Abstract
A low cost three-dimensional (3D) printed clear plastic microfluidic device was fabricated for fast, low cost automated protein detection. The unibody device features three reagent reservoirs, an efficient 3D network for passive mixing, and an optically transparent detection chamber housing a glass capture antibody array for measuring chemiluminescence output with a CCD camera. Sandwich type assays were built onto the glass arrays using a multi-labeled detection antibody-polyHRP (HRP = horseradish peroxidase). Total assay time was ~30 min in a complete automated assay employing a programmable syringe pump so that the protocol required minimal operator intervention. The device was used for multiplexed detection of prostate cancer biomarker proteins prostate specific antigen (PSA) and platelet factor 4 (PF-4). Detection limits of 0.5 pg mL−1 were achieved for these proteins in diluted serum with log dynamic ranges of four orders of magnitude. Good accuracy vs ELISA was validated by analyzing human serum samples. This prototype device holds good promise for further development as a point-of-care cancer diagnostics tool.
Introduction
Three-dimensional (3D) printing is a simple, attractive route for prototyping and fabricating bioanalytical devices that require many modifications along the road to optimization. Fabrication of objects is typically achieved by uploading a computer-aid design (CAD) to the printer, which will construct the desired object layer-by-layer based on “sliced” CAD models.1,2 Recently, 3D printers have become relatively inexpensive due to advances in technology and market competition. Researchers have applied these unique tools to develop microfluidic devices without the need for high end lithography.2–4 While 3D-printer resolution cannot yet rival that of lithography, perfectly functional microfluidic devices can be developed rapidly due to 3D-pritning’s rapid prototyping capability.5,6,7
Stereolithography (SLA) is a 3D printing method that provides low surface roughness8 using acrylate-based resin, an optical transparent plastic.9 SLA prints objects using a laser or digital light processing (DLP) projector to photocure liquid polymer precursor.1–3 A low cost desktop 3D printer with ~300 µm feature resolution can be used to fabricate highly transparent devices after appropriate processing.4,10 Fused deposition modeling (FDM) is an alternative to SLA that print objects by forcing a molten thermoplastic through a heated nozzle onto a moving platform. While the FDM generally cost less than SLA, SLA is superior in terms of resolution, smoother surface finish, and the ability to print transparent objects (Table S1).
Examples of analytical device fabrication by 3D printing2 include devices for monitoring metal ions,11 for cellphone-based food allergen and albumin assays,12,13 for bacteria detection14, and for proteins.15 We recently utilized a SLA printer to fabricate a clear plastic flow sensor cell to measure DNA by electrochemiluminescence (ECL),10 and 3D-printed a supercapacitor-powered electrochemiluminescent (ECL) immunoarray for multiplexed detection of 3 cancer biomarker proteins at fg mL−1 levels.16
Elevated levels of proteins in blood have great potential as biomarkers for early cancer detection and personalized therapy.17–23 Enzyme-linked immunosorbent assays (ELISA) have long served as the gold standard for clinical protein measurements.23,24 Newer commercial techniques such as bead-based optical or electrochemiluminescent (ECL) methods hold great promise for high throughput and multiplexed assays.25–28 Simoa, an advanced nanowell technology for protein detection, has sensitivity in the fg mL−1 range.29 However, reliable lower cost analytical devices are still needed for automated, multiplexed, highly sensitive, and equipped with broad dynamic range for clinical protein detection. Immunoarray devices have been reported that measure small numbers of proteins with accuracy, reliability and in some cases automation.30–35 We developed an amperometric microfluidic system to measure up to four cancer biomarker proteins in serum down to levels 5 fg mL−1.36–38 We also developed ECL arrays with antibody-coated Ru(bpy)32+ doped silica nanoparticles for ultrasensitive automated detection of up to four biomarker proteins as well as 3D-printed gravity flow system.16 Nonetheless, further automation is needed to decrease operator errors and simplify assay protocols while keeping overall cost low.
Detection by chemiluminescence (CL) is simple, low-cost technique offering exquisite sensitivity without a light or power source.39,40 CL signals can be generated by reaction of luminol with hydrogen peroxide in the presence of a peroxidase enzyme. Realizing the possibility to use a simple camera to measure output, the combination of CL with 3D-printed microfluidic devices presents an opportunity for automated, multiplexed detection for cancer biomarker proteins.41,42
In this paper, we use an inexpensive high definition 3D SLA printer to prototype and fabricate an automated device to measure two proteins by simultaneous chemiluminescent detection. Poly-L-lysine glass slides with immobilized capture antibodies were positioned downstream from reagent reservoirs in the device to enable the sandwich immunoassays. We achieved sub pg mL−1 detection limits for prostate cancer biomarker proteins platelet factor 4 (PF-4) and prostate specific antigen (PSA) in diluted calf serum with a wide dynamic range demonstrating good promise as a diagnostic tool for cancer screening in resource-limited settings. Accuracy of the device was confirmed by comparison of assay results on human patient serum samples with ELISA.
