| Male fertile | Male sterile | Male fertile | Male sterile | ||
| Sbms26‐Δ36 | 10a | 0 | 4 | 6 | 0 |
| Sbms26‐Δ78 | 15b | 0 | 10 | 5 | 0 |
SbMs26, sorghum Ms26 ortholog; ZmMs26, maize Ms26 gene.
Includes events #3 and #4.
Includes events #1, #2, and #3.
Figure 5.
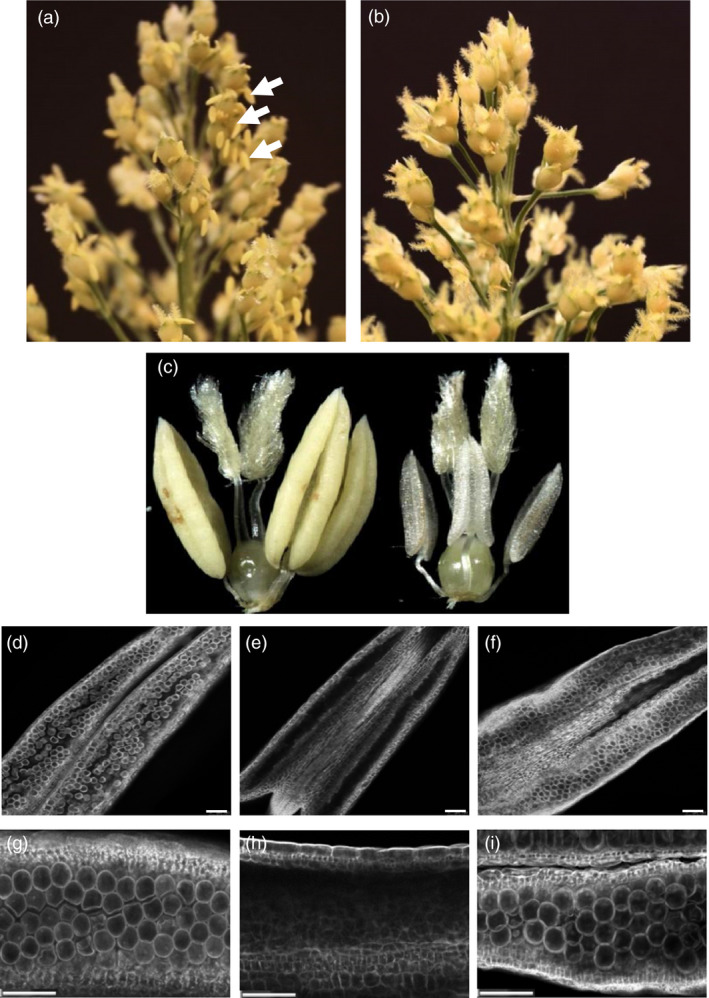
Male sterile sorghum generated from targeted mutagenesis of SbMs26 gene. (a) Panicles from male fertile sorghum plants (SbMs26/Sbms26‐Δ78) with extruded anthers (white arrows). (b) Panicles from male sterile sorghum plants (Sbms26‐Δ78/Sbms2‐Δ78). (c) Floret from male fertile (left) and from male sterile (Sbms26/Sbms26) sorghum plants (right). (d‐i) Images of sorghum anthers derived from wild‐type plants (d and g), homozygous Sbms26/Sbms26 plants (e and h), and Sbms26/Sbms26 plants containing ZmMs26 complementation vector (f and i). Magnification bar = 100 μM.
To further investigate that the male sterile phenotype observed in these sorghum mutant plants was directly due to deletions at SbMs26, restoration of fertility was tested using a copy of the maize Ms26 gene (also referred to as ZmMs26) introduced by transformation. Wild‐type sorghum plants were generated to contain a single copy of the ZmMs26 gene introduced on a T‐DNA expression cassette and crossed with plants carrying either the Sbms26‐Δ36 or the Sbms26‐Δ78 deletion mutation. From these crosses, F1 plants were screened and those that contained both the Sbms26 mutation and ZmMs26 were allowed to self‐pollinate to generate F2 seeds. F2 plants were grown under greenhouse conditions and genotyped for the Sbms26 mutation and presence of ZmMs26. A set of F2 plants homozygous for the Sbms26 mutation but segregated for the ZmMs26 expression cassette were selected. Plants homozygous for Sbms26‐Δ36 or Sbms26‐Δ78 but lacking ZmMs26 were male sterile; anthers did not contain pollen, and plants did not set seed upon self‐pollination (Table 2 and Table S1). However, when ZmMs26 was present, these homozygous Sbms26 mutant plants were male fertile (Table 2) with pollen development comparable to wild type (Figure 5f and i). The requirement of Sb01g045960 for pollen development in sorghum suggests that this gene is the ortholog of the maize male fertility gene Ms26.
