Summary
Wheat is one of the most widely grown cereal crops in the world and is an important food grain source for humans. However, wheat yields can be reduced by many abiotic and biotic stress factors, including powdery mildew disease caused by Blumeria graminis f.sp. tritici (Bgt). Generating resistant varieties is thus a major effort in plant breeding. Here, we took advantage of the non‐transgenic Targeting Induced Lesions IN Genomes (TILLING) technology to select partial loss‐of‐function alleles of TaMlo, the orthologue of the barley Mlo (Mildew resistance locus o) gene. Natural and induced loss‐of‐function alleles (mlo) of barley Mlo are known to confer durable broad‐spectrum powdery mildew resistance, typically at the expense of pleiotropic phenotypes such as premature leaf senescence. We identified 16 missense mutations in the three wheat TaMlo homoeologues, TaMlo‐A1, TaMlo‐B1 and TaMlo‐D1 that each lead to single amino acid exchanges. Using transient gene expression assays in barley single cells, we functionally analysed the different missense mutants and identified the most promising candidates affecting powdery mildew susceptibility. By stacking of selected mutant alleles we generated four independent lines with non‐conservative mutations in each of the three TaMlo homoeologues. Homozygous triple mutant lines and surprisingly also some of the homozygous double mutant lines showed enhanced, yet incomplete, Bgt resistance without the occurrence of discernible pleiotropic phenotypes. These lines thus represent an important step towards the production of commercial non‐transgenic, powdery mildew‐resistant bread wheat varieties.
Keywords: Targeting Induced Local Lesions in Genomes, powdery mildew, Mlo, hexaploid bread wheat, Blumeria graminis, plant disease resistance
Introduction
Bread wheat (Triticum aestivum) is the third largest cultivated crop after maize and rice and the second in terms of dietary intakes. By 2013, global wheat grain production reached 712 million tonnes (http://faostat.fao.org), and by 2050 an estimated increase of a further 60% will be required to meet the demands of our growing population (http://wheatinitiative.org).
Allohexaploid wheat has a genome of 17 Gb in size, of which more than 80% is composed of repetitive transposable elements. Genetically, it has the structure of three independent genomes in one species (AABBDD genome), since meiotic pairing of homoeologous chromosomes is prevented through the action of Ph (pairing homoeologous) genes (IWGSC, 2014).
Yield in wheat can be reduced by abiotic factors, pests or diseases caused by pathogens. A global disease threat is powdery mildew caused by Blumeria graminis f.sp. tritici (Bgt) (Conner et al., 2003; Dean et al., 2012), an obligate biotrophic fungus (ascomycete) of the order Erysiphales (Glawe, 2008). However, more recently, B. graminis f.sp. triticale has also been reported to reproduce on wheat (Menardo et al., 2016). Control of the powdery mildew disease caused by Bgt in wheat is performed mainly by using fungicides and varieties containing R (resistance) genes, the latter typically conferring isolate‐specific protection (Dean et al., 2012). Going forward, the most cost effective solution to control this disease is by exploiting genetic resources to provide enhanced, durable resistance.
In barley, natural and induced loss‐of‐function mutations of the Mildew resistance locus o (Mlo) gene confer broad‐spectrum resistance against most B. graminis f.sp. hordei (Bgh) isolates, an effect that has been shown to last in the field for more than 30 years (Jørgensen, 1992; Lyngkjær et al., 2000). Mlo is a member of an ancient eukaryotic gene family that is conserved throughout the plant kingdom (Kusch et al., 2016), and although its role in powdery mildew resistance has been well studied in various species, the biochemical function of Mlo proteins is still unknown (Acevedo‐Garcia et al., 2014).
On susceptible host plants, once sporelings of the pathogen land on the leaf or stem surface, these germinate and form an appressorium within two hours. The appressorium attempts to penetrate the epidermal cell layer by generating a penetration peg. If the pathogen successfully enters the host cell, in the following hours of infection the penetration peg enlarges to develop the feeding structure known as haustorium. Thereafter, the pathogen will complete its asexual life cycle on the leaf surface with the development of epiphytic hyphae, the production of conidiophores and the release of new spores (reviewed in Glawe, 2008). In the case of resistant mlo plants, a near‐complete arrest of pathogen growth occurs at the penetration stage where the germinating spore is not able to develop a haustorium (Jørgensen and Mortensen, 1977). However, in barley mlo lines resistance is typically associated with pleiotropic phenotypes such as the spontaneous deposition of callose‐containing cell wall appositions, early chlorophyll decay and spontaneous mesophyll cell death, which together lead to chlorotic and necrotic leaf flecking and have been interpreted as signs of premature leaf senescence (Peterhänsel et al., 1997; Piffanelli et al., 2002; Schwarzbach, 1976; Wolter et al., 1993).
In wheat, the three orthologues of barley Mlo, TaMlo‐A1, ‐B1 and ‐D1 (Konishi and Sasanuma, 2010), are located on chromosomes 5AL, 4BL and 4DL (Elliott et al., 2002). In contrast to barley, occurrence of natural wheat mlo mutants has not been reported. This is likely due to its hexaploid nature, which may require mutation of all six gene copies to generate a resistance phenotype that would be detectable within a breeding programme. Previously, Elliott and co‐workers showed that one of the wheat orthologues of barley Mlo, TaMlo‐B1, can complement powdery mildew‐resistant barley mlo mutants at the single‐cell level (Elliott et al., 2002). Furthermore, Várallyay and colleagues used virus‐induced gene silencing (VIGS) to demonstrate that RNAi silencing of the TaMlo homoeologues in wheat results in powdery mildew resistance (Várallyay et al., 2012). Most recently, the teams of Caixia Gao and Jin‐Long Qiu took advantage of the TALEN (transcription activator‐like effector nuclease) genome editing technology to generate transgenic winter wheat plants containing simultaneous knockout lesions in the three TaMlo homoeologues. Compared to the respective parental line, these plants were fully resistant against Bgt infection (Wang et al., 2014).
Targeting Induced Local Lesions IN Genomes (TILLING), first introduced 16 years ago by McCallum and collaborators (McCallum et al., 2000), is a powerful approach that integrates chemical mutagenesis with a high‐throughput detection method to identify single‐nucleotide mutations in a specific region of a gene of interest. In principle, TILLING can be used in different plant species independent of their ploidy. Interestingly, polyploid species can tolerate higher mutation densities than diploids (Slade et al., 2005; Uauy et al., 2009; Wang et al., 2012). This feature translates into a far smaller population size that needs to be screened to reach saturated mutagenesis in a polyploid species (Kurowska et al., 2011). Chemical mutagenesis combined with TILLING provides an ample spectrum of mutations, where a pool of allelic variations can result in a range of weak to strong phenotypes (Slade et al., 2005). To date there are several TILLING resources available for various crop species such as hexaploid and durum wheat, barley, rice, tomato, maize, sorghum, soybean and potato (reviewed in Chen et al., 2014b). Although until recently no TILLING‐derived crop variety has been released commercially, they represent a great advantage for plant breeding (especially in Europe) since these varieties will be considered non‐transgenic (Chen et al., 2014b).
In wheat (diploid, tetraploid or hexaploid), several genes involved in starch synthesis have been targeted by TILLING, for example, Waxy (Rawat et al., 2012; Slade et al., 2005), Starch Branching Enzyme II (SBEII) (Botticella et al., 2011; Sestili et al., 2015; Slade et al., 2012; Uauy et al., 2009) and Starch Synthase II (Sgp‐1/SSII (Dong et al., 2009; Sestili et al., 2009). Additionally, genes involved in other processes such as carotenoid content (Colasuonno et al., 2016), grain width (Simmonds et al., 2016), flowering (Chen et al., 2014a) or vernalization (Chen and Dubcovsky, 2012), have been also targeted by TILLING.
Currently, wheat breeders have a great challenge to select for varieties that on the one hand have a minimal disturbance in growth, development and fertility, thereby maintaining good grain quality and enhancing grain yield, and on the other hand exhibit improved resistance to pests and pathogens. At least in Europe, the deployment of transgenic plants (including those resulting from genome editing approaches) is still socially and politically contentious in plant breeding, agriculture as well as the food, feed and drinks industries. Therefore, methods that are indisputably non‐transgenic are favoured for the development of new varieties.
In this study, we generated hexaploid bread wheat lines with enhanced resistance to the common powdery mildew disease. Our approach made use of the non‐transgenic TILLING technology to select four different lines containing missense mutations in the TaMlo‐A1, TaMlo‐B1 and TaMlo‐D1 homoeologues. Triple and some double mutant lines showed enhanced powdery mildew resistance compared to respective wild‐type (WT) plants. So far these mutant plants did not show discernible abnormalities in growth and development.
