Abstract
Dry root rot (DRR) caused by the fungus Rhizoctonia bataticola (Taub.) Butler, is an emerging disease in chickpea. The disease is often mistaken with other root rots like Fusarium wilt, collar rot and black root rot in chickpea. Therefore, its timely and specific detection is important. Current detection protocols are either based on mycological methods or on protocols involving DNA amplification by polymerase chain reaction (PCR). Here we report the rapid and specific detection of R. bataticola using loop-mediated isothermal amplification (LAMP) assay targeting fungal specific 5.8S rDNA sequence for visual detection of R. bataticola. The reaction was optimized at 63 °C for 75 min using minimum 10 fg of DNA. After adding SYBR Green I in LAMP products, the amplification was found to be highly specific in all the 94 isolates of R. bataticola collected from diverse geographical regions as well as DRR infected plants and sick soil. No reaction was found in other pathogenic fungi infecting chickpea (Fusarium oxysporum f. sp. ciceris, Rhizoctonia solani, Sclerotium rolfsii and Fusarium solani) and pigeonpea (Fusarium udum and Phytophthora cajani). The standardised LAMP assay with its simplicity, rapidity and specificity is very useful for the visual detection of this emerging disease in chickpea.
Dry root rot (DRR) caused by soil borne necrotrophic fungus Rhizoctonia bataticola (Taub.) Butler [Synonyms: Macrophomina phaseolina (Tassi) Goid] is an emerging disease in chickpea (Cicer arietinum L.)1. The DRR is more dominant when the crop is exposed to moisture stress conditions2 and can cause 50 to 100% yield loss under favourable conditions3. In recent years, Rhizoctonia bataticola is becoming more prevalent in agricultural areas where climate change is leading to higher temperatures. It is reported that R. bataticola can infect more than 284 plant species including monocot and dicot plant hosts4. Due to availability of wide range of natural host, R. bataticola can easily sustain in the dry climatic area and persist in soil for prolonged period even after rotation of the crops.
In chickpea, DRR is often mistaken with Fusarium wilt and other root rot diseases (collar rot, black root rot etc.), as the general symptoms of these diseases are nearly similar and visually undistinguishable in field conditions1. In all the cases, affected plants show foliar chlorosis and ultimately cause plant collapse. Therefore, there is a real need of an advance rapid, reliable and easy detection method for diagnosis of R. bataticola for better management of DRR. In recent years, PCR based methods like conventional PCR and real time PCR is being employed to detect fungal species and other microorganism5,6,7, but it is not cost effective and need high-quality DNA due to the effects of inhibitors on PCR sensitivity8,9. Also molecular expertise is required for true diagnosis of pathogens. Now a days, Loop-mediated isothermal amplification (LAMP) has been developed as an alternative and reliable method for the detection of microbial pathogens and diagnosis of plant diseases10,11,12,13,14. The advantages and simplicity of LAMP assay is that the reaction could be easily judged as positive or negative by naked eye through assessing of increased turbidity or colour change15,16, and for that it does not require any expensive instruments like thermal cycler.
The LAMP is highly sensitive, less time-consuming than conventional PCR-based methods, and less prone to inhibition from DNA preparations17. Reliability of primer sets and DNA sequences of interest are the most important factors in development of molecular detection of targeted organisms. The internal transcribed spacer (ITS) region of nuclear rRNA genes is suitable targets for species diversity analysis within the fungal communities18,19,20. The characteristic of high sequence variability within the ITS region makes itself a valuable and ideal target for developing of genus and species specific PCR primers to identify an organism. Since LAMP assay has been reported to be very useful for quick detection and identification of a broad range of microorganisms, including viruses21, bacteria8, and fungi10,11, the present study was proposed to develop highly specific and very sensitive LAMP assay for the detection of R. bataticola from infected plants and soil.
Materials and Methods
Materials studied
Fungal strains
A total 94 isolates of R. bataticola representing different chickpea growing geographical region of India were taken in this study. Other major fungal strains infecting chickpea (e.g. Fusarium oxysporum f. sp. ciceris, Rhizoctonia solani, Sclerotium rolfsii and Fusarium solani) and other legume crop pigeonpea (Fusarium udum and Phytophthora cajani) were taken for validation of studies (Table 1).
Table 1. Details of thee fungal isolates, plant and soil samples used in the LAMP detection assay.
| Samples | Collection site | State | LAMP detection |
Conventional PCR |
||
|---|---|---|---|---|---|---|
| Florescence | Agarose gel | RB_F3/RB_B3 | ITS1/ITS4 | |||
| R. bataticola isolates (chickpea) | ||||||
| RB1 | Kanpur | Uttar Pradesh | + | + | + | + |
| RB2 | Coimbatore | Tamil Nadu | + | + | + | + |
| RB3 | ICRISAT, BIL 01 | Andhra Pradesh | + | + | + | + |
| RB4 | ICRISAT, BIL 01 | Andhra Pradesh | + | + | + | + |
| RB5 | ICRISAT, BP 4 | Andhra Pradesh | + | + | + | + |
| RB6 | ICRISAT, BP 10 | Andhra Pradesh | + | + | + | + |
| RB7 | ICRISAT, BIL 01 | Andhra Pradesh | + | + | + | + |
| RB8 | ICRISAT, BUS 03 | Andhra Pradesh | + | + | + | + |
| RB9 | ICRISAT, BR 05 | Andhra Pradesh | + | + | + | + |
| RB10 | ICRISAT, BIL 02 | Andhra Pradesh | + | + | + | + |
| RB11 | ICRISAT, BIL 02 | Andhra Pradesh | + | + | + | + |
| RB12 | Pati | Andhra Pradesh | + | + | + | + |
| RB13 | ICRISAT, BP 02 | Andhra Pradesh | + | + | + | + |
| RB14 | Jodhpur | Madhya Pradesh | + | + | + | + |
| RB15 | Jabalpur | Madhya Pradesh | + | + | + | + |
| RB16 | Delhi | Delhi | + | + | + | + |
| RB17 | Damoh 1 | Madhya Pradesh | + | + | + | + |
