Summary
The flowering time regulator GIGANTEA (GI) connects networks involved in developmental stage transitions and environmental stress responses in Arabidopsis. However, little is known about the role of GI in growth, development and responses to environmental challenges in the perennial plant poplar. Here, we identified and functionally characterized three GI‐like genes (PagGIa, PagGIb and PagGIc) from poplar (Populus alba × Populus glandulosa). PagGIs are predominantly nuclear localized and their transcripts are rhythmically expressed, with a peak around zeitgeber time 12 under long‐day conditions. Overexpressing PagGIs in wild‐type (WT) Arabidopsis induced early flowering and salt sensitivity, while overexpressing PagGIs in the gi‐2 mutant completely or partially rescued its delayed flowering and enhanced salt tolerance phenotypes. Furthermore, the PagGIs‐PagSOS2 complexes inhibited PagSOS2‐regulated phosphorylation of PagSOS1 in the absence of stress, whereas these inhibitions were eliminated due to the degradation of PagGIs under salt stress. Down‐regulation of PagGIs by RNA interference led to vigorous growth, higher biomass and enhanced salt stress tolerance in transgenic poplar plants. Taken together, these results indicate that several functions of Arabidopsis GI are conserved in its poplar orthologues, and they lay the foundation for developing new approaches to producing salt‐tolerant trees for sustainable development on marginal lands worldwide.
Keywords: poplar, Arabidopsis, PagGI , salt tolerance, RNAi, transgenic plants
Introduction
The worldwide population is expected to reach between 9.6 and 12.3 billion by 2100, which presents challenges to energy security, economic growth and environmental protection (Gerland et al., 2014). Environmental problems such as global warming, drought and salinity severely limit agricultural and forest productivity (Lobell and Gourdji, 2012; Sivakumar et al., 2005). There is an urgent need for plant breeders to develop novel plant varieties with increased growth and tolerance to various environment stresses. Recent studies have revealed that genetic modification of flowering time, or circadian rhythm, represents a useful new strategy for crop and woody plant improvement (Grundy et al., 2015; Jung and Müller, 2009; Ni et al., 2009).
In the annual plant Arabidopsis, GIGANTEA (GI) was originally identified based on its roles in photoperiodic flowering and circadian clock regulation (Fowler et al., 1999; Martin‐Tryon et al., 2007). First, the GI‐CONSTANS (CO)‐Flowering Locus T (FT) regulatory module controls flowering time under long‐day conditions (Takada and Goto, 2003). GI and the FLAVIN‐BING, KELCH REPEAT, F‐BOX 1 protein form a complex that controls daytime CO transcription in a light‐dependent manner by degrading a key CO repressor, CYCLING DOF FACTOR 1 (Sawa et al., 2007). Second, GI interacts with F‐box protein ZEITLUPE (ZTL) through the amino‐terminal flavin‐binding LIGHT, OXYGEN or VOLTAGE domain of ZTL, which is necessary to sustain a normal circadian period by regulating the proteasome‐dependent degradation of the central circadian oscillator, TIMING OF CAB EXPRESSION 1 (Kim et al., 2007). Recent findings indicate that GI also mediates responses to environmental stress. Among abiotic stress factors, salinity affects almost all aspects of plant development, including germination, vegetative growth and reproductive development, which threatens crop production on over 800 million hectares, or one‐quarter to one‐third of all agricultural land on earth (Rengasamy, 2010; Zhu, 2002). The salt stress response is controlled by many genes that function through complex genetic regulatory networks. One of the key responses to salt stress is maintaining cellular ion homoeostasis by restricting the accumulation of toxic sodium (Na+). The Salt Overly Sensitive (SOS) signalling pathway is a well‐defined signalling pathway required for the control of ion homeostasis (Zhu, 2002). The SOS pathway modulates Na+ levels via three known components: calcium‐binding protein SOS3, protein kinase SOS2 and plasma membrane Na+/H+ antiporter SOS1. Under salt stress conditions, the SOS2–SOS3 complex phosphorylates and activates the transport activity of the SOS1 antiporter (Guo et al., 2001; Ji et al., 2013; Zhu, 2000). The SOS pathway is functionally conserved in dicot plants Arabidopsis and Brassica nigra, the monocot plant rice, and the woody plant poplar (Tang et al., 2010; Yang et al., 2015). The role of GI in the salt stress response was recently documented. GI competitively binds to SOS2 kinase and prevents the phosphorylation‐dependent activation of SOS1 under normal conditions. However, in the presence of high salt, GI is degraded by the 26S proteasome. Released SOS2 interacts with SOS3 to form an active SOS2–SOS3 protein kinase complex, which subsequently activates the plasma membrane‐localized Na+/H+ antiporter SOS1. As a result, sodium ions are exported from the cell and salt tolerance is established (Kim et al., 2013a). GI is highly conserved in monocot and dicot plants (Song et al., 2010). However, the nature of the circadian clock gene GI in species other than Arabidopsis is much less well understood.
As perennial plants, forest trees are highly valuable, as they provide raw materials, help maintain biodiversity, protect land and water resources, and help mitigate the effects of climate change (Harfouche et al., 2011; Lambrou and Laub, 2004). Forest trees have many unique characteristics, such as an extended juvenile phase that can last for decades, critical day length originating from different latitudes, and seasonal changes and cycling between periods of growth and dormancy (Bäurle and Dean, 2006; Jansson and Douglas, 2007). Most poplar cultivars are extremely sensitive to saline soils (Chen et al., 2014; Polle and Chen, 2014). Increasing the salt tolerance in crops and woody plants has become a major challenge for modern agriculture. Although much effort has focused on increasing salt tolerance in poplar through conventional breeding and genetic engineering, few studies have successfully produced transgenic poplar plants with enhanced salt tolerance using known abiotic stress‐associated genes from other species (Harfouche et al., 2011; Vinocur and Altman, 2005). Recently, transgenic poplar plants overexpressing the poplar SOS2 gene were generated, which exhibit enhanced tolerance to salt stress (Yang et al., 2015). Since SOS2 is regulated by the photoperiodicity and circadian clock switch GI (Kim et al., 2013a), unravelling the mechanisms underlying the control of flowering time and its correlation with environmental stress tolerance in trees would greatly accelerate current tree‐breeding programmes.
In this study, we identified three GI‐like genes (named PagGIa, PagGIb and PagGIc) from poplar (P. alba × P. glandulosa) and investigated their roles in circadian rhythm and flowering time control in transgenic Arabidopsis plants. In addition, we developed a salt‐tolerant transgenic poplar line by down‐regulating the expression of PagGIs, as these genes negatively regulate salt stress tolerance via an SOS‐mediated pathway. This study provides the first evidence that altering the expression of GI genes through genetic engineering could be used to develop woody plants with enhanced growth and increased salt tolerance.
Results
Cloning and characterization of poplar GIGANTEA (GI)‐like genes
We isolated the cDNA fragments of poplar GI‐like genes (PagGIa, PagGIb and PagGIc) from P. alba × P. glandulosa by homology‐based BLAST searches (DOE JGI, Walnut Creek, CA, USA) against the Joint Genome Institute Phytozome database (www.phytozome.com). Phylogenetic analysis of the deduced plant GI amino acid sequences from 12 species showed that PagGIa, b and c are closely clustered with other dicot GI homologues and belong to the same subgroup as AtGI (Figure 1a). These results suggest that PagGIa, b, c and AtGI share a conserved role.
Figure 1.
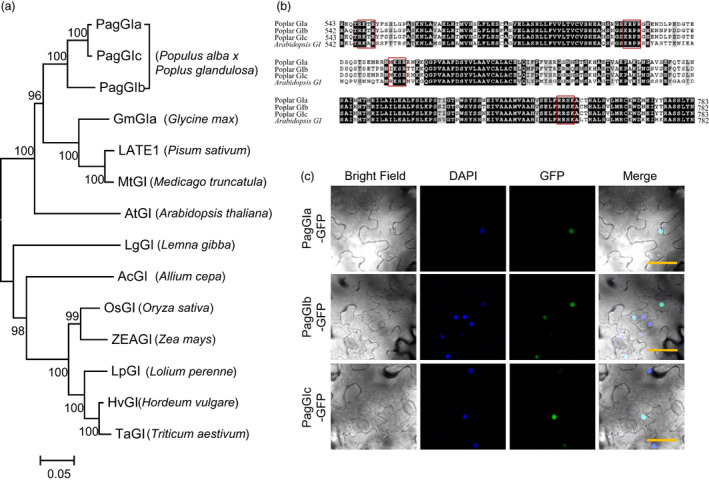
GIGANTEA ‐like Genes in Poplar. (a) Phylogenetic analysis of poplar GIGANTEA‐like genes. (b) Comparison of the amino acid sequences of nuclear localization regions of poplar and Arabidopsis GI. The alignment was performed using BioEdit and BoxShade server. Identical and similar amino acid residues are shaded in black and grey respectively. The nuclear localization sites are indicated by red boxes. (c) Subcellular localization of PagGIa‐, b‐ and c‐GFP fusion proteins. DAPI (second row) and GFP (third row) fluorescence was observed at 3 days after infiltration. Both fluorescence images are merged in the fourth row (merge). Scale bars represent 50 μM.