Ems26+‐targeted mutations in the wheat Ms26 ortholog
As described above, the maize Ms26 polypeptide sequence was used to search the translated hexaploid wheat genome database (Chinese spring) (Consortium, 2014) for homologous sequences. A 4.2‐kb contig derived from Chromosome 4AS was found capable of encoding a 550 amino acid polypeptide having 91% sequence similarity to maize Ms26 protein (Figure S1 and Text S4). Additional BLAST (NCBI, Bethesda, MD, USA) search yielded wheat 4BL and 4DL contigs that only partially overlapped the region corresponding to the 2nd (4DL) and last exons (4BL and 4DL) of the 4AS sequence. To ascertain whether the Ems26+ recognition site was conserved across all three wheat genomes, UNIMS26 primers were used to amplify genomic DNA from the hexaploid spring wheat cultivar Fielder. DNA sequence analysis of these amplified products, when compared to the contigs described above, suggested that the 22‐nt Ems26+ recognition site was conserved. The A‐, B‐ and D‐genomic copies of the wheat Ms26 ortholog (referred to as TaMs26) were assigned to the Fielder genome (Figure S4). Distinguishing features within this amplified region include a six‐nucleotide, two‐amino acid gap; functional relevance of this sequence difference is not yet known, but interestingly is found in a region that is diverse across maize, sorghum, rice and wheat Ms26 protein alignments (Figure S5).
To examine whether the Ems26+ endonuclease was competent to generate targeted mutations at the TaMs26 gene orthologs in hexaploid wheat, Fielder plants containing the ZmMDHpro:Ems26+ T‐DNA were generated by Agrobacterium‐mediated transformation. Immature Fielder embryos were used as transformation targets. Blue‐fluorescing callus sectors indicative of T‐DNA integration were selected to initiate plant regeneration. Young wheat plantlets grown under greenhouse conditions were analysed for ZmMDHpro:Ems26+ T‐DNA copy number by qPCR. Four independently transformed wheat plants containing single‐ or low‐copy ZmMDHpro:Ems26+ T‐DNA were allowed to self‐pollinate and embryos were harvested 15 days postfertilization. Blue‐fluorescing embryos (n = 12) isolated from each plant were either held in the dark at 26 °C (control, n = 4) or incubated at 37 °C for 24 h (treated, n = 8), whereupon the treated embryos were returned to 26 °C and allowed to grow in the dark. As observed for the rice and sorghum experiments, approximately 72 h after treatment, wheat embryos incubated at 37 °C began to develop red‐fluorescing sectors, consistent with the presence of active Ems26+ endonuclease (Figure S6). No red‐fluorescing sectors were detected in control, nontreated wheat embryos. To ascertain whether Ems26+ generated mutations at endogenous TaMs26 sequences, total genomic DNA was isolated from temperature‐treated and control embryos and amplified using UNIMS26 primers. Deep sequencing analysis of these amplicons (approximately 5 million reads per sample) revealed that 1.6% of the amplicon reads from the temperature‐treated embryos contained mutations as compared to <0.007% from control embryos (Table S2 and Figure S7). Callus from embryos treated at 37 °C and containing ZmMDHpro:Ems26+ was used to regenerate 121 plants that were then screened for mutations using the BsiWI digestion assay; ten wheat plants contained BsiWI digestion‐resistant amplicons. DNA sequencing of these resistant PCR products identified seven different mutations within the Ems26+ recognition site consisting of a single‐nucleotide addition and deletions of various sizes; a nonoverlapping subset of these mutations was found within each genome copy of TaMs26 (Figure 6). Plants containing TaMs26 mutations were allowed to self‐pollinate for progeny analysis. Inheritance analysis determined that, with the exception of the single‐C‐nucleotide‐insertion mutant (no seed set), mutations in all three genomes were sexually transmitted to progeny plants. These progeny plants containing single‐genome, homozygous mutations in the A‐, B‐ or D‐genomic copies of TaMs26 were phenotypically normal and male fertile (Table S3). Together these experiments demonstrated that a conditionally expressed Ems26+ was capable of generating targeted mutations at the TaMs26 gene in planta and that single‐genome mutants in this P450 are not sufficient to confer a male sterile phenotype in the spring wheat cultivar Fielder. Studies that genetically combine these different A‐, B‐ and D‐genome mutations are in progress to determine the requirement of Ms26 for pollen development in wheat.
Figure 6.

TaMs26 gene mutations isolated from Fielder wheat A‐, B‐ and D‐genomes via targeted mutagenesis. Ems26+ recognition sequence is highlighted in yellow above the A‐genome sequence. Deleted nucleotide sequence is represented by a hyphen (‘‐’). Inserted nucleotide sequence is in white font and highlighted in black. Sequence differences between the A‐, B‐ and D‐genomes are highlighted in grey. White space indicates a gap in sequence.