| RB18 | Damoh 2 | Madhya Pradesh | + | + | + | + |
| RB19 | Damoh 3 | Madhya Pradesh | + | + | + | + |
| RB20 | Damoh 4 | Madhya Pradesh | + | + | + | + |
| RB21 | Damoh 5 | Madhya Pradesh | + | + | + | + |
| RB22 | Damoh 6 | Madhya Pradesh | + | + | + | + |
| RB23 | Damoh 7 | Madhya Pradesh | + | + | + | + |
| RB24 | ICRISAT, BUS 04 | Andhra Pradesh | + | + | + | + |
| RB25 | ICRISAT, BUS 07 | Andhra Pradesh | + | + | + | + |
| RB26 | ICRISAT, BIL 02 | Andhra Pradesh | + | + | + | + |
| RB27 | ICRISAT, BUS 07 | Andhra Pradesh | + | + | + | + |
| RB28 | ICRISAT, BR 04H | Andhra Pradesh | + | + | + | + |
| RB29 | ICRISAT, BUS 03 | Andhra Pradesh | + | + | + | + |
| RB30 | ICRISAT, BIL 02D | Andhra Pradesh | + | + | + | + |
| RB31 | ICRISAT, BUS 03 | Andhra Pradesh | + | + | + | + |
| RB32 | ICRISAT, BIL 05 | Andhra Pradesh | + | + | + | + |
| RB33 | ICRISAT, BP 04 | Andhra Pradesh | + | + | + | + |
| RB34 | ICRISAT, BR 05D | Andhra Pradesh | + | + | + | + |
| RB35 | ICRISAT, BUS 03 | Andhra Pradesh | + | + | + | + |
| RB36 | ICRISAT, BR 04F | Andhra Pradesh | + | + | + | + |
| RB37 | ICRISAT, BP 04 | Andhra Pradesh | + | + | + | + |
| RB38 | ICRISAT, BR 04F | Andhra Pradesh | + | + | + | + |
| RB39 | ICRISAT, BM 14 | Andhra Pradesh | + | + | + | + |
| RB40 | ICRISAT, BR 04 | Andhra Pradesh | + | + | + | + |
| RB41 | ICRISAT, BP 04 | Andhra Pradesh | + | + | + | + |
| RB42 | ICRISAT, BR 04F | Andhra Pradesh | + | + | + | + |
| RB43 | ICRISAT, BW 01A | Andhra Pradesh | + | + | + | + |
| RB44 | ICRISAT, BIL 01 | Andhra Pradesh | + | + | + | + |
| RB45 | ICRISAT, BW 03 | Andhra Pradesh | + | + | + | + |
| RB46 | ICRISAT, BW 04 | Andhra Pradesh | + | + | + | + |
| RB47 | ICRISAT, BR 04I | Andhra Pradesh | + | + | + | + |
| RB48 | ICRISAT, BR 04H | Andhra Pradesh | + | + | + | + |
| RB49 | Jabalpur 1 | Madhya Pradesh | + | + | + | + |
| RB50 | Jabalpur 5 | Madhya Pradesh | + | + | + | + |
| RB51 | Jabalpur 6 | Madhya Pradesh | + | + | + | + |
| RB52 | Jabalpur 8 | Madhya Pradesh | + | + | + | + |
| RB53 | Brampuri, Damoh 8 | Madhya Pradesh | + | + | + | + |
| RB54 | Brampuri, Damoh 9 | Madhya Pradesh | + | + | + | + |
| RB55 | Katni | Madhya Pradesh | + | + | + | + |
| RB56 | JNKVV, Jabalpur | Madhya Pradesh | + | + | + | + |
| RB57 | Rewa | Madhya Pradesh | + | + | + | + |
| RB58 | Bachara. Satna | Madhya Pradesh | + | + | + | + |
| RB59 | IIPR, Kanpur | Uttar Pradesh | + | + | + | + |
| RB60 | ICRISAT, BIL 02 | Andhra Pradesh | + | + | + | + |
| RB61 | Dhaulakuan | Himachal Pradesh | + | + | + | + |
| RB62 | Ramnagar, Pantnagar | Uttarakhand | + | + | + | + |
| RB63 | BAU, Ranchi | Jharkhand | + | + | + | + |
| RB64 | ICRISAT, BP 02C | Andhra Pradesh | + | + | + | + |
| RB65 | ICRISAT, BP 15 | Andhra Pradesh | + | + | + | + |
| RB66 | ICRISAT, BP 03B | Andhra Pradesh | + | + | + | + |
| RB67 | ICRISAT, BP 03C | Andhra Pradesh | + | + | + | + |
| RB68 | ICRISAT, BP 08A | Andhra Pradesh | + | + | + | + |
| RB69 | ICRISAT, BP 08B | Andhra Pradesh | + | + | + | + |
| RB70 | ICRISAT, BR 05D | Andhra Pradesh | + | + | + | + |
| RB71 | ICRISAT, BR 05B | Andhra Pradesh | + | + | + | + |
| RB72 | ICRISAT, BL 04A | Andhra Pradesh | + | + | + | + |
| RB73 | ICRISAT, BL0 4 | Andhra Pradesh | + | + | + | + |
| RB74 | ICRISAT, BM 13 | Andhra Pradesh | + | + | + | + |
| RB75 | ICRISAT, BM 13 | Andhra Pradesh | + | + | + | + |
| RB76 | ICRISAT, BM 08C | Andhra Pradesh | + | + | + | + |
| RB77 | ICRISAT, BW 02A | Andhra Pradesh | + | + | + | + |
| RB78 | ICRISAT, BW 02B | Andhra Pradesh | + | + | + | + |
| RB79 | ICRISAT, BW 02C | Andhra Pradesh | + | + | + | + |
| RB80 | ICRISAT, BW 04A | Andhra Pradesh | + | + | + | + |
| RB81 | ICRISAT, BW 05A | Andhra Pradesh | + | + | + | + |
| RB82 | ICRISAT, BW 05B | Andhra Pradesh | + | + | + | + |
| RB83 | ICRISAT, BW 08 | Andhra Pradesh | + | + | + | + |
| RB84 | ICRISAT, BIL 01 | Andhra Pradesh | + | + | + | + |
| RB85 | ICRISAT, BIL 01 | Andhra Pradesh | + | + | + | + |
| RB86 | ICRISAT, BIL 01 | Andhra Pradesh | + | + | + | + |
| RB87 | ICRISAT, BIL 03C | Andhra Pradesh | + | + | + | + |
| RB88 | ICRISAT, BIL 03 | Andhra Pradesh | + | + | + | + |
| RB89 | ICRISAT, BIL 04 | Andhra Pradesh | + | + | + | + |
| RB90 | ICRISAT, BIL 05B | Andhra Pradesh | + | + | + | + |
| RB91 | ICRISAT, BIL 05C | Andhra Pradesh | + | + | + | + |
| RB92 | ICRISAT, BIL 05C | Andhra Pradesh | + | + | + | + |
| RB93 | ICRISAT, JM 08B | Andhra Pradesh | + | + | + | + |
| RB94 | ICRISAT, BR 05C | Andhra Pradesh | + | + | + | + |
| DRR infected chickpea plants (field) | ||||||
| Sample 1 | ICRISAT, BIL 02 | Andhra Pradesh | + | + | + | + |
| Sample 2 | ICRISAT, BW 08 | Andhra Pradesh | + | + | + | + |
| Sample 3 | ICRISAT, BM 13 | Andhra Pradesh | + | + | + | + |
| Sample 4 | ICRISAT, BP 15 | Andhra Pradesh | + | + | + | + |
| Sample 5 | ICRISAT, BIL 01 | Andhra Pradesh | + | + | + | + |
| Healthy | ICRISAT, BIL 02 | Andhra Pradesh | − | − | − | − |
| DRR infected chickpea plants (greenhouse) | ||||||
| Sample 1 | Experimental sample | ICRISAT | + | + | + | + |
| Sample 2 | Experimental sample | ICRISAT | + | + | + | + |
| Sample 3 | Experimental sample | ICRISAT | + | + | + | + |
| Sample 4 | Experimental sample | ICRISAT | + | + | + | + |
| Healthy | Experimental sample | ICRISAT | − | − | − | − |
| Sick soil (DRR) | ||||||
| Rhizospheric black soil | ICRISAT, BP 04 | Andhra Pradesh | + | + | + | MBa |
| Rhizospheric red soil | ICRISAT, RL17 | Andhra Pradesh | + | + | + | MB |
| Sick soil (non-rhizospheric) | Experimental sick plot | ICRISAT | + | + | + | + |
| Other fungal pathogens | ||||||
| F. oxysporum f. sp. ciceris (chickpea) | In vitro culture | ICRISAT | − | − | − | + |
| R. solani (chickpea) | In vitro culture | ICRISAT | − | − | − | + |
| S. rolfsii (chickpea) | In vitro culture | ICRISAT | − | − | − | + |
| F. solani (chickpea) | In vitro culture | ICRISAT | − | − | − | + |
| F. udum (pigeonpea) | In vitro culture | ICRISAT | − | − | − | + |
| P. cajani (pigeonpea) | In vitro culture | ICRISAT | − | − | − | + |
aMultiple bands.