Comparison of the deduced amino acid sequences of AtGI and the PagGI genes revealed that the regions for their nuclear localization are conserved among Arabidopsis and poplar GI, which contain four clusters of basic amino acids similar to the clusters identified in established bipartite NLSs in other nuclear proteins (Huq et al., 2000) (Figure 1b). To investigate the subcellular localization of PagGIa, b and c, we transiently transfected N. benthamiana leaves with a 35S:PagGIa/b/c:GFP construct. As shown in Figure 1c, GFP fluorescence produced by PagGIa‐, b‐ and c‐GFP fusion proteins overlapped with blue fluorescence, as revealed by 4′,6‐diamidino‐2‐phenylindole (DAPI) staining. These results suggest that PagGIa, b and c predominantly localize to the nucleus and may play a role in transcriptional regulation. GI function in photoperiodic flowering and circadian rhythms have been extensively studied from monocot to dicot plants (Mouradov et al., 2002), but have not yet been reported in perennial poplar. To verify whether poplar PagGIs are orthologues of Arabidopsis GI, we first characterized its original functions in regulation of circadian clock, flowering time.
PagGIa, b and c are involved in the regulation of circadian rhythm in poplar
To determine whether PagGI mRNA levels fluctuate throughout the day, we examined the expression levels of the PagGI genes every 4 hours for 2 days. As shown in Figure 2a, PagGI (the sum of PagGIa, b and c) transcripts were diurnally regulated, with a peak around zeitgeber 12 (ZT12) (Zerr et al., 1990) in mature leaves. Moreover, PagGI mRNAs were most abundant in young leaves among all tissues examined, including shoot tips, young leaves, mature leaves, bark, xylem and roots (Figure 2b). We then determined whether PagGI transcript levels were affected by salinity stress. As shown in Figure 2a, PagGI mRNA levels were significant up‐regulated throughout the day and in all tissues examined in response to salt stress treatment. We also investigated the transcript levels of PagGIa, b and c separately. Since it is difficult to distinguish between PagGIa and PagGIc due to their high homology, we listed the expression data for PagGIa/c, the sum of PagGIa and PagGIc. As shown in Figure S1a,b, PagGIa/c and PagGIb expression was induced by salt stress, and PagGIb was expressed at higher levels than PagGIa/c throughout the day and in all tissues examined.
Figure 2.
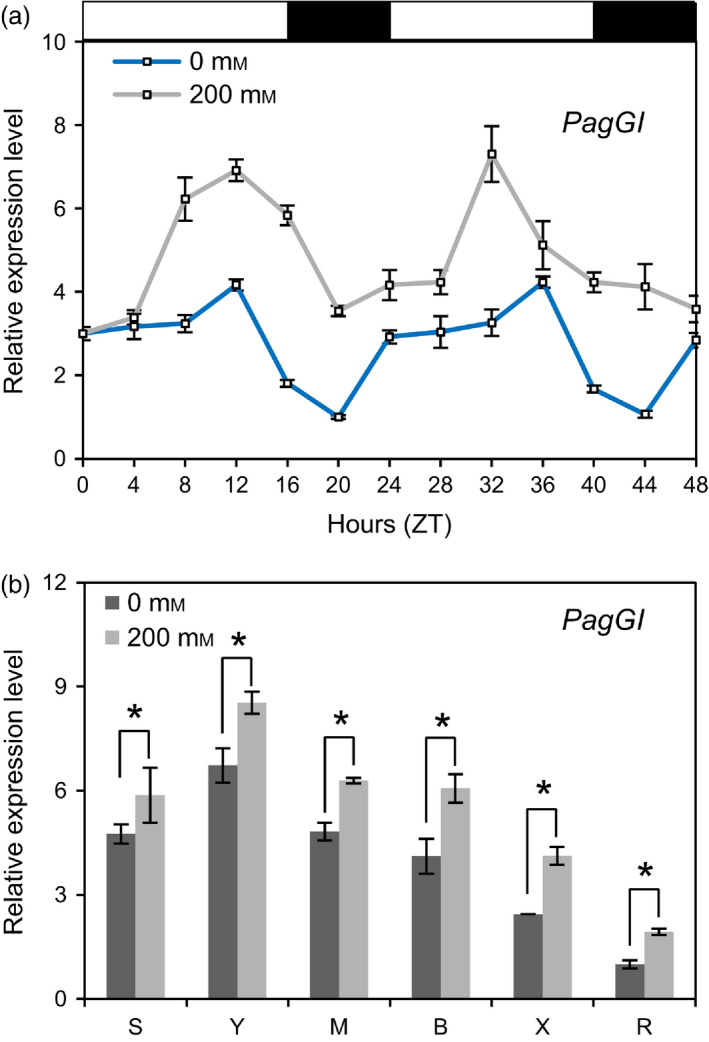
PagGIa, b and c are Involved in the Regulation of Circadian Rhythms. (a) Time course of PagGI expression in mature leaves of poplar plants under LDs. (b) Expression levels of PagGI in different tissues at ZT12. Total RNA samples were collected every 4 h from 2‐month‐old poplar plants entrained in LDs of 48 h with or without 200 mm NaCl treatments. The mRNA abundance was quantified by quantitative RT‐PCR, which was performed in triplicate with three independently harvested samples. Actin expression was used as an internal control. Detected tissues including shoot tip (S), young leaf (Y), mature leaf (M), bark (B), xylem (X) and root (R). White and black bars above the graph indicate day and night periods respectively. Error bars represent SD of three independent experiments. Asterisks indicate significant differences at P < 0.05.
Overexpression of PagGIs induces early flowering in Arabidopsis
To investigate whether PagGIa, b and c play a functionally conserved role in regulating flowering time, we generated transgenic plants overexpressing PagGIa, b and c in the WT (35S:PagGIa/b/c Col‐0) and gi‐2 mutant (35S:PagGIa/b/c gi‐2) backgrounds. We first confirmed the expression levels of PagGIs in transgenic Arabidopsis by RT‐PCR, immunoblotting analysis, and confocal microscopy (Figure S2a–c). Next, we selected transgenic lines overexpressing the PagGI genes (two lines per gene) and subjected them to detailed phenotypic analysis under long‐day conditions (LDs). The 35S:PagGIa/b/c Col‐0 plants flowered earlier than the WT (Figure 3a–c). Arabidopsis gi‐2 mutant plants exhibit a strong late‐flowering phenotype (Fowler et al., 1999). However, this late‐flowering phenotype was partially or completely rescued in 35S:PagGIa/b/c gi‐2 plants. The 35S:PagGIa/b/c Col‐0 and 35S:PagGIa/b/c gi‐2 transgenic lines flowered from 15 to 17.8 days and from 19.7 to 38.2 days of seed sowing at the 3/4 (rosette) stage under LDs, whereas WT and gi‐2 flowered within 19.3 and 41.3 days of seed sowing respectively (Figure 3c). These results suggest that PagGIa, b and c play a role in flowering time regulation in plants.
Figure 3.
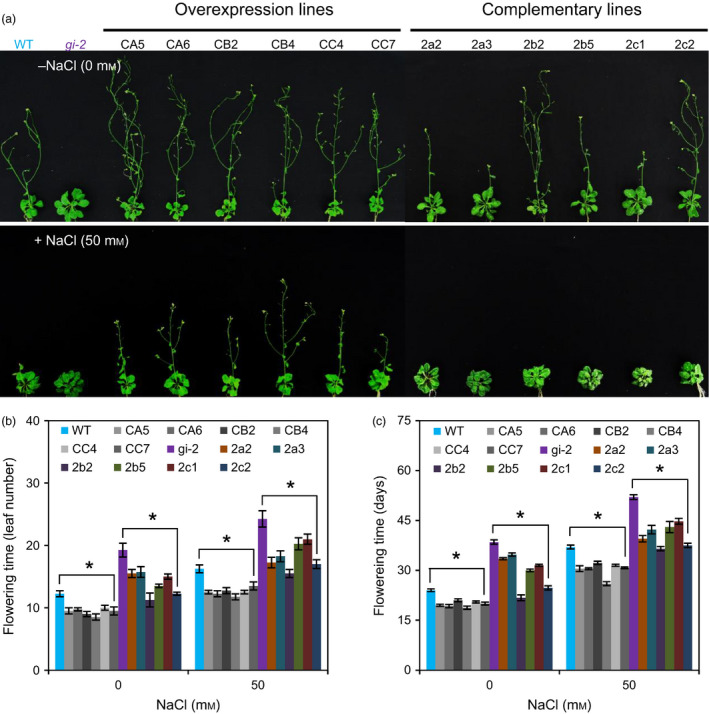
Overexpression of PagGI Genes Induces Early Flowering in Transgenic Arabidopsis (T3 Generation). (a) Flowering phenotype of 35S:PagGIa/b/c Col‐0 (overexpression lines) and 35S:PagGIa/b/c gi‐2 (complemented lines). Three‐day‐old seedlings were transferred from MS basal medium to MS medium without (0 mm) or with 50 mm NaCl. Plants were grown under LDs and photographed at 5 weeks. Flowering time was scored by counting the number of leaves (rosette and cauline) (b) and measuring the days to bolting (c) when bolted main stems were ~1 cm long. More than 10 plants were measured per data point. Error bars represent SD of three independent experiments. Asterisks indicate significant differences at P < 0.05.
