Abstract
Kir2.1 (also known as IRK1) plays key roles in the regulation of resting membrane potential and cell excitability. To achieve its physiological roles, Kir2.1 performs a series of conformational transitions, named as gating. However, the structural basis of gating is still obscure. Here, we combined site-directed mutagenesis, two-electrode voltage clamp with molecular dynamics simulations and determined that H221 regulates the gating process of Kir2.1 by involving a weak interaction network.
Our data show that the H221R mutant accelerates the rundown kinetics and decelerates the reactivation kinetics of Kir2.1. Compared with the WT channel, the H221R mutation strengthens the interaction between the CD- and G-loops (E303-R221) which stabilizes the close state of the G-loop gate and weakens the interactions between C-linker and CD-loop (R221-R189) and the adjacent G-loops (E303-R312) which in turn destabilizes the open state of G-loop gate. Our data indicate that the three pairs of interactions (E303-H221, H221-R189 and E303-R312) precisely regulate the G-loop gate by controlling the conformation of the G-loop.
Keywords: Kir channel, Targeted molecular dynamics, Molecular dynamics, Homology model, gating kinetics, weak interaction
INTRODUCTION
Inward rectifying K+ (Kir) channels are intrinsic membrane proteins that selectively permeate potassium ions across cell membranes with the primary role of regulating the outward directed K+ current. Kir channels regulate many physiological processes, such as pancreatic insulin secretion, vascular tone, cardiomyocytes, neurons, heart rate, fluid balance and K+ homeostasis in the body 1–3.
The Kir family, which includes 16 members, has been classified into seven subfamilies (Kir1-7) on the basis of sequence conservation and biophysical properties 4–9. For the past decade, more and more Kir channel structures (the full-length structures and cytoplasmic domain) have been crystalized 10–20. The crystallographic structures reveal that the Kir channels assemble in tetrameric complexes, which have two different regions in series: a transmembrane domain (TMD) and a cytoplasmic domain (CTD). An expansion linker, named the C-linker, makes the TMD tethered to the CTD. The P loop and transmembrane domain (M1-P-M2 motif) form the “transmembrane pore” and the cytoplasmic N- and C-termini (cytoplasmic domain) form the “cytoplasmic pore”. The cytoplasmic pore is a common architectural characteristic of Kir channels, which extends the length of the canonical transmembrane pore nearly twice. A long ion permeation pathway outlines both the transmembrane and cytoplasmic pores. Three gates are distributed along the permeation pathway in turn: selectivity filter gate, the bundle-crossing gate (both in the transmembrane pore) and the G-loop gate (in the cytoplasmic pore). At the surface of the cytoplasmic domain a cluster of acidic and hydrophobic residues are found to create a favorable environment for a variety of modulators (ATP, H+, G-protein, PIP2, and Na+ and Mg2+) 1, 21, 22 that cause conformational changes of the G-loop, which is coupled to movement of the bundle-crossing gate 8, 18, 23–27. Experimental data combined with crystal structural studies show that the G-loop gate could control potassium ion flow of Kir channels 12, 28.
Kir2 subfamily members are well expressed in skeletal muscle, brain, heart and cardiovascular and nervous systems. 1–3, 7, 29. Loss-of-function Kir2.1 mutations, mostly situated in the cytoplasmic domain, cause Andersen’s syndrome in humans, which is characterized by periodic paralysis, abnormal electrocardiogram and ventricular arrhythmias 12, 30, 31. It has been suggested that conformational changes of the G-loop are accompanied by transformation of a network of weak interactions in the cytoplasmic domain 18, 32, 33. The E303K and V302M mutations in Kir2.1 channel can change the weak interactions between amino acids of the cytoplasmic domain that are able to affect the conformational changes of the G-loop 28, 34. However, the key residues near the G-loop of the Kir2.1 channel that precisely regulate the gating of the G-loop are far from being clear.
Our earlier studies of the Kir2.1 wild-type and mutant channel demonstrated that weak interactions (E303-H221 and H221-R189) are important for controlling the gating of the G-loop 24, 33. Previous results showed that the residue E303 is essential in maintaining the function of the Kir2.1 channel 33. To investigate if the H221 is a key residue during gating, we carried out measurements using a combination of site-directed mutagenesis, electrophysiology with molecular dynamics (MD) and targeted MD simulations. Our data show that the H221R mutant accelerates the rundown kinetics, and decelerates reactivation kinetics relative to the WT upon change of the three pairs of weak interactions.
MATERIALS AND METHODS
Homology Modeling
The full-length mouse Kir2.1 channel was used as a model template. Chain A of 3SPI (chicken Kir2.2 in the closed state) and 3SYQ (mouse Kir3.2 in the open state), were obtained from the Protein Data Bank (PDB). The homology models were generated using the SWISS-MODEL server 35–37. Due to high sequence homology (about 77% and 53% sequence identity to cKir2.2 and mKir3.2 respectively), the QMEAN4 score 38 was 0.566 and 0.545 respectively and QMEAN Z-score 39 was -3.36 and -3.66 respectively. The H221R mutant was generated with the VMD software 40. The H221R model was constructed by substituting the His sidechain with the Arg’s. Compared to its template models, the main chain geometry in experimental models had no change.
Conventional Molecular Dynamics
For simulation of four experimental structures, the structures of wild type (WT) and H221R in the closed and open states were embedded in an explicit palmitoyloleoyl-phosphatidylcholine (POPC) bilayer using a membrane package that is a plugin of VMD 40. The two binding sites (S1 and S3) of the selectivity filter were occupied by K+ ion, and the other two (S2 and S4) by water. All molecular dynamics (MD) simulations with explicit solvent and ions were performed in ~0.15 M KCL which was placed randomly to neutralize the system. A TIP3P water model 41 was employed for the solvent. The solvated systems covered altogether 24–34 ns (including 14-ns equilibration of the lipid and solution with protein released and 10–20 ns production run until reaching equilibrium). Each Targeted Molecular Dynamic was performed on the equilibrated system.