Plant and soil samples
Healthy and infected DRR chickpea plant samples were collected from greenhouse and experimental fields of International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru, Telangana State, India. The soil samples taken in this study were collected from screening plot for R. bataticola and from rhizosphere of DRR infected chickpea plants in field (Table 1).
DNA extraction
Total genomic DNA (gDNA) was isolated from all the fungal isolates and DRR infected plants using PureLink Plant Total DNA Purification kit (Invitogen, USA) as per manufacturer’s protocol. About 100 mg of frozen mycelial tissue/plant tissue was grounded in liquid N2 and resuspended in 250 μL Resuspension Buffer (supplied in the kit). Total gDNA was eluted in 50 μL of nuclease free water and stored at −20 °C for further downstream application. The soil DNA was extracted from 100 mg of R. bataticola sick soil and DRR infected chickpea rhizospheric soil using SoilMaster™ DNA Extraction Kit (Epicentre, USA) according to the manufacturer’s protocol. The obtained DNA was suspended in 200 μL of elution buffer. The purified DNA was evaluated in 0.8% agarose gel as well as by UV spectrophotometry.
Primer design
As R. bataticola is an important plant pathogen with a broad host range and causes disease in diverse commercial crops, primers for the LAMP assay were designed from the conserved region in the partial ITS and 5.8S rRNA sequences of R. bataticola and M. phaseolina identified by multiple sequence alignment of representative isolates from different crops (Table 2 and Fig. 1). A set of six primers for LAMP assay, comprising two outer primers (RB_F3 and RB_B3), two innermost primers (RB_FIP and RB_BIP), and two loop primers (RB_LoopF and RB_LoopB) were designed manually by following the directions of Torres et al.22. The FIP was made with the complementary sequence of F1 (F1c) and F2, and the BIP made with the complementary sequence of R1 (B1c) and B2 (Table 3). The organization and position of the LAMP primers and their complementarity to target DNA used in this study are shown in Fig. 1. The designed primers were nBLAST searched on NCBI for analysing the sequence specificity and to get chance of cross reaction with other sibling species if environmental samples will assay in LAMP reaction (Table 4). To avoid the false positive reaction by cross reactivity within the primers, the secondary structures (hairpin, self-dimer and hetero-dimer) of the primers and their corresponding stability with ΔG were reviewed23,24 in OligoAnalyzer 3.1 tool (http://eu.idtdna.com/calc/analyzer) and were determined (Table 5). The universal primer pair ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) and RB_F3 and RB_B3 were used for conventional PCR.
Table 2. Nucleotide sequence analysis of representative Rhizoctonia bataticola/Macrophomina phaseolina isolates infecting economically important plants and their disease list.
| Pathogen | Host |
Disease | Acc. No. | Country source | Sequence alignment analysis for Acc. No. HQ392814 (QC/ID in per cent)* |
||
|---|---|---|---|---|---|---|---|
| Common name | Scientific name | Complete sequence | Sequence fragment taken for primer design | ||||
| Rhizoctonia bataticola | Chickpea | Cicer arientinum | Dry root rot | HQ392814 | India | 100/99 | 100/100 |
| Pigeonpea | Cajanus cajan | Dry root rot | KJ629078 | India | 95/99 | 100/100 | |
| Linseed | Linum usitatissimum | Wilt | KM247370 | India | 95/98 | 100/99 | |
| Spider lily | Crinum asiaticum | Leaf disease | KX447538 | Malaysia | 99/98 | 100/100 | |
| Macrophomina phaseolina | Impatiens | Impatiens sp. | Root disease complex | KU726237 | USA | 98/99 | 100/100 |
| Sorghum | Sorghum bicolor | Charcoal rot | KU856652 | Denmark | 98/99 | 100/100 | |
| Olive | Olea europea | Die-back | KU863545 | Tunisia | 98/99 | 100/100 | |
| Sunflower | Helianthus annuus | Charcoal rot | KT862032 | Mongolia | 95/99 | 100/100 | |
| Potato | Solanum tuberosum | Charcoal rot/tuber blemishes | KU721993 | South Africa | 85/100 | 100/100 | |
| Common bean | Phaseolus vulgaris | Charcoal rot/leaf blight | KU831500 | Tunisia | 94/99 | 100/100 | |
| Indian jasmine | Jasminum multiflorum | Root rot | KT768135 | India | 98/99 | 100/100 | |
| Cowpea | Vigna unguiculata | Charcoal rot | KF951783 | Senegal | 92/99 | 100/100 | |
| Peanut | Arachis hypogae | Dry root rot/charcoal rot | KF951759 | Senegal | 96/99 | 100/100 | |
| Lady’s fingers | Abelmoschus esculentas | Dry root and foot rot | KF951754 | Senegal | 97/99 | 100/100 | |
| Roselle | Hibiscus sabdarifa | Charcoal rot | KF951701 | Louga, Senegal | |||
| 97/99 | 100/100 | ||||||
| Pearl millet | Pennisetum glaucum | Dry root rot | KF951691 | Niger | 92/99 | 100/100 | |
| Black gram | Phaseolus mungo | Charcoal rot | KF951637 | Denmark | 93/99 | 100/100 | |
| Chrysanthemum | Chrysanthemum sp. | Charcoal rot | KF951633 | USA | 92/99 | 100/100 | |
| Sugarcane | Saccharum officinarum | Charcoal rot | KF951631 | India | 97/99 | 100/100 | |
| Pigeonpea | Cajanus indicus | Dry root rot | KF951628 | Sri Lanka | 93/99 | 100/100 | |
| Maize | Zea mays | Charcoal rot | KF951627 | Palestine | 92/99 | 100/100 | |
| Sesamum | Sesamum indicum | Charcoal rot | KF951624 | Uganda | 93/99 | 100/100 | |
| Derris legume | Derris elliptica | Charcoal rot | KF951623 | Malaysia | 93/99 | 100/100 | |
| Black cottonwood | Populus trichocarpa | Charcoal rot | KF428466 | USA | 97/99 | 100/100 | |
| Cumin | Cuminum cyminum | Charcoal rot | KF453968 | Turkey | 95/99 | 100/100 | |
| Strawberry | Fragaria sp. | Crown rot | KC822431 | China | 98/99 | 100/100 | |
| Pectilis orchid | Pectilis susannae | Charcoal rot | KC920477 | India | 78/100 | 100/100 | |
| Soybean | Glycin max | Charcoal rot | KC202823 | Iran | 93/99 | 100/100 | |
| Sweet potato | Ipomoea batatas | Charcoal rot | JX945170 | USA | 98/99 | 100/100 | |
| Turmeric | Carcuma longa | Charcoal rot | JX535007 | India | 96/99 | 100/100 | |
| Mungbean | Vigna radiata | Root rot and leaf blight | HQ660594 | China | 98/99 | 100/100 | |
| Golden samphire | Inula crithmoides | Root rot | HQ649832 | Spain | 93/99 | 100/100 | |
*Values of QC (quarry coverage) and identity (ID) scores of all sequence segments retrieve from NCBI nucleotide sequence database that matched the query sequence (HQ392814).