To our knowledge, this is the first report of non-PDMS 3D-printed channels with mixer and detection chamber integrated into an unibody immunoarray to measure protein biomarkers. The current system incorporates a reusable 3D-printed microfluidic device with sensitive CL detection into a unique immuno-diagnostic tool for multiplexed detection at low cost. The novelty lies in the integration of automated assay features in a valve-free, low cost device. Such features are essential for future translation of multi-protein detection into the cancer clinic. With appropriately spotted protein arrays, more than four proteins can be measured to enable low cost multiplexed protein detection.
Experimental
Chemicals
Clear resin (GCPL02) was from Formlabs (Somerville, MA). Antibody pairs and protein standards were obtained from Prostate specific antigen (PSA) DuoSet (catalog # DY1344) and platelet factor-4 (PF-4) DuoSet (catalog # DY795) from R&D Systems, Inc. Streptavidin Poly-HRP and 1% casein was from Fitzgerald, Inc. Tween-20 was from Sigma Aldrich, Inc. FemtoWest chemiluminescence reagent was from ThermoFisher. Immunoreagents were dissolved in pH 7.4 PBS buffer (0.01 Min phosphate, 0.14 M NaCl, 2.7 mM KCl) unless otherwise noted. Protein standards were prepared in 500× diluted calf serum. Single protein ELISA kits for PSA (RAB0331) and PF-4 (RAB0402) were from Sigma Aldrich.16 Normal patient serum sample was from Capital Biosciences, and prostate cancer patient serum samples were provided by George Washington University Hospital.
3D-printed device fabrication
A desktop 3D stereolithographic printer, Formlab Form 1+, and clear transparent methacrylate-based resin (FormLab GCPL02) was used to fabricate the fluidic device. Computer aided design (CAD) was generated using the software 123D Design (AutoDesk) and converted to 3D printer format (Figure 1). The model was imported into the Formlab software, PreForm, where printer settings were optimized to add extra supports to eliminate deformation. Layer heights were set to 50 µm to balance print speed while maintaining resolution. Uncured resin within the fluidic device was removed by forcing isopropanol through the channels using a syringe equipped with 23-guage needle followed by submerging the device in an isopropanol bath for 15 min on a shaker. Supports were snapped off the device, which was then rinsed with water and dried. To enhance transparency, devices were spray-coated with clear acrylic top coat (Krylon, Cleveland, OH).
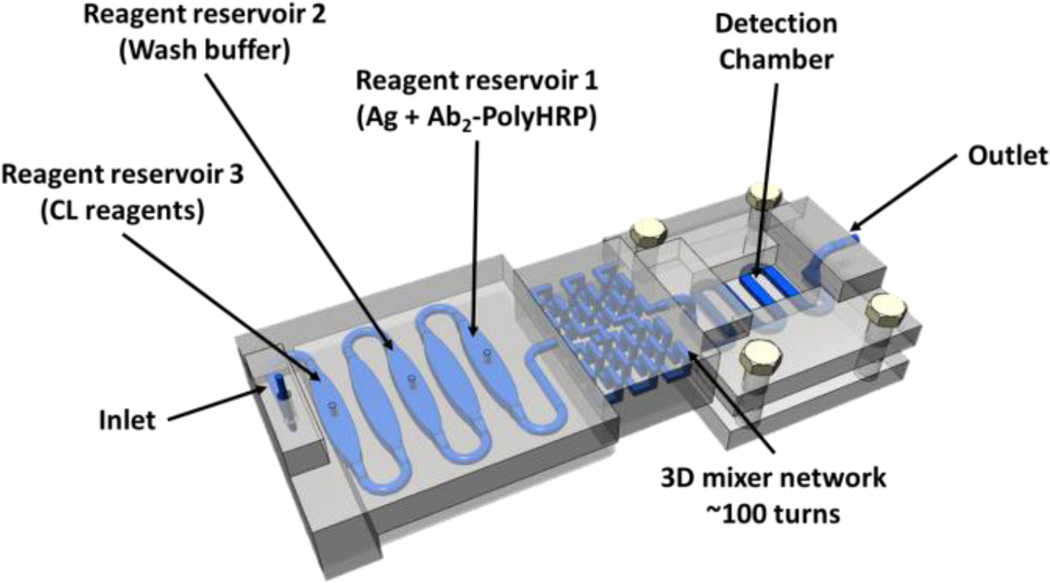
Fig. 1.
Model of 3D-printed unibody immunoarray for automated detection of cancer biomarker proteins (Ag).
