Abstract
Secreted frizzled-related proteins (SFRPs), containing five family members (SFRPs 1–5) are putative extracellular Wnt inhibitors. Given their abilities to inhibit Wnt signalling, as well as the loss of SFRP1 in many cancers, this family is generally considered to be tumour suppressive. In this study we analyzed gene expression, promoter methylation and survival data from over 8000 tumour and normal samples from 29 cancers in order to map the context-specific associations of SFRPs 1–5 with patient survival, gene silencing and gene expression signatures. We show that only SFRP1 associates consistently with tumour suppressive functions, and that SFRP2 and SFRP4 typically associate with a poor prognosis concomitant with the expression of genes associated with epithelial-to-mesenchymal transition. Moreover, our results indicate that while SFRP1 is lost in cancer cells via the process of DNA methylation, SFRP2 and 4 are likely derived from the tumour stroma, and thus tend to increase in tumours as compared to normal tissues. This in-depth analysis highlights the need to study each SFRP as a separate entity and suggests that SFRP2 and SFRP4 should be approached as complex matricellular proteins with functions that extend far beyond their putative Wnt antagonistic ability.
Secreted Frizzled-related proteins (SFRPs) were initially described as tumour suppressor genes when SFRP1 was found to be downregulated by loss of heterozygosity or promoter methylation in breast and colorectal cancer cell lines1,2. Given their crucial role in Wnt signalling, and in development, the SFRP gene family was quickly recognized for its potential to modulate tumourigenic behaviour.
SFRPs 1–5 are secreted glycoproteins of ~300 amino acids in length, which fold into two independent domains: (1) a N-terminal cysteine-rich domain (CRD), and (2) a C-terminal netrin-like domain (NTR)3. The cysteine-rich domain shares considerable sequence homology with Fzd receptors, and due to this molecular mimicry, SFRPs were immediately recognized for their potential to sequester Wnt ligands away from receptor complexes and ultimately antagonize Wnt signalling4,5. Wnt signalling pathways have been shown to play central roles in cell survival, proliferation, fate determination, polarity, and tissue patterning. Unsurprisingly, dysregulation of Wnt-associated pathways is a key event in the development of many types of cancer. In general, constitutive activation of Wnt signalling (eg. through stabilizing beta-catenin mutations) is recognized to contribute to tumourigenesis6. Thus, due to their ability to antagonize Wnt signalling, and their frequent epigenetic silencing, SFRPs were initially designated as tumour suppressor genes and many studies have gone on to support this proposed functional role (reviewed in ref. 7).
However, accumulating evidence suggests that they may also promote tumourigenesis in certain contexts. One instance is canine mammary gland tumours, where SFRP2 was found to be overexpressed and induces cancerous transformation in normal mammary epithelial cells. In this case, SFRP2 associated with a fibronectin-integrin extracellular matrix protein complex, and this interaction mediated cell adhesion and blocked apoptosis8,9,10. In metastatic renal cell carcinoma, SFRP1 was found to be upregulated, concomitant with a hypomethylated promoter region. Functionally, knocking down SFRP1 resulted in increased apoptosis and decreased invasive potential11. Furthermore, in renal cancer, SFRP2 was also shown to have oncogenic potential; SFRP2 promoted both in vitro cellular proliferation and in vivo tumour growth12. Deciphering the complex effects of SFRPs on tumour progression is likely complicated by the local context of Wnt signalling components, differences between SFRP family members and the unknown impact of NTR domain interactions.
Recent transcriptional and genomic profiling of thousands of patient tumour samples by The Cancer Genome Atlas (TCGA) has enabled a thorough investigation of the functions of the SFRP gene family across different types of cancers. In this study, we investigate the context-specific associations of SFRP1–5 expression in over 8000 tumour and normal samples from 29 different cancers. We show that the putative tumour suppressor function is not consistent between members of the SFRP family and that specific SFRPs behave in a cancer type-dependent manner. We found that SFRP1 is the only family member whose expression is consistently decreased in primary cancer samples as compared to associated normal tissues. Moreover, this loss of SFRP1 expression correlates with gene promoter methylation. Despite these abstruse associations, SFRP2 and 4 expression consistently clusters together, suggesting a common gene program. We found that SFRP2 and 4 expression is tightly correlated to stromal content, and forms part of a common epithelial-to-mesenchymal transition (EMT) gene program that is expressed in a multitude of different cancers.