Figure 1. Nucleotide sequence alignment of partial ITS and 5.8S rDNA region of Rhizoctonia bataticola and Macrophomina phaseolina of different legumes.
The shades sequences in different colour indicate the regions of primer development for LAMP assay. The primer sequences are specified by red colour and arrows are the direction of amplification.
Table 3. Information of the primers used for the LAMP reaction.
| Primers name | Sequences (5′-3′) | Type | Primer position (nt) | Length (bp) | GC (%) | Tm |
|---|---|---|---|---|---|---|
| RB_F3 | CCTCCCACCCTTTGTATACCTACC | Forward outer | 1–24 | 24 | 54.2 | 58.4 |
| RB_B3 | CGCAAAGGACGGTGCCCAA | Backward outer | 336–354 | 19 | 63.2 | 61.4 |
| RB_FIP (F1c+F2) | CTGCAACGTTTACTGACTGGAGTTTG-CCGATTTTGGGGGGTGGCTAGT | Forward inner | (104–129) + (65–86) | 26 + 22 | 46.2–59.1 | 58.2–61.7 |
| RB_BIP (B1c + B2) | CGGATCTCTTGGTTCTGGCATCG-GAAATGACGCTCGAACAGGCATG | Backward inner | (168–190) + (291–313) | 23 + 23 | 54.2–52.2 | 60.9–58.8 |
| RB_LoopF | TCCTCTGGCGGGCACTAG | Forward loop forming | 82–99 | 18 | 66.7 | 59.4 |
| RB_LoopB | ACATTGCGCCCCTTGGGATT | Backward loop forming | 263–282 | 20 | 55 | 60.2 |
Table 4. In silico specificity and cross reactivity analysis of LAMP primers through NCBI database and details of 100 hits generated in nBLAST.
| List of primers |
R. bataticola/M. phaseolina |
Other microbes |
|||
|---|---|---|---|---|---|
| Identity (%) | Number of hits | Identity (%) | Number of hits | Name of the dominant microbes hit in primer blast (numbers in the parentheses denote the number of hits) | |
| RB_F3 | 100 | 96 | 100 | 4 | Uncultured Helotiales sp. (4) |
| RB_B3 | 100 | 40 | 100 | 60 | Botryosphaeria dothidea (51), Uncultured fungal strains (7), Fusicoccum fabicercianum (2) |
| RB_F1c | 100 | 100 | — | — | — |
| RB_F2 | 100 | 100 | — | — | — |
| RB_B1c | — | ?* | 100 | 100 | Uncultured fungal strains (22), Coniella spp. (14), Uncultured Helotiales sp. (7), Fusarium spp. (6), Oidiodendron sp. (5), Xylariaceae sp. (5), Botryosphaeria dothidea (4), Nemania sp. (4) and remaining hits are of different microbes with single entry |
| RB_B2 | — | ?* | 100 | 100 | Coniella sp. (60), Uncultured fungal strains (9), Oidiodendron sp. (9), Fusarium spp. (6), Xylariaceae sp. (5), Microsporum gypseum (4), Nemania sp. (4) and remaining hits are of different microbes with single entry |
| RB_Loop_F | 100 | 89 | 100 | 11 | Botryosphaeria mamane (11) |
| RB_Loop_B | 100 | 3 | 100 | 6 | Dothideomycetes sp. (1), Hormonema sp. (1), Saccharomyces cerevisiae (1), Ampelomyces quisqualis (1), Uncultured Heyderia (1), Uncultured Helotiales (1) |
| — | — | 85–95 | 91 | Uncultured fungal strains (36), Lasiodiplodia theobromae, (10), Russula sp. (4), Mesorhizobium (3), Uncultured Helotiales sp (2) and remaining hits are of different microbes with single entry | |
Table 5. In silico hairpin, self-dimer or hetero-dimer analysis of primers used in LAMP assay.
| List of primers | ΔG for hetero-dimer |
ΔG for self-dimer | Hairpin |
|||||
|---|---|---|---|---|---|---|---|---|
| RB_B3 | RB_FIP | RB_BIP | RB_LoopF | RB_LoopB | ΔG | Tm | ||
| RB_F3 | −10.51 | −12.51 | −5.02 | −6.14 | −9.67 | −6.08 | 0.23 | 22.3 |
| RB_B3 | — | −10.04 | −8.16 | −12.57 | −10.65 | −5.09 | −0.59 | 31.6 |
| RB_FIP | — | — | −9.73 | −8.16 | −9.21 | −10.18 | −1.75 | 43.0 |
| RB_BIP | — | — | — | −6.75 | −6.75 | −6.76 | −1.97 | 36.5 |
| RB_Loop_F | — | — | — | — | −9.82 | −4.16 | 0.3 | 17.1 |
| RB_Loop_B | — | — | — | — | — | −9.89 | −1.69 | 50.5 |
LAMP reaction
The constituents of the LAMP assay were optimized using total gDNA extracted from R. bataticola fungal culture (positive control) and two negative controls (without DNA and DNA from a healthy chickpea plant). The LAMP reaction was performed in a 25 μL volume contained 2.0 μL primer mixture (20 μM each of FIP, BIP, Loop F, and Loop B primers, and 2.5 μM each of F3 and B3 primers), 1 mM dNTPs, 4 mM MgCl2, 2.5 μL of 10X ThermoPol Reaction Buffer (1X reaction buffer (pH 8.8) contained 20 mM Tris–HCl, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100), 8 U of Bst DNA polymerase (NEB, UK), 1.5 μL target DNA (about 100 ng). After completing the isothermal amplification 1 μL of SYBR Green I (Invitrogen, USA) was added for visual assay of the amplification. The reaction was carried out at 63 °C for 60 min followed by incubation at 80 °C for 10 min to inactivate the Bst DNA polymerase. The reactions were performed in a 0.5 mL microcentrifuge in a water bath for temperature control.