Salt stress delays the floral transition in Arabidopsis (Kim et al., 2013a; Li et al., 2007). To determine whether PagGIa, b and c transcript levels affect flowering time under salt stress, we observed the flowering time of WT and transgenic Arabidopsis plants under salt stress. As shown in Figure 3a–c, salt‐induced late flowering was fully suppressed in 35S:PagGIa/b/c Col‐0 and partially suppressed in 35S:PagGIa/b/c gi‐2 plants, whereas WT displayed a late‐flowering phenotype under salt stress.
Overexpression of PagGI genes confers salt sensitivity in Arabidopsis
To determine whether PagGIa, b and c play a functionally conserved role in negatively regulating the salt stress response, we evaluated the roles of PagGIa, b and c in transgenic Arabidopsis under salt stress condition. To examine the sensitivity of transgenic Arabidopsis seedlings to salt stress, we planted WT, 35S:PagGIa/b/c Col‐0 and 35S:PagGIa/b/c gi‐2 seeds on MS medium with or without the indicted concentration of NaCl. In the absence of NaCl, all lines exhibited similar seed germination rates. However, in the presence of 100 mm NaCl, 35S:PagGIa/b/c Col‐0 and 35S:PagGIa/b/c gi‐2 seeds exhibited delayed germination compared to WT and gi‐2 seeds respectively (Figure S3a,b). We next examined the root growth phenotype of salt stress‐treated plants. Root elongation was significantly suppressed in 35S:PagGIa/b/c Col‐0 and 35S:PagGIa/b/c gi‐2 compared to WT and gi‐2 plants, respectively, on MS medium containing 125 mm NaCl. The 35S:PagGIa/b/c Col‐0 and 35S:PagGIa/b/c gi‐2 transgenic plants lost more root fresh weight (78%–85.9% and 47.6%–68%) than WT (59.7%) and gi‐2 (38.3%) plants respectively (Figure S3c,d).
We also evaluated the salt sensitivity of the transgenic Arabidopsis plants grown in soil. Compared to WT plants, 35S:PagGIa/b/c Col‐0 transgenic lines had reduced tolerance to NaCl‐induced osmotic stress (Figure 4a,b). In addition, the increased salt tolerance of gi‐2 mutant plants was fully or partially reversed in the 35S:PagGIa/b/c gi‐2 transgenic lines (Figure 4a,b). These results suggest that PagGIa, b and c also function as negative regulators of the salt stress signalling pathway.
Figure 4.
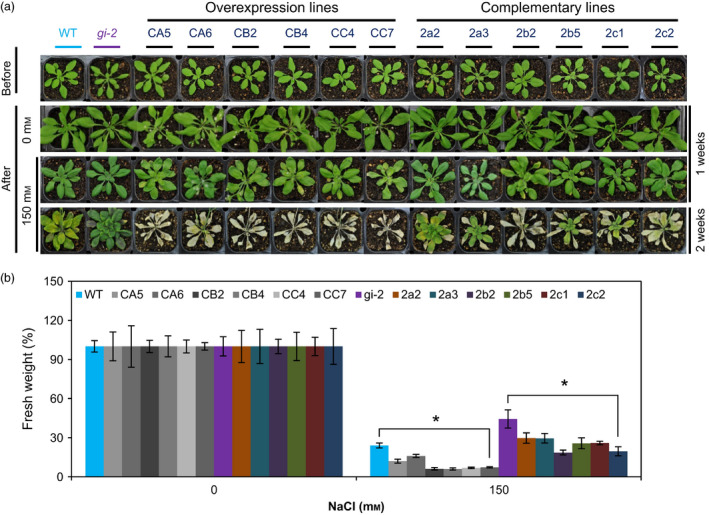
Overexpression of PagGI Genes Confers Salt Sensitivity in Transgenic Arabidopsis. (a) 35S:PagGIa/b/c Col‐0 (overexpression lines) and 35S:PagGIa/b/c gi‐2 (complemented lines) plants were grown in soil under LDs for 3 weeks (first row, before) and then (after) watered with water (0 mm NaCl) for 1 week (second row) or 150 mm NaCl solution for 1 week (third row) or 2 weeks (fourth row), and (b) relative fresh weights at the end of the treatments (shown in a) were measured. More than 10 plants were measured per data point. Error bars represent SD of three independent experiments. Asterisks indicate significant differences at P < 0.05.
PagGIs inhibit PagSOS2‐dependent phosphorylation of PagSOS1
We next examined how PagGIa, b, and c participate in the regulation of the salinity stress response. GI appears to be involved in salt stress tolerance through the SOS pathway in Arabidopsis (Kim et al., 2013a). To determine if PagGIs are also involved in the SOS pathway, we isolated genes encoding components of the SOS pathway in poplar, that is, PagSOS1 and PagSOS2 (Figure S4 and S5). We next examined the interaction between PagGIs and PagSOS2 via His pull‐down assays in vitro. His‐PagGIs and GST‐PagSOS2C fusion proteins were purified as described in Supporting information. His‐PagGIa, ‐b and ‐c were incubated with GST‐PagSOS2C, and the pull‐down extracts were analysed by immunoblotting using the respective anti‐GST antibody. GST‐SOS2 was pulled down with His‐PagGIa, ‐b and ‐c (Figure 5a), suggesting that PagGIa, b and c interact with PagSOS2 in vitro. To test the interaction between PagGI proteins and PagSOS2 in plant cells, we used BIFC assays. Cell suspensions of Agrobacterium tumefaciens carrying N‐terminal Venus fluorescent protein PagGIa‐, b‐ and c‐VN and C‐terminal Venus fluorescent protein PagSOS2C‐VC constructs were infiltrated into N. benthamiana leaves respectively. We detected strong Venus fluorescence in the nucleus when combinations of PagGIa‐, b‐ and c‐VN/PagSOS2C‐VC were used, indicating that PagGI proteins interact with PagSOS2, predominantly in the nucleus (Figure 5b and Figure S6).
Figure 5.
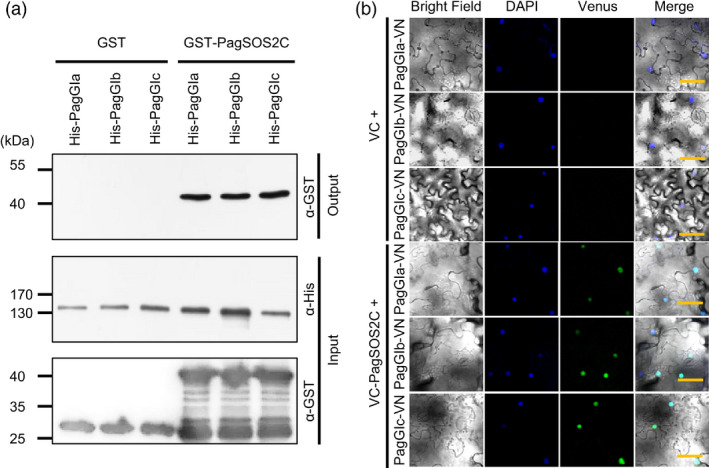
PagGI Proteins Negatively Regulate Salinity Stress Tolerance. (a) His pull‐down assay to examine the association between PagGI proteins and PagSOS2 in vitro. For input, the protein extracts were analysed by SDS‐PAGE, followed by western blotting with the indicated antibodies (α‐His, second row; α‐GST, third row). For output, the protein extracts were incubated overnight with His‐agarose beads. After extensive washing, bound proteins were analysed as above with antibodies against GST‐tag (α‐GST, first row). (b) BIFC assay to monitor the interaction between PagGI proteins and PagSOS2 in vivo. DAPI (second row) and Venus (third row) fluorescence was observed at 3 days after infiltration. Both fluorescence images are merged in the fourth row (merge). Scale bars represent 50 μM.
To determine whether the PagGIs‐PagSOS2 complex affects phosphorylation of PagSOS1 by PagSOS2, we performed an in vitro kinase assay using a constitutively active form of PagSOS2 (PagSOS2TD), in which Thr168 is changed to Asp. A C‐terminal fragment of SOS1 (PagSOS1C) was used as the substrate. PagSOS2TD showed strong autophosphorylation activity in vitro, and it constitutively activated PagSOS1 (Figure S7), whereas when purified recombinant PagGIa, b and c were included in the kinase reaction, the phosphorylation level of PagSOS1 was greatly reduced (Figure 6a). Moreover, we also tested the effects of PagGIs‐PagSOS2 complex on the phosphorylation of PagSOS1 by reconstitution of poplar SOS pathway in yeast cells. As shown in Figure 6b, auto‐phosphorylated PagSOS2 (PagSOS2TD) was sufficient to activate the PagSOS1, however, when coordinate expression of PagSOS2TD and PagGIs, the level of phosphorylated/activated PagSOS1 were greatly reduced in the Yeast AXT3K cells (lack the Na+ efflux proteins ENA1‐4 and NHA1, and the vacuolar Na+/H+ antiporter). These results indicate that PagGI proteins negatively regulate salt stress tolerance by inhibiting PagSOS2‐dependent phosphorylation of PagSOS1 via PagGIs‐PagSOS2 interaction.
Figure 6.