Three reservoirs with volume 125 ± 5 µL were printed, with equal volume empty chambers in between to separate the solutions and prevent mixing. A 3D network passive mixer (210 ± 10 µL) was placed after the reservoirs. A 30 ± 2 µL detection chamber with a serpentine channel was located downstream. The detection chamber houses a poly-L-lysine coated glass slide decorated with capture antibodies. An open window was placed directly on top of the PDMS base detection chamber for maximum light transmittance.
Antibody Array Fabrication
Capture antibodies (Ab1) were diluted to 100 µg mL−1 from stock solution, and 1 µL of the respective antibody solution was then spotted onto the poly-L-lysine-coated glass slide using a multichannel pipette with 4.5 mm centre-to-centre pitch spacing vertically and 2 mm lateral spacing. After spotting, slides were incubated overnight in a humidity chamber at 4°C allowing the antibodies to be immobilized onto the glass substrate. The protein arrays were than rinsed with PBS and blocked with 1% casein in PBS for 1 hr to minimize non-specific binding. Arrays were then washed with PBS and used immediately. Unused arrays were stored at −20°C under vacuum for up to 14 days.
Sample and Reagent Loading
A mixture of 1:80 detection antibodies (Ab2) and 1:200 polyHRP diluted in PBS with 1% casein and 0.05% Tween-20 from their respective stock solutions were premixed. and loaded into reservoir 1 (Fig. 2). A wash buffer containing 0.5% casein and 0.05% Tween-20 in PBS was loaded into the reservoir 2 (Fig. 2) followed by 1:1 mixture of the chemiluminescent reagent in the reservoir 3 (Fig. 2). Then, 10 µL of a mixture of protein standards in 500-fold diluted calf serum were pipetted into reservoir 1 previously loaded with antibodies and polyHRP allowing the immuno-reagents to react with corresponding protein partner as the mixture moves through the 3D mixer network. For patient serum samples, 2 µL of each sample was first diluted 500-fold in PBS prior to loading into reservoir 1.
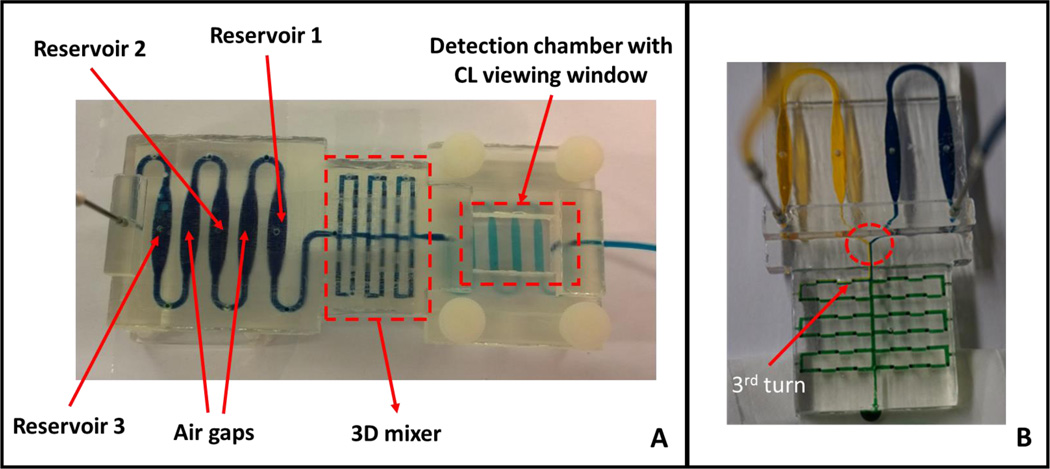
Fig. 2.
Completed 3D-printed unibody CL immunoarray device. (A) Upstream reagent reservoirs are separated by empty air chambers, followed by the 3D mixing network in the center and a detection chamber downstream housing the antibody array. Blue dye solution indicates the fluidic path of the device. (B) A modified 3D-printed device to demonstrate the mixing ability of the passive 3D mixing network. The mixer contains with 96 0.8 mm× 0.8 mm × 0.8 mm 90° turns in a 2 cm × 3.5 cm space. Yellow and blue dyes were pumped at 50 µL min−1 into the mixer network with solution mixing by the third turn (red arrow) demonstrating excellent mixing efficiency of this passive mixer. There was a clear separation of two dyes before the dyes entered the mixer (red circle).
Assay Protocol
After the capture antibody array was placed within the detection chamber, a syringe pump (Chemyx Fusion 400) was programmed through its touchscreen interface to follow a sequence of start and stop flow and change in flow rates during different stages of the assay (see SI file for details).