Discussion
The ability to perform targeted mutagenesis in plants is a valuable tool for gene functional analysis and generation of genetic diversity. In this report, targeted mutagenesis of the monocot male fertility gene, Ms26, was accomplished in three different plant species using either a constitutively or conditionally expressed custom designed I‐CreI endonuclease, Ems26+. In rice, approximately 7% of the primary plants contained mutations across the highly conserved Ems26+ target site; these mutations ranged from single‐nucleotide deletions to large insertions. Similar to mutations identified by classical mutagenesis of maize and rice, targeted mutations to the rice Ms26 ortholog also resulted in complete male sterility by preventing the formation of pollen in homozygous recessive mutant plants. During these studies, it was determined that plants carrying a constitutively expressed Ems26+ endonuclease did not promote mutagenesis in subsequent generations of plants suggesting that mutations occurred early in the plant transformation process and functional endonucleases were not stably transmittable. In contrast, when Ems26+ was introduced into transformed plants and placed under the transcriptional control of a conditional promoter, the endonuclease was functional in progeny sorghum and wheat plants.
In previous reports, the low frequency of stable event recovery has been suggested to be an indicator of cytotoxicity due to off‐target recognition for ZFNs in human cell lines and plants (Cornu et al., 2007; Szczepek et al., 2007; Townsend et al., 2009). This suggestion is supported by the observation that in maize and rice experiments, in contrast to control experiments that did not introduce UBIpro:Ems26+, event recovery was lower when Ems26+ was delivered by biolistic transformation. Although it cannot be ruled out that reduced transformation frequency in these experiments was simply a consequence of the high concentration of vectors containing Ems26+ delivered by particle bombardment when compared to Agrobacterium transformation, differences in delivery and regulation of Ems26+ were important design considerations for these targeted mutagenesis experiments. The establishment of pre‐integrated and conditionally regulated endonuclease was one strategy that mitigated potential toxicity by taking advantage of a promoter capable of induction at higher temperatures and the temperature‐dependent biochemical activity of this class of endonucleases (Wang et al., 1997). In our studies, Ems26+ demonstrated increased nuclease activity at 37 °C both in vitro and in planta. Thus, short pulses at elevated temperatures during tissue culture or plant growth resulted in both increased transcription and nucleolytic activity of the Ems26+ gene and protein, respectively. This dual switch was an effective method for controlling DSB activity in these experiments and has been exploited as a means to rapidly generate large collections of allelic variants for gene discovery.
Pollen formation in plants is highly conserved at the biochemical and genetic level (for review see Shi et al., 2015). A number of pollen‐ and anther‐specific genes have been identified whose function is conserved across plants. The maize Ms26 and rice OsCYP704B2 genes encode a cytochrome P450 that plays a critical role in male fertility. In this study, conservation of sequence within these orthologous genes was used to introduce targeted mutations in rice, sorghum and wheat. Importantly, this is the first isolation and functional characterization of a male fertility gene in sorghum, a close relative to maize. As in rice, sorghum plants containing homozygous recessive mutations in the ortholog of the maize Ms26 gene were male sterile. Moreover, a transformed copy of the maize Ms26 gene was able to restore fertility in these sorghum mutant plants. The ability to restore fertility in these sorghum mutants using a maize Ms26 gene further supports biochemical conservation of P450 function.
This observed functional conservation is particularly relevant to the foundational experiments described in this report where the orthologs of the maize Ms26 gene were targeted in wheat. To date, there are only two recessive male fertility mutants that have been described in Triticum: Ms1 and Ms5 (Whitford et al., 2013). Given the high sequence similarity across maize, sorghum and rice, it is likely that orthologs of Ms26 in wheat are also required for pollen development. In contrast to these diploid species, wheat is an allohexaploid. In this study, it was observed that single‐genome homozygous TaMs26 mutations in hexaploid wheat did not confer a male sterility phenotype. These results suggest that no single TaMs26 genomic copy from the A‐, B‐ or D‐genome is essential to fertility in wheat, as the other wild‐type TaMs26 copies present in these plants are likely to function to maintain pollen development and a male fertility phenotype. It is interesting to speculate that despite the high amino acid sequence conservation across all three wheat genome copies, functional differences may be revealed and associated with relatively minor sequence changes, in particular within the regions of amino acid diversity (Figure S5). Thus, combining these TaMs26 gene mutations via genetic crosses will allow examination of the effects of various combinations of homozygous TaMs26 mutations in the three genomes of hexaploid wheat (i.e. in one, two or all three single genomes) on male fertility.