ALL MD simulations with a total of ~140,000 atoms were carried out using the NAMD2 program (http://www.ks.uiuc.edu/Research/namd/) 42 and the CHARMM 27 force filed 43–45 with NBFIX correction 46–48. The simulations had similar characteristics as in a reference24. Constant pressure (1 atm) and constant temperature (310 K) were controlled by Nose’-Hoover-Langevin piston and Langevin dynamics49, 50. Long-range electrostatic interactions, which were calculated using the particle-mesh Ewald (PME) 51, 52, had 120 × 120 × 180 grid points with a grid density of 1 Å3. Short-range interactions were calculated with a cutoff radius of 12 Å. A smooth switching function was employed at 10 Å and distance of pair list at 13.5 Å. A frame was saved every 4 ps. Simulation analysis and structural diagrams were treated by simulaid and VMD. The Plots were produced by the XMGrace progam (http://plasma-gate.weizmann.ac.il/Grace). Simulations were performed on a 64-processor Linux cluster.
Targeted Molecular Dynamics (Targeted MD)
Targeted MD simulation algorithm 53, 54 which was developed by Schlitter et al. was a simple method for producing large-scale conformational change in large proteins within a short simulation time. The conformational change is induced by the steering force which is dependent on the RMSD value. The force is given by the gradient of the potential.
where the force constant is represented by k, the number of atoms (N) used to calculate the RMSD from the target structure are set as targeted structure, RMSD(t) is the RMSD of the current structure to the target structure at time t, and RMSD*(t) is the prescribed target RMSD value at time t. The value of target RMSD is decreased monotonically from the initial RMSD to the target structure until it reached 0 Å at the end of Targeted MD simulation, which showed that the Targeted MD is accomplished 55.
Molecular Biology
All mouse Kir2.1 channels were subcloned into the pGEMHE expression vector and used as described. H221R mutantion was generated using a Quickchange kit (Stratagene). DNA sequencing was used to confirm Sequences. As previously described, recombinant wild-type Kir2.1, H221R and Ci-VSP were used for Xenopus laevis oocyte expression 56, 57. cRNA was prepared with T7 RNA polymerase using a kit from Promega. According to the functional expression level, cRNAs of the Kir2.1, H221R and of Ci-VSP were microinjected into Xenopus oocytes (0.5–10 ng/oocyte).
Electrophysiology
Whole-cell currents were recorded from Xenopus laevis oocytes 1–2 days after cRNA injection, using conventional two-electrode voltage-clamp (TEVC) with a GeneClamp 500 amplifier (Molecular Devices, CA). Microelectrodes were filled with 3 M KCl dissolved in 1% agarose to prevent leakage of KCl into the oocytes. Pipettes had a tip resistance of less than 1 MΩ. The bath solution with a high-potassium (ND96K) contained 96 mM KCl, 1 mM NaCl, 1.8 mM CaCl2, 1 mM MgCl2, 5 mM HEPES; the