Optimization of LAMP reaction condition
To optimize the LAMP reaction conditions, the reaction was carried out at different temperatures, 57 °C, 60 °C, 63 °C, 66 °C and 69 °C using a gradient thermo cycler (ARKTIK Thermal Cycler, Thermo Scientific, USA). The LAMP reaction mixture was incubated at the different temperatures for different time periods, 15 min, 30 min, 45 min, 60 min, 75 min and 90 min to optimize the temperature and time/duration for the LAMP reaction. The reactions were terminated by heat inactivation at 80 °C for 10 min. The LAMP reaction was assessed visually based on colour change after adding SYBR Green I and under UV light. The LAMP amplified products were then analysed by 2% agarose gel electrophoresis.
Specificity of the LAMP
To determine the specificity of LAMP assay, the reaction was carried out with the extracted DNA from other legume infecting pathogenic fungi discussed previously in fungal strain section. The LAMP assay was done as described earlier at optimized temperature and time duration. The assays were visualized based on SYBR Green I colour change and then by 2% agarose gel electrophoresis. Individual fungal sample was tested by three replications and the experiment was repeated three times.
Sensitivity of the LAMP method
The sensitivity of the LAMP assay was determined using 10 fold serially diluted R. bataticola DNA ranged from 10 ng to 0.1 fg. Reaction mixture without DNA template was included as a negative control. The LAMP amplification products were analysed visually by addition of 1 μL SYBR Green I under UV light or by 2% agarose gel electrophoresis. To compare the sensitivities and specificities between LAMP assay and PCR; PCR was performed with the extracted DNA from all fungal isolates using ITS primers (Table 2). The PCR reaction was carried using the protocol detailed in Ghosh et al. (2015). The PCR amplified products were then analysed in 1% agarose gel stained with ethidium bromide. The experiment was repeated thrice. The detection limit in LAMP assay was defined as the last dilution with positive reaction.
Validation of the LAMP assay
To validate the LAMP assay, the DNA from DRR infected chickpea plants and soil samples were tested for the presence of R. bataticola. Based on colour change and gel electrophoresis as described above, the LAMP reaction was visualised. The experiment was carried out in triplicate and samples were considered as positive if two of the replicates showed positive response.
Results
Selection of LAMP primers
All R. bataticola isolates were identified by amplifying and sequencing of approximately 544 bp of partial ITS and 5.8S region using universal primers, ITS1 and ITS4 (Table 1). For designing of LAMP assay primers for R. bataticola, the conserve region of the partial ITS and 5.8S sequences was identified by multiple sequence alignment of corresponding nucleotide sequences of representative isolates in different crops (Table 2) and a set of six primers were designed (Table 3). The primers were analysed in silico against the nBLAST search tool using the NCBI sequence database. The BLAST results revealed that except both parts (B1c and B2) of BIP primer all primers were significantly hits the target sequences of R. bataticola and M. phaseolina with 100% identity (Table 4), although the sequence of BIP primer is 100% similar with the target sequences (Fig. 1). The hit also resulted in 100% similarity with other non-pathogenic microbes of chickpea (Table 4). During the design of LAMP primers, ΔG values were determined and the values were fixed around −9 kcal/mole or more positive than −9 kcal/mole (Table 5). For hairpins, the Tm of the hairpin was kept below of 50 °C and lower than the annealing temperature for the LAMP reaction as the strongest hairpin should be at least 50 °C (Table 5).
Optimization of LAMP assay
To determine the optimum temperature and reaction time of LAMP assay, the assay was tested within a wide range of temperatures (57 °C–69 °C) and time (15 min–90 min) using pure DNA of R. bataticola culture. The assay showed positive reaction at all temperatures, whether assessment was based on visual fluorescence detection or in gel electrophoresis. The characteristic ladder like bands were evident in the gel when the reaction was positive, but not if the reaction was negative. However, the high intense ladder-like band pattern was obtained in gel electrophoresis at 63 °C (Fig. 2a). Fluorescence detection result was consistent with the results from 2% agarose gel electrophoresis. For optimizing time, the LAMP reaction was conducted at 63 °C for various time duration mentioned above. The positive reaction was found in all the time duration, but strong band pattern was observed after 75 min of reaction (Fig. 2b). Therefore, 63 °C for 75 min is the optimized temperature and time for LAMP reaction for detection of DRR pathogen.
Figure 2. Optimization of LAMP reaction for detection of Rhizoctonia bataticola.
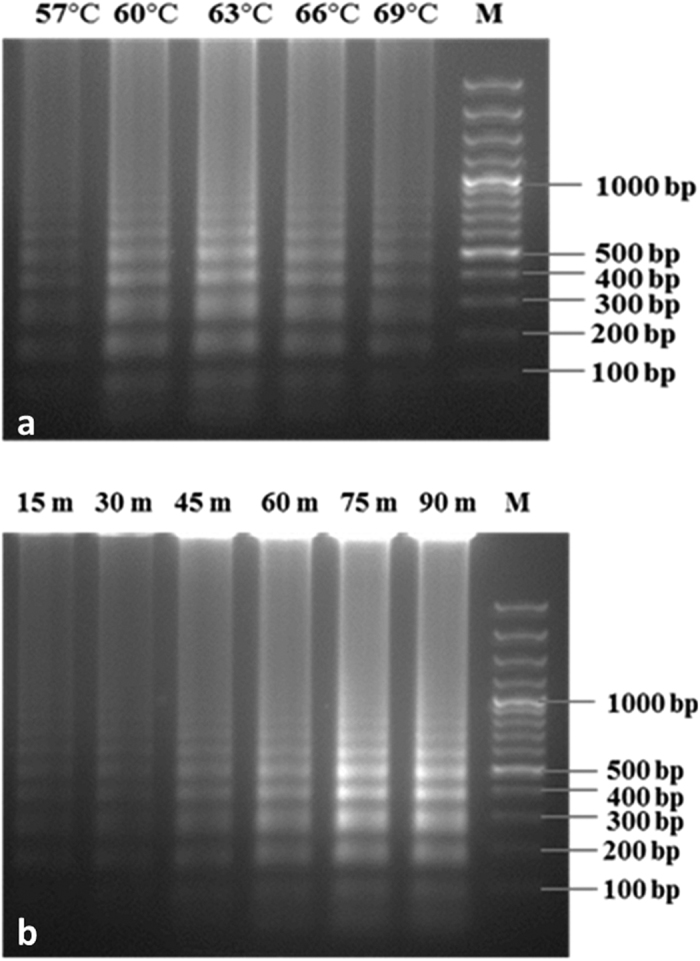
(a) Optimization of temperature of LAMP. M GeneRulerTM 100 bp Plus DNA Ladder; lanes 57 °C–69 °C indicated the reaction temperatures of LAMP. (b) Optimization of time duration of LAMP. M GeneRulerTM 100 bp Plus DNA Ladder; lanes 15 m–90 m indicated the reaction time duration of LAMP. All the products were detected on the basis of 2% agarose gel electrophoresis.