In this report, targeted mutagenesis of the monocot male fertility gene, Ms26, was made possible by advances of genomics, gene editing and plant transformation that overcome two barriers in crop improvement. First, having the ability to specifically target and modify fertility genes would extend SPT systems to crops having perfect flowers enabling yield gains by heterosis not yet fully realized. Second, gene editing, when combined with advanced plant transformation methods, would allow for precision breeding of elite germplasm reducing the time needed for introgression of induced sequence diversity by backcrossing. Classical methods for introducing genetic diversity into plants include mutagenesis with chemical and physical agents or transposition. These nontargeted methods result in random and often additional and undesired changes in the genome. These approaches are both time‐consuming and resource‐intensive requiring multiple generations of backcrossing and selection to convert back to the original state of elite genetics for maximal crop yields. In contrast to these nontargeted approaches that slow discovery and crop improvement, adoption of the precision breeding innovations described above is essential to sustaining agriculture as population increases and as protection of natural resources becomes more challenging (Tian et al., 2016; Tilman et al., 2011; West et al., 2014). Targeted mutagenesis of the Ms26 gene is a good example of how advanced technologies could be deployed to rapidly and precisely generate mutant fertility alleles in elite germplasm of maize, rice, sorghum and potentially wheat. While not exclusively considered a replacement for classical breeding methods, these advances could have a positive impact and change modern agriculture. As with classical mutagenesis methods, should undesired genome modifications occur (i.e. random integration of plasmid DNA or off‐target mutations), these changes could be easily bred away by genetic crosses to elite parental lines, which would eliminate unwanted genome alterations yet maintain targeted sequence variation. In addition, high‐resolution sequencing is particularly useful in the design and testing of nucleases with unique recognition and cutting specificity, thus further improving and advancing gene editing as an accelerated breeding tool.
Experimental procedures
DNA methods
Standard DNA techniques as described in Sambrook et al. (1989) were used for vector construction. The Ems26+ was maize codon‐optimized and designed to include an amino‐terminal nuclear localization signal SV40 (MAPKKKRKV, nucleotide position +1–30) and the potato ST‐LS1 intron [Accession number X04753 (nucleotides 2837–2892)] at nucleotide position +403–591 (Text S5). The chemically synthesized Ems26+, contained on a 1.280‐kb NcoI‐KpnI DNA fragment, was subcloned into PHP17720 (Sabelli et al., 2009) digested with NcoI‐KpnI that links Ems26+ to a maize constitutive ubiquitin‐1 promoter, including the first intron [−899 to +1092 (Christensen et al., 1992)], while transcription is terminated by the addition of the 3′ sequences from the potato proteinase inhibitor II gene (PinII) [nucleotides 2–310 (An et al., 1989)] to generate UBIpro:Ems26+ vector. To generate ZmENDpro:PAT RFP fusion plasmid, the maize END2 promoter [Endosperm‐ and embryo‐expressed promoter from maize; Chromosome 8, nt 171120325‐171119383 Maize (B73) Public Genome Assembly (AGP_v3.8)] was fused to the MoPAT‐DsRED (a translational fusion of the bialophos resistance gene, phosphinothricin‐N‐acetyl‐transferase, and the red fluorescent protein DsRED) expression cassette previously described in Ananiev et al. (2009) with transcription terminated by the PinII terminator described above. To enhance expression of this expression cassette, a 485‐bp fragment of the CaMV 35S promoter was inserted upstream of the ZmEnd2 promoter as described in Unger et al. (2001).
To generate a conditionally expressed Ems26+, a 1048‐bp fragment of temperature‐regulated promoter from the maize mannitol dehydrogenase gene [MDH; temperature‐inducible maize promoter; Chromosome 2, nt 8783228–8784275 Maize (B73) Public Genome Assembly (AGP_v3.2)] was modified to contain an RcaI restriction site at the 3′ end of the sequence (Svitashev et al., 2015) to accommodate translational fusion to the maize‐optimized Ems26+, producing the ZmMDHpro:Ems26+ expression cassette. To construct the stably integrated Ems26+ expression cassette, a T‐DNA vector containing ZmMDHpro:Ems26+ was generated and was identical to that described in Svitashev et al. (2015) with the exception that the ZmMDHpro:Ems26+ present on a 3.339‐kb HindIII‐PacI fragment replaced the UBIpro:Cas9 portion of this vector. In addition, the duplicated DsRED fragments in this gene were separated by a 136‐bp spacer that contained sequences for recognition by the Ems26+ endonuclease (Text S2). This plasmid was introduced into Agrobacterium strain LBA4404 (Komari et al., 1996) by electroporation using a Gene Pulser II (Bio‐Rad Laboratories, Inc., Hercules, CA, USA) as described by Gao et al. (2010) to generate Agrobacterium strains for plant transformation.
To generate the ms26 fertility complementation T‐DNA vector, the Ms45 gene in PHP24490 (Wu et al., 2015) was replaced by a 3.9‐kb DNA fragment [Chromosome 1, nt 4509689–14506399 Maize (B73) RefGen_v2 (MGSC)], which contained the wild‐type Ms26 gene. The LTP2 promoter present in this T‐DNA was also replaced by the maize END2 promoter as described above to transcriptionally regulate the RFP gene in this plasmid. The final T‐DNA cassette, pPG47::Bt1:ZmAA1//Ms26//35SEN‐pEND2::DsRed2, was introduced into Agrobacterium strain LBA4404 as described above.