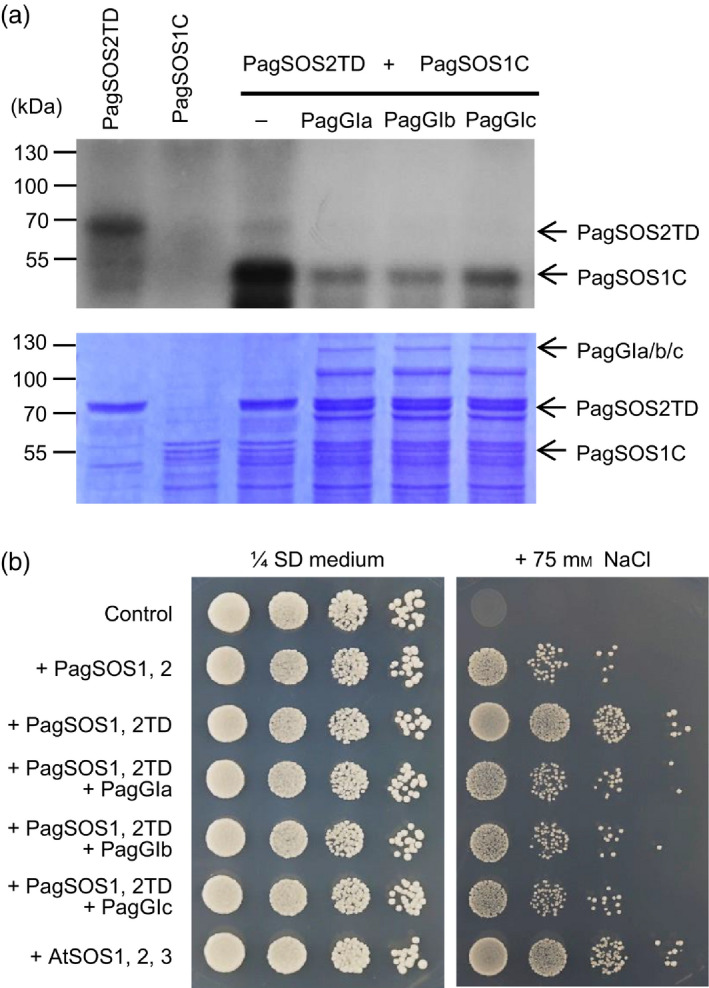
PagGI proteins negatively regulate salinity stress tolerance. (a) PagGIa/b/c inhibit PagSOS2‐mediated PagSOS1 phosphorylation in vitro. An in vitro kinase assay was performed using purified bacterially GST‐PagSOS1C, GST‐PagSOS2TD and His‐PagGI a, b, and c proteins in the indicated combinations. Shown are autoradiogram (top panel) and CBB staining (bottom panel) of a gel containing resolved reactions. (b) PagGIs decrease the NaCl tolerance of yeast expressing the auto‐phosphorylated PagSOS2 (PagSOS2TD)and PagSOS1 ion transporter. Yeast strain AXT3K cells transformed with an empty vector (control) or indicated combination of genes were grown overnight in liquid selective medium. Five microlitres of serial decimal dilutions were spotted onto plates of the same medium or supplemented with 75 mm NaCl. Plates were incubated at 28 °C and photographed after 4 days.
In Arabidopsis, AtGI is degraded upon exposure to salt, thus causing the release of PagSOS2, which subsequently regulates the salt stress response (Kim et al., 2013a). Therefore, we investigated the protein stability of PagGIa, b and c under salt stress. As shown in Figure 7, PagGIa, b and c protein levels were reduced upon exposure to salt stress, but these decreases were blocked by the proteasome inhibitor MG132, suggesting that PagGIs are unstable proteins that are targeted for 26S proteasome‐dependent proteolysis. Taken together, these results indicate that poplar PagGIa, b and c play a functionally conserved role as negative factors in salt stress tolerance.
Figure 7.
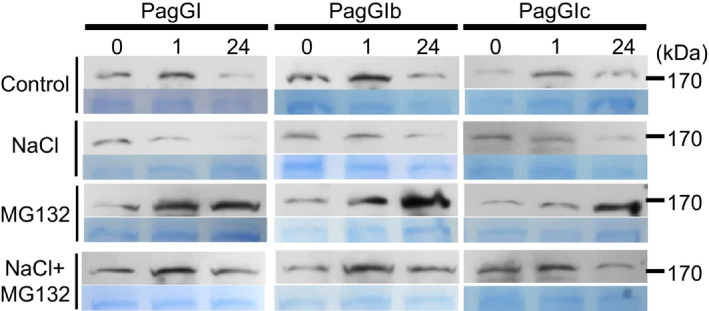
PagGIa, b and c are degraded upon exposure to salt in a proteasome‐dependent manner. Whole 3‐week‐old, soil‐grown 35S:PagGIa/b/c Col‐0 plants were treated with NaCl (100 mm), MG132 (100 mm) or NaCl plus MG132 at ZT1. PagGIa, b and c protein levels were evaluated after 0, 12 and 24 h treatments via immunoblot analysis with anti‐GFP antibody. Coomassie Brilliant blue (CBB)‐stained blots are shown as a loading control. Molecular weight markers in kDa.
Generation of transgenic poplar with down‐regulated expression of PagGIs
The above results encouraged us to generate transgenic poplar plants with improved growth and salt tolerance by down‐regulating PagGI genes. An RNA interference (RNAi) construct was produced and introduced into poplar plants (P. alba × P. glandulosa; referred to as RB) using Agrobacterium‐mediated transformation (Figure 8a). Nine independent RB plants were confirmed by genome PCR analysis with Int F/Nos R‐ and Bar‐specific primers, and three lines (RB5, RB16 and RB32) were selected by RT‐PCR and Southern blot analysis for further characterization (Figure S8a–c). To evaluate the morphological phenotypes of non‐transgenic (NT) and RB plants, poplar plants grown on rooting medium (RM) for 1 month were transferred to pots. Two‐month‐old pot‐cultured RB plants exhibited elongated petioles and large leaves with downward curvature (Figure 8b,c). Moreover, RB plants had thick stems and increased biomass (1.29–1.58‐fold higher biomass than NT plants), but no significant difference in height compared to NT plants (Figure 8d–i). Plant viability and developmental processes, including cell division, cell growth and differentiation, are precisely monitored and determined by auxin (Dhonukshe et al., 2008). Auxin regulates the expression of numerous auxin‐responsive genes, such as Aux/IAA genes (Hagen and Guilfoyle, 2002; Kim et al., 2011). Interestingly, the transcript levels of early auxin‐response genes including IAA1, IAA2, IAA4 and IAA5 were significantly higher in all RB plants under normal growth conditions compared to the control (Figure 8j). These results indicate that the phenotypes of RB plants are positively correlated with the transcript levels of auxin‐response genes.
Figure 8.
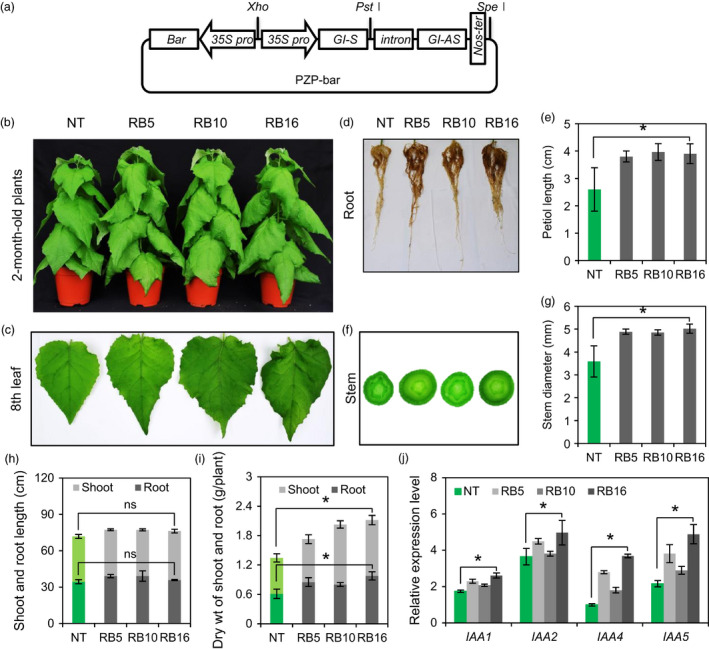
Morphological Phenotypes of RB Poplar Plants. (a) Schematic diagram of the constructs. (b) Plant phenotype of non‐transgenic (NT) and RB plants after 2 months of growth in pots. (c) The 8th leaf, (d) root morphology, (e) petiole length and (f, g) Stem diameter of 2‐month‐old NT and RB plants. Petiole length was determined in the 10th leaves. Stem diameter was measured from the position ~20 cm below the terminal bud. (h) The height and (i) dry weight of shoots and roots of 2‐month‐old poplar plants. (j) Transcript levels of auxin‐responsive IAA genes ( IAA1, IAA2, IAA4 and IAA5) were measured by quantitative RT‐PCR in the 10th leaves of 2‐month‐old NT and RB plants. The expression levels of IAA genes were normalized to the poplar actin gene as an internal control. Data represent three independent experiments. Asterisks and ns indicate significant and nonsignificant differences at P < 0.05 respectively.