Results
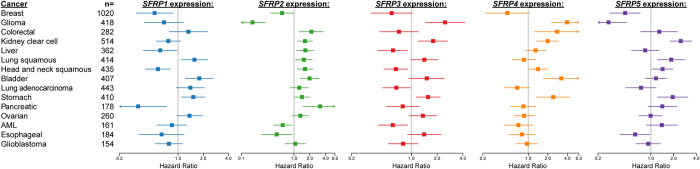
Association of SFRPs with patient survival reveals strong correlations, but inconsistency between family members and cancer type
We were first interested in determining if SFRP expression was associated with favourable patient outcomes, as suggested by their proposed tumour suppressor function. We looked at primary tumours from fifteen different cancer types (Supplementary Fig. 1) and dichotimized SFRP expression into high and low expressors by ROC curve. Univariate Cox regression analysis was conducted and Kaplan-Meier curves were constructed based on overall survival (Fig. 1, Supplementary Fig. 2). To determine the robustness of these associations, re-sampling analysis was conducted (Supplementary Fig. 3). In general, we found that SFRP expression was frequently significantly associated with patient outcomes. However, the direction of that association varied with regards to the particular SFRP isoform queried and the cancer type. Despite this, we observed select consistent cancer-specific or gene-specific effects: For example, high expression of any SFRP was associated with poor prognosis in stomach cancer; and high expression of SFRP4 only associated with poor outcomes (p < 0.05). In colorectal cancer, where promoter SFRP methylation and functional studies in cell lines have implicated their role as tumour suppressive13,14, we found that high expression of SFRP2 and SFRP4 is associated with poor patient outcomes (SFRP2: HR = 2.14 [1.27–3.58], p = 0.004; SFRP4: HR = 2.76 [1.25–6.08], p = 0.01).
Figure 1. Association of SFRP expression with patient survival across different cancer types.
Hazard ratios and 95% confidence intervals for overall survival, by cancer type. The forest plot shows the overall survival advantage or disadvantage of increased SFRP expression (high versus low as stratified by ROC curve) by cancer type, unadjusted for other covariates. The vertical line represents a hazard ratio of one, where there are no survival differences between the two groups.
Expression of SFRP1 and 5, but not other SFRPs, is lost in primary tumours
One of the defining features of SFRP expression during tumourigenesis is a decrease in gene expression. While much of this work has been conducted on normal and cancer cell lines, we investigated the expression levels of all five genes in over 8,000 primary tumours, and 780 associated normal tissues from 29 different cancers (Fig. 2, Supplementary Tables 1 and 2). SFRP1 and 5 consistently have lower expression in primary tumour tissue compared to normal tissue. However, expression of SFRP2, SFRP3 and SFRP4 were often unchanged or even increased in tumour tissue, indicating that they do not undergo the same silencing process as SFRP1 and 5.
Figure 2. Expression levels of SFRPs in normal and cancerous tissue types.
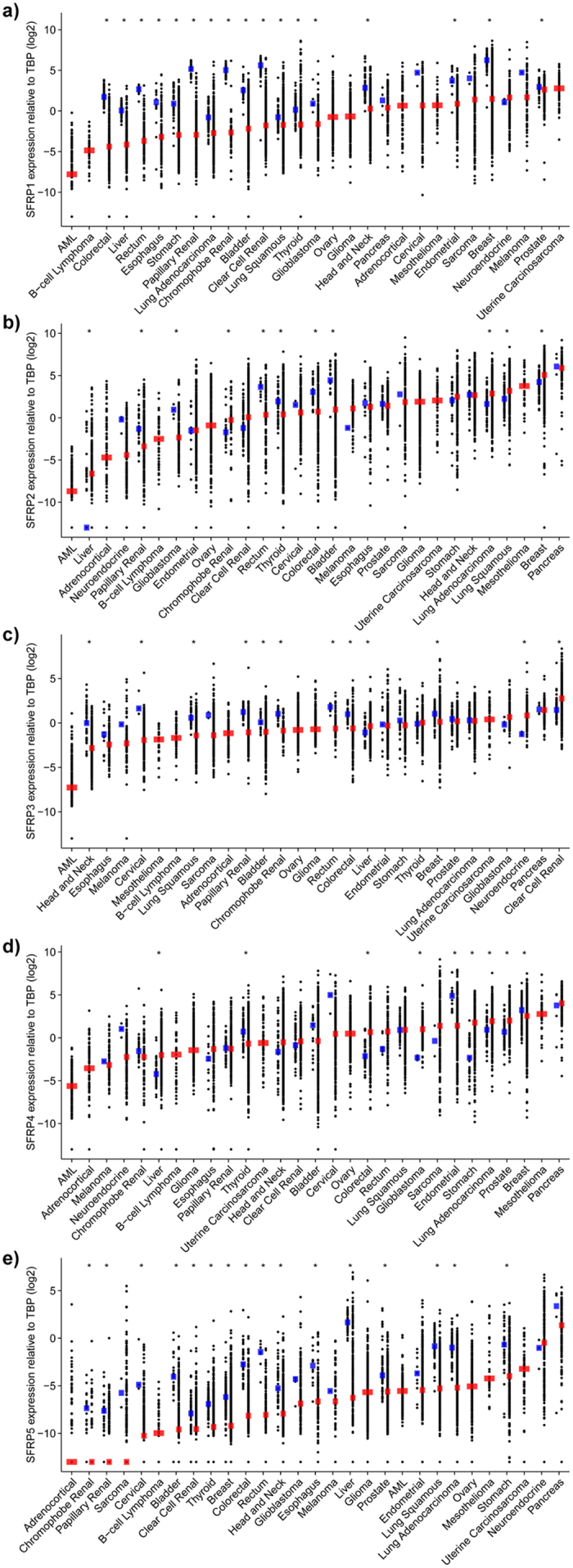
Expression levels of (a) SFRP1, (b) SFRP2, (c) SFRP3, (d) SFRP4, and (e) SFRP5 in patient samples. Each data point represents the SFRP expression levels (log2[RSEM normalized values relative to TBP]) of one tumour or normal sample. Horizontal bars indicate median expression values for normal (blue) or primary tumour (red) samples. Zero value SFRP expressors are plotted at the bottom of the y axis. Comparisons between the normal and tumour expression values were performed using the Mann-Whitney U test to determine significance (*p < 0.05).