Results
Cloning of genomic sequences of the wheat TaMlo‐A1, TaMlo‐B1 and TaMlo‐D1 homoeologues
An essential requirement to perform a TILLING screening is the development of primers for gene‐specific polymerase chain reaction (PCR) amplification. A limiting factor in hexaploid wheat in this respect is the very high nucleotide similarity between the coding sequences of its homoeologous genes, which often constrains primer design. This is also the case for TaMlo, which exhibits 95%, 96% and 97% nucleotide sequence identity between the coding sequences of the A and B, A and D, and B and D genomes, respectively (File S1). To overcome this limitation, homoeologue‐specific PCR primers are frequently designed on the basis of intron sequences, which typically show a higher degree of nucleotide sequence polymorphisms than the exonic coding sequences. We thus first opted to obtain genomic DNA sequence information for the three TaMlo homoeologues. Based on the available known cDNA sequences in NCBI: (i) TaMlo‐A1: AF361933 and AX063298; (ii) TaMlo‐B1: AF361932, AX063294, AF384145; and (iii) TaMlo‐D1: AX063296, we designed common oligonucleotide primers, predicted to equally bind to all three TaMlo homoeologues, and used them to amplify and clone several overlapping genomic TaMlo fragments from DNA obtained from the hexaploid spring wheat cultivar (cv.) Cadenza. Following in silico assembly of the amplicons and also taking information from the known cDNA sequences into account, we obtained almost complete genomic assemblies representing the three TaMlo homoeologues. These were reconciled with the manually annotated sequences generated on the basis of the available wheat genome survey data to finally produce tentative genomic TaMlo consensus sequences (cv. Cadenza and cv. Chinese Spring hybrid sequences; File S2). The determined exon/intron structure of the TaMlo homoeologues is similar to the one of barley Mlo (Büschges et al., 1997) and comprises 11 exons each (Figure 1). The deduced TaMlo protein sequences are each one amino acid longer (534 amino acids) than barley Mlo (533 amino acids); consequently, as a result of small gaps in the alignment between wheat and barley Mlo proteins, the amino acid numbering after position 115 is shifted by +1 in the former compared to the latter.
Figure 1.
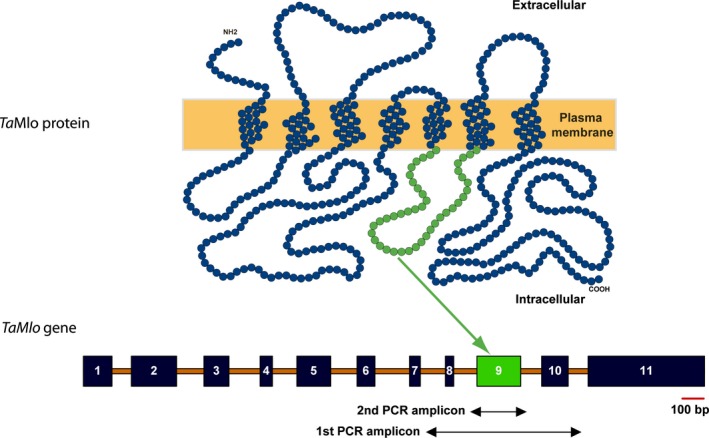
Exon 9 of TaMlo is the target for the TILLING screen. The scheme illustrates the predicted topology of the 7 transmembrane domain TaMlo proteins (top) and the experimentally determined common exon‐intron structure of the corresponding TaMlo homoeologues (bottom). The 11 exons are indicated by dark boxes, introns by orange lines. The third cytoplasmic loop of TaMlo, depicted in green and encoded by exon 9, was the target for TILLING. Arrows below the TaMlo gene model specify the amplicons of the 1st and 2nd round PCR. A scale bar, shown in red, is given below exon 11 (100 bp).
TILLING target and primer design
Previously, we identified the second and third cytoplasmic loop of the barley Mlo protein as relevant regions for its powdery mildew susceptibility‐conferring activity. Mutations in these parts of the Mlo protein that lead to a single amino acid exchange (missense mutations) often result in a loss‐of‐function of the protein and thus resistance against the powdery mildew pathogen (Reinstädler et al., 2010). Hence, we selected the third cytoplasmic loop, encoded by exon 9 of the TaMlo homoeologues, as the target region for the TILLING screening in wheat (Figure 1). We developed homoeologue‐specific primers for a 1st round PCR to amplify a product (between 750 and 1700 bp in size, depending on the TaMlo homoeologue) that contains the target exon (Figure 1 and Table S1). In addition, as High‐Resolution Melt (HRM) analysis is most accurate on DNA fragments up to ~300 bp in size (Reed and Wittwer, 2004), homoeologue‐specific primers were designed for a 2nd round PCR with a maximum amplicon size of 310 bp (Figure 1 and Table S1). Primer specificity was confirmed by amplification from genomic template DNA of cv. Cadenza followed by direct amplicon sequencing.
Identification of Tamlo mutant candidates by TILLING
The ethyl methane sulfonate (EMS)‐mutagenized population of spring bread wheat cv. Cadenza has been previously described (Rakszegi et al., 2010). Genomic DNA samples derived from ~2020 M2 individuals were pooled twofold and employed as templates for 1st round PCR amplification in the target region using the validated homoeologue‐specific primer pairs. The primary PCR product was used as template for a 2nd round of PCR and the resulting ~300 bp amplicons were subjected to HRM analysis to detect mutations in the target exon of the TaMlo homoeologues.
After screening the entire population, a total of 76 candidate mutants were identified (28, 27 and 21 in TaMlo‐A1, TaMlo‐B1 and TaMlo‐D1, respectively, Table 1). Twenty of these candidate mutations could not be confirmed in a secondary round of analysis; these were considered false positives and discarded from further study. Among the remaining 56 mutants, 14 mutations were located in intron regions flanking exon 9, but did not affect the consensus GT/AG splice junctions, and 26 were silent (synonymous mutations, Table 1), resulting in an unaltered TaMlo amino acid sequence. The remaining 16 mutants each represented missense mutations (non‐synonymous mutations, Table 2); nonsense mutations were not found in this screen. At the nucleotide level, the identified missense mutations were all transitions of the type C → T or G → A, as expected for EMS mutagenesis (Table 2). Furthermore, the mutations covered a broad spectrum of amino acid exchanges, ranging from substitutions of amino acids with similar biophysical properties (e.g. S315N, V323I, A354V) to exchanges with dramatic alterations in biophysical properties (e.g. R313W, P335L, G296E). Three of the mutations (G319R, A320T and T345M) were found in the TaMlo‐B1 as well as in the TaMlo‐D1 genome (Table 2). Interestingly, mutations G319R (TaMlo‐B1 and TaMlo‐D1) and P335L (TaMlo‐D1) were previously identified as sequence variants with the characteristic resistance phenotype in barley plants (mlo‐38 and mlo‐29, respectively; Lundqvist et al., 1991; Piffanelli et al., 2002; Müller et al., 2005; Reinstädler et al., 2010). Furthermore, a similar mutation to Tamlo‐A1 (P325L) was studied by transient gene expression in barley (P324A), showing a partial resistance phenotype (Reinstädler et al., 2010). Analysis with the online tool SIFT (Sorting Intolerant From Tolerant; Kumar et al., 2009) revealed in each genome at least one mutation predicted to affect protein function (Table 2). Taken together, we identified 16 missense mutations, distributed between all three homoeologues that would potentially affect TaMlo function. Since nonsense mutations were not identified (Table 1), we further functionally characterized the missense mutations (Table 2) to select the most suitable candidates for stacking of the alleles in single wheat lines.
Table 1.
Mutations in the region of exon 9 of TaMlo identified by TILLING
| Target | Total candidates | False positives | Intron | Silent | Missense | Non sense |
|---|---|---|---|---|---|---|
| TaMlo‐A1 | 28 | 9 | 7 | 10 | 2 | 0 |
| TaMlo‐B1 | 27 | 4 | 5 | 11 | 7 | 0 |
| TaMlo‐D1 | 21 | 7 | 2 | 5 | 7 | 0 |
Table 2.
TaMlo exon 9 missense mutants identified by TILLING
| Gene | EMS line | SNP | Amino acid exchange | Zygositya | SIFT scoreb | Previously described in barley | |
|---|---|---|---|---|---|---|---|
| WT | Mutant | ||||||
| TaMlo‐A1 | CAD1‐29‐A8 | C | T | P325L | Het | 0.20 | mlo P324A Reinstädler et al. (2010) |
| TaMlo‐A1 | CAD‐22‐D12 | C | T | A354V | Hom | 0.00* | No |
| TaMlo‐B1 | CAD1‐2‐H9 | G | A | G296E | Het | 0.00* | No |
| TaMlo‐B1 | CAD2‐18‐H1 | C | T | T297I | Het | 0.04 | No |
| TaMlo‐B1 | CAD2‐22‐E11 | C | T | R313W | Het | 0.01* | No |
| TaMlo‐B1 | CAD2‐1‐A3 | G | A | S315N | Het | 0.49 | No |
| TaMlo‐B1 | CAD2‐21‐D7 | G | A | G319R | Hom | 0.00* | mlo‐38 Lundqvist et al. (1991), Reinstädler et al. (2010) |
| TaMlo‐B1 | CAD1‐4‐H10 | G | A | A320T | Het | 0.97 | No |
| TaMlo‐B1 | CAD1‐2‐G9 | C | T | T345M | Het | 0.09 | No |
| TaMlo‐D1 | CAD2‐1‐A2 | G | A | A314T | Het | 0.65 | No |
| TaMlo‐D1 | CAD2‐19‐C9 | G | A | G319R | Het | 0.01* | mlo‐38 Lundqvist et al. (1991), Reinstädler et al. (2010) |
| TaMlo‐D1 | CAD2‐23‐A2 | G | A | A320T | Het | 1.00 | No |
| TaMlo‐D1 | CAD2‐24‐C8 | C | T | P321S | Het | 0.25 | No |
| TaMlo‐D1 | CAD2‐19‐H4 | G | A | V323I | Hom | 0.13 | No |
| TaMlo‐D1 | CAD2‐3‐B6 | C | T | P335L | Hom | 0.00* | mlo‐29 Piffanelli et al. (2002), Müller et al. (2005) |
| TaMlo‐D1 | CAD2‐29‐A1 | C | T | T345M | Hom | 0.09 | No |
Hom, Homozygous; Het, Heterozygous.