Chemiluminescence Measurement
Chemiluminescence (CL) measurements were obtained using a G:Box bioimaging system and GeneSnap software (Syngene, Cambridge, U.K.). CL intensities for images captured by the CCD camera were determined by integrating CL signal over the area of the individual capture antibody spot using GeneTools software (Syngene, Cambridge, U.K.). CL images obtained using the CCD camera were recolorized using ImageJ software.
Results
Mixing Efficiency of the 3D-Printed Passive Mixing Network
Mixing was achieved by using an optimally configured three dimensional network of channels by testing different geometries (planar and non-planar) of the channels (Figure S2, ESI). Effective mixing of yellow and blue dyes was achieved when flow rates in the mixer was set at 50 µL min−1 (Figure S3, ESI). There was a clear separation of the two dyes before entering the 3D mixer (circled in Fig. 2B) followed by complete mixing by the third turn in the mixer as indicated by the formation of green-colored dye (red arrow, Fig. 2B, also see Fig. S1, ESI). The mixture continued to mix throughout the entire length of the mixer network until it reaches the detection chamber.
Stability of the Capture Antibody Array
The stability of the capture antibody array prepared as described above was tested by storing the arrays at −20°C under vacuum after blocking with 1% casein and dried under gentle N2. The CL responses generated for 50 pg mL−1 of PSA and PF4 antigen standards in 500× diluted calf serum were monitored over 2 weeks. The arrays were found to be stable with minimal change in signals after 2 weeks (Fig. S4, ESI).
Assay Development
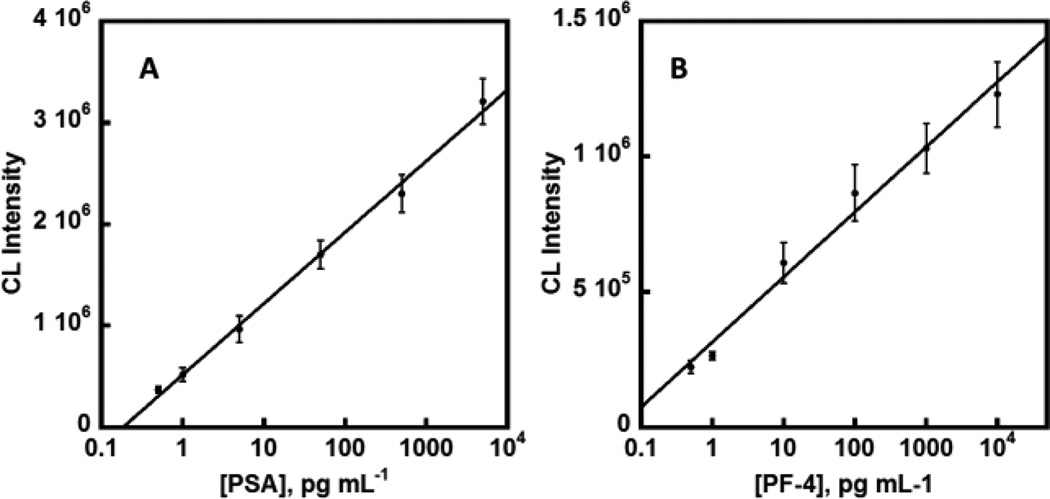
The entire assay protocol was automated using a programmable syringe pump once reagents were loaded into their respective reservoirs. The device was placed into a G:Box bioimaging system equipped with a CCD camera.. The syringe pump was started at 50 µL min−1 forcing the immuno-reagents to enter the 3D network mixer and allow antibodies to capture their respective specific antigen and also bind to the polyHRP label. After 6.5 min inside the mixer and delivery to the detection chamber, the flow was stopped for 15 min to allow the Ab2-polyHRP-protein bioconjugates to be captured by capture antibodies (Ab1) immobilized on the Ab1-patterned glass slide. Flow was resumed after the incubation, flowing wash buffer to remove excess reagents, followed by introduction of the chemiluminescent (CL) reagent. The CL image of the protein array within the detection chamber was then recorded by integrated CCD camera measurement for 60 s immediately after the detection chamber was filled with CL reagent (Figure S5, ESI). The entire assay takes 30 min with the ability to scale up with multiple devices running concurrently. The device was rinsed with 0.05% Tween-20 in PBS and PBS before drying with air using a syringe to eliminate sample carry over and any non-specific binding before the next run. Detection limits of 0.5 pg mL−1 (as 3SD + control) were achieved for PSA and PF-4. A wide dynamic range of 4 orders of log concentration between 0.5 pg mL−1 to 10 ng mL−1 for PF-4 and 0.5 pg mL−1 to 5 ng mL−1 for PSA, which eliminate the need of multiple serial dilutions (Figure 3).
Fig. 3.