LAMP assay specificity
LAMP specificity was examined using DNA templates extracted from total 94 R. bataticola isolates collected from diverse geographical region in India. Six pathogenic wilt and root rot pathogens viz., Fusarium oxysporum f. sp. ciceris, Rhizoctonia solani, Sclerotium rolfsii, Fusarium solani, Fusarium udum and Phytophthora cajani infecting chickpea and pigeonpea were also taken for testing specificity (Table 1). DNA template of R. bataticola isolates gave positive reaction, whereas no amplification was observed for the other fungal species after incubation at 63 °C for 75 min. The LAMP reaction was assessed using 2% agarose (Fig. 3) and SYBR Green I visualisation. The result indicated that the LAMP assay developed in this study is highly specific to R. bataticola. Furthermore, to confirm the specificity of the LAMP primers, a PCR was carried out using primer pair, RB_F3 and RB_B3. All the R. bataticola isolates were amplified with a unique DNA fragment of 354 bp. However, these primers did not produce any band when DNA templates from other pathogenic fungi were used. However, primer pair ITS1 and ITS4 gave amplification of expected size of DNA fragment (Table 1).
Figure 3. Specificity of LAMP assay for detection of Rhizoctonia bataticola.
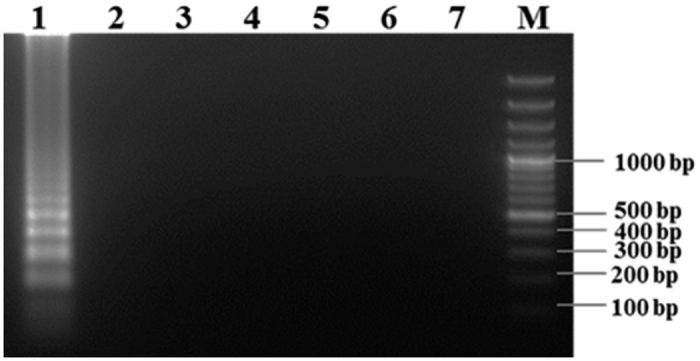
Approximately 10 ng of fungal genomic DNA was used in LAMP reaction. M GeneRulerTM 100 bp Plus DNA Ladder; lane 1–7 DNA of Rhizoctonia bataticola, Fusarium oxysporum f. sp. ciceris, Rhizoctonia solani, Sclerotium rolfsii, Fusarium solani, Fusarium udum and Phytophthora cajani, respectively.
Sensitivity of LAMP assay
To determine the detection limit, the sensitivity of the LAMP reaction was assessed using 10-fold serially diluted R. bataticola DNA template. The amplicons were detected in both, by visual assessment using SYBR Green I (Fig. 4a) and 2% agarose gel electrophoresis (Fig. 4b). No positive signal was produced when less than 10 fg DNA of R. bataticola was used in LAMP reaction, indicating the potential detection limit of R. bataticola up to 10 fg of DNA. It was noticed that the amplified DNA fragments were slightly faint in 10 fg than those produced by a less diluted DNA (>10 fg). On the other hand, when same amount of DNA was used in conventional PCR, no such amplification was obtained after 1.0 pg of dilution (Fig. 4c). Results of visual detection correlated with agarose gel electrophoresis.
Figure 4. Sensitivity of LAMP assay vs. conventional PCR for detection of Rhizoctonia bataticola using similar concentration of DNA template.
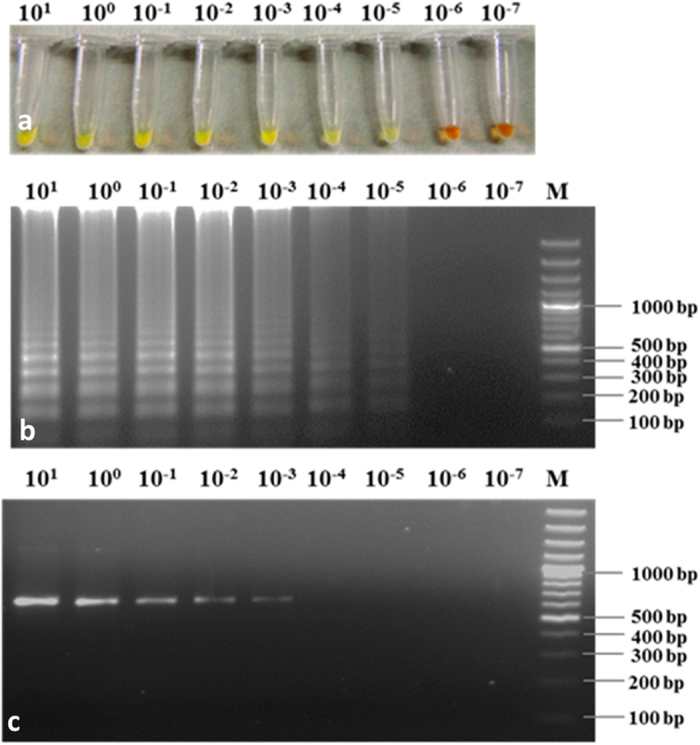
(a) Visual assessment of LAMP assay using SYBR Green I. (b) LAMP assay on the basis of 2% agarose gel electrophoresis. (c) Result of conventional PCR using ITS1 and ITS4 primers. M GeneRulerTM 100 bp Plus DNA Ladder; lane 101–10−7 indicated the DNA concentration in LAMP reaction starting from 10 ng (101 ng) to subsequent 10 fold diluted DNA up to 0.1 fg (10−7 ng), respectively.
Evaluation of the LAMP assays with plant and soil sample
To validate the applicability of the LAMP assay at field level, the developed LAMP assay was evaluated with R. bataticola infested chickpea plants and soil samples as well. The plants showing typical symptom of DRR were collected from different experimental fields of ICRISAT. The LAMP assay was carried out at optimised condition to detect the presence of R. bataticola in tested samples. The positive LAMP reaction was found when DNA templates from the R. bataticola infected chickpea plants were assessed, and the products turned green in colour with SYBR Green I. Moreover, none of the DNA template from healthy plants gave positive signals, and remained orange in colour. Furthermore, the LAMP assay conducted with the DNA from rhizospheric soil of DRR infected chickpea plants as well as R. bataticola inoculated sick soil was also found to be positive (Fig. 5). The LAMP result was consistent when the assays were repeated. Results were consistent with PCR method.
Figure 5. Evaluation of the LAMP assay for detection of Rhizoctonia bataticola in optimized conditions (63 °C for 75 min) using DNA template from infected chickpea plants and rhizospheric soil.
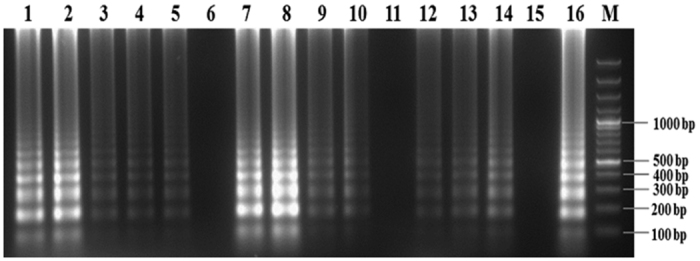
M GeneRulerTM 100 bp Plus DNA Ladder; lane 1–5 Dry root rot infected chickpea plants sampled from different fields; 6 Healthy chickpea plant from field; 7–10R. bataticola inoculated chickpea plants sampled from green house; 11 healthy chickpea plant from greenhouse; 12 Rhizospheric black soil from field; 13 Rhizospheric red soil from field; 14 Sick soil from greenhouse; 15 Negative control without any DNA template; 16 Positive control with genomic DNA of R. bataticola.