SIFT scores ≤0.05 indicate mutations that are predicted to affect protein functions (indicated by an asterisk), while SIFT scores >0.05 indicate mutations that are predicted to be tolerated.
Validation of mutant candidates by transient gene expression
Previously, we demonstrated that TaMlo‐B1 is able to complement a powdery mildew‐resistant barley mlo mutant at the single‐cell level in leaf epidermal tissue (Elliott et al., 2002). Heterologous complementation is possible because of the high level of sequence identity between the wheat and barley Mlo proteins (~89%). We took advantage of this particle bombardment‐based barley transient expression assay to experimentally evaluate the potential of the TaMlo variants with amino acid substitutions to alter the powdery mildew infection phenotype.
Full‐length cDNAs of TaMlo‐A1, TaMlo‐B1 and TaMlo‐D1 were cloned as expression constructs driven by the maize Ubiquitin1 promoter (pUbiGATE) and used as a template to recreate each of the 16 missense mutations by PCR‐based, site‐directed mutagenesis. In addition, WT barley Mlo cDNA (also driven by the Ubiquitin1 promoter) was used to generate the mutant variants P324A, mlo‐29 (P334L) and mlo‐38 (G318R), which were included as additional controls.
Detached leaves of the barley mlo‐3 null mutant were co‐bombarded with the various TaMlo or Mlo constructs and a plasmid expressing the β‐glucuronidase (GUS) reporter gene. As a negative control we employed the GUS plasmid only, and as a positive control we used barley Mlo. Twenty‐four hours after bombardment the treated leaves were inoculated with the barley powdery mildew pathogen Bgh. All three WT wheat TaMlo genes showed a complementation efficiency similar to barley Mlo (~80% host cell entry, median value, Figure 2). Transient expression of mutants Tamlo‐A1 (P325L), its barley counterpart mlo (P324A), Tamlo‐B1 (R313W) and Tamlo‐D1 (A314T), (P321S) and (T345M) resulted in ~40%–60% host cell entry (median value, Figure 2). Expression of mutants Tamlo‐B1 (G296E) and Tamlo‐B1, Tamlo‐D1 (G319R) led to an even lower host cell entry rate of ~15%–20% (Figure 2). Similar results were obtained with the corresponding G318R barley mutant (mlo‐38, Figure 2). Interestingly, the entry rates obtained by expression of Tamlo‐D1 (P335L) and the corresponding barley mutant variant (mlo‐29) were close to background levels (GUS only, Figure 2), suggesting a near‐complete loss‐of‐function in the case of these amino acid substitutions. Expression of the remaining seven wheat mutant variants showed host cell entry rates not considerably different from the respective TaMlo WT versions (Figure 2). Overall there was a good correlation between the results of the in silico SIFT analysis and the experimental data. Exceptions were mutations Tamlo‐A1 (A354V), Tamlo‐B1 (T297I) and Tamlo‐B1 (R313W), which were predicted to have a significant impact on protein function but were only moderately affected regarding host cell entry rates (Figure 2 and Table 2).
Figure 2.
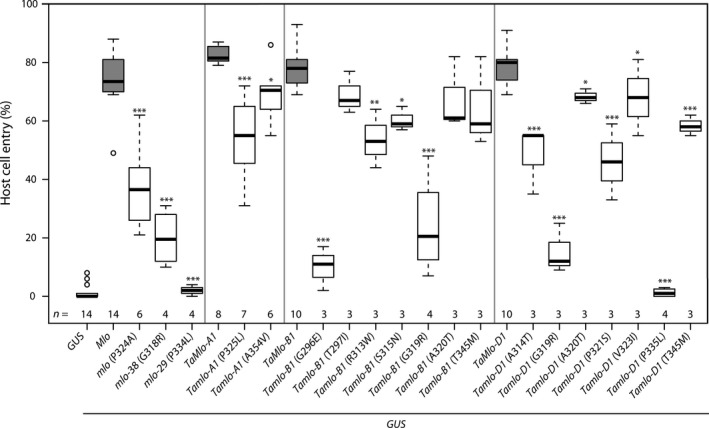
TILLING‐derived variants of Tamlo homoeologues exhibit different levels of functionality in a transient gene expression assay. Detached 8‐day‐old leaves of the powdery mildew‐resistant barley mlo‐3 mutant were co‐bombarded with a GUS reporter plasmid and a plasmid encoding the indicated TaMlo protein variant (WT or mutant version under transcriptional control of the maize Ubiquitin1 promoter). Expression of GUS alone and GUS plus WT barley Mlo, driven by the maize Ubiquitin1 promoter, were used as negative and positive controls, respectively, for restoration of Bgh susceptibility. Host cell entry was scored at 48 hours post inoculation (h p.i.) in GUS‐stained cells attacked by powdery mildew sporelings and results visualized as box plots. Centre lines show the medians; upper and lower box limits indicate the 25th and 75th percentiles respectively; upper and lower whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, respectively, and outliers are represented by dots. Numbers at the bottom of the boxplots indicate the number of biological replicates per sample (n). One biological replicate was typically composed of six leaves with 150 scored cells. Asterisks indicate a statistically significant difference to the respective WT (barley Mlo or TaMlo) with ***P < 0.001, **P < 0.01 and *P < 0.5 as determined by a Generalized Linear Model (GLM) test. Statistics were performed and boxplots generated with R software.
Barley mutants mlo‐29 and mlo‐38 as a case study to compare results from transient gene expression assays and resistance in planta
Since we found wheat equivalents of barley mutants mlo‐29 (P335L) and mlo‐38 (G319R) in our wheat TILLING screening, we compared the powdery mildew host cell entry rate in planta (which has not been assessed before; Figure 3) with the one obtained in transient expression experiments (Figure 2). The respective WT barley parental lines cv. Bonus, Kristina and Sultan showed high susceptibility to Bgh (Figure 3a) with ~85% host cell entry (Figure 3b). By contrast, the mutants mlo‐38 (two lines: SR59, background cv. Bonus and SR65, background cv. Kristina) and mlo‐29 (background cv. Sultan) were highly resistant to the pathogen (Figure 3a) with a host cell entry rate of less than 6% (Figure 3b). At least in the case of mlo‐38, this value is substantially lower than the one obtained upon transient expression of the mlo‐38 variant (~20% entry rate, median value; Figure 2). These results suggest that overexpression of mlo mutant variants may result in an overestimation of the residual protein function (i.e. artificially high protein levels may partly compensate for the defect in the protein). Consequently, for a given mlo mutant allele the resulting level of resistance in planta can be higher (i.e. respective plants are more resistant) than indicated by the results of the single‐cell transient gene expression assays.
Figure 3.
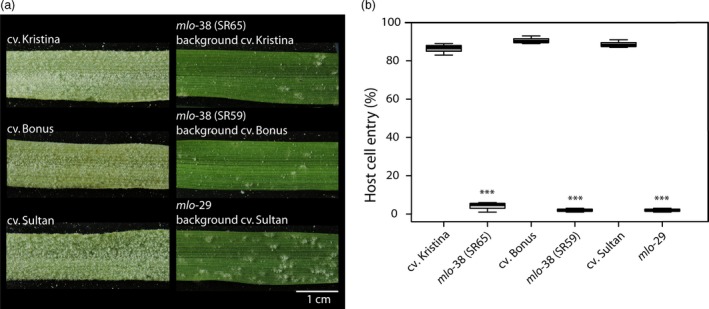
Bgh infection phenotypes of barley mutants mlo‐38 and mlo‐29 and their respective parental lines. Seven‐day‐old leaves were inoculated with Bgh isolate K1. (a) The macroscopic phenotype was recorded at 7 days post inoculation (d.p.i.). A scale bar shown in white is given in the lower right corner (1 cm). (b) Host cell entry was scored at 48 h p.i. Centre lines show the medians; upper and lower box limits indicate the 25th and 75th percentiles respectively; upper and lower whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, respectively. Shown are data of n = 3 independent biological replicates with 3–4 leaves (typically 200 scored interaction sites per leaf) per replicate. Asterisks indicate a statistically significant difference to the respective parental background with ***P < 0.001 as determined by a GLM test. Statistics were performed and boxplots established by R software.