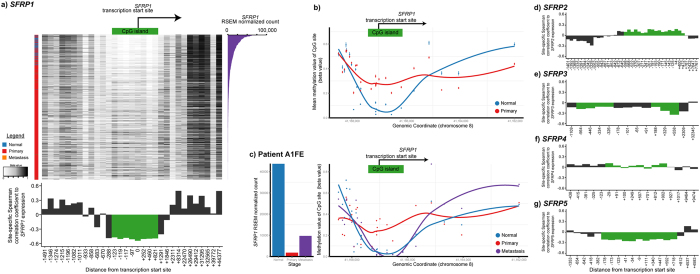
Regulation of SFRP gene expression is commonly observed at the epigenetic level, through DNA methylation, but has also been observed at the genetic level, through loss of heterozygosity1,2,15,16. We investigated these possibilities in breast cancer by looking at how copy number events and CpG site methylation correlates with SFRP expression levels. We found that there were no copy number alterations that consistently associated with gene levels (Supplementary Fig. 4). At the epigenetic level, methylation of the SFRP1 promoter region was strongly inversely correlated with SFRP1 gene expression (Fig. 3a). Methylation of this region in primary tumours was also observed to increase as compared to matched normal tissues at both a cancer-specific and a patient-specific level (Fig. 3b,c). Gene expression did not correlate with CpG methylation for any other SFRPs, indicating that their expression is regulated at a different level (Fig. 3d–g).
Figure 3. SFRP1 promoter methylation correlates to SFRP1 gene expression in breast cancer.
(a) Heatmap depiction of SFRP1 methylation levels of individual CpG sites (beta values: black, methylated; white, unmethylated) in breast tumour samples ordered from high SFRP1 expression levels (top) to low SFRP1 expression levels (bottom). Left bar indicates sample source: blue, normal samples; red, primary sample; orange, metastatic sample. Right barplot depicts SFRP1 expression levels (RSEM normalized values). Bottom barplot depicts Spearman’s correlation coefficients for site-specific methylation levels (beta values) to SFRP1 RSEM normalized values. (b) Scatterplot depicting average CpG methylation values +/− SEM of the SFRP1 genomic locus of normal (blue) and primary (red) breast cancer tumour samples. Curves represent loess smoothed data. (c) Barplot of SFRP1 expression levels (RSEM normalized values) in normal, primary tumour and metastatic tumour samples from the same patient. Scatterplot of CpG methylation values of the SFRP1 genomic locus in normal, primary and metastatic tumour samples from the same patient. Barplot of Spearman’s correlation coefficients for site-specific methylation beta levels to (d) SFRP2, (e) SFRP3, (f) SFRP4, and (g) SFRP5 RSEM normalized values.
SFRP2 and 4 expression in tumours is likely contributed by stroma
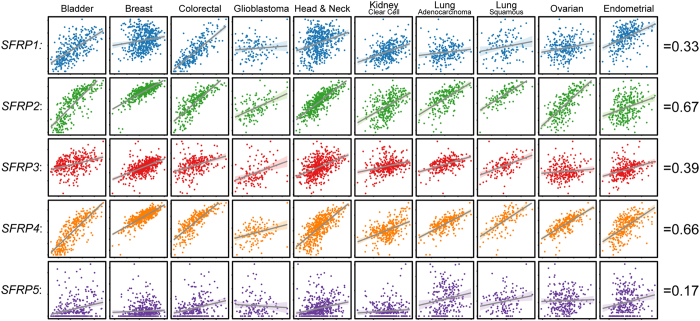
A possible alternative mechanism governing alterations in SFRP expression in tumours is that SFRPs are differentially expressed by various cell types in the tumour microenvironment. Several studies have indicated that SFRPs may be expressed by tissue stroma17,18,19 and we investigated that possibility by correlating SFRP expression to tumour-specific Stromal Scores (as determined by the ESTIMATE algorithm, Fig. 4). We found that SFRP2 and SFRP4 strongly correlated with Stromal Scores in the fourteen cancer types investigated, with an average Spearman’s correlation coefficient of 0.67 and 0.66, respectively. Furthermore, single cell RNA-sequencing in breast cancer suggests that SFRP2 and SFRP4 expression is restricted to cells identified as stromal (Supplementary Fig. 5). By contrast, expression of SFRP1, 3, and 5 only weakly or conditionally associate with Stromal Scores: for example, SFRP1 expression strongly correlates with Stromal Scores in colorectal cancer, a cancer where SFRP1 promoter methylation has been demonstrated to occur in both cell lines and patient samples13,14.