Calibration data in 500× diluted calf serum showing influence of biomarker proteins concentrations on camera-imaged CL signals for (A) PSA and (B) PF-4. (n=3)
Validation of Accuracy
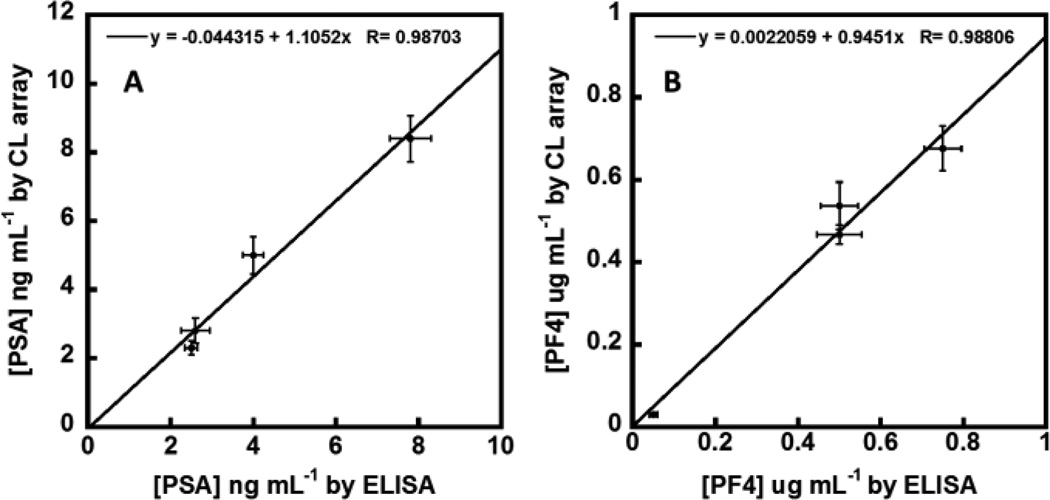
Three serum samples from prostate cancer patients and one serum sample from a cancer-free patient were analyzed and compared with results from single-protein ELISA (Figure S6, ESI). These samples were diluted 500-fold in PBS to bring the CL response into the linear dynamic ranges of the calibrations. Concentrations of PF-4 and PSA were found to be within the detection limits of their respective ELISAs. Linear correlation plots of the ELISA vs. immunoarray data gave slopes of 1.10 ± 0.13 for PSA and 0.95 ± 0.10 for PF-4. Intercepts of these plots were near zero, i.e. −0.05 ± 0.60 for PSA and 0.00 ± 0.05 for PF-4 (Figure 4). These results demonstrate excellent correlation of the automated CL array assay with standard ELISA while confirming the high selectivity and specificity of the assay for the two proteins in the presence of the hundreds of other proteins in human serum.43
Fig. 4.
Linear correlation plots of 3D-printed CL immunoarray vs. ELISA results for 4 human serum samples for (A) PSA and (B) PF-4. Error bars are standard deviations for the CL arrays (n = 3) and ELISA (n = 3).
Discussion
Results above show that stereolithographic 3D printing represents an excellent method for rapid prototyping, optimizing, and fabricating all-in-one unibody microfluidic immunoarrays with three dimensional features. When combined with an easily prepared immunoarray slide enabling chemiluminescent detection, the 3D-printed immunoarray measured two proteins simultaneously in an automated fashion when interfaced with a programmable syringe pump. The 3D-printed device is fully reusable, and the programmable pump controls the flow of reagents within device, allowing the automated addition of the necessary reagents into the detection chamber. Utilizing the pump’s touchscreen interface, the start and stop of flow can be timed in accordance to user’s specifications. Flow rates can also be programed to control the mixing speed within the 3D mixer to control the time to complete the assay.
Assay time was ~30 min starting from the time serum protein samples are loaded until the chemiluminescent signal was captured by the CCD camera. Good reproducibility was found between different arrays for protein standards with RSD ≤ ± 8% for the 2-proteins detection. Loading of reagents into the reservoirs can be done using inexpensive disposable fixed volume pipettes capable of precisely delivering µL volumes. The automated CL immunoarray presented here costs less and is capable of performing multiplexed detection in less time with similar or better analytical performance in comparison to traditional ELISA (Table S2).
PSA is clinically used as a serum biomarker for prostate cancer and has been used as an early screening tool.44 A serum PSA concentration of 4–10 ng mL−1 suggests the possibility of early stage prostate cancer, while normal levels are typically 0.5–2 ng mL−1 and late stage prostate cancer is characterized by values above 100 ng mL−1.45 However, serum PSA test has prediction success of ~70% since PSA levels can be elevated in several benign prostate diseases.46 Therefore, it has been recommended to measure a panel of several biomarkers to improve the predictive power of serum protein tests and provide information for therapeutic monitoring.23,47 PF-4 is another useful protein marker that is elevated in invasive and metastatic prostate cancer with normal serum level of 2–10 ng mL−1 and much higher levels in prostate cancer patients..48,49 Detection limits of 0.5 pg mL−1 for both PSA and PF4 with dynamic ranges from 0.5 pg mL−1 to 5–10 ng mL−1, make the immunoarray relevant to clinical ranges of both these proteins after 500-fold dilution, eliminating the need for serial dilutions.