Generation of triple homozygous Tamlo lines
Based on the data obtained with transient gene expression experiments (Figure 2), mutant candidates that showed reduced host cell entry compared to the respective TaMlo WT genes were selected for allele stacking (Table 3). While several suitable mutants with moderately or strongly reduced TaMlo function were available for the wheat genomes B and D, only two mutants were available for the A genome, which conferred moderately (P325L) and slightly (A354V) reduced host cell entry in the transient gene expression assay (Figure 2 and Table 3). Due to the lack of alternatives for genome A, mutant P325L was used in all crossings. With the aim of balancing the degree of powdery mildew resistance and potential pleiotropic phenotypes associated with the loss of Mlo function (Hückelhoven et al., 2013), we generated four wheat lines containing different mutant allele combinations that ranged from predicted weak to strong effects on powdery mildew susceptibility (Table 3). Depending on the zygosity of the available single mutants identified in the TILLING screen we followed a crossing scheme similar to the one shown in Figure S1 to obtain ideally all possible mutant combinations. To identify the respective mutations in the progeny of the crossings we developed cleaved amplified polymorphic sequence (CAPS) markers (Table S2). Genotype confirmation was subsequently performed by DNA sequencing of the 2nd round PCR products of the particular TaMlo homoeologues.
Table 3.
Triple mutant wheat lines obtained by stacking of Tamlo mutant alleles
| Line No. | Tamlo‐A1 | Tamlo‐B1 | Tamlo‐D1 | Current status | |||
|---|---|---|---|---|---|---|---|
| Mutation | Effecta | Mutation | Effecta | Mutation | Effecta | ||
| 1 | P325L | Medium | G319R | Strong | P335L | Very strong | aabbdd |
| 2 | P325L | Medium | G319R | Strong | G319R | Strong | aabbdd |
| 3 | P325L | Medium | G296E | Strong | P321S | Medium | AaBbDd |
| 4 | P325L | Medium | T297I | Weak | P321S | Medium | AaBbDd |
Effect on powdery mildew susceptibility based on data from transient gene expression assays (Figure 2).
Currently, we have produced four different Tamlo lines that bear mutations in all three TaMlo homoeologues. Lines 1 and 2 (Table 3) are already homozygous and are referred as Tamlo‐aabbdd. We also obtained two of the three possible double mutant combinations (Tamlo‐aabbDD and Tamlo‐AAbbdd). Lines 3 and 4 (Table 3) are presently triple heterozygous plants in self‐fertilization stage.
Mutations in TaMlo homoeologues confer enhanced Bgt resistance
We next aimed to test the powdery mildew infection phenotype with the set of available single, double and triple mutants. We inoculated 10‐day‐old leaves with a Bgt field isolate and estimated microscopically the host cell entry rate at 72 h p.i. (hours post inoculation; Figure 4a). Furthermore, we evaluated the macroscopic phenotype of these mutants at 6 h p.i. (hours post inoculation; Figure 4b).
Figure 4.
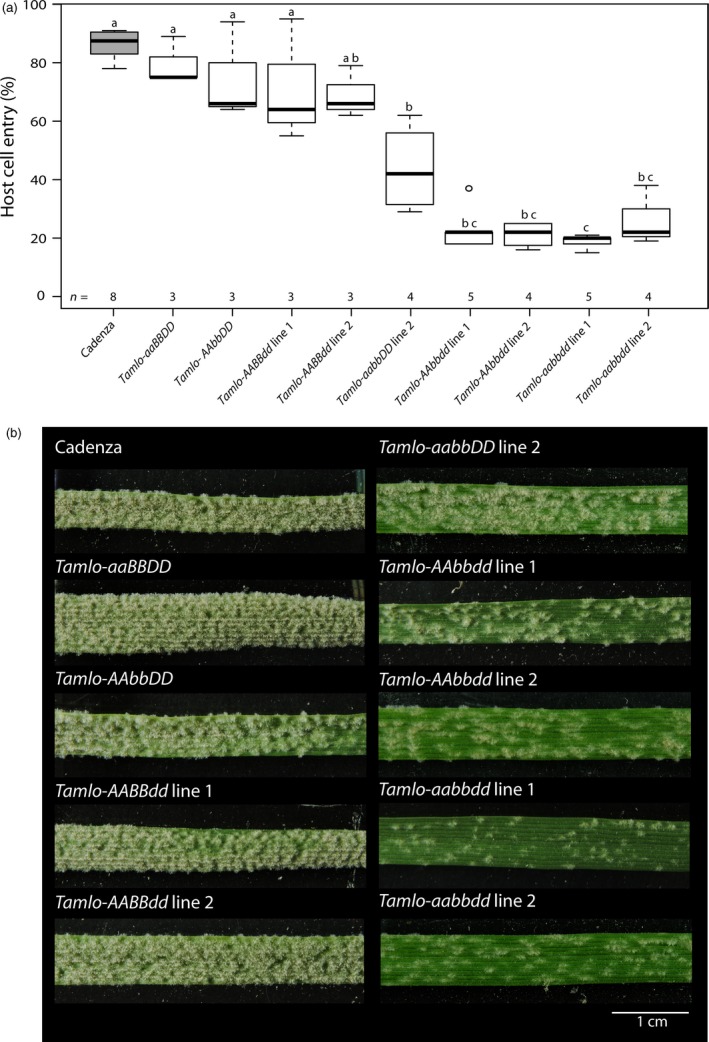
Bgt infection phenotypes of wheat WT (cv. Cadenza), single, double and triple Tamlo mutants. Ten‐day‐old leaves were inoculated with Bgt isolate JA82. (a) Host cell entry was scored at 72 h p.i. Centre lines show the medians; upper and lower box limits indicate the 25th and 75th percentiles respectively; upper and lower whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, respectively, and outliers are represented by dots. Numbers at the bottom of the boxplots indicate the number of biological replicates per sample (n). One biological replicate was typically composed of four leaves with 200 scored cells. Letters indicate genotypes whose data are significantly (P < 0.05) different from genotypes labelled with other letters, as determined by pair‐wise testing with a Games–Howell post hoc test. Statistics were performed and boxplots generated with R software. (b) First leaves of germinated seedlings were fixed with surgical tape to a polycarbonate platform and inoculated with Bgt conidiospores. The macroscopic phenotype was recorded at 6 d.p.i. A scale bar shown in white is given in the lower right corner (1 cm).
The cv. Cadenza exhibited a Bgt penetration rate of ~85%. The various single homozygous mutants showed a slight reduction in entry rate (~65%–75%, not significantly different from cv. Cadenza), with the tendency of Tamlo‐AAbbDD and Tamlo‐AABBdd to be lower than Tamlo‐aaBBDD; Figure 4a). Interestingly, both Tamlo‐AAbbdd double mutant lines showed a lower penetration rate than the Tamlo‐aabbDD double mutant line (between 14% and 22% for Tamlo‐AAbbdd versus 42% for Tamlo‐aabbDD, Figure 4a and S2a; note that Tamlo‐aabbDD was obtained from segregating progeny in line 2, but is based on the same allele combination as in line 1). Furthermore, both Tamlo‐aabbdd triple mutant lines exhibited enhanced resistance compared with WT, with host cell entry values of ~20%, similar to the Tamlo‐AAbbdd double mutant (Figures 4a and S2a). This result suggests that the contribution of the Tamlo‐A1 allele to resistance in the triple homozygous line is lower than the contribution of the Tamlo‐B1 and ‐D1 alleles, as was predicted by the transient expression data (Figure 2).
The evaluation of the macroscopic phenotype upon Bgt inoculation was largely consistent with the microscopic assessment (Figures 4b and S2b). The four single mutants exhibited an infection phenotype similar to WT, while the Tamlo‐aabbDD and Tamlo‐AAbbdd double mutant lines and the two Tamlo‐aabbdd triple mutant lines showed enhanced resistance compared to the WT (Figures 4b and S2b). However, in contrast to the host cell entry results, at the level of the macroscopic infection phenotype the triple Tamlo‐aabbdd mutants seem to be more resistant than the Tamlo‐AAbbdd and Tamlo‐aabbDD double mutants, as exemplified by fewer colonies on the leaf surface (Figures 4b and S2b). This observation suggests that the Tamlo‐aabbdd lines might have an additional post‐penetration effect in powdery mildew infection. Nevertheless, resistance of each mutant was clearly weaker than seen for the triple knockout mutant obtained by TALENs (Wang et al., 2014; Figures 4b and S2b).
Although resistance of the TILLING mutants in the field has not yet been scored, plants were naturally and spontaneously infected by Bgt in the greenhouse. Throughout the entire wheat growth cycle we observed considerably fewer powdery mildew symptoms on the Tamlo‐aabbdd lines compared to the WT or heterozygous individuals within segregating populations (Figure S3).