Figure 4. Correlation of SFRP expression values to Stromal Scores in various cancers.
Scatterplot representations of SFRP RSEM expression values (y axes) to Stromal Scores (x axes, as determined by the ESTIMATE algorithm) in bladder cancer, breast cancer, colorectal cancer, glioblastoma, head and neck cancer, clear cell kidney cancer, lung adenocarcinoma, lung squamous cell carcinoma, ovarian cancer and endometrial cancer. Lines indicate linear regression lines; shading indicates confidence interval. Values (right) indicate average Spearman’s correlation coefficients of the individual SFRP family members in the described cancers.
Common pan-cancer gene program associated with SFRP2 and 4 expression
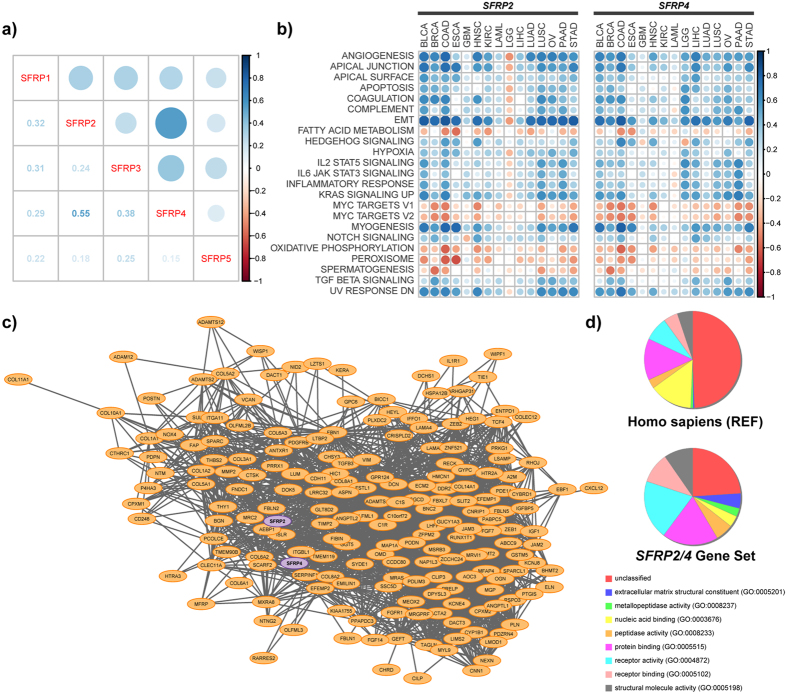
When expression levels of the various SFRPs were correlated to each other (Supplementary Fig. 6) and averaged across cancers, we found SFRP2 and 4 expression to be tightly correlated (Fig. 5a). This suggests that those two SFRPs share a common gene program. Furthermore, the pattern of correlation of SFRP2 and 4 expression to gene set enrichments are highly concordant, with tight correlations to EMT and angiogenesis gene sets (Supplementary Fig. 7, Fig. 5b). Correlation network analysis of SFRP2 and 4 reveals a common gene program of 180 genes that are tightly correlated (r > 0.5) across multiple cancers (Fig. 5c). This program includes previously identified key EMT proteins such as ZEB1, ZEB2, VIM, and MMP2. Gene ontology reveals an enrichment of extracellular matrix constituents and metallopeptidase activity ontologies in the identified SFRP2/4 gene set (Fig. 5D). This correlation network analysis and gene ontology strengthens the notion that SFRP2 and 4 have stromal functions — distinct from their proposed Wnt antagonistic activity, and that they may promote processes such as cellular invasion and metastasis.
Figure 5. SFRP2 and SFRP4 expression strongly associates with an EMT signature.