For PCR and DNA sequence analysis, DNA was extracted from a small amount (0.5 cm in diameter) of callus tissue or two leaf punches (four leaf punches for wheat) as described in Gao et al. (2010). PCR was performed using REDExtract‐N‐Amp™ PCR ReadyMix (Sigma‐Aldrich Co., St. Louis, MO, USA) or Phusion® High‐Fidelity PCR Master Mix (New England Biolabs Inc., Ipswich, MA, USA) according to the manufacturer's recommendations. Ms26 target site mutations were identified by amplification of the region by PCR using the primer pair UNIMS26 5′‐2 (5′‐GACGTGGTGCTCAACTTCGTGAT‐3′) and UNIMS26 3′‐1 (5′‐GCCATGGAGAGGATGGTCATCAT‐3′) and digestion of the amplified products with BsiWI, the DNA restriction enzyme that recognizes the sequence 5′‐CGTACG‐3′, as described in Djukanovic et al. (2013). Products of these reactions were electrophoresed on 1% agarose gels and screened for BsiWI digestion‐resistant bands indicative of mutations at the Ms26 target site. PCR‐amplified fragments were also subcloned for DNA sequence analysis using the pCR2.1‐TOPO cloning vector (Thermo Fisher Scientific, Waltham, MA, USA).
Deep sequencing
Total genomic DNA was extracted from the temperature‐treated and nontreated calli transformed with the Ems26+ homing endonuclease using a modified CTAB method (Unger et al., 2001). The region surrounding the Ms26 target site was PCR‐amplified with Phusion® High‐Fidelity PCR Master Mix (New England Biolabs) adding on the sequences necessary for amplicon‐specific barcodes and Illumina sequencing using ‘tailed’ primers through two rounds of PCR. The primers used in the primary PCR Ms26 reaction were Ms26F (5′‐CTACACTCTTTCCCTACACGACGCTCTTCCGATCTAACCCGCGGAGGACGACGTGCTC‐3′) and Ms26R (5′‐CAAGCAGAAGACGGCATACGAGCTCTTCCGATCTCGTCGGGGCCCCAGTTGTAC‐3′), while the primers used in the secondary PCR reaction were F2 (5′‐AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACG‐3′) and R2 (5′‐CAAGCAGAAGACGGCATA‐3′). Genomic DNA extracted from leaves of untransformed Fielder plants served as a negative control. The resulting PCR amplifications were concentrated using a Qiagen MinElute® PCR purification spin column (QIAGEN, Inc., Venlo, Netherlands), electrophoresed on a 2% agarose gel, and amplifications were then excised and purified with a Qiagen gel extraction spin column. The concentration of the gel‐purified amplifications was measured with a Hoechst dye‐based fluorometric assay, combined in an equimolar ratio, and single‐read 100‐nt‐length deep sequencing was performed on an Illumina Genome Analyzer IIx (ELIM Biopharmaceuticals, Inc., Hayward, CA, USA) with a 30%–40% (v/v) spike of PhiX control v3 (Illumina, Inc., San Diego, CA, USA) to off‐set sequence bias. Only those reads with a ≥1 nucleotide indel arising within a 6‐nt window centred over the expected site of cleavage and not found in a similar level in the negative control were classified as NHEJ mutations. The total numbers of NHEJ mutations were then used to calculate the % mutant reads based on the total number of reads of an appropriate length containing a perfect match to the barcode and forward primer.
Plant methods
Rice, sorghum and wheat plant transformation and tissue culture details are outlined in Data S1.
Microscopy
Sorghum samples were dissected to remove the glumes and expose the anthers, and fixed in 2% paraformaldehyde and 4% glutaraldehyde overnight under vacuum (10 psi). After fixation, the samples were rinsed three times in 1× PBS (Phosphate Buffer Solution). Samples were then cleared in graded TDE (2, 2′‐Thiodiethanol) series (10%, 25%, 50% and 97%), and the 97% was repeated to assure optimal clearing. Samples were mounted in 97% TDE and the coverslip was sealed with nail polish. Confocal images were taken with the Leica (Wetzlar, Germany) TCS SPE using the solid‐state 405 nm with the DAPI setting (emission: 430–480); the 488 nm with the GFP setting (emission: 500–600); and the 561 nm with the DRED setting (emission: 650–700) laser lines and merged using the Leica LAS X software. Final image adjustments were accomplished with Adobe Systems Photoshop CS6 (San Jose, CA).
Conflict of interest
AMC and JY are inventors on pending applications on this work and are current employees of DuPont Pioneer who owns the pending patent applications. MS, GB, LF, MK, M‐JC, NS and SS are current or former employees of DuPont Pioneer.
Supporting information
Figure S1 Ms26 protein sequence is highly conserved across crop species.
Figure S2 BsiWI restriction enzyme resistance assay screen for OsMs26 mutations.
Figure S3 SbMs26 gene mutations in sorghum achieved via targeted mutagenesis.
Figure S4 DNA sequences corresponding to the IWGSC contigs containing the wheat 4AS, 4BL and 4DL genomic regions orthologous to maize Ms26 gene and aligned with DNA sequences from Fielder.
Figure S5 Predicted protein translation products of amplicons derived from UNIMS26 primers.
Figure S6 Harvested Fielder wheat embryo containing Ems26+ under transcriptional control of a maize temperature‐regulated promoter.