Differential transcript accumulation of the TaMlo1 homoeologues
Since we observed a higher reduction in the Bgt penetration rate and less sporulation on the leaf surface of the Tamlo‐AAbbdd double mutant compared to the Tamlo‐aabbDD double mutant (Figure 4) we reasoned that this difference could be based (i) on the particular mutant allele combinations; (ii) on potential unequal functional redundancy of the TaMlo homoeologues; or (iii) on differential expression of the three TaMlo homoeologues. To explore the latter possibility, we made use of a data set of RNA‐seq samples derived from several tissues (root, leaf, stem, spike and grain) at different developmental stages of bread wheat cv. Chinese Spring (Choulet et al., 2014). The paired‐end RNA‐seq reads were mapped to a transcriptome reference containing the coding sequences of TaMlo obtained in this study (File S1) together with non‐redundant cDNA sequences from The International Wheat Genome Sequencing Consortium (IWGSC, 2014). The respective mean fragments per kilobase (kb) per million mapped reads (FPKM) values revealed that the TaMlo homoeologues are highly expressed in leaves compared to the other tissues, especially at Zadoks stage 71 (Zadoks et al., 1974; Figure 5 and Table S3). Furthermore, the FPKM values suggest that in leaves the three homoeologues have differential expression levels, with TaMlo‐D1 showing the highest level of expression, followed by TaMlo‐B1, while the level of TaMlo‐A1 transcripts seems strikingly lower (Figure 5).
Figure 5.
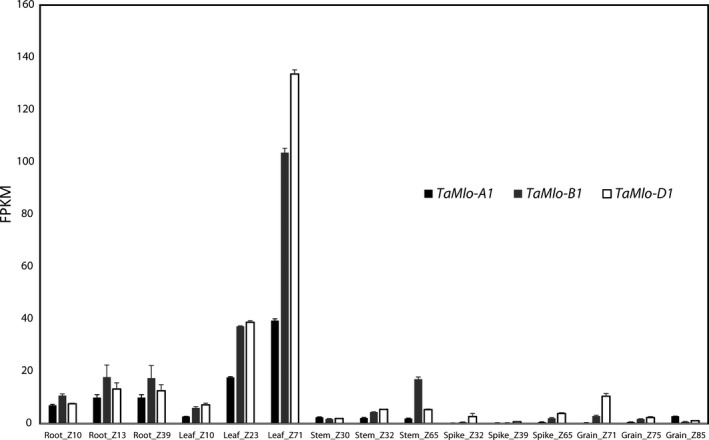
Transcript accumulation of TaMlo homoeologues in five bread wheat tissues at different developmental stages. Transcript levels of each homoeologue were summed up and then averaged across replicates, ± standard errors of the mean. FPKM: Fragments per kb per million mapped reads; Z: Zadoks developmental scale (Zadoks et al., 1974).
To further test the notion that TaMlo‐A1 shows lower transcript levels in leaf tissue compared to TaMlo‐B1 and ‐D1, we followed an entirely independent experimental route. We PCR‐amplified and cloned a TaMlo cDNA fragment using oligonucleotide primers that have a complete match in all three TaMlo homoeologues. This cDNA fragment was further chosen to contain several homoeologue‐specific single‐nucleotide polymorphisms (SNPs) that distinguish them unequivocally (Figure S4). We randomly picked and sequenced 28 clones representing the various TaMlo cDNA fragments. Among these, we found 14, 10 and 4 clones corresponding to TaMlo‐B1, TaMlo‐D1 and TaMlo‐A1, respectively. Together the results of these two approaches suggest that TaMlo‐A1 is the homoeologue with the lowest transcript levels in leaves.
TILLING‐derived Tamlo triple mutant lines lack obvious pleiotropic phenotypes
In our conditions, the Tamlo‐aabbdd lines did not show any obvious and macroscopically discernible differences regarding overall growth habit, height, the number of spikes or seed number compared to cv. Cadenza WT plants. We also did not notice any signs of early leaf chlorosis in the TILLING mutants compared to the WT. This is in stark contrast to the pronounced leaf chlorosis observed under our growth conditions in case of the TALEN‐derived Tamlo‐aabbdd line, which represents a full Tamlo knockout mutant (Wang et al., 2014, Figure S5).
Discussion
In this study, we used the non‐transgenic TILLING technology as a strategy to identify mutant candidates in the TaMlo genes from an EMS‐mutagenized population of spring wheat cv. Cadenza. TILLING has been previously suggested as a means to create mutants in ‘susceptibility genes’ such as Mlo in a targeted manner (Hückelhoven et al., 2013; van Schie and Takken, 2014). By using transient gene expression assays in barley and an adapted powdery mildew species for the bioassay we were able to test the obtained missense mutant wheat variants and to select candidates that confer different degrees of reduction in powdery mildew host cell entry (Figure 2). This experimental assessment of TaMlo protein functionality broadly agreed with the results of in silico prediction via SIFT analysis (Figure 2 and Table 2). However, the observed exceptions [Tamlo‐A1 (A354V), Tamlo‐B1 (T297I) and Tamlo‐B1 (R313W)] highlight the need for validation of mutant variants by experimentation. Stacking of the selected mutant alleles resulted in four different wheat lines carrying a missense mutation in each of the three TaMlo homoeologues (Table 3). The two triple homozygous Tamlo‐aabbdd mutant lines (1 and 2) showed enhanced powdery mildew disease resistance, exemplified by a considerable reduction in Bgt host cell entry [from ~85% in WT to ~20% (median values) in the triple mutant] and a substantial decrease in macroscopically visible Bgt colonies (Figures 4 and S2). Notably, the Tamlo‐AAbbdd double mutants showed a similar level of enhanced Bgt penetration resistance.
In our TILLING screening we targeted TaMlo exon 9, which encodes the third cytoplasmic loop of the TaMlo protein. Excluding the false positives, we found a total of 56 mutants within an approximate population of 2020 individuals (Table 1), which represents an average density of 1 mutation per 32 kb screened. These results are comparable to the ones obtained by Botticella and co‐workers after screening the same TILLING population for mutants in the SbIIa genes (Botticella et al., 2011). These authors identified an average of 60 candidate mutants per exon and a density of approximately 1 mutation per 40 kb screened. As in our screening, the authors did not find nonsense mutations for each exon assessed. However, in contrast to our results, the number of identified missense mutations was higher than the silent ones. The lack of identification of nonsense mutations in our study is somewhat surprising because exon 9 has six glutamine (Q; CAG triplet) or tryptophan (W; TGG triplet) codons that could potentially have been mutated to stop codons.
Using transient gene expression experiments in single barley epidermal cells we demonstrated that cDNAs from each of the three TaMlo homoeologues are able to complement the barley mlo resistance phenotype and can restore host cell entry to WT‐like levels (Figure 2). Of the 16 identified missense mutations, the non‐conservative changes had a more dramatic effect on susceptibility to powdery mildew, especially G296E, G319R and P335L (Figure 2 and Table 3). These data are consistent with previous site‐directed mutagenesis studies performed in barley, further reinforcing the relevance of the third cytoplasmic loop of Mlo as an important domain in the susceptibility‐conferring activity of the protein (Reinstädler et al., 2010). Therefore, the application of transient gene expression assays was an efficient tool to select suitable mutant alleles for subsequent stacking.
Surprisingly, not only the Tamlo‐aabbdd triple mutants but also the double mutants Tamlo‐aabbDD and Tamlo‐AAbbdd exhibited significantly enhanced Bgt resistance compared to the WT (Figures 4 and S2). These results are in stark contrast with the ones obtained by Wang and co‐workers, where only the triple knockout mutant exhibited a notable difference in the powdery mildew infection phenotype (Wang et al., 2014). However, the Tamlo‐AAbbdd double mutant line was missing in the set of tested TALEN plants (Wang et al., 2014); therefore, a comparison between this double mutant and ours generated by TILLING is not possible. It is conceivable that some of the discrepancies between the two data sets are due to the type of evaluation, since we scored host cell entry while Wang and co‐workers evaluated the number of microcolonies per germinated spores (Wang et al., 2014). Additionally, second‐site mutations present in our TILLING lines, which are derived from EMS mutagenesis, might lead to differences in the powdery mildew infection phenotype. Furthermore, the TILLING lines are based on a spring wheat cultivar (cv. Cadenza), while the TALEN lines are derived from a winter wheat. Together, our results challenge the hypothesis of strict functional redundancy between wheat TaMlo homoeologues proposed by Borrill et al. (2015) and suggest instead that the powdery mildew infection phenotype can be the result of gene dosage effects or differential expression of the respective TaMlo homoeologues. Evidence to support this notion is provided not only by the differences in host cell entry and macroscopic infection phenotypes observed between the double mutants Tamlo‐aabbDD and Tamlo‐AAbbdd compared to WT (Figure 4), but also the seemingly lower transcript levels of TaMlo‐A1 compared to TaMlo‐B1 and ‐D1 (Figure 5). Tamlo double mutants with enhanced powdery mildew resistance might be advantageous in future breeding programmes, since only two mutant alleles must be followed in segregating populations.
The incomplete Bgt resistance found in our Tamlo‐aabbdd triple mutants is likely based on the partial loss‐of‐function alleles used in these lines (Figure 2; Table 3). Deployment of yet to be identified stronger EMS‐induced alleles (e.g. nonsense mutations) may, similar to the TALEN‐derived line (Wang et al., 2014), result in a TILLING‐based non‐transgenic triple mutant with full Bgt resistance. However, the incomplete resistance phenotype in our generated Tamlo‐aabbdd TILLING lines might in fact represent an advantage in several different ways, namely, reduced pleiotropic effects, more durable resistance and an opportunity for breeders since products of induced mutagenesis are free of government regulation.