(a) Correlation matrix of the expression levels of different SFRP family members (log2[RSEM normalized values + 1]) in primary tumours. Average of Pearson’s correlation coefficient of gene-gene pairs from 15 different cancers. (b) Correlation matrix of SFRP2 and SFRP4 expression values (log2[RSEM normalized values + 1]) with individual gene sets (GAGE determined enrichment values for MSigDb Hallmark gene sets) in 15 different cancers. Gene sets were included if they had an absolute correlation coefficient of over 0.5 with SFRP2 or 4 expression values. (c) Correlation network analysis of SFRP2 and 4 reveals a common gene set. The network displayed genes whose expression strongly correlates (r > 0.5) with SFRP2 and 4 in over 50% of cancers where they are a poor prognostic factor. Nodes colours indicate SFRP member location (purple, SFRPs; orange, others); edges represent average correlation coefficient >0.8 in the STAD dataset. (d) SFRP2 and 4 correlated genes (n = 180) were classified using PANTHER Classification System based on molecular function.
Discussion
This is the first study of its kind to systematically investigate the survival associations and context-specific interactions of secreted Frizzled-related protein family members. Our results contradict the notion that SFRPs are tumour suppressive across all cancers. Indeed, in many cancers, high expression is associated with poor patient outcomes. We show that while SFRP1 is frequently silenced in many cancers through methylation, the other SFRPs do not undergo this silencing event. In fact, SFRP2 and SFRP4 expression is often increased in tumours and are tightly correlated to Stromal Scores, suggesting that their expression is produced by the tumour stroma. Gene ontology strengths this observation, with both genes contributing to a common pan-cancer gene set that is tightly associated with EMT. Based on these discoveries, we anticipate that SFRP2 and SFRP4 function as complex matricellular proteins, and that they may have functions that extend far beyond the regulation of Wnt signalling.
SFRP1 was the first of the gene family found to be altered during tumourigenesis. It was discovered in this manner when it was found to be silenced by methylation or loss of heterozygosity in colorectal and breast cancers1,2. Following these initial reports, others went on to show similar trends and to tie the effects of SFRP1 loss to increased Wnt-related signalling in a variety of other cancers. We further validated the essence of these studies with our pancancer analysis by showing that in 17 different cancers, SFRP1 expression is significantly downregulated in tumourous tissues compared to normal counterparts. In addition, we confirmed that in breast cancer the downregulation of SFRP1 associated with promoter methylation. Several groups have proposed the use of SFRP1 promoter methylation as a cancer biomarker, and our study provides further support for this concept20,21,22,23,24.
However, our investigation into the other SFRPs did not lead to the same conclusions. We found that high expression of SFRP2 and 4 most often was associated with poor patient prognoses. Furthermore, expression of SFRP2 and 4 often increased in primary tumours compared to their normal counterparts, and this expression tightly correlated to tumour stromal content. Given that SFRPs are secreted proteins, expression from any cell type has the potential to modulate the tumour microenvironment, and affect tumourigenesis. Since many of the initial studies into SFRP2 and 4 were done using cancer cell lines, which are composed solely of tumour cells, this nuance has likely been largely overlooked. The effects of stromal-derived SFRPs could be completely different from tumour cell-derived SFRPs. For example, only tumour cell-derived, not stromal-derived, MMP-2 and MMP-13 correlate with poor patient outcomes and aggressive tumour phenotypes in ovarian or breast cancer, respectively25,26. This is in line with multiple studies that are beginning to appreciate and characterize the complexity involved in biomarker generation27,28,29. Moreover, the effects of SFRPs on three-dimensional tumours composed of a plethora of cell types is likely entirely distinct from their effects on cancer cell lines. This theory is strengthened by a recent study of SFRP2 in melanoma30. Kaur et al. found that SFRP2 expressed by aged fibroblasts drove melanoma angiogenesis and metastasis. This study was unique in that much of the work was conducted in models that incorporated alternative cells types, such as skin reconstructions or transfer of conditioned media. We suggest that future studies investigating the role of SFRP2 or 4 in tumourigenesis consider possible contribution by the tumour microenvironment and incorporate this into model choice.
Despite production by tumour stroma, SFRP2 or 4 promoter gene methylation may still show promise as a cancer biomarker. Kalmar et al. found that despite an increase in SFRP2 expression in colorectal cancer compared to associated normals, the SFRP2 promoter region became hypermethylated in the cancerous tissues31. They used laser-capture microdissection of colonic epithelial cells to show that this increase was not due to expression within the tumour cells themselves. Moreover, the tumour cells had hypermethylation in the SFRP2 promoter region. This has also been observed in various cell lines, where the SFRP2 promoter region of normal cell line derivatives are unmethylated, but that region is hypermethylated in tumour cell lines16,32,33. Therefore, despite the stromal contribution of SFRP2 and/or SFRP4, hypermethylation within the tumour cell compartment may still show utility as a clinical biomarker. Given that the robustness of cancer biomarkers is often an issue, the utilization of SFRPs with other biomarkers (individual or signature-based) would likely improve accuracy.