pH was adjusted with KOH to 7.4 at room temperature. A low-potassium solution (ND96) composed of (in mM) 96 NaCl, 1 KCl, 1.8 CaCl2, 1 MgCl2, 5 HEPES at a pH of 7.4 with NaOH. ND96 solution was made in some experiments to inhibit most of the Kir2.1 currents at –80 mV. The voltage-pulse protocol for activation of Ci-VSP was carried out on TEVC oocyte recordings: 1s-sweeps contained a 170 ms stepped depolarizations from -80 to +80 mV and an 830 ms step to +80 mV. To deactivate Ci-VSP, the holding potential was -80mV on the oocytes. Sweeps were applied till the resulting currents reached steady state. Current amplitudes were elicited by 1s pulses applied to potentials ranging from +80 mV to –80 mV. Results are reported as mean ± S.E.M (n = number of oocytes). Each experiment shown or described was performed on 5–7 oocytes from the same frog (batch) for TEVC recording. A minimum of 3–4 batches of oocytes was tested for each experiment shown. Data acquisition and analysis were done with pClamp9.2 (Molecular Devices, CA) and Origin 7.5 (Microcal, MA) software.
RESULTS AND DISCUSSION
Construction, equilibration and Targeted MD simulation
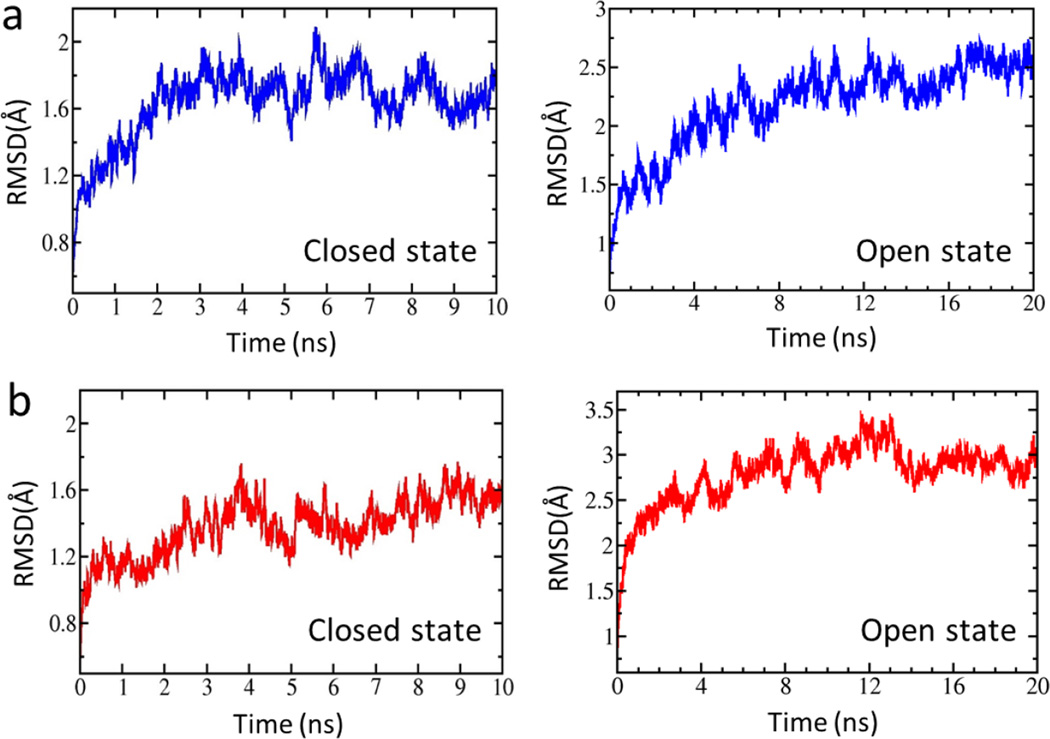
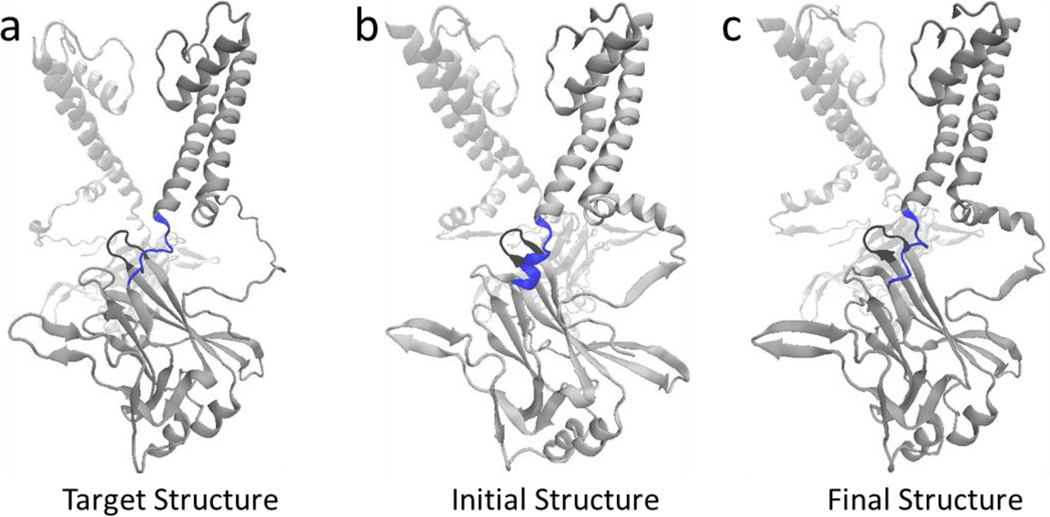
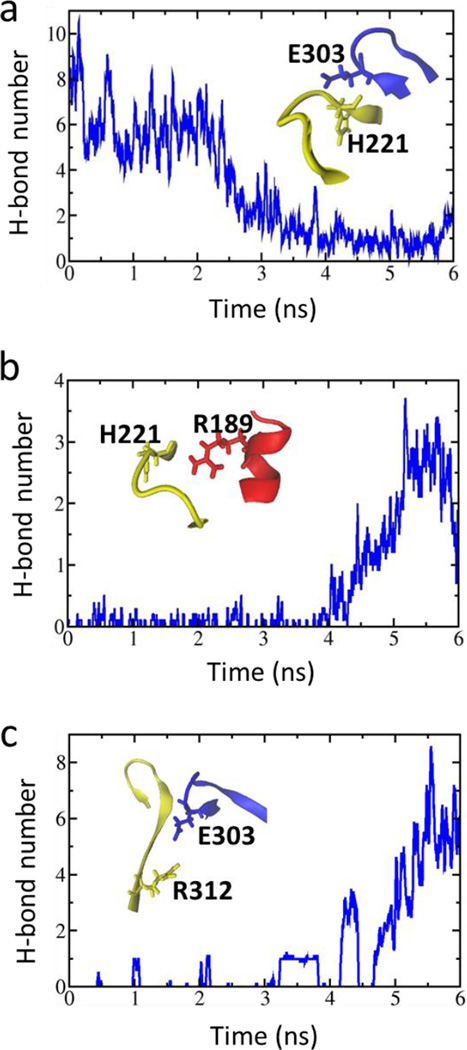
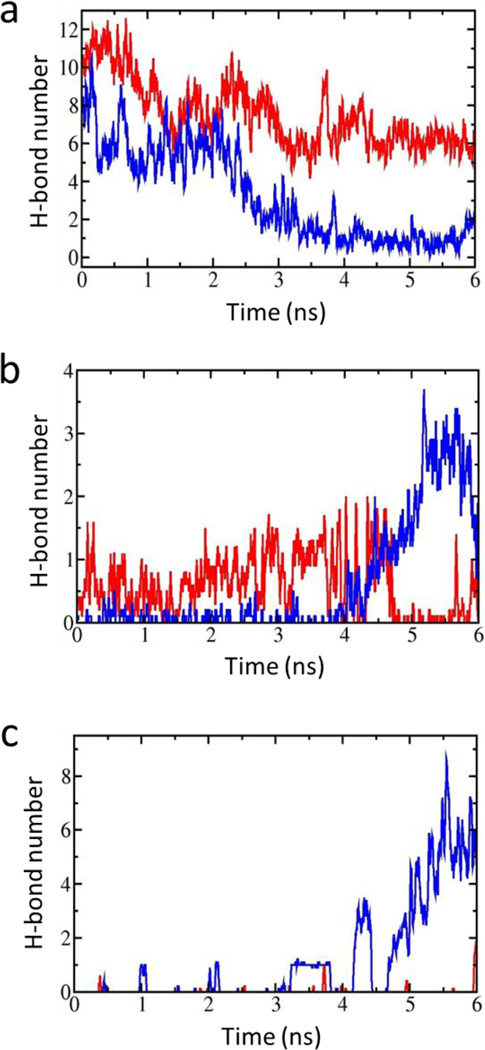
In the present study, four full-length structures of the Kir2.1 (WT) and the Kir2.1 (H221R) channels in the closed and open states were constructed using the crystal structures of chicken Kir2.2 (PDB ID: 3SPI) and mouse Kir3.2 (PDB ID: 3SYQ) as the templates. The sequences identities are 77% and 53%, respectively. Based on the radius of the pore, we named them closed and open states of Kir2.1, respectively. During 10–20 ns free MD simulations, root mean square deviation (RMSD) values of the four structures were no more than 3 Å which means that these systems reached their equilibration states (Fig. 1). We performed a Targeted MD simulation on the equilibrated closed Kir2.1 WT channel with the targeted structure in the open state (Fig. 2a, b). Figure 2c showed the final structure. During this Targeted MD simulation, an external force (force constant: 500 kcal/mol/Å2) was put on the backbone atoms of the PIP2 binding sites (Arg80, Trp81, Arg82, Lys182, Lys185 to Thr192 and Arg218) to make the specific domain reached targeted structure. The simulation showed that three pairs of weak interactions controlled the G-loop gate (Fig. 2, black domain), which is located at the CTD along the pore (Fig. 3). One pair of weak interaction between G- and CD-loop (E303-H221) was decreased, which led the closed G-loop gate to become unstable, making the G-loop gate easy to open (Fig. 3a). The other two pairs of weak interactions between the CD-loop and the C-linker (H221-R189) and between the adjacent G-loops (E303-R312) were increased (Fig. 3b-c), which are thought to maintain stability of the G-loop gate in the open state.