Discussion
In this report, we have demonstrated and optimized the LAMP for the detection of R. bataticola, the fungus that cause DRR disease in chickpea. LAMP assay reported here is rapid, highly sensitive, less time-consuming than conventional PCR-based DNA amplification method. It has been applied for the detection of a wide range of microorganisms including viral13,25, bacterial26,27, phytoplasma28, mycoplasma29, fungal10,11,12 and parasitic agents30.
In our study, we used SYBR Green I in LAMP assay as florescent dye which is non-mutagenic and eco-friendly in nature, as the replacement of other potential human mutagen florescent dyes like ethidium bromide. Subsequently, uses of Bst DNA polymerase in LAMP reaction, it permits to strand displacement DNA synthesis and the reactions can be performed under isothermal conditions using a simple incubator, such as a water bath or heating block. With taking these advantages, LAMP detection technique can be employed in diagnosis of disease in field level even also in remote area where the laboratories are not well equipped.
We validated our developed LAMP assay using the primers generated from partial ITS and 5.8S rDNA gene for quick detection of R. bataticola from varied samples viz. fungal culture, diseased plants and soil samples. The highly sensitive and variable ITS region is idle target rather than other single-copy genes in genomic DNA to sufficient discriminate of some closely related fungal species31,32 because of its presence in 100 or more copies in the fungal genome and has competence to give amplification from a very small number of micro-organisms33. Then again, to avoid secondary structures in primers with abundant G-C bonds e.g. hairpins, self-dimer and heterodimer, the ΔG of the primers were kept more positive than −9 kcal/mole or very less negative than the same value23,24 as those can give false positive reaction in assay. When the LAMP primers were analysed in silico for its specificity, it showed significant hit with our target organisms R. bataticola and M. phaseolina in nBLAST. For confirming the specificity of our developed LAMP primers, we assayed those primers with DNA isolated from 94 R. bataticola isolates along with six other plant pathogenic fungi. During this LAMP assay, except all R. bataticola DNA, no colour change was obtained with DNA of other six pathogenic fungus. Thus, this result indicated that the designed primers and LAMP assay were highly specific for R. bataticola, as it correctly distinguished between R. bataticola and the other pathogens. The reaction mixture without DNA also showed no change in colour during LAMP assay. Previously, the primers from rDNA-ITS gene have been used to successfully detect Pythium aphanidermatum in infected tomatoes34 and Phytophthora capsici in infected peppers, tomatoes, and other agronomic and ornamental crops of the Solanaceae and Cucurbitaceae families35.
The detection limit in our study was found to be 10 fg DNA of R. bataticola, below this level no colour change was noticed. This detection limit was lower than previously reported LAMP methods used to detect other fungal pathogens e.g. Sclerotinia sclerotiorum12, Phytophthora sojae36, P. ramorum and P. kernoviae37, indicating greater sensitivity. Comparison of LAMP assay with the conventional PCR showed the LAMP assay using SYBR Green I dye significantly improved the detection efficiency of R. bataticola. This result of LAMP assay was significant and concordant with the reports published previously for the detection of some plant pathogens12,36,37.
Positive reaction in LAMP assay with the DNA isolated from R. bataticola infected chickpea plants sampled from field, further validated our results. DNA from healthy plants gave no reaction. It has previously been used in detection/screening of plant pathogens like Phytophthora capsici35, P. ramorum38, Pythium aphanidermatum34, Meloidogyne enterolobii39 from infected plants in field and soil samples. In this study, we have designed LAMP primers using sequence of R. bataticola/M. phaseolina isolates infecting chickpea as well as other crops from worldwide and in silico analyses of ITS sequences showed low genetic diversity within the Indian and global isolates, hence the utility of our developed LAMP assay will be equally useful for detection of R. bataticola isolates in any other crops globally. In future, LAMP diagnostic kit will be very useful for monitoring the disease complex in the fields, further helpful in developing the timely management strategies.
Additional Information
How to cite this article: Ghosh, R. et al. Rapid and sensitive diagnoses of dry root rot pathogen of chickpea (Rhizoctonia bataticola (Taub.) Butler) using loop-mediated isothermal amplification assay. Sci. Rep. 7, 42737; doi: 10.1038/srep42737 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
This work was supported by grant from Department of Science and Technology-Strategic Programmes Large Initiatives & Coordinated Action Enabler (SPLICE) and Science and Engineering Research Board (SERB), Govt. of India.
Footnotes
The authors declare no competing financial interests.
Author Contributions R.G. and M.S. conceived, designed and initiated the study. A.T. and R.G. were responsible for conducting, analyzing and interpretation of results and initial drafting of the manuscript. M.S. edited the manuscript and provided critical inputs at various stages of the study. All authors read and approved the manuscript.