A major advantage of 3D-printing is its ability to construct three dimensional features within the device in an unibody design. This allows the fluidic device to maintain channel integrity and eliminates leaking as compared to multi-layer devices that can suffer from leakage if not properly assembled. For the device we describe here, flow rates as high as 1000 µL min−1 were achieved without any leakage. Reproducibility of reservoir volumes of 125 ± 10 µL suggests that some uncured photopolymer resin might have been left within the channels even after vigorous removal by flowing isopropyl alcohol with a syringe pump for 1 min and curing on a shaker table while submersed in isopropyl alcohol for additional 15 min.10,50 To eliminate volume errors, a calibrated pipette was used to load the reservoirs with precise volumes of diluted sample and reagents.
A major challenge in designing microfluidic devices for immuno-diagnostics is efficient mixing in microchannels to promote necessary reaction of the immuno-reagents.51–53 It is essential to design microfluidic devices that enable turbulent mixing, while maintaining overall small dimensions to realize the benefits of miniaturization. Turbulent mixing can be achieved actively or passively. Typically, active mixing strategies are implemented by stirring or agitation of the fluids using external forces such as acoustic waves, electric field, and heat, within the microfluidic device.52 However, these mixing methods require additional parts that increase cost and size of the devices. Passive mixers promote mixing by moving fluids through specified microchannel designs and are more suitable for integrated diagnostics since they require no additional energy or moving parts. In this paper, a passive mixer consisting of three-dimensional serpentine structures was integrated into the microfluidic device to promote turbulent mixing of immuno-reagents. As compared to planar serpentine designs housed in the same dimension (20 × 31 × 10 mm), more than 3 times (31 turns vs 96 turns) the number of turns and >3 times the length were incorporated, ensuring efficient mixing of immuno-reagents before entering the detection chamber.
The 3D-printed CL microfluidic array is an excellent candidate for inexpensive, low cost diagnostics in a low or moderate resource setting. The 3D-printed components cost ~$5.00 in materials. Considering these parts are reusable by replacing the antibody array with a fresh one, a single immunoassay to measure two biomarker proteins costs ~$2.00 (~$0.33 per replicate) for the reagents including the antibody array. Furthermore, coupling of the device with a microcontrolled micropump16 and a less expensive camera would further miniaturize and automate this diagnostic tool for resource-limited settings.
Conclusions
Results above demonstrate the use of stereolithographic 3D-printing to prototype and fabricate an all-in-one automated chemiluminescence immuno-device in an unibody fashion. The overall assay cost for two biomarker proteins is ~$2.00 in assay time 30 min. While other techniques for protein detection may offer competitive or better sensitivity and detection limits15,21–32,54,55 the CL immunoarray described here can provide fast quality results at very low cost, which is crucial to future diagnostic applications of multiprotein analyses in the clinic. Additional improvements such as microprocessor/micropump control, data acquisition and reporting could lead to a low cost point-of-care device suitable for widespread use in cancer diagnostics.
Supplementary Material
Acknowledgments
This work was supported financially by grants Nos. EB016707 and EB014586 from the National Institute of Biomedical Imaging and Bioengineering (NIBIB), NIH.