Figure 1.
RMSD variations for four systems (a, closed and open states of wild type. b, closed and open states of H221R.) throughout the simulation. RMSDs were calculated based on all Cα atoms of the channel.
Figure 2.
(a) The schematic diagram of a targeted structure, which is in the open state. (b) The schematic diagram of the initial structure, which is in the closed state. The target residues, to which the targeted force is applied (residues Lys185-Thr192 and PIP2 binding sites), are colored blue. The G-loop is highlighted in black. (c) The schematic diagram of the final conformation is one which is achieved by our last Targeted MD simulation.
Figure 3.
(a) The time course of the interaction between G-loop (blue) and CD-loop (yellow) through an intra-subunit E303-H221 interaction; (b) Variation of the strengthened interactions between the CD-loop (yellow) and C-linker (red) through H221-R189 and between adjacent G-loops through E303-R312 (c) as a function of simulation time. (a-c) are taken the average of H-bond number every 40ps.
H221R mutant affects the gating kinetics of Kir2.1 channel
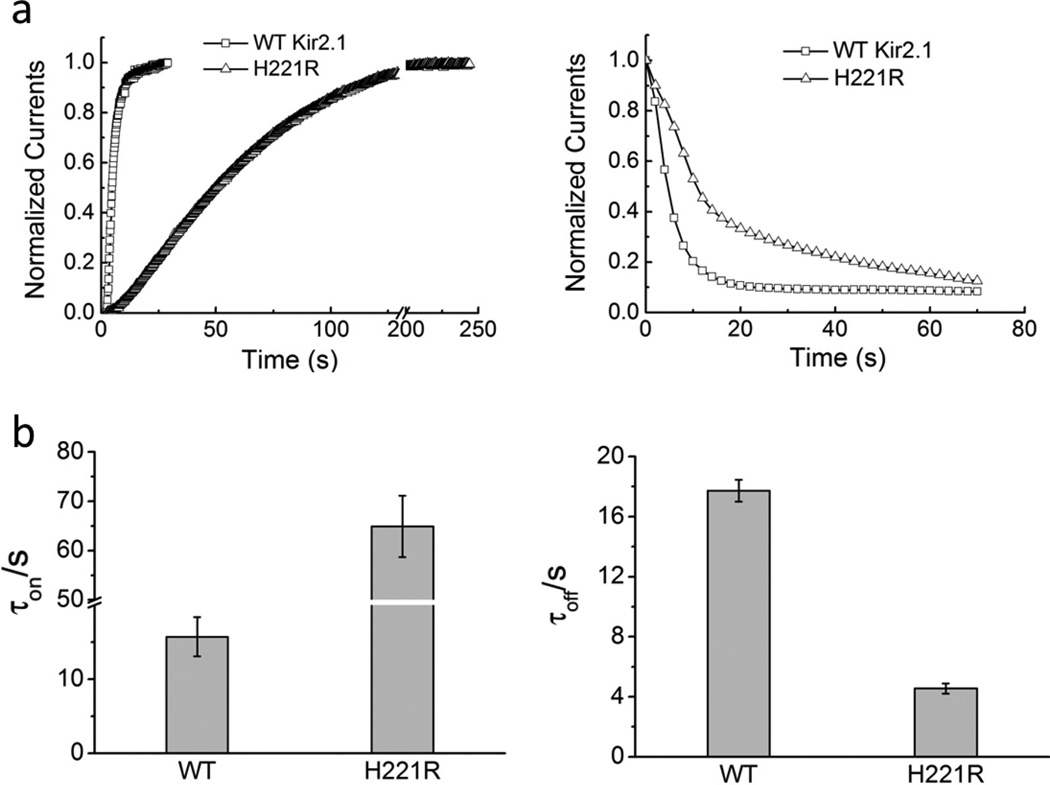
To illustrate how the H221 residue is involved in the gating of Kir2.1, we designed H221R mutation and measured the gating kinetics of the Kir2.1 H221R channel, using the two-electrode voltage clamp (TEVC) technique. The Ciona intestinalis voltage-sensitive phosphatase (Ci-VSP) was used to control the membrane PIP2 levels 58, 59 in intact cells. Ci-VSP co-expression with WT and H221R mutant channel was activated by the voltage protocol, which was able to achieve rapid and reversible PIP2 depletion. PIP2 de-phosphorylation and re-phosphorylation in turn produced the gating kinetics of Kir2 channel during the closing and opening process.
The kinetics of current recovery (τon) and inhibition (τoff) of H221R through inhibition (left panels) or activation (right panels) of Ci-VSP was four-fold effect of WT (τon for H221R: 64.90 ± 6.20s and for WT: 15.73 ± 2.66s; τoff for H221R: 4.55 ± 0.34s and for WT: 17.73 ± 0.72s) (Fig. 4a, b). The data show that the H221R mutant decelerated the gating kinetics of channel reactivation by PIP2 rephosphorylation, and accelerated by inhibition by PIP2 dephosphorylation compared to WT.
Figure 4. H221R mutation impacts the gating kinetics of Kir2.1.