Figure S7 Top 10 amplicon sequences containing mutations from genomic DNA derived from Fielder wheat callus transformed with ZmMDHpro:Ems26+.
Table S1 Restoration of male fertility to homozygous Sbms26/Sbms26 plants with a transformed copy of maize ZmMs26 was achieved using four nonidentical events in two different Sbms26 mutant backgrounds.
Table S2 Percent (%) mutant reads at the wheat Ems26+ target locus identified by deep sequencing.
Table S3 Male fertility phenotype for wheat TaMs26 mutant lines.
Text S1 4.194‐kb region containing wheat Ms26 ortholog IWGSC 4AS.
Text S2 Spacer sequence (136 bp) containing multiple nuclease target sites.
Text S3 Nucleotide sequence of Ems26+ target site (underlined) present in Chromosome 7 in Sorghum bicolor Tx430.
Text S4 Predicted 550 amino acid sequence encoded by wheat Ms26 ortholog IWGSC 4AS coding region.
Text S5 Maize‐optimized Ems26+ coding region.
Data S1 Experimental procedures detailing plant transformation and tissue culture methods.
Acknowledgements
The authors thank Marc Albertsen and Peggy Ozias‐Akins for reviewing the manuscript and providing comments. The authors also thank Katherine Thilges for her assistance with the sorghum microscopy and special thanks to Tracey Fisher for her assistance during manuscript preparation and for valuable comments during review. Authors also thank anonymous reviewers for insightful input.
References
- An, G. , Mitra, A. , Choi, H.K. , Costa, M.A. , An, K. , Thornburg, R.W. and Ryan, C.A. (1989) Functional analysis of the 3′ control region of the potato wound‐inducible proteinase inhibitor II gene. Plant Cell, 1, 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev, E. , Wu, C. , Chamberlin, M. , Svitashev, S. , Schwartz, C. , Gordon‐Kamm, W. and Tingey, S. (2009) Artificial chromosome formation in maize (Zea mays L.). Chromosoma, 118, 157–177. [DOI] [PubMed] [Google Scholar]
- Casas, A.M. , Kononowicz, A.K. , Haan, T.G. , Zhang, L. , Tomes, D.T. , Bressan, R.A. and Hasegawa, P.M. (1997) Transgenic sorghum plants obtained after microprojectile bombardment of immature inflorescences. In Vitro Cellular & Developmental Biology‐Plant, 33, 92–100. [Google Scholar]
- Christensen, A.H. , Sharrock, R.A. and Quail, P.H. (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 18, 675–689. [DOI] [PubMed] [Google Scholar]
- Cigan, A.M. , Unger, E. , Xu, R.‐J. , Kendall, T.L. and Fox, T.W. (2001) Phenotypic complementation of ms45 maize requires tapetal expression of MS45. Sex. Plant Reprod. 14, 135–142. [Google Scholar]
- Cigan, A.M. , Unger‐Wallace, E. and Haug‐Collet, K. (2005) Transcriptional gene silencing as a tool for uncovering gene function in maize. Plant J. 43, 929–940. [DOI] [PubMed] [Google Scholar]
- Consortium, I.W.G.S. (2014) A chromosome‐based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science, 345, 1251788. [DOI] [PubMed] [Google Scholar]
- Cornu, T.I. , Thibodeau‐Beganny, S. , Guhl, E. , Alwin, S. , Eichtinger, M. , Joung, J.K. and Cathomen, T. (2007) DNA‐binding specificity is a major determinant of the activity and toxicity of zinc‐finger nucleases. Mol. Ther. 16, 352–358. [DOI] [PubMed] [Google Scholar]
- Djukanovic, V. , Smith, J. , Lowe, K. , Yang, M. , Gao, H. , Jones, S. , Nicholson, M.G. et al. (2013) Male‐sterile maize plants produced by targeted mutagenesis of the cytochrome P450‐like gene (MS26) using a re‐designed I‐CreI homing endonuclease. Plant J. 76, 888–899. [DOI] [PubMed] [Google Scholar]
- Duvick, D.N. (1999) Heterosis: feeding people and protecting natural resources. In: The Genetics and Exploitation of Heterosis in Crops ( Corrs, J.G. , Pandey, S. and Gerdes, J.T. eds), pp. 19–29. Madison, WI: ASA‐CSSA‐SSSA. [Google Scholar]
- Dwivedi, S. , Perotti, E. and Ortiz, R. (2008) Towards molecular breeding of reproductive traits in cereal crops. Plant Biotechnol. J. 6, 529–559. [DOI] [PubMed] [Google Scholar]
- Gao, H. , Smith, J. , Yang, M. , Jones, S. , Djukanovic, V. , Nicholson, M.G. , West, A. et al. (2010) Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 61, 176–187. [DOI] [PubMed] [Google Scholar]
- Guo, J.‐X. and Liu, Y.‐G. (2012) Molecular control of male reproductive development and pollen fertility in rice. J. Integr. Plant Biol. 54, 967–978. [DOI] [PubMed] [Google Scholar]