Pleiotropic effects such as early leaf senescence associated with mlo mutants have been better studied in barley where lines carrying weak resistance alleles, permitting residual fungal growth, exhibit fewer pleiotropic phenotypes than mlo mutants harbouring strong alleles, which confer complete immunity (Hentrich, 1979; Piffanelli et al., 2002). TILLING has thus been suggested as a means to find partial loss‐of‐function mutants of ‘susceptibility genes’ that show mild pleiotropy (Hückelhoven et al., 2013). During the first cycle of propagation we did not observe differences in growth, development and fertility between our Tamlo‐aabbdd TILLING lines (partial loss‐of‐function alleles) and the WT, nor did we observe enhanced or premature leaf senescence (Figure S5). These results are in contrast to the noticeable leaf chlorosis in TALEN‐based mutants (complete loss‐of‐function alleles) in comparison to its parental line KN199 (Figure S5). Nevertheless, this is only a first qualitative assessment of these plants and quantitative measurements of photosynthetic performance and chlorophyll levels under various growth conditions are needed to substantiate these observations in future experiments.
The most common allele introduced in the spring barley varieties cultivated in Europe is mlo‐11 (Brown and Rant, 2013; Kokina et al., 2007; Piffanelli et al., 2004). Barley plants carrying this natural partial loss‐of‐function allele are not fully resistant and allow the growth of powdery mildew colonies at a low level (Piffanelli et al., 2004). The barley mlo‐11 plants nevertheless confer broad‐spectrum resistance to the pathogen, which has been durable in the field for more than 30 years (reviewed in Acevedo‐Garcia et al., 2014). One may speculate that the lower selection pressure exerted on powdery mildew populations by partial resistance might have contributed to this durability under agricultural conditions. Our Tamlo‐aabbdd lines are also partially resistant, allowing a low level of Bgt sporulation (Figure 4). This feature may contribute to the robustness of the trait once mlo‐based resistance is used in commercial wheat crops. Finally, our non‐transgenic wheat Tamlo‐aabbdd plants represent an excellent alternative for breeders and farmers in regions where growth of genetically‐modified organisms (GMOs) is banned and where regulations for the cultivation of genome‐edited crops are still under discussion (Chen et al., 2014b; Huang et al., 2016).
This study reports the first step towards the generation of new commercial bread wheat varieties with an enhanced resistance phenotype to the globally important powdery mildew disease, generated by TILLING technology targeting the TaMlo gene. Currently, our Tamlo‐aabbdd plants are being backcrossed to remove excess of EMS mutations that may affect additional traits. It will be interesting to determine the performance of the backcrossed plants in the field at the agronomic level and their response to other pathogens and pests that attack wheat crops.
Experimental procedures
Plant and fungal material
The EMS‐mutagenized population of the spring wheat (Triticum aestivum) cv. Cadenza has been previously described (Rakszegi et al., 2010). Barley (Hordeum vulgare) seeds NGB14661.2 (cv. Kristina), NGB14682.1 (cv. Bonus) NGB119178.1 (SR59, mlo‐38, background cv. Bonus), NGB119185.1 (SR65, mlo‐38, background cv. Kristina) were originally reported in Lundqvist et al. (1991) and obtained from NordGen (Nordic Genetic Resource Centre) in Sweden. The mlo‐29 mutant and its parent cv. Sultan were described previously (Piffanelli et al., 2002). The TALEN‐derived Tamlo‐aabbdd line and its parental line KN199 (Wang et al., 2014) were kindly provided by Prof. Caixia Gao and Prof. Jin‐Long Qiu from the State Key Laboratories (Chinese Academy of Sciences, Beijing, China). The Bgt isolate, termed JA82, was obtained from plant material spontaneously infected in the greenhouse, latitude: 50.7784, longitude: 6.04863. It was propagated weekly on 10‐day‐old seedlings comprising a mixture of summer wheat cv. Munk and commercially available seeds of unknown identity. Bgh isolate K1 was propagated weekly on 7‐day‐old barley seedlings cv. Margret.
TILLING screening
M2 wheat genomic DNA samples (~2020) of the TILLING library developed at Rothamsted Research were twofold pooled in 96‐well plates at 25 ng/μL. The amplicons for HRM were produced by a nested PCR strategy. The 1st round PCR was carried out in 10 μL final volume using 1 μL of pooled template DNA, 5 μL of HotShot Diamond™ Master Mix (Clent Life Sciences, Stourbridge, UK) and 0.5 μm primers. The PCR program was: 98 °C, 5 min; (97 °C, 30 s; 66–67 °C, 30 s; 72 °C 1 min) × 35 cycles; 72 °C, 5 min; 10 °C until end.
For HRM, the 1st round PCR reaction was diluted 100‐fold and 2 μL were used as template for the 2nd round PCR. The 2nd round PCR was prepared in FrameStar 96‐well skirted plates, black frame with white wells (4titude, Surrey, UK). The reaction contained 2 μL of diluted DNA template, 0.5 μm of each primer, 5 μL HotShot Diamond Mastermix, and 1 μL of LCGreen Plus (Clent Life Sciences, Stourbridge, UK) in a total volume of 10 μL; 10 μL of mineral oil (M5904, Sigma, Deisenhofen, Germany) was added to prevent evaporation. The PCR program was similar to the primary PCR, with annealing at 66 °C and after final extension, an additional denaturation step at 98 °C, 3 min, followed by cooling to 10 °C at 0.2 °C/s.
High‐Resolution Melting analysis
The 2nd round PCR plates were used for HRM analysis using the LightScanner instrument (Idaho Technology Inc., Utah) and with the following temperatures: start, 82 °C; end 98 °C and a hold of 78 °C. The analysis was performed with the software provided with the scanner and as described in Botticella et al. (2011). Negative samples were repeated and positive candidates were selected from the independent plate. A twofold pool of DNA from candidate lines together with cv. Cadenza DNA was generated for the 2nd round PCR and screening was repeated. The amplicon from putative positive candidates was re‐amplified and sent for sequencing. DNA sequencing was performed by Eurofins MWG Operon (Ebersberg, Germany).
Transient gene expression in barley leaf epidermal cells
Ballistic transformation of 8‐day‐old detached barley mlo‐3 (background cv. Ingrid) leaves was performed as previously described (Reinstädler et al., 2010). Inoculation with Bgh isolate K1 was carried out at 24 h after bombardment and histochemical staining with X‐Gluc (5‐Bromo‐4‐chloro‐3‐indolyl‐ß‐D‐glucoronide; Carl Roth GmbH, Karlsruhe, Germany) for β‐glucoronidase (GUS) activity was performed at 48 h p.i. Fungal structures were stained with Coomassie Brilliant Blue R‐250 (Carl Roth GmbH, Karlsruhe, Germany). The host cell entry rate was calculated as: (number of transformed, Bgh‐attacked cells with haustoria/transformed and Bgh‐attacked cells)*100. At least six leaves were evaluated per mutant with a minimum of 150–200 cells per experiment and at least three independent biological replicates.
For further experimental details, see Files S3 and S4.
Supporting information
Figure S1 Crossing scheme to obtain the Tamlo‐aabbdd line 1. The procedure started with the propagation of M2 seeds and the selection of single homozygous mutants for each TaMlo homoeologue (Tamlo‐aaBBDD, Tamlo‐AAbbDD and Tamlo‐AABBdd). The homozygous single mutant lines Tamlo‐AAbbDD and Tamlo‐AABBdd were crossed, the resulting heterozygous F1 progeny (genotype Tamlo‐AABbDd) self‐fertilized, and homozygous Tamlo‐AAbbdd double mutant plants selected in the F2 generation. These were finally crossed with the homozygous Tamlo‐aaBBDD single mutant, the resulting heterozygous F1 progeny (genotype Tamlo‐AaBbDd) self‐fertilized, and homozygous Tamlo‐aabbdd triple mutant plants selected in the F2 generation. A crossing is represented with an x and self‐pollination is represented by an x inside a circle. Similar crossing schemes were adapted for the other allele combinations.
Figure S2 Bgt infection phenotypes of additional independent Tamlo mutant lines and the TALEN‐derived Tamlo‐aabbdd mutant with its respective parent. Ten‐day‐old leaves were inoculated with Bgt conidiospores. (a) Host cell entry was scored at 72 h p.i. Centre lines show the medians; upper and lower box limits indicate the 25th and 75th percentiles, respectively; upper and lower whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles respectively. Numbers at the bottom of the boxplots indicate the number of biological replicates per sample (n). One biological replicate was typically composed of four leaves with 200 scored cells. Letters indicate genotypes whose data are significantly (P < 0.001) different from genotypes labelled with other letters, as determined by pair‐wise testing with a Games–Howell post hoc test. Statistics were performed and boxplots generated with R software. (b) First leaves of germinated seedlings were fixed with surgical tape to a polycarbonate platform and inoculated with Bgt conidiospores. The macroscopic phenotype was recorded at 6 d.p.i. A scale bar shown in white is given in the lower right corner (1 cm).
Figure S3 Spontaneous powdery mildew infection of wheat plants growing under greenhouse conditions. (a) cv. Cadenza. (b) Heterozygous plant of segregating population, genotype Tamlo‐AaBBdd. (c–d) Tamlo‐aabbdd line 1. (a and c) 2‐month‐old, (b and d) 3‐month‐old plants.