(a) represents the time course of reactivation (PIP2 re-phosphorylation, a 4-fold effect) and inhibition (PIP2 de-phosphorylation induced by Ci-VSP in a 4-fold effect) of WT-Kir2.1 and H221R; (b) Bars in b are the time constants (mean ± SEM of at least six experiments) corresponding to a.
Three pairs of weak interactions that control the G-loop gate
To understand how the H221 residue affects the gating kinetics of the Kir channel, we performed molecular dynamics (MD) and targeted MD simulation on the H221R Kir2.1 mutant. Compared with the WT channel, the H221R mutation strengthened the interaction between the CD- and G-loops (E303-R221), which stabilized the close state of the G-loop gate (Fig. 5a). This weakened the two pairs of weak interactions between the CD-loop and the C-linker (R221-R189) and between the adjacent G-loops (E303-R312), which could not maintain the G-loop gate in the open state (Fig. 5b, c).
Figure 5. The three interactions that controls the G-loop gate.
(a) The time course of the interaction between G-loop and CD-loop through intra-subunit E303-R221. (b) and (c) show the interaction between CD-loop and C-linker through R221-R189 (b) and between adjacent G-loops through E303-R312 (c). The red and blue lines are shown the average of the H-bond number every 40 ps of H221R (red) and WT-Kir2.1 (blue) as a function of simulation time, respectively.
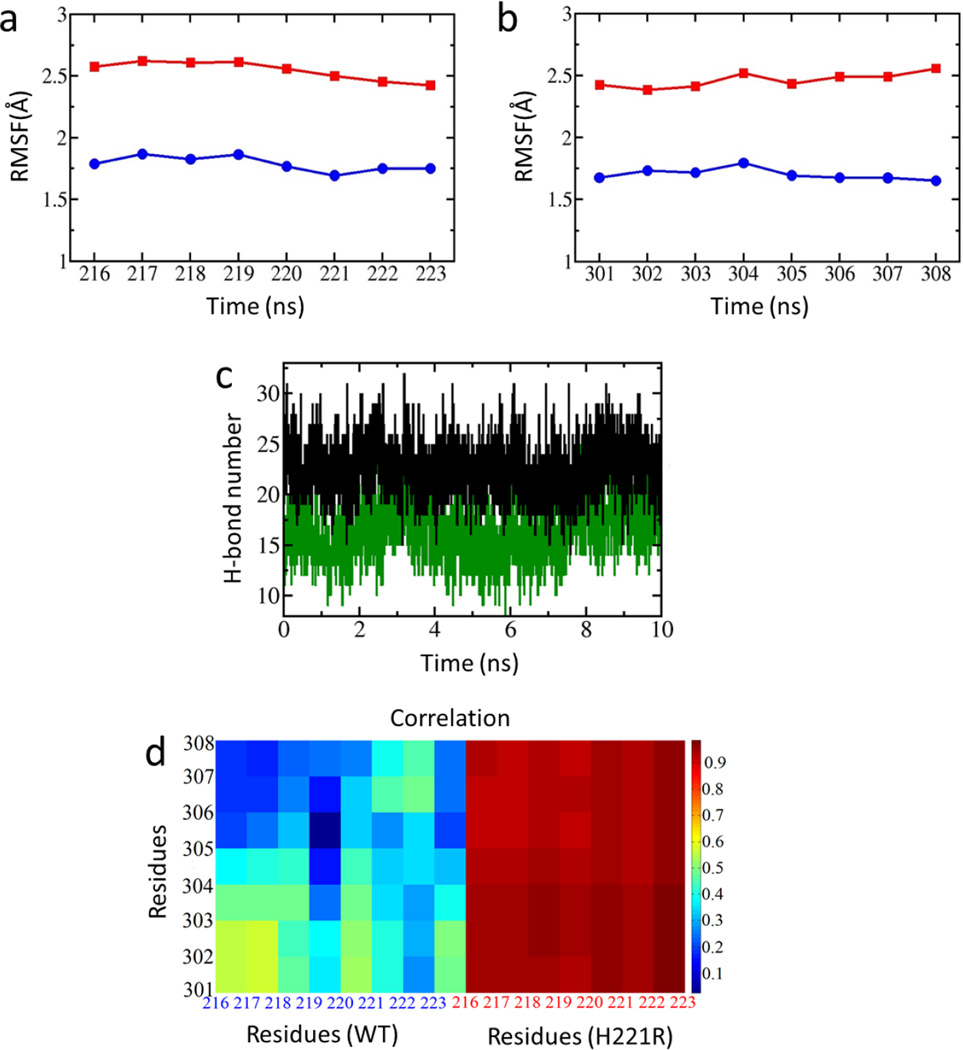
To explore the reason why the mutation at Kir2.1 221 position (H221R) led the weak interactions to change, we analyzed the RMSF (root mean square fluctuation) and relative movements between the CD- and G-loops. The RMSF is a standard of the flexibility of CD- and G-loop for the WT and the H221R mutant. The H221R mutation increased the flexibility of the CD-loop (residues 216 to 223) and G-loop (residues 301 to 308) compared to the WT (Fig. 6a, b). From the free MD simulations the Kir2.1 221 position from His to Arg could increase the interaction and correlation between the CD- and G-loops (Fig. 6c, d), which may cause the enhanced flexibility that could be correlated with its effects on the gating kinetics 60.
Figure 6.
RMSF (root mean square fluctuation) analysis of the CD-loop (residues 216 to 223) (a) and G-loop (residues 301 to 308) (b) of Kir2.1 and H221R. (c) The time course of the interactions between CD-loop and G-loop of H221R (black) and WT-Kir2.1 (green). (d) Correlation of movements among CD-loop and G-loop of WT and H221R.