In all solid cancers investigated, SFRP2 and 4 showed high concordance in terms of associations with survival, correlation to one another and association with enriched gene sets. The exception to this was brain cancers, where these two proteins appear to act independently of one another. In glioma, high SFRP4 expression is strongly associated with poor outcomes (HR = 3.93 [2.27–6.81], p < 0.0001), while high SFRP2 expression is strongly associated with favourable outcomes (HR = 0.16 [0.09–0.28], p < 0.0001). In addition, in glioma, SFRP4 is strongly associated with EMT and angiogenesis gene sets, like in all other cancers. On the other hand, SFRP2 in glioma is an exception: It is the only cancer where SFRP2 expression negatively correlates with EMT and angiogenesis gene sets. This disparity may be driven by tissue of origin effects — SFRP2 has been shown to be highly expressed in the developing neural system whereas SFRP4 is not expressed in those structures34,35,36. Further studies are needed to clarify the details of this divergence. However, it may provide a unique opportunity to elucidate some of the distinct molecular mechanisms of these two proteins.
Conclusions
This study is the first of its kind to systematically analyze the SFRP gene family in multiple cancers. We focused on gene expression and patient survival data and sought to identify if all SFRPs are associated with tumour suppression. We determined that SFRP1, the prototypical family member, is downregulated during tumour formation, silenced by methylation and likely follows much of the dogma surrounding this gene family. On the other hand, SFRP2 and 4 expression levels often increase during tumourigenesis, likely as a result of increased production by the tumour stroma. These two SFRPs, which are often associated with poor patient outcomes, form part of a common gene program that is expressed across many cancers and is associated with EMT. We anticipate that as more studies are conducted, new functions will be discovered for these complex matricellular proteins that extend far beyond their putative Wnt antagonistic ability.
Methods
Datasets
RNA-seq expression, patient clinical and methylation data were retrieved from The Cancer Genome Atlas Data Matrix on 17 August 2014. For gene expression analysis of normals and primaries, RNASeqV2 (HiSeq) data was used for ACC, BLCA, BRCA, CESC, COAD, DLBC, GBM, HNSC, KICH, KIRC, KIRP, LAML, LGG, LIHC, LUAD, LUSC, MESO, OV, PAAD, PCPG, PRAD, READ, SARC, SKCM, THCA, and UCS. RNASeqV2 (GA) data was used for UCEC. RNASeq (HiSeq) level data was used for ESCA and STAD cancers. SFRP expression values (RSEM or RPKM) were normalized to the expression of TBP for each tumour sample. Comparisons between the normal and tumour RNA-seq-derived expression values were performed using the Mann-Whitney U test to determine significance. Breast cancer (BRCA) SFRP copy number status was accessed using the cBioPortal on 17 May 2015 (http://www.cbioportal.org/). Single cell RNA-sequencing data was accessed on 7 December 2015 for 51 breast cancer cells from tumour BC02 profiled by the Gene Expression Omnibus (GEO) series, GSE75688.
Survival Analysis
To include the most recently released patient samples, additional RNASeqV2 (HiSeq) and clinical data were downloaded on 20 August 2015 for COAD, LIHC, BLCA, STAD, and ESCA cancers. These data were used in downstream survival analyses. SFRP expression values (RSEM normalized values) were dichotomized by receiver operating characteristics (ROC) curves and the Youden index J method was used to determine the optimal cutoff. Survival curves for overall survival (OS) were constructed using the Kaplan-Meier method and significance was determined by log-rank test. The associations between SFRP expression (high versus low) and OS were tested in univariate Cox regression models. Re-sampling Cox regression analysis was conducted by calculating hazard ratios on SFRP expression on 70% of the dataset and re-sampled 500 times.
Methylation analysis
Breast cancer sample associations between individual CpG site methylation (beta values) and SFRP expression (log2[RSEM + 1]) were calculated using Pearson’s correlation in normal, primary and metastatic samples.
Tumour purity
Stromal Scores were defined for tumours through the use of the ESTIMATE (Estimation of STromal and Immune cells in MAlignant Tumour tissues using Expression data; original publication37) algorithm using RNASeqV2 data or accessed through the MD Anderson Bioinformatics Portal on 15 May 2015 (http://bioinformatics.mdanderson.org/estimate/). Spearman’s correlation coefficient was used to calculate the association of specific genes to Stromal Scores.
Gene set enrichment analysis
Enrichment for SFRP associated gene sets was conducted using Generally Applicable Gene-set Enrichment (GAGE, v2.12.3). Hallmark gene sets were downloaded from the Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb) v5.0 on 10 August 201538. Enrichment was calculated against a formulated sample composed of the mean expression values for each gene and sample-specific test statistics were correlated to SFRP expression values (log2[RSEM + 1]) using Spearman’s rank correlation on a cancer-specific level.