Figure S4 Nucleotide sequences of the TaMlo cDNA fragment chosen for indirect TaMlo expression analysis by PCR product cloning. The sequence represents the PCR fragment obtained with oligonucleotide primers TaMlo‐ABD‐F and TaMlo‐ABD‐R (Table S1), indicated with bold lines. Shown in yellow are SNPs for TaMlo‐A1, in blue SNPs for TaMlo‐B1, in green SNPs for TaMlo‐D1 and in grey a SNP for all three homoeologues.
Figure S5 Early leaf senescence phenotype of the TALEN‐derived Tamlo‐aabbdd line. (a) Transgenic winter wheat TALEN line (genotype Tamlo‐aabbdd; left) and its parental line WT KN199 (genotype Tamlo‐AABBDD; right). (b) TILLING‐derived spring wheat line 1 (genotype Tamlo‐aabbdd; left) and its WT parent cv. Cadenza (genotype Tamlo‐AABBDD; right). (c) TILLING‐derived spring wheat line 2 (genotype Tamlo‐aabbdd; left) and its WT parent cv. Cadenza (genotype Tamlo‐AABBDD; right). All plants were around 3 months old when images were taken.
Table S1 Sequences of oligonucleotide primers used in this study.
Table S2 CAPS markers developed for genotyping Tamlo mutants.
Table S3 Homoeologue‐specific TaMlo gene transcript levels in wheat tissues.
File S1 Multiple sequence alignment of TaMlo coding sequences.
File S2 TaMlo consensus genomic sequences.
File S3 Supplementary Materials and Methods.
File S4 R script for the Games–Howell post hoc test.
Acknowledgements
We are thankful to Prof. Caixia Gao and Prof. Jin‐Long Qiu from the State Key Laboratories (Chinese Academy of Sciences, Beijing, China) for providing seeds of the transgenic TALEN‐derived Tamlo‐aabbdd mutant and its parental line KN199, and to Prof. Hans Thordal‐Christensen (University of Copenhagen, Denmark) for facilitating the delivery of the seeds. We acknowledge Stefan Kusch for his advice regarding statistical analysis. This work was funded by a shared grant of the German Federal Ministry of Food and Agriculture (BMEL; project management via the Specialist Agency for Renewable Resources; grant number 22030411) and the Germany Society for the Advancement of Plant Innovation (GFPi; grant number G 135/12 NR) respectively. AP and KHK are funded by the 20:20 Wheat Institute Strategic Programme Grant to Rothamsted Research from the Biotechnology and Biological Sciences Research Council of the U.K (grant number BB/J/00426X/1).
References
- Acevedo‐Garcia, J. , Kusch, S. and Panstruga, R. (2014) Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 204, 273–281. [DOI] [PubMed] [Google Scholar]
- Borrill, P. , Adamski, N. and Uauy, C. (2015) Genomics as the key to unlocking the polyploid potential of wheat. New Phytol. 208, 1008–1022. [DOI] [PubMed] [Google Scholar]
- Botticella, E. , Sestili, F. , Hernandez‐Lopez, A. , Phillips, A. and Lafiandra, D. (2011) High resolution melting analysis for the detection of EMS induced mutations in wheat SBEIIa genes. BMC Plant Biol. 11, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.K.M. and Rant, J.C. (2013) Fitness costs and trade‐offs of disease resistance and their consequences for breeding arable crops. Plant. Pathol. 62, 83–95. [Google Scholar]
- Büschges, R. , Hollricher, K. , Panstruga, R. , Simons, G. , Wolter, M. , Frijters, A. , van Daelen, R. et al (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell, 88, 695–705. [DOI] [PubMed] [Google Scholar]
- Chen, A. and Dubcovsky, J. (2012) Wheat TILLING mutants show that the vernalization gene VRN1 down‐regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet. 8, e1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, A. , Li, C. , Hu, W. , Lau, M.Y. , Lin, H. , Rockwell, N.C. , Martin, S.S. et al (2014a) PHYTOCHROME C plays a major role in the acceleration of wheat flowering under long‐day photoperiod. Proc. Natl Acad. Sci. USA, 111, 10037–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Hao, L. , Parry, M.A.J. , Phillips, A.L. and Hu, Y.‐G. (2014b) Progress in TILLING as a tool for functional genomics and improvement of crops. J. Integr. Plant Biol. 56, 425–443. [DOI] [PubMed] [Google Scholar]
- Choulet, F. , Alberti, A. , Theil, S. , Glover, N. , Barbe, V. , Daron, J. , Pingault, L. et al (2014) Structural and functional partitioning of bread wheat chromosome 3B. Science, 345, 1249721. [DOI] [PubMed] [Google Scholar]
- Colasuonno, P. , Incerti, O. , Lozito, M.L. , Simeone, R. , Gadaleta, A. and Blanco, A. (2016) DHPLC technology for high‐throughput detection of mutations in a durum wheat TILLING population. BMC Genet. 17, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner, R.L. , Kuzyk, A.D. and Su, H. (2003) Impact of powdery mildew on the yield of soft white spring wheat cultivars. Can. J. Plant Sci. 83, 725–728. [Google Scholar]
- Dean, R. , van Kan, J.A.L. , Pretorius, Z.A. , Hammond‐Kosack, K. , Pietro, A.Di , Spanu, P.D. , Rudd, J.J. et al (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C. , Vincent, K. and Sharp, P. (2009) Simultaneous mutation detection of three homoeologous genes in wheat by High Resolution Melting analysis and Mutation Surveyor® . BMC Plant Biol. 9, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, C. , Zhou, F. , Spielmeyer, W. , Panstruga, R. and Schulze‐Lefert, P. (2002) Functional conservation of wheat and rice Mlo orthologs in defense modulation to the powdery mildew fungus. Mol. Plant Microbe Interact. 15, 1069–1077. [DOI] [PubMed] [Google Scholar]
- Glawe, D.A. (2008) The powdery mildews: a review of the world's most familiar (yet poorly known) plant pathogens. Ann. Rev. Phytopathol. 46, 27–51. [DOI] [PubMed] [Google Scholar]
- Hentrich, W. (1979) Multiple Allelie, Pleiotropie und züchterische Nutzung mehltauresistenter Mutanten des mlo‐Locus der Gerste. Tag‐Ber. Akad. Landwirtsch‐Wiss. 175, 191–202. [Google Scholar]
- Huang, S. , Weigel, D. , Beachy, R.N. and Li, J. (2016) A proposed regulatory framework for genome‐edited crops. Nat. Genet. 48, 109–111. [DOI] [PubMed] [Google Scholar]
- Hückelhoven, R. , Eichmann, R. , Weis, C. , Hoefle, C. and Proels, R.K. (2013) Genetic loss of susceptibility: a costly route to disease resistance? Plant. Pathol. 62, 56–62. [Google Scholar]
- IWGSC . (2014) A chromosome‐based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science, 345, 1251788. [DOI] [PubMed] [Google Scholar]
- Jørgensen, J.H. (1992) Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica, 63, 141–152. [Google Scholar]
- Jørgensen, J.H. and Mortensen, K. (1977) Primary infection by Erysiphe graminis f. sp. hordei of barley mutants with resistance genes in the ml‐o locus. Phytopathology, 5, 678–685. [Google Scholar]
- Kokina, A. , Legzdiņa, L. , Bērziņa, I. , Bleidere, M. , Rashal, I. and Rostoks, N. (2007) Molecular marker‐based characterization of barley powdery mildew Mlo resistance locus in European varieties and breeding lines. Lat. J. Agron. 11, 77–83. [Google Scholar]
- Konishi, S. and Sasanuma, T. (2010) Identification of novel Mlo family members in wheat and their genetic characterization. Genes Genet. Syst. 85, 167–175. [DOI] [PubMed] [Google Scholar]
- Kumar, P. , Henikoff, S. and Ng, P.C. (2009) Predicting the effects of coding non‐synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Kurowska, M. , Daszkowska‐Golec, A. , Gruszka, D. , Marzec, M. , Szurman, M. , Szarejko, I. and Maluszynski, M. (2011) TILLING – a shortcut in functional genomics. J. App. Genet. 52, 371–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch, S. , Pesch, L. and Panstruga, R. (2016) Comprehensive phylogenetic analysis sheds light on the diversity and origin of the MLO family of integral membrane proteins. Genome Biol. Evol. 8, 878–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundqvist, U. , Meyer, J. and Lundqvist, A. (1991) Mutagen specificity for 71 lines resistant to barley powdery mildew race D1 and isolated in four highbred barley varieties. Hereditas, 115, 227–239. [DOI] [PubMed] [Google Scholar]
- Lyngkjær, M.F. , Newton, A.C. , Atzema, J.L. and Baker, S.J. (2000) The barley mlo‐gene: an important powdery mildew resistance source. Agronomie, 20, 745–756. [Google Scholar]
- McCallum, C.M. , Comai, L. , Greene, E.A. and Henikoff, S. (2000) Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant. Physiol. 123, 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menardo, F. , Praz, C. , Wyder, S. , BenDavid, R. , Bourras, S.A. , Matsumae, H. , McNally, K.E. et al (2016) Hybridization of powdery mildew strains gives raise to pathogens on novel agricultural crop species. Nat. Genet. 48, 201–205. [DOI] [PubMed] [Google Scholar]