RB plants exhibit increased tolerance to salt stress
To evaluate the salt stress tolerance of RB plants, we cultured poplar stem cuttings (NT, RB5, RB10 and RB16) with double nodes in RM tubes supplemented with or without the indicated concentration of NaCl. In the presence of 50 mm NaCl, RB plants exhibited less loss (33.2%–44.1%) of root fresh weight than NT plants (74.7%), whereas very little rooting of NT plants occurred in the presence of 75 mm NaCl after 1 month of treatment (Figure S9a,b). Furthermore, we subjected 2‐month‐old pot‐cultured NT and RB plants of the same health status to 200 mm NaCl for 6 days. NT plants displayed faster wilting and more chlorosis than RB plants under salt stress conditions. The differences in wilting symptoms between NT and RB plants became even more pronounced after normal irrigation was resumed (Figure 9a). As expected, reduced levels of PagGI transcripts were detected in RB plants (Figure 9b). RB plants maintained higher photosystem II efficiency (Fv/Fm) and higher levels of chlorophyll compared to NT plants under salt stress, which is consistent with their salt‐resistant phenotypes (Figure 9c). All of these results indicate that down‐regulating PagGI genes confers tolerance to salt stress.
Figure 9.
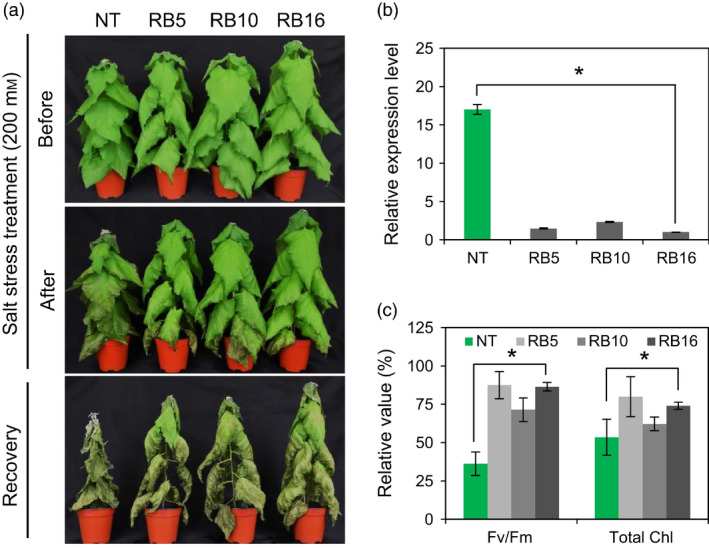
Down‐Regulation of PagGI Genes Confers Salt Tolerance in Poplar. (a) Effect of salt stress on RB poplar plants. Two‐month‐old non‐transgenic and RB plants were subjected to 200 mm NaCl treatment for 6 day and recovery for 15 day. (b) Transcript levels of PagGIs in three individual RB poplar plants. The 10th leaves from 1‐month‐old plants entrained in LDs were harvested at ZT12. The mRNA abundance was quantified by quantitative RT‐PCR and normalized to the level of poplar actin transcript. (c) Fv/Fm and total Chl contents in the leaves (10th) of poplar plants were determined at 4 day after treatment. Data represent means ± SD of three independent experiments. Asterisks indicate significant differences at P < 0.05 respectively.
Discussion
PagGIs are functional orthologues of Arabidopsis GI
GIGANTEA (GI) regulates several signalling pathways, but only its role in photoperiodic flowering and circadian rhythms have been extensively studied (Mouradov et al., 2002). Poplar differs from annual Arabidopsis plant in many ways (Bäurle and Dean, 2006; Jansson and Douglas, 2007). Elucidating how perennial plants adapt their growth and development in response to perceived environmental stress at the molecular level would greatly accelerate current tree‐breeding programmes.
The GI gene has undergone several intraspecific gene duplications: soybean has at least four GI paralogs (GmGI 1α, GmGI 1β, GmGI 2 and GmGI 3), and onion has two GI‐like genes (AcGIa and AcGIb) involved in flowering promotion (Taylor et al., 2010; Watanabe et al., 2011). In this study, we identified three GI homologs (paralogs) in poplar. PagGIa and PagGIc are predicted to be localized to chromosome 5, while PagGIb is found on chromosome 2 (data not shown). Although the overall expression profiles of PagGIa, b and c were similar, slight but interesting differences were observed among the three GI paralogues. The peak transcript levels were higher for PagGIb than the total PagGIa and PagGIc transcript levels throughout the day and in all investigated tissues (Figure S1a,b). Gene duplication initially generates two identical copies of a gene that are functionally redundant. The original function may be conserved in both genes, providing extra amounts of protein or RNA products, or one gene may evolve freely without much pressure from natural selection until it becomes a non‐functional pseudogene or acquires a novel function (Teichmann and Babu, 2004; Zhang, 2003). Although we detected no significant differences in the physiological functions of these genes, such as the induction of early flowering and salt sensitivity in transgenic Arabidopsis (Figure 3a and 4a), the GI paralogues appear to be differentially regulated in poplar.
Several lines of evidence indicate that PagGIa, b and c are AtGI orthologues in poplar (Figure 10). PagGIs regulate circadian rhythms via direct protein–protein interaction with PagZTLs (orthologues of Arabidopsis ZTL; Figure S10 and S11), which is necessary to sustain a normal circadian period by controlling the proteasome‐dependent degradation of the central clock protein TIMING OF CAB EXPRESSION 1 (TOC1) (Kim et al., 2007, 2013b). PagGIs appear to regulate flowering (at least in part) by influencing PagCO2 (orthologue of Arabidopsis CO; Figure S12 and S13), and PagGI‐PagCO‐PagFT (in the photoperiodic pathway) might play a role in regulating both flowering time and the timing of growth cessation (Böhlenius et al., 2006). PagGIs negatively regulate salt stress tolerance by caging PagSOS2 to the nucleoplasm and cytoplasm, which might influence the activities of the plasma membrane Na+/H+ antiporter PagSOS1 (Figure 6). These results provide strong evidence that these three poplar genes are (at least in part) functionally conserved between the herbaceous plant Arabidopsis and the woody plant poplar.
Figure 10.
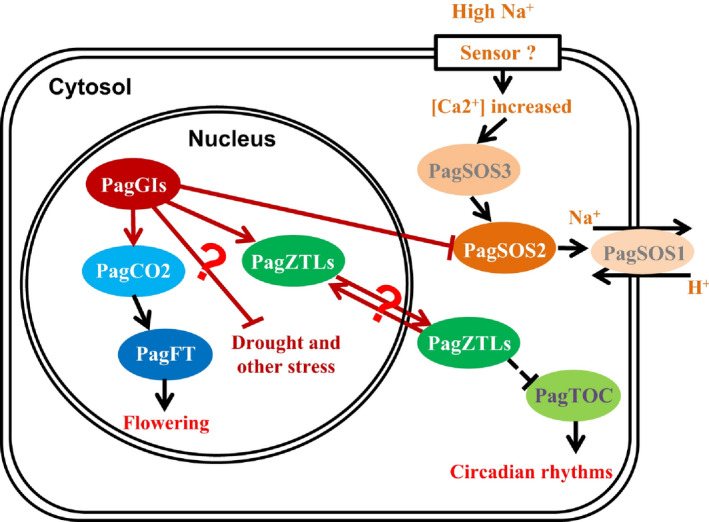
Model of the Roles of PagGI Genes in Regulating Circadian Rhythms, Flowering Time and the Salt Stress Response.
PagGIs play the negative regulatory roles in the salt tolerance response
Among abiotic factors, soil salinity represents an increasing global agricultural and environmental challenge, particularly in irrigated lands (Zhu, 2002). High salt stress disrupts the homoeostasis of water potential and ion distribution. Maintaining ion homoeostasis is critical for plants combating high salinity stress (Zhu, 2000). The initiation of flowering involves components of the circadian clock and is precisely monitored by multiple environmental cues (Andrés and Coupland, 2012; McClung, 2006). A number of components of the circadian clock are transcription factors, which play dual roles in ensuring the functioning of the central oscillator and controlling the rhythmic expression of downstream genes, such as abiotic stress‐responsive genes (Grundy et al., 2015). As GIs are large proteins, they possess various functional domains and can participate in multiple signalling pathways. In Arabidopsis, AtGI negatively regulates the salt stress response by caging SOS2 to the nucleoplasm and cytoplasm under normal growth conditions, but is degraded in response to salt stress (Kim et al., 2013a). Released SOS2 then interacts with SOS3 to form an active SOS2‐SOS3 protein complex, which regulates the expression and/or activities of various ion transporters, including the plasma membrane Na+/H+ antiporter SOS1, thereby promoting salt stress tolerance (Guo et al., 2004; Zhu, 2000, 2002). The SOS pathway, which functions in the salt stress response, is conserved in poplar: PtSOS1, PtSOS2 and PtSOS3 are functional homologues of their Arabidopsis counterparts, reconstitution of poplar SOS pathway in yeast cells revealed that PtSOS2 and PtSOS3 acted coordinately to activate PtSOS1 (Tang et al., 2010). Transgenic poplar plants overexpressing PtSOS2 or PtSOS2TD (constitutively active form of PtSOS2) have improved salt tolerance (Yang et al., 2015; Zhou et al., 2014). Although the exact mechanism in poplar remains unknown, our findings suggest that PagGIs function in the regulation of the salt stress response. Wild‐type Arabidopsis plants overexpressing PagGIa, b and c exhibited early flowering and enhanced salt sensitivity, while overexpressing these genes in the gi‐2 Arabidopsis mutant rescued the delayed flowering phenotype and increased salt tolerance (Figure 3a, b, Figure 4a, b). His pull‐down assays and BIFC experiments revealed that PagGI proteins interact with PagSOS2 in poplar. PagGI‐PagSOS2 complexes inhibit the PagSOS2‐based phosphorylation of PagSOS1 (Figure 5 and Figure 6a), which subsequently weakens Na+/H+ ion exchange under normal growth conditions (Figure 6b). PagGIs might play negative roles in salt response either by interfering with the PagSOS2‐regulated phosphorylation of PagSOS1 in cytosol or regulating cytosolic amount of PagSOS2 proteins by sequestering PagSOS2 to the nuclear (Figure 5 and Figure S6). However, it remains to be elucidated. These inhibitions were counteracted by the degradation of PagGI proteins in a proteasome‐dependent manner under salt stress (Figure 7). Down‐regulation of PagGI genes confers enhanced salt tolerance in poplar, further confirming that a link exists between salt stress responses and flowering control, and that PagGI genes may play pivotal roles in this process.