Phosphatidylinositol 4,5-bisphosphate (PIP2) is known to regulate the activity of all Kir family members, including the Kir2.1 channel, through interactions with the positively charged amino acids, which exposed to the surface of interface between CTD and TMD 16, 56, 61, 62. These binding sites include the residues H53, R67, R82, K182, K185, K187, K188, R189, R218, K219, K228, and R312 (numbers correspond to the Kir2.1 channel). The gating of Kir channels is controlled by the inner weak interactions network of the CTD, when PIP2 is bound at its binding sites 16, 24, 32. Following our previous study, the interactions networks between CD- and G-loop and C-linker play a vital role in the PIP2-induced Kir channels gating 24. One pair of weak interactions (E303-H221) between the CD- and G-loops may be bound up with the gating of the Kir2.1 channel (Fig. 3a). The Kir2.1 E303D mutation disrupted the weak interaction within the CTD and rendered Kir2.1 dysfunctional 33. Three pairs of interactions (E303-H221, H221-R189 and E303-R312) precisely regulated the G-loop gate 24, 33.
CONCLUSIONS
To explore the mechanism by which H221 controlled channel function, we used the combined approaches of computational chemistry and electrophysiology. Experimental data show that H221R was able to affect the gating kinetics of the Kir2.1 channel (accelerating the inhibition kinetics-τoff, and decelerating reactivation kinetics-τon relative to the WT). The simulation data show that the H221R mutation compared to the WT enhanced E303-R221 interaction (between CD- and G-loops), which delayed the closed time of the G-loop gate, and weakened the H221-R189 and E303-R312 interactions (between the C-linker and CD-loop in adjacent G-loops), which decreased the open time of the G-loop gate. These three pairs of interactions control the gating kinetics of Kir2.1 channel. These results can be helpful for targeted drug design of Kir channelopathies.
Acknowledgments
Grant sponsor: Natural Science Fund for Distinguished Young Scholars of Hebei Province of China; Grant numbers: C2015202340 and BJ2014014; Grant sponsor: Fund for Outstanding Talents of Hebei Province of China; Grant number: C201400305; Grant sponsor: Hebei Province Science and Technology Program Self Financing Project; Grant number: 15271704; Grant sponsor: Scientific Innovation Grant for Excellent Young Scientists of Hebei University of Technology; Grant number: 2015010; Grant sponor: Foundation for the Science and Technology Program of Higher Education Institutions of Hebei Province, China: Grant number: QN2016113; Grant sponsor: Natural Science Fund of Hebei Province; Grant number: C2013202244; Grant sponsor: National Natural Science Fund of China; Grant numbers: 11247010, 11475053, 51505123, 11347017, and 31400711. DEL grant number R01HL059949-19; Grant sponsor: National Institutes of Health, Heart Lung and Blood Institute.
Abbreviations
- Kir
Inwardly rectifying K+
- TMD
transmembrane domain
- CTD
cytoplasmic domain
- MD
molecular dynamics
- PDB
Protein Data Bank
- PME
particle-mesh Ewald
- PIP2
Phosphatidylinositol 4,5-bisphosphate
- Targeted MD
Targeted Molecular Dynamics
- TEVC
two-electrode voltage-clamp
- POPC
palmitoyloleoyl-phosphatidylcholine
- RMSD
root mean square deviation
- Ci-VSP
Ciona intestinalis voltage-sensitive phosphatase
- VMD
Visual Molecular Dynamics
Footnotes
Author contributions
JL, YZ and HA designed the project and wrote the main manuscript text. JL and HA did most computation and data analysis and prepared all figures. XX, HZ and CP did most of the computational work and data analysis. SX and SL helped with the preparation of some figures. DEL, SL, HLZ, YZ and HA revised the manuscript. All authors reviewed the manuscript.
Competing financial interests: The authors declare no competing financial interests.
REFERENCES
- 1.Hibino H, Inanobe A, Furutani K, Murakami S, Findlay I, Kurachi Y. Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol Rev. 2010;90:291–366. doi: 10.1152/physrev.00021.2009. [DOI] [PubMed] [Google Scholar]
- 2.Pruss H, Derst C, Lommel R, Veh RW. Differential distribution of individual subunits of strongly inwardly rectifying potassium channels (Kir2 family) in rat brain. Brain Res Mol Brain Res. 2005;139:63–79. doi: 10.1016/j.molbrainres.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Miyashita T, Kubo Y. Localization and developmental changes of the expression of two inward rectifying K(+)-channel proteins in the rat brain. Brain Res. 1997;750:251–263. doi: 10.1016/s0006-8993(96)01365-0. [DOI] [PubMed] [Google Scholar]
- 4.Kubo Y, Baldwin TJ, Jan YN, Jan LY. Primary structure and functional expression of a mouse inward rectifier potassium channel. Nature. 1993;362:127–133. doi: 10.1038/362127a0. [DOI] [PubMed] [Google Scholar]
- 5.Doupnik CA, Davidson N, Lester HA. The inward rectifier potassium channel family. Curr Opin Neurobiol. 1995;5:268–277. doi: 10.1016/0959-4388(95)80038-7. [DOI] [PubMed] [Google Scholar]
- 6.Fakler B, Schultz JH, Yang J, Schulte U, Brandle U, Zenner HP, Jan LY, Ruppersberg JP. Identification of a titratable lysine residue that determines sensitivity of kidney potassium channels (ROMK) to intracellular pH. EMBO J. 1996;15:4093–4099. [PMC free article] [PubMed] [Google Scholar]
- 7.Nichols CG, Lopatin AN. Inward rectifier potassium channels. Annu Rev Physiol. 1997;59:171–191. doi: 10.1146/annurev.physiol.59.1.171. [DOI] [PubMed] [Google Scholar]
- 8.Bichet D, Haass FA, Jan LY. Merging functional studies with structures of inward-rectifier K(+) channels. Nat Rev Neurosci. 2003;4:957–967. doi: 10.1038/nrn1244. [DOI] [PubMed] [Google Scholar]
- 9.Kubo Y, Adelman JP, Clapham DE, Jan LY, Karschin A, Kurachi Y, Lazdunski M, Nichols CG, Seino S, Vandenberg CA. International Union of Pharmacology. LIV. Nomenclature and molecular relationships of inwardly rectifying potassium channels. Pharmacol Rev. 2005;57:509–526. doi: 10.1124/pr.57.4.11. [DOI] [PubMed] [Google Scholar]