Correlation network analysis
Correlation network analysis was conducted to determine the context in which SFRPs are expressed. Pearson’s correlation coefficient were determined for SFRPs (log2[RSEM + 1]) to all genes. Correlations were determined for cancers where SFRP2 is associated with poor prognosis (BLCA, COAD, HNSC, KIRC, LIHC, LUSC, PAAD, and STAD) and for cancers where SFRP4 is associated with poor prognosis (BLCA, COAD, HNSC, KIRC, LGG, and STAD). Genes were included in the network if their expression values (log2[RSEM + 1]) were strongly correlated (Pearson’s correlation coefficient >0.5) in at least 50% of the cancers analyzed for correlation with SFRP2 and 50% of the cancers analyzed for correlation with SFRP4. Cytoscape (v3.2.1) was used to visualize the constructed network39. Edges depict an average gene-gene expression correlation coefficient of >0.8 in the STAD dataset.
Gene ontology
The identified SFRP2/4 gene set (n = 180) was classified using the PANTHER Classification System (http://www.pantherdb.org, version 10.0, released 2015-05-15)40. The genes were classified based on their molecular function and a Statistical Overrepresentation Test was performed on these genes to examine enrichment of GO terms.
Statistical analysis
We conducted all analyses and visualizations in the RStudio programming environment (v0.98.501). R/Bioconductor packages ggplot2, corrplot, plyr, gplots, matrixStats, survival, and GAGE were used where appropriate.
Additional Information
How to cite this article: Vincent, K. M. and Postovit, L.-M. A pan-cancer analysis of secreted Frizzled-related proteins: re-examining their proposed tumour suppressive function. Sci. Rep. 7, 42719; doi: 10.1038/srep42719 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by an Alberta Innovates Health Solutions Translational Health Chair in cancer and a Canadian Breast Cancer Foundation operating grant awarded to LMP. LMP was the recipient of the Peter-Lougheed Premier New Investigator Award from the Canadian Institutes of Health Research. KMV is a Vanier Scholar.
Footnotes
The authors declare no competing financial interests.
Author Contributions K.M.V. and L.-M.P. conceived of the analysis. K.M.V. prepared the data and performed all data analysis. All authors participated in interpreting results. K.M.V. wrote the manuscript. L.-M.P. participated in revising the manuscript. All authors have read and approve the final manuscript.
References
- Ugolini F. et al. Differential expression assay of chromosome arm 8p genes identifies Frizzled-related (FRP1/FRZB) and Fibroblast Growth Factor Receptor 1 (FGFR1) as candidate breast cancer genes. Oncogene. 18, 1903–1910 (1999). [DOI] [PubMed] [Google Scholar]
- Suzuki H. et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 31, 141–149 (2002). [DOI] [PubMed] [Google Scholar]
- Chong J. M., Uren A., Rubin J. S. & Speicher D. W. Disulfide bond assignments of secreted Frizzled-related protein-1 provide insights about Frizzled homology and netrin modules. J Biol Chem. 277, 5134–5144 (2002). [DOI] [PubMed] [Google Scholar]
- Wang S., Krinks M., Lin K., Luyten F. P. & Moos M. Frzb, a secreted protein expressed in the Spemann organizer, binds and inhibits Wnt-8. Cell. 88, 757–766 (1997). [DOI] [PubMed] [Google Scholar]
- Leyns L., Bouwmeester T., Kim S. H., Piccolo S. & De Robertis E. M. Frzb-1 is a secreted antagonist of Wnt signalling expressed in the Spemann organizer. Cell. 88, 747–756 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. & Nusse R. Wnt/B-Catenin Signalling and Disease. Cell 149, 1192–1205 (2012). [DOI] [PubMed] [Google Scholar]
- Surana R. et al. Secreted frizzled related proteins: Implications in cancers. Biochim Biophys Acta. 1845, 53–65 (2014). [DOI] [PubMed] [Google Scholar]
- Lee J.-L., Chang C.-J., Wu S.-Y., Sargan D. R. & Lin C.-T. Secreted frizzled-related protein 2 (SFRP2) is highly expressed in canine mammary gland tumours but not in normal mammary glands. Breast Cancer Res Treat. 84, 139–149 (2004). [DOI] [PubMed] [Google Scholar]
- Lee J.-L., Lin C.-T., Chueh L.-L. & Chang C.-J. Autocrine/paracrine secreted Frizzled-related protein 2 induces cellular resistance to apoptosis: a possible mechanism of mammary tumourigenesis. J Biol Chem. 279, 14602–14609 (2004). [DOI] [PubMed] [Google Scholar]
- Lee J.-L., Chang C.-J., Chueh L.-L. & Lin C.-T. Secreted frizzled related protein 2 (sFRP2) decreases susceptibility to UV-induced apoptosis in primary culture of canine mammary gland tumours by NF-κB activation or JNK suppression. Breast Cancer Res Treat. 100, 49–58 (2006). [DOI] [PubMed] [Google Scholar]
- Saini S. et al. Functional significance of secreted Frizzled-related protein 1 in metastatic renal cell carcinomas. Cancer Res. 69, 6815–6822 (2009). [DOI] [PubMed] [Google Scholar]