Footnotes
Electronic Supplementary Information (ESI) available: See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Gross BC, Erkal JL, Lockwood SY, Chen C, Spence DM. Anal. Chem. 2014;86:3240–3253. doi: 10.1021/ac403397r. [DOI] [PubMed] [Google Scholar]
- 2.Bishop GW, Satterwhite-Warden JE, Kadimisetty K, Rusling JF. Nanotechnology. 2016;27:284002. doi: 10.1088/0957-4484/27/28/284002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Neill PF, Azouz AB, Vázquez M, Liu J, Marczak S, Slouka Z, Chang HC, Diamond D, Brabazon D. Biomicrofluidics. 2014;8:052112. doi: 10.1063/1.4898632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng C, Ho B, Ng SH, Ho cK, Li H, Yoon Y-J. Lab Chip. 2015;15:3627–3637. doi: 10.1039/c5lc00685f. [DOI] [PubMed] [Google Scholar]
- 5.Au AK, Bhattacarjee N, Horowitz LF, Chang TC, Folch A. Lab Chip. 2015;15:1934–1941. doi: 10.1039/c5lc00126a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharjee N, Urrios A, Kang S, Folch A. Lab Chip. 2016;16:1720–1742. doi: 10.1039/c6lc00163g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waheed S, Gabot JM, Macdonald NP, Lewis T, Guijt RM, Paull B, Breadmore MC. Lab Chip. 2016;16:1993–2013. doi: 10.1039/c6lc00284f. [DOI] [PubMed] [Google Scholar]
- 8.Comina G, Suska A, Filippini D. Lab Chip. 2014;14:2978–2982. doi: 10.1039/c4lc00394b. [DOI] [PubMed] [Google Scholar]
- 9.Au AK, Lee W, Folch A. Lab Chip. 2014;14:1294–1301. doi: 10.1039/c3lc51360b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop GW, Satterwhite-Warden JE, Bist I, Chen E, Rusling JF. ACS Sens. 2016;1:197–202. doi: 10.1021/acssensors.5b00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su C, Hsia S, Sun Y. Anal. Chim. Acta. 2014;838:58–63. doi: 10.1016/j.aca.2014.06.037. [DOI] [PubMed] [Google Scholar]
- 12.Coskun AF, Nagi R, Sadeghi K, Phillips S, Ozcan A. Lab Chip. 2013;13:4231–4238. doi: 10.1039/c3lc50785h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coskun AF, Wong J, Khodadadi D, Nagi R, Tey A, Ozcan A. Lab Chip. 2013;13:636–640. doi: 10.1039/c2lc41152k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee W, Kwon D, Choi W, Jung GY, Jeon S. Sci. Rep. 2015;5:7717. doi: 10.1038/srep07717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixit CK, Kadimisetty K, Otieno B, Tang CK, Malla S, Krause CE, Rusling JF. Analyst. 2016;141:536–547. doi: 10.1039/c5an01829c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadimisetty K, Mosa IM, Malla S, Satterwhite-Warden JE, Kuhns TM, Faria RC, Lee NH, Rusling JF. Biosens. Bioelectron. 2016:188–193. doi: 10.1016/j.bios.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giljohan DA, Mirkin CA. Nature. 2009;426:461–464. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulasingam V, Diamandis EP. Nat. Clin. Prac.: Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 19.Hanash SM, Baik CS, Kallioniemi O. Nat. Rev. Clin. Oncol. 2011;8:142–150. doi: 10.1038/nrclinonc.2010.220. [DOI] [PubMed] [Google Scholar]
- 20.Kingsmore SF. Nat. Rev. Drug Discov. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rusling JF, Kumar CV, Gutkind JS, Patel V. Analyst. 2010;135:2496–2511. doi: 10.1039/c0an00204f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rusling JF, Bishop GW, Doan N, Papadimitrakopoulos F. J Mater. Chem. B. 2014;2:12–30. doi: 10.1039/C3TB21323D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rusling JF. Anal. Chem. 2013;85:5304–5310. doi: 10.1021/ac401058v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J. Biosens. Bioelectron. 2006;21:1887–1892. doi: 10.1016/j.bios.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 25.Beveridge JS, Stephens JR, Williams ME. Annu. Rev. Anal. Chem. 2011;4:251–273. doi: 10.1146/annurev-anchem-061010-114041. [DOI] [PubMed] [Google Scholar]
- 26.Roche Diagnostics. [Accessed December 2015]; http://www.roche.com. [Google Scholar]
- 27.Meso Scale Diagnostics. [Accessed December 2015]; http://www.mesoscale.com. [Google Scholar]
- 28.Perkin Elmer. [Accessed December 2015]; http://www.perkinelmer.com/catalog/cate-gory/id/alphatech. [Google Scholar]