- 10.Nishida M, MacKinnon R. Structural basis of inward rectification: cytoplasmic pore of the G protein-gated inward rectifier GIRK1 at 1.8 A resolution. Cell. 2002;111:957–965. doi: 10.1016/s0092-8674(02)01227-8. [DOI] [PubMed] [Google Scholar]
- 11.Kuo A, Gulbis JM, Antcliff JF, Rahman T, Lowe ED, Zimmer J, Cuthbertson J, Ashcroft FM, Ezaki T, Doyle DA. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 2003;300:1922–1926. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- 12.Pegan S, Arrabit C, Zhou W, Kwiatkowski W, Collins A, Slesinger PA, Choe S. Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification. Nat Neurosci. 2005;8:279–287. doi: 10.1038/nn1411. [DOI] [PubMed] [Google Scholar]
- 13.Nishida M, Cadene M, Chait BT, MacKinnon R. Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. EMBO J. 2007;26:4005–4015. doi: 10.1038/sj.emboj.7601828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao X, Avalos JL, Chen J, MacKinnon R. Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science. 2009;326:1668–1674. doi: 10.1126/science.1180310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke OB, Caputo AT, Hill AP, Vandenberg JI, Smith BJ, Gulbis JM. Domain reorientation and rotation of an intracellular assembly regulate conduction in Kir potassium channels. Cell. 2010;141:1018–1029. doi: 10.1016/j.cell.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Hansen SB, Tao X, MacKinnon R. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature. 2011;477:495–498. doi: 10.1038/nature10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whorton MR, MacKinnon R. Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell. 2011;147:199–208. doi: 10.1016/j.cell.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bavro VN, De Zorzi R, Schmidt MR, Muniz JR, Zubcevic L, Sansom MS, Venien-Bryan C, Tucker SJ. Structure of a KirBac potassium channel with an open bundle crossing indicates a mechanism of channel gating. Nat Struct Mol Biol. 2012;19:158–163. doi: 10.1038/nsmb.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whorton MR, MacKinnon R. X-ray structure of the mammalian GIRK2-betagamma G-protein complex. Nature. 2013;498:190–197. doi: 10.1038/nature12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zubcevic L, Bavro VN, Muniz JR, Schmidt MR, Wang S, De Zorzi R, Venien-Bryan C, Sansom MS, Nichols CG, Tucker SJ. Control of KirBac3.1 potassium channel gating at the interface between cytoplasmic domains. J Biol Chem. 2014;289:143–151. doi: 10.1074/jbc.M113.501833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie LH, John SA, Ribalet B, Weiss JN. Activation of inwardly rectifying potassium (Kir) channels by phosphatidylinositol-4,5-bisphosphate (PIP2): interaction with other regulatory ligands. Prog Biophys Mol Biol. 2007;94:320–335. doi: 10.1016/j.pbiomolbio.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Xie X, Liu J, Yu H, Zhang S, Zhan Y, Zhang H, Logothetis DE, An H. Lack of negatively charged residues at the external mouth of kir2.2 channels enable the voltage-dependent block by external mg2+ PLoS One. 2014;9:e111372. doi: 10.1371/journal.pone.0111372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SJ, Wang S, Borschel W, Heyman S, Gyore J, Nichols CG. Secondary anionic phospholipid binding site and gating mechanism in Kir2.1 inward rectifier channels. Nat Commun. 2013;4:2786. doi: 10.1038/ncomms3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Lü S, Liu Y, Pang C, Chen Y, Zhang S, Yu H, Long M, Zhang H, Logothetis DE, Zhan Y, An H. Identification of the Conformational transition pathway in PIP2 Opening Kir Channels. Sci Rep. 2015;5:11289. doi: 10.1038/srep11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swartz KJ. Towards a structural view of gating in potassium channels. Nat Rev Neurosci. 2004;5:905–916. doi: 10.1038/nrn1559. [DOI] [PubMed] [Google Scholar]
- 26.Kuo A, Domene C, Johnson LN, Doyle DA, Venien-Bryan C. Two different conformational states of the KirBac3.1 potassium channel revealed by electron crystallography. Structure. 2005;13:1463–1472. doi: 10.1016/j.str.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Domene C, Doyle DA, Venien-Bryan C. Modeling of an ion channel in its open conformation. Biophys J. 2005;89:L01–L03. doi: 10.1529/biophysj.105.060855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma D, Tang XD, Rogers TB, Welling PA. An andersen-Tawil syndrome mutation in Kir2.1 (V302M) alters the G-loop cytoplasmic K+ conduction pathway. J Biol Chem. 2007;282:5781–5789. doi: 10.1074/jbc.M608776200. [DOI] [PubMed] [Google Scholar]
- 29.Lujan R. Organisation of potassium channels on the neuronal surface. J Chem Neuroanat. 2010;40:1–20. doi: 10.1016/j.jchemneu.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Lopes CM, Zhang H, Rohacs T, Jin T, Yang J, Logothetis DE. Alterations in conserved Kir channel-PIP2 interactions underlie channelopathies. Neuron. 2002;34:933–944. doi: 10.1016/s0896-6273(02)00725-0. [DOI] [PubMed] [Google Scholar]
- 31.Donaldson MR, Jensen JL, Tristani-Firouzi M, Tawil R, Bendahhou S, Suarez WA, Cobo AM, Poza JJ, Behr E, Wagstaff J, Szepetowski P, Pereira S, Mozaffar T, Escolar DM, Fu YH, Ptacek LJ. PIP2 binding residues of Kir2.1 are common targets of mutations causing Andersen syndrome. Neurology. 2003;60:1811–1816. doi: 10.1212/01.wnl.0000072261.14060.47. [DOI] [PubMed] [Google Scholar]
- 32.Xuan-Yu Meng H-XZ. The Molecular Mechanism by which PIP2 Opens the Intracellular G-Loop Gate of a Kir3.1 Channel. Biophys J. 2012;102:2049–2059. doi: 10.1016/j.bpj.2012.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J-W, Xiao S-Y, Xie X-X, Yu H, Zhang H-L, Zhan Y, An H-L. Identification of Three Interactions to Determine the Conformation Change and to Maintain the Function of Kir2.1 Channel Protein. Chin Phys Lett. 2015;32:028702. [Google Scholar]