Down‐regulation of PagGIs by RNA interference led to vigorous growth, higher biomass and enhanced salt stress tolerance in transgenic poplar plants
RNA interference (RNAi) is an effective tool used to manipulate gene expression experimentally and to probe gene function on a genome‐wide scale in both plants and animals (Hannon, 2002). Altering flowering time regulation by overexpressing or suppressing gene activity can be used in crop breeding (Jung and Müller, 2009). Suppressing flowering time‐related gene activity by RNAi has been used to delay bolting and flowering in Arabidopsis, rice, and wheat (Curtis et al., 2002; Peng et al., 2008; Yong et al., 2003). The development of salt‐tolerant crops is urgently needed to sustain agricultural production. However, conventional tree‐breeding techniques alone are no longer sufficient to meet the increasing demand for forest products or environmental requirements. Genetic engineering of trees would accelerate the domestication of forest trees (Harfouche et al., 2011; Lambrou and Laub, 2004). We previously focused on developing environmental stress‐tolerant poplar plants using known abiotic stress‐related genes from other species (Ke et al., 2015; Kim et al., 2011). In the current study, we generated transgenic poplar plants with down‐regulated expression of PagGI genes (RB plants), and RB plants exhibited increased biomass and tolerance to salt stress (Figure 8a–i). Interestingly, the RB plants displayed auxin‐overproduction morphological phenotypes associated with increased transcript levels of early auxin‐response genes including IAA1, IAA2, IAA4 and IAA5 in all RB plants under normal growth conditions (Figure 8j). Furtermore, GI regulates phytochrome A‐mediated photo‐morphogenesis in Arabidopsis (Karina et al. 2007). Recent studies have shown that the phytochrome light receptors impose a strong influence on auxin levels in planta, by regulating SUPERROOT 2 (SUR2), a suppressor, and TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1), an enhancer of IAA biosynthesis (Halliday et al. 2009). Changed phenotypes of transgenic poplar plants might be achieved by light modulation of auxin signalling systems. However, the relationship between down‐regulation of GI genes and increased biomass remains to be elucidated.
Modern‐day plants evolved from their early plant ancestors through long‐term adaption to multiple environmental changes (Niklas and Kutschera, 2010; Tufto, 2000; Zhu, 2002). Perennial woody plants differ from annual plants in several ways and are thought to have evolved a complicated regulatory mechanism in response to environmental cues and seasonal changes (Nicotra et al., 2010). In light of the increasingly serious global environmental problems, developing abiotic stress‐tolerant poplar would be quite useful for maintaining biodiversity, moderating climatic conditions and combating desertification (Harfouche et al., 2011; Lambrou and Laub, 2004). Our data demonstrate that the link between developmental stage transitions and environmental cues mediated by GI is conserved between the model plant Arabidopsis and the perennial woody plant poplar. Specifically, we developed transgenic poplar plants with enhanced growth and tolerance to salt stress by down‐regulating PagGI genes. Although the performance of these transgenic poplar plants must be further characterized under field conditions, PagGIs represent promising candidate genes for use in genetic engineering of salt‐tolerant trees. Therefore, this study provides a new approach for sustainable production of salt‐tolerant forest products on global marginal lands.
Experimental procedures
Plant materials and growth conditions
The Arabidopsis thaliana ecotype Colombia‐0 (Col‐0) was used as the WT for all experiments. The gi‐2 mutant (Col‐0 background), which was described previously (Fowler et al., 1999), was kindly provided by Dr. Dae‐Jin Yun (Gyeongsang National University, South Korea). Arabidopsis seeds were stratified at 4 °C in darkness for 3 day and transferred to a growth chamber at 22 °C under a 16 h light/8 h dark photoperiod (light intensity ~120 μmol/m2/s), unless otherwise specified.
A hybrid poplar clone (P. alba × P. glandulosa) was used in this study. The plants were sub‐cultured monthly by aseptically transferring shoot apices with double nodes to fresh RM comprising 1× Murashige and Skoog medium (MS, Murashige and Skoog, 1962) containing 0.2 mg/L indole‐3‐butyric acid. After 1 month culture in RM, the rooted plantlets were transplanted to pots and grown in a growth chamber at 25 °C with 60% relative humidity under a light intensity of 150 μmol/m/s and a 16/8 h (light/dark) photoperiod.
Gene cloning
The poplar GIGANTEA (PagGI) and ZEITLUPE (PagZTL) genes were identified by sequence comparison of Arabidopsis GI and ZTL, respectively, to the Populous trichocarpa genomic database. Poplar CO2 (PagCO2) and SOS2 (PagSOS2) were identified based on the cDNA sequences of PtCO2 and PtSOS2 respectively. The coding sequences of PagGIa, PagGIb, PagGIc, PagZTL1, PagZTL2, PagCO2 and PagSOS2 were amplified from cDNA from P. alba × P. glandulosa leaves with Pfu‐X DNA polymerase (Solgent, Daejeon, Korea) using the primer pairs listed in Table S1. GenBank accession numbers are shown in Table S2.
Phylogenic analysis
For details, see Supporting information.
Plasmid construction
For details, see Supporting information.
RNA preparation and analysis of gene expression
Total RNA was extracted from the indicated plant tissues using a GeneAll Ribospin Plant™ kit (GeneAll, Seoul, Korea) according to the manufacturer's instructions. For cDNA production, 2 μg of total RNA was reverse transcribed using an RT‐PCR kit (Enzynomics, Daejeon, Korea). The reaction mixture was diluted to 100 μL with sterilized water, and 2 μL was used for real‐time quantitative RT‐PCR. Gene‐specific primers are listed in Table S1. All quantitative RT‐PCR analysis was performed with a CFX real‐time PCR system and CFX system software (Bio‐Rad, CA) using Ever‐Green 20 fluorescent dye (BioFACT, Daejeon, Korea). The following program was used for PCR: an initial denaturation step for 5 min at 95 °C, followed by 44 cycles of 95 °C for 20 s, 60 °C for 20 s and 72 °C for 20 s. There were at least three biological repeats and three technical repeats per data point.
Arabidopsis and poplar transformation
The PGWB5‐35S:PagGIa/b/c‐GFP constructs were introduced into Agrobacterium tumefaciens strain GV3101 by the freeze‐thaw method (Wise et al., 2006) and transferred into WT and gi‐2 mutant plants by floral dip infiltration (Clough and Bent, 1998). For each gene, two independent transgenic lines (T3 generation), which were homozygous for single‐copy insertions of transgene T‐DNAs in the WT or gi‐2 mutant background, were used for the experiments.
The indicated construct PZP‐35S‐(GI‐S)‐Intron‐(GI‐AS)‐Nos was introduced into poplar plants by Agrobacterium tumefaciens (strain EHA105)‐mediated transformation, as described by Choi et al. (2005). The transformed calli were selected on callus induction medium (CIM) comprising 1× MS medium containing 1.0 mg/L 2,4‐dichlorophenoxyacetic (2,4‐D), 0.1 mg/L benzylaminopurine (BAP), 0.01 mg/L 1‐naphthylacetic acid (NAA) and the appropriate antibiotics (0.5 mg/L Phosphinothricin [PPT]). PPT‐resistant shoots were regenerated from calli by transferring to shoot induction medium (SIM) comprising 1× Woody Plant Medium (Lloyd and McCown, 1980) containing 1.0 mg/L zeatin, 0.1 mg/L BAP and 0.01 mg/L NAA. Regenerated shoots were transferred to fresh RM for rooting, and the rooted plantlets were used for in‐tube or in‐pot growth assays.
Salt stress treatment
For details, see Supporting information.