- Yamamura S. et al. Oncogenic functions of secreted Frizzled-related protein 2 in human renal cancer. Mol Cancer Ther. 9, 1680–1687 (2010). [DOI] [PubMed] [Google Scholar]
- Caldwell G. M. et al. The Wnt antagonist sFRP1 in colorectal tumourigenesis. Cancer Res. 64, 883–888 (2004). [DOI] [PubMed] [Google Scholar]
- Suzuki H. et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signalling in colorectal cancer. Nat Genet. 36, 417–422 (2004). [DOI] [PubMed] [Google Scholar]
- Veeck J. et al. Aberrant methylation of the Wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene. 25, 3479–3488 (2006). [DOI] [PubMed] [Google Scholar]
- Suzuki H. et al. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 98, 1147–1156 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Jawdeh G. et al. Differential expression of frpHE: a novel human stromal protein of the secreted frizzled gene family, during the endometrial cycle and malignancy. Lab Invest. 79, 439–447 (1999). [PubMed] [Google Scholar]
- Carmon K. S. & Loose D. S. Secreted Frizzled-Related Protein 4 Regulates Two Wnt7a Signalling Pathways and Inhibits Proliferation in Endometrial Cancer Cells. Mol Cancer Res. 6, 1017–1028 (2008). [DOI] [PubMed] [Google Scholar]
- Sun Y. et al. SFRP2 augments WNT16B signalling to promote therapeutic resistance in the damaged tumour microenvironment. Oncogene, doi: 10.1038/onc.2015.494 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Wang D. & Li H. Combination analysis of hypermethylated SFRP1 and SFRP2 gene in fecal as a novel epigenetic biomarker panel for colorectal cancer screening. J Nanjing Med Univ. 22, 96–101 (2008). [Google Scholar]
- Wang D.-R. & Tang D. Hypermethylated SFRP2 gene in fecal DNA is a high potential biomarker for colorectal cancer noninvasive screening. World J Gastroentero. 14, 524–531 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. et al. Circulating SFRP1 promoter methylation status in gastric adenocarcinoma and esophageal square cell carcinoma. Biomed Rep. 3, 123–127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. W. et al. Transcriptional inactivation of secreted frizzled-related protein 1 by promoter hypermethylation as a potential biomarker for non-small cell lung cancer. Neoplasma. 57, 228–233 (2010). [DOI] [PubMed] [Google Scholar]
- Urakami S. Combination Analysis of Hypermethylated Wnt-Antagonist Family Genes as a Novel Epigenetic Biomarker Panel for Bladder Cancer Detection. Clin Cancer Res. 12, 2109–2116 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang B. et al. Tumour-derived matrix metalloproteinase-13 (MMP-13) correlates with poor prognoses of invasive breast cancer. BMC Cancer. 8, 83 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z. et al. The expression of tumour-derived and stromal-derived matrix metalloproteinase 2 predicted prognosis of ovarian cancer. Int J Gynecol Cancer. 25, 356–362 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 513, 382–387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell. 166, 755–765 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat Comms. 1, 34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A. et al. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature. 532, 250–254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmár A. et al. DNA hypermethylation and decreased mRNA expression of MAL, PRIMA1, PTGDR and SFRP1 in colorectal adenoma and cancer. BMC Cancer. 15, 736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeck J. et al. Promoter hypermethylation of the SFRP2 gene is a high-frequent alteration and tumour-specific epigenetic marker in human breast cancer. Mol Cancer. 7, 83 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H. et al. Frequent epigenetic inactivation of SFRP genes in hepatocellular carcinoma. J Gastroenterol. 43, 378–389 (2008). [DOI] [PubMed] [Google Scholar]
- Leimeister C., Bach A. & Gessler M. Developmental expression patterns of mouse sFRP genes encoding members of the secreted frizzled related protein family. Mech Dev. 75, 29–42 (1998). [DOI] [PubMed] [Google Scholar]
- Terry K., Magan H., Baranski M. & Burrus L. W. Sfrp-1 and sfrp-2 are expressed in overlapping and distinct domains during chick development. Mech Dev. 97, 177–182 (2000). [DOI] [PubMed] [Google Scholar]
- Lin C.-T., Lin Y.-T. & Kuo T.-F. Investigation of mRNA expression for secreted frizzled-related protein 2 (sFRP2) in chick embryos. J Reprod Dev. 53, 801–810 (2007). [DOI] [PubMed] [Google Scholar]
- Yoshihara K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Comms. 4, 2612 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Poudel S., Muruganujan A., Casagrande J. T. & Thomas P. D. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 44, D336–42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.