- 34.Pegan S, Arrabit C, Slesinger PA, Choe S. Andersen's syndrome mutation effects on the structure and assembly of the cytoplasmic domains of Kir2.1. Biochemistry. 2006;45:8599–8606. doi: 10.1021/bi060653d. [DOI] [PubMed] [Google Scholar]
- 35.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 36.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 37.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benkert P, Tosatto SC, Schomburg D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins. 2008;71:261–277. doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- 39.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 41.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926. [Google Scholar]
- 42.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacKerell AD, Bashford D, Bellott M, Dunbrack R, Evanseck J, Field MJ, Fischer S, Gao J, Guo H, Ha Sa. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 44.Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 45.Feller SE, Yin D, Pastor RW, MacKerell AD., Jr Molecular dynamics simulation of unsaturated lipid bilayers at low hydration: parameterization and comparison with diffraction studies. Biophys J. 1997;73:2269. doi: 10.1016/S0006-3495(97)78259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roux B, Bernèche S. On the potential functions used in molecular dynamics simulations of ion channels. Biophys J. 2002;82:1681. doi: 10.1016/S0006-3495(02)75520-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noskov SY, Roux B. Control of ion selectivity in LeuT: two Na+ binding sites with two different mechanisms. J Mol Biol. 2008;377:804–818. doi: 10.1016/j.jmb.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caplan DA, Subbotina JO, Noskov SY. Molecular mechanism of ion-ion and ion-substrate coupling in the Na+-dependent leucine transporter LeuT. Biophys J. 2008;95:4613–4621. doi: 10.1529/biophysj.108.139741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. The Journal of Chemical Physics. 1994;101:4177. [Google Scholar]
- 50.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: the Langevin piston method. The Journal of Chemical Physics. 1995;103:4613. [Google Scholar]
- 51.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. Journal of Chemical Physics. 1995;103:8577–8593. [Google Scholar]
- 52.Darden T, York D, Pedersen L. Particle mesh Ewald: An N· log (N) method for Ewald sums in large systems. The Journal of chemical physics. 1993;98:10089. [Google Scholar]
- 53.Schlitter J, Engels M, Kruger P. Targeted molecular dynamics: a new approach for searching pathways of conformational transitions. J Mol Graph. 1994;12:84–89. doi: 10.1016/0263-7855(94)80072-3. [DOI] [PubMed] [Google Scholar]
- 54.Schlitter J, Engels M, Krüger P, Jacoby E, Wollmer A. Targeted molecular dynamics simulation of conformational change-application to the T↔ R transition in insulin. Molecular Simulation. 1993;10:291–308. [Google Scholar]
- 55.Salo-Ahen OM, Wade RC. The active-inactive transition of human thymidylate synthase: targeted molecular dynamics simulations. Proteins. 2011;79:2886–2899. doi: 10.1002/prot.23123. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H, He C, Yan X, Mirshahi T, Logothetis DE. Activation of inwardly rectifying K+ channels by distinct PtdIns(4,5)P2 interactions. Nat Cell Biol. 1999;1:183–188. doi: 10.1038/11103. [DOI] [PubMed] [Google Scholar]
- 57.Rodriguez-Menchaca AA, Adney SK, Tang QY, Meng XY, Rosenhouse-Dantsker A, Cui M, Logothetis DE. PIP2 controls voltage-sensor movement and pore opening of Kv channels through the S4-S5 linker. Proc Natl Acad Sci U S A. 2012;109:E2399–E2408. doi: 10.1073/pnas.1207901109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horn R. Electrifying Phosphatases. Sci STKE. 2005;307:50. doi: 10.1126/stke.3072005pe50. [DOI] [PubMed] [Google Scholar]
- 59.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 60.An HL, Lu SQ, Li JW, Meng XY, Zhan Y, Cui M, Long M, Zhang HL, Logothetis DE. The cytosolic GH loop regulates the phosphatidylinositol 4,5-bisphosphate-induced gating kinetics of Kir2 channels. J Biol Chem. 2012;287:42278–42287. doi: 10.1074/jbc.M112.418640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang C-L, Feng S, Hilgemann DW. Direct activation of inward rectifier potassium channels by PIP2 and its stabilization by Gβγ. Nature. 1998;391:803–806. doi: 10.1038/35882. [DOI] [PubMed] [Google Scholar]
- 62.Suh BC, Hille B. Regulation of ion channels by phosphatidylinositol 4,5-bisphosphate. Curr Opin Neurobiol. 2005;15:370–378. doi: 10.1016/j.conb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 63.Davies NP, Imbrici P, Fialho D, Herd C, Bilsland LG, Weber A, Mueller R, Hilton-Jones D, Ealing J, Boothman BR, Giunti P, Parsons LM, Thomas M, Manzur AY, Jurkat-Rott K, Lehmann-Horn F, Chinnery PF, Rose M, Kullmann DM, Hanna MG. Andersen-Tawil syndrome: new potassium channel mutations and possible phenotypic variation. Neurology. 2005;65:1083–1089. doi: 10.1212/01.wnl.0000178888.03767.74. [DOI] [PubMed] [Google Scholar]
- 64.Sansone V, Tawil R. Management and treatment of Andersen-Tawil syndrome (ATS) Neurotherapeutics. 2007;4:233–237. doi: 10.1016/j.nurt.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 65.Zitron E, Kiesecker C, Luck S, Kathofer S, Thomas D, Kreye VA, Kiehn J, Katus HA, Schoels W, Karle CA. Human cardiac inwardly rectifying current IKir2.2 is upregulated by activation of protein kinase A. Cardiovasc Res. 2004;63:520–527. doi: 10.1016/j.cardiores.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 66.Soom M, Schonherr R, Kubo Y, Kirsch C, Klinger R, Heinemann SH. Multiple PIP2 binding sites in Kir2.1 inwardly rectifying potassium channels. FEBS Lett. 2001;490:49–53. doi: 10.1016/s0014-5793(01)02136-6. [DOI] [PubMed] [Google Scholar]