Transient expression in N. benthamiana and confocal microscopy
Agrobacterium tumefaciens GV3101 strains carrying the indicated constructs and P19‐silencing plasmid were grown in YEP medium supplemented with the appropriate antibiotics overnight. Cultures were spun down and resuspended in infiltration solution (10 mm MES, 10 mm MgCl2 and 100 μM acetosyringone). Agrobacterium carrying the indicated construct or combinations of constructs and P19 were co‐infiltrated into 2‐week‐old N. benthamiana leaves for subcellular localization analysis. The cultures were grown in infiltration solution to a final OD600 = 0.5. After 3 days of growth in a greenhouse at 25 °C under LDs, the infiltrated parts of leaves were cut and immersed in DAPI solution for nuclear staining and subjected to fluorescent signal detection under a Leica TCs SP2 confocal microscope (Leica Microsystems, Heidelberg, Germany) with proper filter sets, as described by Gehl et al. (2009).
Protein expression and purification
For details, see Supporting information.
Immunoblot analysis
Total protein was extracted from whole plants of 3‐week‐old soil‐grown PagGIa/b/c‐OX plants, which were treated with NaCl (100 mm), MG132 (100 mm) or NaCl plus MG132 at the indicated time point. The extraction solution contained 50 mm Tris‐HCl (pH 7.5), 5 mm EDTA, 5 mm EGTA, 10 mm DTT and one protease inhibitor tablet (Roche, Mannheim, Germany). The extracts were separated by SDS‐PAGE. Immunoblot analysis was performed with anti‐GFP antibody.
His pull‐down assay
First, 5–10 μg of GST or GST fusion protein (GST‐PagSOS2F or GST‐PagSOS2C) was incubated with 30–50 μg of His fusion protein (His‐PagGIa, His‐PagGIb or His‐PagGIc) in lysis buffer (50 mm Tris‐HCl [pH 7.5], 250 mm NaCl, 5 mm EDTA, 0.1% Tween20, 1 mm DTT and one tablet of protease inhibitor) at 4 °C for 4 h. Next, 50–100 μL of Ni‐NTA agarose was added to the sample, followed by incubation overnight at 4 °C. The beads were washed three times with lysis buffer. Bound proteins were eluted from the beads by boiling in SDS sample buffer and analysed by SDS‐PAGE, followed by staining with Coomassie Brilliant Blue or Western blotting using the appropriate antibodies.
Kinase assay
For the in vitro kinase assay, kinase reactions were performed in 20 μL of kinase buffer (20 mm Tris‐HCl (pH 7.5), 10 mm MgCl2, 5 mm EGTA, 100 mm NaCl, and 1 mm DTT) containing 5 μg of fusion protein with 1 μL of [γ‐32P] ATP at room temperature for 1 h. The reactions were stopped by adding 4× SDS loading buffer. The phosphorylation of fusion proteins was analysed by autoradiography after separation by 10% SDS‐PAGE.
Yeast experiments
The AXT3K (▵ena1::HIS3::ena4, ▵nha1::LEU2, ▵nhx1::KanMX4), which was described previously (Quintero et al., 2002), was kindly provided by Dr. Dae‐Jin Yun (Gyeongsang National University, South Korea). The ion uptake experiment was performed as described previously (Ardie et al., 2009). Yeast AXT3K cells transformed with an empty vector or expressing combinations of PagSOS1, PagSOS2, PagSOS2TD and PagGIa/b/c were incubated in the 1/4 strength SD plate supplemented with or without 75 mm NaCl. Plates were incubated at 28 °C and photographed after 4 days.
Measurement of flowering time
Flowering time was measured by counting the number of rosette and cauline leaves or days to bolting when floral buds were visible (1 cm long) at the centre of the rosette. Flowering time was measured at least twice with similar results.
Analysis of photosynthetic activity and chlorophyll contents
Photosynthetic activity in leaves was estimated based on chlorophyll fluorescence determination of photochemical yield (Fv/Fm), which represents the maximal yield of the photochemical reaction in photosystem II (PSII), using a portable chlorophyll fluorescence meter (Handy PEA, Hansatech, England) after 30 min of dark adaption. Chlorophyll contents were measured with a portable chlorophyll meter (SPAD‐502; Konica Minolta, Japan). Total chlorophyll contents after stress treatment were compared with those under normal conditions. Both of these values were detected using the 5th–10th intact, fully expanded leaves (counting from the shoot apical meristem) of individual plants.
Statistical analysis
Data were statistically analysed with Statistical Package for the Social Sciences (SPSS 12.0, SPSS Inc., Chicago, IL). Means were separated using Duncan's multiple range test at P = 0.05.
Supporting information
Figure S1 PagGI genes are involved in the regulation of circadian rhythms
Figure S2 Generation of transgenic Arabidopsis plants overexpressing PagGI genes
Figure S3 Sensitivity of transgenic Arabidopsis seedlings to salt stress
Figure S4 SOS1‐like gene in poplar
Figure S5 SOS2‐like gene in poplar
Figure S6 Nuclear and cytosolic distribution of PagGI proteins
Figure S7 PagSOS2 phosphorylates PagSOS1 in vitro
Figure S8 Generation of transgenic poplar plants by down‐regulating PagGI genes
Figure S9 Effects of salt stress on NT and RB plants grown under in‐tube conditions
Figure S10 ZEITLUPE (ZTL)‐like genes in poplar
Figure S11 The interaction between PagGI proteins and PagZTL proteins in vivo
Figure S12 CONSTANS(CO)‐like gene in poplar
Figure S13 The interaction between PagGI proteins and PagCO2 in vivo
Table S1 Gene‐specific primers used in this study.
Table S2 GenBank accession numbers of genes described in this study.
Acknowledgements
This work was supported by grants from the KRIBB Initiative Program, the Korea‐China International Collaboration Project, National Research Foundation of Korea (NRF), Korea and the 111 Project of the Ministry of Education, China (no. B12007).
References
- Andrés, F. and Coupland, G. (2012) The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. [DOI] [PubMed] [Google Scholar]
- Ardie, S.W. , Xie, L. , Takahashi, R. , Liu, S. and Takano, T. (2009) Cloning of a high‐affinity K+ transporter gene PutHKT2; 1 from Puccinellia tenuiflora and its functional comparison with OsHKT2; 1 from rice in yeast and Arabidopsis . J. Exp. Bot. 184, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäurle, I. and Dean, C. (2006) The timing of developmental transitions in plants. Cell, 125, 655–664. [DOI] [PubMed] [Google Scholar]
- Böhlenius, H. , Huang, T. , Charbonnel‐Campaa, L. , Brunner, A.M. , Jansson, S. , Strauss, S.H. and Nilsson, O. (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science, 312, 1040–1043. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Hawighorst, P. , Sun, J. and Polle, A. (2014) Salt tolerance in Populus: significance of stress signaling networks, mycorrhization, and soil amendments for cellular and whole‐plant nutrition. Environ. Exp. Bot. 107, 113–124. [Google Scholar]
- Choi, Y.I. , Noh, E.W. , Lee, H.S. , Han, M.S. , Lee, J.S. and Choi, K.S. (2005) An efficient and novel plant selectable marker based on organomercurial resistance. J. Plant Biol. 48, 351–355. [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis, I.S. , Nam, H.G. , Yun, J.Y. and Seo, K.H. (2002) Expression of an antisense GIGANTEA (GI) gene fragment in transgenic radish causes delayed bolting and flowering. Transgenic Res. 11, 249–256. [DOI] [PubMed] [Google Scholar]
- Dhonukshe, P. , Tanaka, H. , Goh, T. , Ebine, K. , Mähönen, A.P. , Prasad, K. , Blilou, I. et al. (2008) Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature, 456, 962–966. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fowler, S. , Lee, K. , Onouchi, H. , Samach, A. , Richardson, K. , Morris, B. , Coupland, G. et al. (1999) GIGANTEA: a circadian clock‐controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane‐spanning domains. EMBO J. 18, 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl, C. , Waadt, R. , Kudla, J. , Mendel, R.R. and Hänsch, R. (2009) New GATEWAY vectors for high throughput analyses of protein‐protein interactions by bimolecular fluorescence complementation. Mol. Plant, 2, 1051–1058. [DOI] [PubMed] [Google Scholar]
- Gerland, P. , Raftery, A.E. , Ševčíková, H. , Li, N. , Gu, D. , Spoorenberg, T. , Alkema, L. et al. (2014) World population stabilization unlikely this century. Science, 346, 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy, J. , Stoker, C. and Carré, I.A. (2015) Circadian regulation of abiotic stress tolerance in plants. Front Plant Sci. 6, 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y. , Halfter, U. , Ishitani, M. and Zhu, J.K. (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell, 13, 1383–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y. , Qiu, Q.S. , Quintero, F.J. , Pardo, J.M. , Ohta, M. , Zhang, C. , Schumaker, K.S. et al. (2004) Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C‐terminal regulatory domain for salt tolerance in Arabidopsis thaliana . Plant Cell, 16, 435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen, G. and Guilfoyle, T. (2002) Auxin‐responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. [PubMed] [Google Scholar]