- Hervé, P. and Kayano, T. (2006) Japonica rice varieties (Oryza sativa, Nipponbare, and others). In: Agrobacterium Protocols ( Wang, K. ed) pp. 213–222. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Hobo, T. , Suwabe, K. , Aya, K. , Suzuki, G. , Yano, K. , Ishimizu, T. , Fujita, M. et al. (2008) Various spatiotemporal expression profiles of anther‐expressed genes in rice. Plant Cell Physiol. 49, 1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komari, T. , Hiei, Y. , Saito, Y. , Murai, N. and Kumashiro, T. (1996) Vectors carrying two separate T‐DNAs for co‐transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J. 10, 165–174. [DOI] [PubMed] [Google Scholar]
- Kumar, V. and Jain, M. (2015) The CRISPR–Cas system for plant genome editing: advances and opportunities. J. Exp. Bot. 66, 47–57. [DOI] [PubMed] [Google Scholar]
- Li, J. and Yuan, L. (1999) Hybrid rice: genetics, breeding and seed production. In Plant Breeding Reviews ( Janick, J. , ed), pp. 15–158. Oxford, U.K.: John Wiley & Sons Inc. [Google Scholar]
- Li, H. , Pinot, F. , Sauveplane, V. , Werck‐Reichhart, D. , Diehl, P. , Schreiber, L. , Franke, R. et al. (2010) Cytochrome P450 family member CYP704B2 catalyzes the ω‐hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. The Plant Cell Online, 22, 173–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukides, C.A. , Broadwater, A.H. and Bedinger, P.A. (1995) Two new male‐sterile mutants of Zea mays (Poaceae) with abnormal tapetal cell morphology. Am. J. Bot. 82, 1017–1023. [Google Scholar]
- Melchinger, A.E. (1999) Genetic diversity and heterosis. In: The Genetics and Exploitation of Heterosis in Crops ( Coors, J.G. and Pandy, S. eds), pp. 99–118. Madison, WI: CSSA. [Google Scholar]
- Miller, F.R. (1984) Registration of RTx430 sorghum parental line. Crop Sci. 24, 1224. [Google Scholar]
- Neuffer, M.G. , Coe, E.H. and Wessler, S.R. (1997) Mutants of Maize. Plainview, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Osakabe, Y. and Osakabe, K. (2015) Genome editing with engineered nucleases in plants. Plant Cell Physiol. 56, 389–400. [DOI] [PubMed] [Google Scholar]
- Parry, M.A.J. and Hawkesford, M.J. (2012) An integrated approach to crop genetic improvement. J. Integr. Plant Biol. 54, 250–259. [DOI] [PubMed] [Google Scholar]
- Puchta, H. and Hohn, B. (2012) In planta somatic homologous recombination assay revisited: a successful and versatile, but delicate tool. Plant Cell, 24, 4324–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, D.K. , Mueller, N.D. , West, P.C. and Foley, J.A. (2013) Yield trends are insufficient to double global crop production by 2050. PLoS One, 8, e66428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, B.V.S. , Ramesh, S. and Ortiz, R. (2005) Genetic and cytoplasmic‐nuclear male sterility in sorghum. In Plant Breeding Reviews ( Janick, J. , ed), pp. 139–172. Oxford, U.K.: John Wiley & Sons Inc. [Google Scholar]
- Sabelli, P.A. , Hoerster, G. , Lizarraga, L.E. , Brown, S.W. , Gordon‐Kamm, W.J. and Larkins, B.A. (2009) Positive regulation of minichromosome maintenance gene expression, DNA replication, and cell transformation by a plant retinoblastoma gene. Proc. Natl Acad. Sci. 106, 4042–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Shi, J. , Cui, M. , Yang, L. , Kim, Y.J. and Zhang, D. (2015) Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci. 20, 741–753. [DOI] [PubMed] [Google Scholar]
- Southern, E.M. (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98, 503–517. [DOI] [PubMed] [Google Scholar]
- Svitashev, S. , Young, J.K. , Schwartz, C. , Gao, H. , Falco, S.C. and Cigan, A.M. (2015) Targeted mutagenesis, precise gene editing, and site‐specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169, 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepek, M. , Brondani, V. , Buchel, J. , Serrano, L. , Segal, D.J. and Cathomen, T. (2007) Structure‐based redesign of the dimerization interface reduces the toxicity of zinc‐finger nucleases. Nat. Biotechnol. 25, 786–793. [DOI] [PubMed] [Google Scholar]
- Tester, M. and Landrige, P. (2010) Breeding technologies to increase crop production in a changing world. Science, 327, 818–822. [DOI] [PubMed] [Google Scholar]
- Tian, H. , Lu, C. , Ciais, P. , Michalak, A.M. , Canadell, J.G. , Saikawa, E. , Huntzinger, D.N. et al. (2016) The terrestrial biosphere as a net source of greenhouse gases to the atmosphere. Nature, 531, 225–228. [DOI] [PubMed] [Google Scholar]