References
- Sharma M., Ghosh R. & Pande S. Dry root rot (Rhizoctonia bataticola (Taub.) Butler): an emerging disease of chickpea–where do we stand? Arch. Phytopath. Plant Prot. 48, 1–16 (2015). [Google Scholar]
- Sharma M. & Pande S. Unravelling effects of temperature and soil moisture stress response on development of dry root rot [Rhizoctonia bataticola (Taub.)] Butler in Chickpea. Am. J. Plant Sci. 4(3), 584–589 (2013). [Google Scholar]
- Ghosh R., Pande S., Telengare R. D. & Sharma M. Participatory varietal selection of chickpea in rainfed rice fallow lands of Chhattisgarh and Madhya Pradesh in India for sustainable crop production. Int. J. Plant Prod. 8, 243–254 (2014). [Google Scholar]
- Farr D. F., Bills G. F., Chamuris G. P. & Rossman A. Y. Fungi on plants and plant products in the United States (2nd ed) (St Paul, MN: APS Press; 1995). [Google Scholar]
- Tarafdar A., Godara S., Diwedi S. &Biswas K. K. Characterization of Citrus tristeza virus and determination of genetic variability in Northeast and South India. Indian Phytopathol. 66(3), 302–307 (2013). [Google Scholar]
- Meng K. et al. Rapid detection and quantification of zearalenone-producing Fusarium species by targeting the zearalenone synthase gene PKS4. Food Control 21, 207–211(2010). [Google Scholar]
- Nicolaisen M. et al. Real-time PCR for quantification of eleven individual Fusarium species in cereals. J. Microbiol. Meth. 76, 234–240 (2009). [DOI] [PubMed] [Google Scholar]
- Pan W. et al. Development and application of the novel visual loop-mediated isothermal amplification of Omp25 sequence for rapid detection of Brucella sp. J. Anim. Vet. Adv. 10, 2120–2126 (2011). [Google Scholar]
- Opel K. L., Chung D. & McCord B. R. A study of PCR inhibition mechanisms using real time PCR. J. Forensic Sci. 55, 25–33(2010). [DOI] [PubMed] [Google Scholar]
- Chen X., Ma L., Qiang S. & Ma D. Development of a loop-mediated isothermal amplification method for the rapid diagnosis of Aschochyta rabiei L. in chickpeas. Scientific Reports 6, 25688 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R., Nagavardhini A., Sengupta A. & Sharma M. Development of Loop-mediated isothermal amplification (LAMP) assay for rapid detection of Fusarium oxysporum f. sp. ciceris - wilt pathogen of chickpea. BMC Res. Notes 8, 40 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y. B., Ge C. Y., Zhang X. K., Wang J. X. & Zhou M. G. A rapid detection method for the plant pathogen Sclerotinia sclerotiorum based on loop-mediated isothermal amplification (LAMP). Australas. Plant Pathol. 43, 61–66 (2014). [Google Scholar]
- Li R. & Ling K. Development of reverse transcription loop-mediated isothermal amplification assay for rapid detection of an emerging potyvirus: Tomato necrotic stunt virus. J. Virol. Methods 200, 35–40 (2014). [DOI] [PubMed] [Google Scholar]
- Lu C., Dai T., Zhang H., Wang Y. & Zheng X. Development of a Loop-mediated isothermal amplification assay to detect Fusarium oxysporum. J. Phytopathol. 163, 63–66 (2013). [Google Scholar]
- Curtis K. A., Rudolph D. L. & Owen S. M. Rapid detection of HIV-1 by reverse- transcription, loop-mediated isothermal amplification (RT-LAMP). J. Virol. Methods 151, 264–270 (2008). [DOI] [PubMed] [Google Scholar]
- Hill J., Beriwal S., Chandra I., Paul V. K. & Kapil A. Loop-mediated isothermal amplification assay for rapid detection of common strains of Escherichia coli. J. Clin. Microbiol. 46, 2800–2804 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y. & Notomi T. Loop-mediated isothermal amplification (LAMP): A rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 15, 62–69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge P. & Spooner B. Soil fungi: diversity and detection. Plant Soil 232, 147–154 (2001). [Google Scholar]
- Heinonsalo J., Jorgensen K. S. & Sen R. Microcosm based analyses of Scots pine seedling growth, ectomycorrhizal fungal community structure and bacterial carbon utilization profiles in boreal forest humus and underlying illuvial mineral horizons. FEMS Microbiol. Ecol. 36, 73–84 (2001). [DOI] [PubMed] [Google Scholar]
- Grades M. & Bruns T. D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 110–118 (1993). [DOI] [PubMed] [Google Scholar]
- Parida M., Posadas G., Inoue S., Hasebe F. & Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 42, 257–263 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres C. et al. LAVA: An open-source approach to designing LAMP (Loop-Mediated Isothermal Amplification) DNA signatures. BMC Bioinfo. 12, 240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallone P. M. & Butler J. M. AutoDimer: A screening tool for primer-dimer and hairpin structures. BioTechniques 37, 226–231 (2004). [DOI] [PubMed] [Google Scholar]
- Rychlik W. Selection of primers for polymerase chain reaction. Mol. Biotech. 3, 129–134 (1995). [DOI] [PubMed] [Google Scholar]
- Almasi Md. A. et al. Detection of coat protein gene of the Potato leafroll virus by reverse transcription Loop-mediated isothermal amplification. J. Plant Pathol. Microb. 4, 188 (2013). [Google Scholar]
- Hong M. et al. A modified visual loop-mediated isothermal amplification method for diagnosis and differentiation of main pathogens from Mycobacterium tuberculosis complex. World J. Microb. Biot. 28, 523–531(2012). [DOI] [PubMed] [Google Scholar]
- Ravindran A., Levy J., Pierson E. & Gross D. C. Development of a loop-Mediated isothermal amplification procedure as a sensitive and rapid method for detection of ‘Candidatus Liberibacter solanacearum’ in potatoes and psyllids. Phytopathology 102, 899–907 (2012). [DOI] [PubMed] [Google Scholar]
- Nair S., Manimekalai R., Raj P. G. & Hegde V. Loop mediated isothermal amplification (LAMP) assay for detection of coconut root wilt disease and arecanut yellow leaf disease phytoplasma. World J. Microb. Biot. 32, 108 (2016). [DOI] [PubMed] [Google Scholar]
- Li J. et al. Loop-mediated isothermal amplification for rapid and convenient detection of Mycoplasma hyopneumoniae. World J. Microb. Biot. 29, 607–616 (2013). [DOI] [PubMed] [Google Scholar]
- Kuboki N. et al. Loop-mediated isothermal amplification for detection of African trypanosomes. J. Clin. Microbiol. 41, 5517–5524 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern E., Siefring S., Paar J., Doolittle M. & Haugland R. Comparison of quantitative PCR assays for Escherichia coli targeting ribosomal RNA and single copy genes. Lett. Appl. Microbiol. 52, 298–306 (2011). [DOI] [PubMed] [Google Scholar]
- YongYang G., Nan W., GuanPeng G. & Wei W. Triplex PCR detection of Cladosporium cucumerinum, Fusarium oxysporum f. sp. niveum and Mycosphaerella melonis in infected plant tissues. Acta Phytopathol. Sin. 40, 343–350 (2010). [Google Scholar]
- Sandhu G. S., Kline B. C., Stockman L. & Roberts G. D. Molecular probes for diagnosis of fungal infections. J. Clin. Microbiol. 33(11), 2913–2919 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuta S. et al. Detection of Pythium aphanidermatum in tomato using loop-mediated isothermal amplification (LAMP) with species-specific primers. Eur. J. Plant Pathol. 136, 689–701(2013). [Google Scholar]
- Dong Z. et al. Loop-mediated isothermal amplification assay for sensitive and rapid detection of Phytophthora capsici. Can. J. Plant Pathol. 37, 485–494 (2015). [Google Scholar]
- Dai T. T. et al. Development of a loop-mediated isothermal amplification assay for detection of Phytophthora sojae. FEMS Microbiol. Lett. 334, 27–34 (2012). [DOI] [PubMed] [Google Scholar]
- Tomlinson J., Dickinson M. & Boonham N. Rapid detection of Phytophthora ramorum and P. kernoviae by two-minute DNA extraction followed by isothermal amplification and amplicon detection by generic lateral flow device. Phytopathology 100, 143–149 (2010). [DOI] [PubMed] [Google Scholar]
- Tomlinson J., Barker I. & Boonham N. Faster, simpler, more-specific methods for improved molecular detection of Phytophthora ramorum in the field. Appl. Environ. Microb. 73, 4040–4047 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J. H. et al. Evaluation of loop-mediated isothermal amplification (LAMP) assays based on 5S rDNA-IGS2 regions for detecting Meloidogyne enterolobii. Plant Pathol. 61, 809–819 (2012). [Google Scholar]