- Halliday, K.J. , Martínez‐García, J.F. , Josse, E.M. (2009) Integration of light and auxin signaling. CSH Perspect Biol. 1, a001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon, G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- Harfouche, A. , Meilan, R. and Altman, A. (2011) Tree genetic engineering and applications to sustainable forestry and biomass production. Trends Biotechnol. 29, 9–17. [DOI] [PubMed] [Google Scholar]
- Huq, E. , Tepperman, J.M. and Quail, P.H. (2000) GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis . Proc. Natl Acad. Sci. USA, 97, 9789–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson, S. and Douglas, C.J. (2007) Populus: a model system for plant biology. Annu. Rev. Plant Biol. 58, 435–458. [DOI] [PubMed] [Google Scholar]
- Ji, H. , Pardo, J.M. , Batelli, G. , Van Oosten, M.J. , Bressan, R.A. and Li, X. (2013) The Salt Overly Sensitive (SOS) pathway: established and emerging roles. Mol. Plant, 6, 275–286. [DOI] [PubMed] [Google Scholar]
- Jung, C. and Müller, A.E. (2009) Flowering time control and applications in plant breeding. Trends Plant Sci. 14, 563–573. [DOI] [PubMed] [Google Scholar]
- Ke, Q. , Wang, Z. , Ji, C.Y. , Jeong, J.C. , Lee, H.S. , Li, H. , Xu, B. et al. (2015) Transgenic poplar expressing Arabidopsis YUCCA6 exhibits auxin‐overproduction phenotypes and increased tolerance to abiotic stress. Plant Physiol. Biochem. 94, 19–27. [DOI] [PubMed] [Google Scholar]
- Kim, W.Y. , Fujiwara, S. , Suh, S.S. , Kim, J. , Kim, Y. , Han, L. , David, K. et al. (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature, 449, 356–360. [DOI] [PubMed] [Google Scholar]
- Kim, Y.H. , Kim, M.D. , Choi, Y.I. , Park, S.C. , Yun, D.J. , Noh, E.W. , Lee, H.S. et al. (2011) Transgenic poplar expressing Arabidopsis NDPK2 enhances growth as well as oxidative stress tolerance. Plant Biotechnol. J. 9, 334–347. [DOI] [PubMed] [Google Scholar]
- Kim, W.Y. , Ali, Z. , Park, H.J. , Park, S.J. , Cha, J.Y. , Perez‐Hormaeche, J. , Quintero, F.J. et al. (2013a) Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis . Nat. Commun. 4, 1352. [DOI] [PubMed] [Google Scholar]
- Kim, J. , Geng, R. , Gallenstein, R.A. and Somers, D.E. (2013b) The F‐box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development, 140, 4060–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrou, Y. , Laub, R. (2004) Gender perspectives on the conventions on biodiversity, climate change and desertification. United Nations, FAO: Gender and Population Division. [Google Scholar]
- Li, K. , Wang, Y. , Han, C. , Zhang, W. , Jia, H. and Li, X. (2007) GA signaling and CO/FT regulatory module mediate salt‐induced late flowering in Arabidopsis thaliana . Plant Growth Regul. 53, 195–206. [Google Scholar]
- Lloyd, G. and McCown, B. (1980) Commercially‐feasible micropropagation of mountain laurel, Kalmia latifolia, by use of shoot‐tip culture. Comb. Proc. Int. Plant Prop. Soc. 30, 421–427. [Google Scholar]
- Lobell, D.B. and Gourdji, S.M. (2012) The influence of climate change on global crop productivity. Plant Physiol. 160, 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Tryon, E.L. , Kreps, J.A. and Harmer, S.L. (2007) GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 143, 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung, C.R. (2006) Plant circadian rhythms. Plant Cell, 18, 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov, A. , Cremer, F. and Coupland, G. (2002) Control of flowering time interacting pathways as a basis for diversity. Plant Cell, 14, S111–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T. and Skoog, F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant, 15, 473–497. [Google Scholar]
- Ni, Z. , Kim, E.D. , Ha, M. , Lackey, E. , Liu, J. , Zhang, Y. , Sun, Q. et al. (2009) Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature, 457, 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra, A.B. , Atkin, O.K. , Bonser, S.P. , Davidson, A.M. , Finnegan, E. , Mathesius, U. , Poot, P. et al. (2010) Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15, 684–692. [DOI] [PubMed] [Google Scholar]
- Niklas, K.J. and Kutschera, U. (2010) The evolution of the land plant life cycle. New Phytol. 185, 27–41. [DOI] [PubMed] [Google Scholar]
- Peng, L.T. , Shi, Z.Y. , Li, L. , Shen, G.Z. and Zhang, J.L. (2008) Overexpression of transcription factor OsLFL1 delays flowering time in Oryza sativa . J. Plant Physiol. 165, 876–885. [DOI] [PubMed] [Google Scholar]
- Polle, A. and Chen, S. (2014) On the salty side of life: molecular, physiological and anatomical adaptation and acclimation of trees to extreme habitats. Plant, Cell Environ. 38, 1794–1816. [DOI] [PubMed] [Google Scholar]
- Quintero, F.J. , Ohta, M. , Shi, H. , Zhu, J.K. , Pardo, J.M. , (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl Acad. Sci. USA, 99, 9061–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengasamy, P. (2010) Soil processes affecting crop production in salt‐affected soils. Funct. Plant Biol. 37, 613–620. [Google Scholar]
- Sivakumar, M. , Das, H. and Brunini, O. (2005) Impacts of present and future climate variability and change on agriculture and forestry in the arid and semi‐arid tropics. Clim. Change, 70, 31–72. [Google Scholar]
- Song, Y.H. , Ito, S. and Imaizumi, T. (2010) Similarities in the circadian clock and photoperiodism in plants. Curr. Opin. Plant Biol. 13, 594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S. , Goto, K. (2003) TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell, 15, 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, R.J. , Liu, H. , Bao, Y. , Lv, Q.D. , Yang, L. and Zhang, H.X. (2010) The woody plant poplar has a functionally conserved salt overly sensitive pathway in response to salinity stress. Plant Mol. Biol. 74, 367–380. [DOI] [PubMed] [Google Scholar]
- Taylor, A. , Massiah, A.J. and Thomas, B. (2010) Conservation of Arabidopsis thaliana photoperiodic flowering time genes in onion (Allium cepa L.) . Plant Cell Physiol. 51, 1638–1647. [DOI] [PubMed] [Google Scholar]
- Teichmann, S.A. and Babu, M.M. (2004) Gene regulatory network growth by duplication. Nat. Genet. 36, 492–496. [DOI] [PubMed] [Google Scholar]
- Tufto, J. (2000) The evolution of plasticity and nonplastic spatial and temporal adaptations in the presence of imperfect environmental cues. Am. Nat. 156, 121–130. [DOI] [PubMed] [Google Scholar]
- Vinocur, B. and Altman, A. (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr. Opin. Biotechnol. 16, 123–132. [DOI] [PubMed] [Google Scholar]
- Watanabe, S. , Xia, Z. , Hideshima, R. , Tsubokura, Y. , Sato, S. , Yamanaka, N. , Takahashi, R. et al. (2011) A map‐based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics, 188, 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, A.A. , Liu, Z. and Binns, A.N. (2006) Three methods for the introduction of foreign DNA into Agrobacterium . Agrobacterium Protoc. 343, 43–54. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Tang, R.J. , Jiang, C.M. , Li, B. , Kang, T. , Liu, H. , Zhao, N. et al. (2015) Overexpression of the PtSOS2 gene improves tolerance to salt stress in transgenic poplar plants. Plant Biotechnol. J. 13, 962–973. [DOI] [PubMed] [Google Scholar]
- Yong, W.D. , Xu, Y.Y. , Xu, W.Z. , Wang, X. , Li, N. , Wu, J.S. , Liang, T.B. et al. (2003) Vernalization‐induced flowering in wheat is mediated by a lectin‐like gene VER2 . Planta, 217, 261–270. [DOI] [PubMed] [Google Scholar]
- Zerr, D. , Hall, J. , Rosbash, M. and Siwicki, K. (1990) Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J. Neurosci. 10, 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. (2003) Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298. [Google Scholar]
- Zhou, J. , Wang, J. , Bi, Y. , Wang, L. , Tang, L.Z. , Yu, X. , Ohtani, M. et al. (2014) Overexpression of PtSOS2 enhances salt tolerance in transgenic poplars. Plant Mol. Biol. Rep. 32, 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.K. (2000) Genetic analysis of plant salt tolerance using Arabidopsis . Plant Physiol. 124, 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.K. (2002) Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 PagGI genes are involved in the regulation of circadian rhythms
Figure S2 Generation of transgenic Arabidopsis plants overexpressing PagGI genes
Figure S3 Sensitivity of transgenic Arabidopsis seedlings to salt stress
Figure S4 SOS1‐like gene in poplar
Figure S5 SOS2‐like gene in poplar
Figure S6 Nuclear and cytosolic distribution of PagGI proteins
Figure S7 PagSOS2 phosphorylates PagSOS1 in vitro
Figure S8 Generation of transgenic poplar plants by down‐regulating PagGI genes
Figure S9 Effects of salt stress on NT and RB plants grown under in‐tube conditions
Figure S10 ZEITLUPE (ZTL)‐like genes in poplar
Figure S11 The interaction between PagGI proteins and PagZTL proteins in vivo
Figure S12 CONSTANS(CO)‐like gene in poplar
Figure S13 The interaction between PagGI proteins and PagCO2 in vivo
Table S1 Gene‐specific primers used in this study.
Table S2 GenBank accession numbers of genes described in this study.


