Abstract
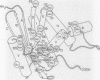
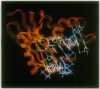
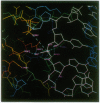
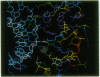
The mechanism of RNase H substrate recognition is proposed from a model of a chemically modified DNA.RNA hybrid Escherichia coli RNase H complex. Site-directed mutagenesis of the enzyme and substrate titration observed by heteronuclear two-dimensional NMR spectra have been carried out. A model complex has been built, based on free structures of the enzyme and the substrate independently determined by x-ray crystallography and NMR distance geometry, respectively. In addition to steric and electrostatic complementarities between the molecular surfaces of the enzyme and the minor groove of the hybrid in the model, putative hydrogen bonds between the polar groups in the enzyme and 2'-oxygens of the RNA strand of the hybrid fix the hybrid close to the active site of the enzyme. The enzymatic activities of the mutant proteins and the changes in NMR spectra during the course of substrate titration are consistent with the present model. Moreover, the specific cleavage of the RNA strand in DNA.RNA hybrids can be explained as well as cleavage modes in modified heteroduplexes. A mechanism of enzymatic action is proposed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. E., Ptashne M., Harrison S. C. Structure of the repressor-operator complex of bacteriophage 434. 1987 Apr 30-May 6Nature. 326(6116):846–852. doi: 10.1038/326846a0. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Millane R. P., Park H. S. DNA-RNA hybrid secondary structures. J Mol Biol. 1986 Apr 20;188(4):631–640. doi: 10.1016/s0022-2836(86)80011-0. [DOI] [PubMed] [Google Scholar]
- Benevides J. M., Thomas G. J., Jr A solution structure for poly(rA).poly(dT) with different furanose pucker and backbone geometry in rA and dT strands and intrastrand hydrogen bonding of adenine 8CH. Biochemistry. 1988 May 17;27(10):3868–3873. doi: 10.1021/bi00410a051. [DOI] [PubMed] [Google Scholar]
- Chou S. H., Flynn P., Reid B. High-resolution NMR study of a synthetic DNA-RNA hybrid dodecamer containing the consensus pribnow promoter sequence: d(CGTTATAATGCG).r(CGCAUUAUAACG). Biochemistry. 1989 Mar 21;28(6):2435–2443. doi: 10.1021/bi00432a014. [DOI] [PubMed] [Google Scholar]
- Davies J. F., 2nd, Hostomska Z., Hostomsky Z., Jordan S. R., Matthews D. A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991 Apr 5;252(5002):88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Gupta G., Sarma M. H., Sarma R. H. Secondary structure of the hybrid poly(rA).poly(dT) in solution. Studies involving NOE at 500 MHz and stereochemical modelling within the constraints of NOE data. J Mol Biol. 1985 Nov 20;186(2):463–469. doi: 10.1016/0022-2836(85)90118-4. [DOI] [PubMed] [Google Scholar]
- Inoue H., Hayase Y., Iwai S., Ohtsuka E. Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett. 1987 May 11;215(2):327–330. doi: 10.1016/0014-5793(87)80171-0. [DOI] [PubMed] [Google Scholar]
- Kanaya S., Kohara A., Miura Y., Sekiguchi A., Iwai S., Inoue H., Ohtsuka E., Ikehara M. Identification of the amino acid residues involved in an active site of Escherichia coli ribonuclease H by site-directed mutagenesis. J Biol Chem. 1990 Mar 15;265(8):4615–4621. [PubMed] [Google Scholar]
- Kanaya S., Kohara A., Miyagawa M., Matsuzaki T., Morikawa K., Ikehara M. Overproduction and preliminary crystallographic study of ribonuclease H from Escherichia coli. J Biol Chem. 1989 Jul 15;264(20):11546–11549. [PubMed] [Google Scholar]
- Kane C. M. Renaturase and ribonuclease H: a novel mechanism that influences transcript displacement by RNA polymerase II in vitro. Biochemistry. 1988 May 3;27(9):3187–3196. doi: 10.1021/bi00409a010. [DOI] [PubMed] [Google Scholar]
- Katayanagi K., Miyagawa M., Matsushima M., Ishikawa M., Kanaya S., Ikehara M., Matsuzaki T., Morikawa K. Three-dimensional structure of ribonuclease H from E. coli. Nature. 1990 Sep 20;347(6290):306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- Metelev V. G., Zayakina G. V., Ryabushenko I. L., Krynetskaya N. F., Romanova E. A., Oretskaya T. S., Shabarova Z. A. Influence of probe structure on unique (regiospecific) cleavage of RNA by RNase H. FEBS Lett. 1988 Jan 4;226(2):232–234. doi: 10.1016/0014-5793(88)81429-7. [DOI] [PubMed] [Google Scholar]
- Nagayama K., Yamazaki T., Yoshida M., Kanaya S., Nakamura H. Combination of heteronuclear 1H-15N and 1H-13C three-dimensional nuclear magnetic resonance experiments for amide-directed sequential assignment in larger proteins. J Biochem. 1990 Aug;108(2):149–152. doi: 10.1093/oxfordjournals.jbchem.a123173. [DOI] [PubMed] [Google Scholar]
- Nakamura H., Katayanagi K., Morikawa K., Ikehara M. Structural models of ribonuclease H domains in reverse transcriptases from retroviruses. Nucleic Acids Res. 1991 Apr 25;19(8):1817–1823. doi: 10.1093/nar/19.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo H., Matsumoto U. Direct evidence for a bimorphic structure of a DNA-RNA hybrid, poly(rA).poly(dT), at high relative humidity. J Biol Chem. 1984 Jul 25;259(14):8682–8684. [PubMed] [Google Scholar]
- Suck D., Lahm A., Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988 Mar 31;332(6163):464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- Suck D., Oefner C. Structure of DNase I at 2.0 A resolution suggests a mechanism for binding to and cutting DNA. Nature. 1986 Jun 5;321(6070):620–625. doi: 10.1038/321620a0. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Fujii S., van Boom J. H., van der Marel G. A., van Boeckel S. A., Rich A. Molecular structure of r(GCG)d(TATACGC): a DNA--RNA hybrid helix joined to double helical DNA. Nature. 1982 Oct 14;299(5884):601–604. doi: 10.1038/299601a0. [DOI] [PubMed] [Google Scholar]
- Wyatt J. R., Walker G. T. Deoxynucleotide-containing oligoribonucleotide duplexes: stability and susceptibility to RNase V1 and RNase H. Nucleic Acids Res. 1989 Oct 11;17(19):7833–7842. doi: 10.1093/nar/17.19.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T., Yoshida M., Kanaya S., Nakamura H., Nagayama K. Assignments of backbone 1H, 13C, and 15N resonances and secondary structure of ribonuclease H from Escherichia coli by heteronuclear three-dimensional NMR spectroscopy. Biochemistry. 1991 Jun 18;30(24):6036–6047. doi: 10.1021/bi00238a030. [DOI] [PubMed] [Google Scholar]
- Yang W., Hendrickson W. A., Crouch R. J., Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990 Sep 21;249(4975):1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Pheiffer B. H. A RNA.DNA hybrid that can adopt two conformations: an x-ray diffraction study of poly(rA).poly(dT) in concentrated solution or in fibers. Proc Natl Acad Sci U S A. 1981 Jan;78(1):78–82. doi: 10.1073/pnas.78.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]