- 29.Meissner EG, Decalf J, Casrouge A, Masur H, Kottilil S, Albert ML, Duffy D. PLoS ONE. 2015;10(7):e0133236. doi: 10.1371/journal.pone.0133236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, Wang J, Moore H, Rouse R, Umvilligihozo G, Karitia E, Mwamarangewe L, Braunstein S, van de Wijgert J, Sahabo R, Justman J, El-Sadr W, Sia SK. Nat. Med. 2011;17:1015–1019. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 31.Chin CD, Linder V, Sia SK. Lab Chip. 2012;12:2118–2134. doi: 10.1039/c2lc21204h. [DOI] [PubMed] [Google Scholar]
- 32.Yu ZT, Guan H, Cheung MK, McHugh WM, Cornell TT, Shanley TP, Kurabayashi K, Fu J. Sci. Rep. 2015;5:11339. doi: 10.1038/srep11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laksanasopin T, Guo TW, Nayak S, Sridhara AA, Xie S, Olowookere OO, Cadinu P, Meng F, Chee NH, Kim J, Chin CD, Munyazesa E, Mugwaneza P, Rai AJ, Mugisha V, Castro AR, Steinmiller D, Linder V, Justman JE, Nsanzimana S, Sia SK. Sci. Transl. Med. 2015;7:273re1–273re1. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- 34.Kadimisetty K, Malla S, Sardesai NP, Joshi AA, Faria RC, Lee NH, Rusling JF. Anal. Chem. 2015;87:4472–4478. doi: 10.1021/acs.analchem.5b00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fragoso A, Latta D, Laboria N, von Germar F, Hansen-Hagge TE, Kemmer W, Gartner C, Klemm R, Drese KS, O’Sullivan CK. Lab Chip. 2011;11:625–631. doi: 10.1039/c0lc00398k. [DOI] [PubMed] [Google Scholar]
- 36.Malhotra R, Patel V, Chikkaveeraiah BV, Munge BS, Cheong SC, Zain RB, Abraham MT, Dey DK, Gutkind JS, Rusling JF. Anal. Chem. 2012;84:6249–6255. doi: 10.1021/ac301392g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krause CE, Otieno BA, Bishop GW, Choquette PG, Lalla RV, Peterson DE, Rusling JF. Anal. Bioanal. Chem. 2015;407:7239–7243. doi: 10.1007/s00216-015-8873-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krause CE, Otieno BA, Latus A, Faria RC, Patel V, Gutkind JS, Rusling JF. ChemistryOpen. 2013;2:141–145. doi: 10.1002/open.201300018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Road A, Mirasoli M, Dolci LS, Buragina A, Bonvicini F, Simon P, Guardigi M. Anal. Chem. 2011;83:3178–3185. doi: 10.1021/ac200360k. [DOI] [PubMed] [Google Scholar]
- 40.Zhao LX, Sun L, Chu XG. TrAC, Trends Anal. Chem. 2009;28:404–415. [Google Scholar]
- 41.Roda A, Michelini E, Cevenini L, Calabria D, Calabretta MM, Simoni P. Anal. Chem. 2014;86:7299–7304. doi: 10.1021/ac502137s. [DOI] [PubMed] [Google Scholar]
- 42.Roda A, Guardigli M, Calabria D, Calabretta MM, Cevenini L, Michelini E. Analyst. 2014;139:6494–6501. doi: 10.1039/c4an01612b. [DOI] [PubMed] [Google Scholar]
- 43.Pieper R, Gatlin CL, Makusky AJ, Russo PS, Schatz CR, Miller SS, Su Q, McGrath AM, Estock MA, Parmar PP, Zhao M, Huang S, Zhou J, Wang F, Esquer-Blasco R, Anderson NL, Taylor J, Steiner S. Proteomics. 2003;3:1345–1364. doi: 10.1002/pmic.200300449. [DOI] [PubMed] [Google Scholar]
- 44.Smith RA, Cokkinides V, Brawley OW. CA Cancer J. Clin. 2012;62:129–142. doi: 10.3322/caac.20143. [DOI] [PubMed] [Google Scholar]
- 45.Kulasingam V, Diamandis EP. Nat. Clin. Pract. Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 46.Ward AM, Catto JWF, Hamdy FC. Ann. Clin. Biochem. 2001;38:633–651. doi: 10.1258/0004563011901055. [DOI] [PubMed] [Google Scholar]
- 47.Hanash SM, Baik CS, Kalliioiemi O. Nat. Rev. Clin. Oncol. 2011;8:142–150. doi: 10.1038/nrclinonc.2010.220. [DOI] [PubMed] [Google Scholar]
- 48.Wu Q, Dhir R, Wells A. Mol. Cancer. 2012;11:3–18. doi: 10.1186/1476-4598-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chikkaveeraiah BV, Bhirde A, Malhotra R, Patel V, Gutkind JS, Rusling JF. Anal. Chem. 2009;81:9129–9134. doi: 10.1021/ac9018022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shallan AI, Smejkal P, Corban M, Guijt RM, Breadmore MC. Anal. Chem. 2014;86:3124–3130. doi: 10.1021/ac4041857. [DOI] [PubMed] [Google Scholar]
- 51.Jeong GS, Chung S, Kim CB, Lee SH. Analyst. 2010;135:460–473. doi: 10.1039/b921430e. [DOI] [PubMed] [Google Scholar]
- 52.Lee CY, Chang CL, Wang YN, Fu LM. Int. J. Mol. Sci. 2011;12:3263–3287. doi: 10.3390/ijms12053263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Deng Y, Zhang P, Liu Z, Wu Y. J Micromech. Microeng. 2013;2013:075002. [Google Scholar]
- 54.Walt DR. Lab Chip. 2014;14:3195–3200. doi: 10.1039/c4lc00277f. [DOI] [PubMed] [Google Scholar]
- 55.Kelley SO, Mirkin CA, Walt DR, Ismagilov RF, Toner M, Sargent EH. Nat. Nanotech. 2014;9:969–980. doi: 10.1038/nnano.2014.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.