- Müller, J. , Piffanelli, P. , Devoto, A. , Miklis, M. , Elliott, C. , Ortmann, B. , Schulze‐Lefert, P. et al (2005) Conserved ERAD‐like quality control of a plant polytopic membrane protein. Plant. Cell, 17, 149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhänsel, C. , Freialdenhoven, A. , Kurth, J. , Kolsch, R. and Schulze‐Lefert, P. (1997) Interaction analyses of genes required for resistance responses to powdery mildew in barley reveal distinct pathways leading to leaf cell death. Plant. Cell, 9, 1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli, P. , Zhou, F. , Casais, C. , Orme, J. , Jarosch, B. , Schaffrath, U. , Collins, N.C. et al (2002) The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant. Physiol. 129, 1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli, P. , Ramsay, L. , Waugh, R. , Benabdelmouna, A. , D'Hont, A. , Hollricher, K. , Jørgensen, J.H. et al (2004) A barley cultivation‐associated polymorphism conveys resistance to powdery mildew. Nature, 430, 887–891. [DOI] [PubMed] [Google Scholar]
- Rakszegi, M. , Kisgyörgy, B.N. , Tearall, K. , Shewry, P.R. , Láng, L. , Phillips, A. and Bedő, Z. (2010) Diversity of agronomic and morphological traits in a mutant population of bread wheat studied in the Healthgrain program. Euphytica, 174, 409–421. [Google Scholar]
- Rawat, N. , Sehgal, S.K. , Joshi, A. , Rothe, N. , Wilson, D.L. , McGraw, N. , Vadlani, P.V. et al (2012) A diploid wheat TILLING resource for wheat functional genomics. BMC Plant. Biol. 12, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, G.H. and Wittwer, C.T. (2004) Sensitivity and specificity of single‐nucleotide polymorphism scanning by high‐resolution melting analysis. Clin. Chem. 50, 1748–1754. [DOI] [PubMed] [Google Scholar]
- Reinstädler, A. , Müller, J. , Czembor, J.H. , Piffanelli, P. and Panstruga, R. (2010) Novel induced mlo mutant alleles in combination with site‐directed mutagenesis reveal functionally important domains in the heptahelical barley Mlo protein. BMC Plant. Biol. 10, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie, C.C.N. and Takken, F.L.W. (2014) Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581. [DOI] [PubMed] [Google Scholar]
- Schwarzbach, E. (1976) The pleiotropic effects of the ml‐o gene and their implications in breeding In Barley Genetics III (Gaul H., ed), pp. 440–445. Munich: Karl Thiemig Verlag. [Google Scholar]
- Sestili, F. , Botticella, E. , Bedo, Z. , Phillips, A. and Lafiandra, D. (2009) Production of novel allelic variation for genes involved in starch biosynthesis through mutagenesis. Mol. Breed. 25, 145–154. [Google Scholar]
- Sestili, F. , Palombieri, S. , Botticella, E. , Mantovani, P. , Bovina, R. and Lafiandra, D. (2015) TILLING mutants of durum wheat result in a high amylose phenotype and provide information on alternative splicing mechanisms. Plant. Sci. 9116, 1–7. [DOI] [PubMed] [Google Scholar]
- Simmonds, J. , Scott, P. , Brinton, J. , Mestre, T.C. , Bush, M. , del Blanco, A. and Dubcovsky, J. , et al (2016) A splice acceptor site mutation in TaGW2‐A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theor. Appl. Genet. 129, 1099–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade, A.J. , Fuerstenberg, S.I. , Loeffler, D. , Steine, M.N. and Facciotti, D. (2005) A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat. Biotechnol. 23, 75–81. [DOI] [PubMed] [Google Scholar]
- Slade, A.J. , McGuire, C. , Loeffler, D. , Mullenberg, J. , Skinner, W. , Fazio, G. , Holm, A. et al (2012) Development of high amylose wheat through TILLING. BMC Plant. Biol. 12, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy, C. , Paraiso, F. , Colasuonno, P. , Tran, R.K. , Tsai, H. , Berardi, S. , Comai, L. et al (2009) A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant. Biol. 9, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várallyay, É. , Giczey, G. and Burgyán, J. (2012) Virus‐induced gene silencing of Mlo genes induces powdery mildew resistance in Triticum aestivum . Arch. Virol. 157, 1345–1350. [DOI] [PubMed] [Google Scholar]
- Wang, T.L. , Uauy, C. , Robson, F. and Till, B. (2012) TILLING in extremis . Plant. Biotechnol. J. 10, 761–772. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Cheng, X. , Shan, Q. , Zhang, Y. , Liu, J. , Gao, C. and Qiu, J.L. (2014) Simultaneous editing of three homoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Wolter, M. , Hollricher, K. , Salamini, F. and Schulze‐Lefert, P. (1993) The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol. Gen. Genet. 239, 122–128. [DOI] [PubMed] [Google Scholar]
- Zadoks, J.C. , Chang, T.T. and Konzak, C.F. (1974) A decimal code for the growth stages of cereals. Weed Res. 14, 415–421. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Crossing scheme to obtain the Tamlo‐aabbdd line 1. The procedure started with the propagation of M2 seeds and the selection of single homozygous mutants for each TaMlo homoeologue (Tamlo‐aaBBDD, Tamlo‐AAbbDD and Tamlo‐AABBdd). The homozygous single mutant lines Tamlo‐AAbbDD and Tamlo‐AABBdd were crossed, the resulting heterozygous F1 progeny (genotype Tamlo‐AABbDd) self‐fertilized, and homozygous Tamlo‐AAbbdd double mutant plants selected in the F2 generation. These were finally crossed with the homozygous Tamlo‐aaBBDD single mutant, the resulting heterozygous F1 progeny (genotype Tamlo‐AaBbDd) self‐fertilized, and homozygous Tamlo‐aabbdd triple mutant plants selected in the F2 generation. A crossing is represented with an x and self‐pollination is represented by an x inside a circle. Similar crossing schemes were adapted for the other allele combinations.
Figure S2 Bgt infection phenotypes of additional independent Tamlo mutant lines and the TALEN‐derived Tamlo‐aabbdd mutant with its respective parent. Ten‐day‐old leaves were inoculated with Bgt conidiospores. (a) Host cell entry was scored at 72 h p.i. Centre lines show the medians; upper and lower box limits indicate the 25th and 75th percentiles, respectively; upper and lower whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles respectively. Numbers at the bottom of the boxplots indicate the number of biological replicates per sample (n). One biological replicate was typically composed of four leaves with 200 scored cells. Letters indicate genotypes whose data are significantly (P < 0.001) different from genotypes labelled with other letters, as determined by pair‐wise testing with a Games–Howell post hoc test. Statistics were performed and boxplots generated with R software. (b) First leaves of germinated seedlings were fixed with surgical tape to a polycarbonate platform and inoculated with Bgt conidiospores. The macroscopic phenotype was recorded at 6 d.p.i. A scale bar shown in white is given in the lower right corner (1 cm).
Figure S3 Spontaneous powdery mildew infection of wheat plants growing under greenhouse conditions. (a) cv. Cadenza. (b) Heterozygous plant of segregating population, genotype Tamlo‐AaBBdd. (c–d) Tamlo‐aabbdd line 1. (a and c) 2‐month‐old, (b and d) 3‐month‐old plants.
Figure S4 Nucleotide sequences of the TaMlo cDNA fragment chosen for indirect TaMlo expression analysis by PCR product cloning. The sequence represents the PCR fragment obtained with oligonucleotide primers TaMlo‐ABD‐F and TaMlo‐ABD‐R (Table S1), indicated with bold lines. Shown in yellow are SNPs for TaMlo‐A1, in blue SNPs for TaMlo‐B1, in green SNPs for TaMlo‐D1 and in grey a SNP for all three homoeologues.
Figure S5 Early leaf senescence phenotype of the TALEN‐derived Tamlo‐aabbdd line. (a) Transgenic winter wheat TALEN line (genotype Tamlo‐aabbdd; left) and its parental line WT KN199 (genotype Tamlo‐AABBDD; right). (b) TILLING‐derived spring wheat line 1 (genotype Tamlo‐aabbdd; left) and its WT parent cv. Cadenza (genotype Tamlo‐AABBDD; right). (c) TILLING‐derived spring wheat line 2 (genotype Tamlo‐aabbdd; left) and its WT parent cv. Cadenza (genotype Tamlo‐AABBDD; right). All plants were around 3 months old when images were taken.
Table S1 Sequences of oligonucleotide primers used in this study.
Table S2 CAPS markers developed for genotyping Tamlo mutants.
Table S3 Homoeologue‐specific TaMlo gene transcript levels in wheat tissues.
File S1 Multiple sequence alignment of TaMlo coding sequences.
File S2 TaMlo consensus genomic sequences.
File S3 Supplementary Materials and Methods.
File S4 R script for the Games–Howell post hoc test.