- Tilman, D. , Balzer, C. , Hill, J. and Befort, B.L. (2011) Global food demand and the sustainable intensification of agriculture. Proc. Natl Acad. Sci. USA, 108, 20260–20264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki, S. (1997) Rapid and efficient Agrobacterium‐mediated transformation in rice. Plant Mol. Biol. Rep. 15, 16–21. [Google Scholar]
- Townsend, J.A. , Wright, D.A. , Winfrey, R.J. , Fu, F. , Maeder, M.L. , Joung, J.K. and Voytas, D.F. (2009) High‐frequency modification of plant genes using engineered zinc‐finger nucleases. Nature, 459, 442–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger, E. , Betz, S. , Xu, R. and Cigan, A.M. (2001) Selection and orientation of adjacent genes influences DAM‐mediated male sterility in transformed maize. Transgenic Res. 10, 409–422. [DOI] [PubMed] [Google Scholar]
- Voytas, D.F. and Gao, C. (2014) Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 12, e1001877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Kim, H. , Yuan, X. and Herrin, D. (1997) Purification, biochemical characterization and protein‐DNA interactions of the I‐CreI endonuclease produced in Escherichia coli . 10.1093/nar/25.19.3767. Nucleic Acids Res. 25, 3767–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck‐Reichhart, D. and Feyereisen, R. (2000) Cytochromes P450: a success story. Genome Biol. 1, 3003.1–3003.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, P.C. , Gerber, J.S. , Engstrom, P.M. , Mueller, N.D. , Brauman, K.A. , Carlson, K.M. , Cassidy, E.S. et al. (2014) Leverage points for improving global food security and the environment. Science, 345, 325–328. [DOI] [PubMed] [Google Scholar]
- Whitford, R. , Fleury, D. , Reif, J.C. , Garcia, M. , Okada, T. , Korzun, V. and Langridge, P. (2013) Hybrid breeding in wheat: technologies to improve hybrid wheat seed production. J. Exp. Bot. 64, 5411–5428. [DOI] [PubMed] [Google Scholar]
- Wu, Y. , Fox, T.W. , Trimnell, M.R. , Wang, L. , Xu, R.J. , Cigan, A.M. , Huffman, G.A. et al. (2015) Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross‐pollinating crops. Plant Biotechnol. J. 14, 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata, Y. , Doi, K. , Yasui, H. and Yoshimura, A. (2007) Identification of mutants for abnormal pollen development in rice. Breeding Science, 57, 331–337. [Google Scholar]
- Zhao, Z.‐Y. , Cai, T. , Tagliani, L. , Miller, M. , Wang, N. , Pang, H. , Rudert, M. et al. (2000) Agrobacterium‐mediated sorghum transformation. Plant Mol. Biol. 44, 789–798. [DOI] [PubMed] [Google Scholar]
- Zhu, H. , Jeoung, J.M. , Liang, G.H. , Muthukrishnan, S. , Krishnaveni, S. and Wilde, G. (1998) Biolistic transformation of sorghum using a rice chitinase gene [Sorghum bicolor (L.) Moench‐Oryza sativa L.]. Journal of Genetics & Breeding (Italy), 52, 243–252. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Ms26 protein sequence is highly conserved across crop species.
Figure S2 BsiWI restriction enzyme resistance assay screen for OsMs26 mutations.
Figure S3 SbMs26 gene mutations in sorghum achieved via targeted mutagenesis.
Figure S4 DNA sequences corresponding to the IWGSC contigs containing the wheat 4AS, 4BL and 4DL genomic regions orthologous to maize Ms26 gene and aligned with DNA sequences from Fielder.
Figure S5 Predicted protein translation products of amplicons derived from UNIMS26 primers.
Figure S6 Harvested Fielder wheat embryo containing Ems26+ under transcriptional control of a maize temperature‐regulated promoter.
Figure S7 Top 10 amplicon sequences containing mutations from genomic DNA derived from Fielder wheat callus transformed with ZmMDHpro:Ems26+.
Table S1 Restoration of male fertility to homozygous Sbms26/Sbms26 plants with a transformed copy of maize ZmMs26 was achieved using four nonidentical events in two different Sbms26 mutant backgrounds.
Table S2 Percent (%) mutant reads at the wheat Ems26+ target locus identified by deep sequencing.
Table S3 Male fertility phenotype for wheat TaMs26 mutant lines.
Text S1 4.194‐kb region containing wheat Ms26 ortholog IWGSC 4AS.
Text S2 Spacer sequence (136 bp) containing multiple nuclease target sites.
Text S3 Nucleotide sequence of Ems26+ target site (underlined) present in Chromosome 7 in Sorghum bicolor Tx430.
Text S4 Predicted 550 amino acid sequence encoded by wheat Ms26 ortholog IWGSC 4AS coding region.
Text S5 Maize‐optimized Ems26+ coding region.
Data S1 Experimental procedures detailing plant transformation and tissue culture methods.


