Abstract
Ambient ionization mass spectrometry imaging (MSI) has been increasingly used to investigate the molecular distribution of biological tissue samples. Here, we report the integration and optimization of desorption electrospray ionization (DESI) and liquid-microjunction surface sampling probe (LMJ-SSP) with a chip-based high-field asymmetric waveform ion mobility spectrometry (FAIMS) device to image metabolites, lipids, and proteins in biological tissue samples. Optimized FAIMS parameters for specific molecular classes enabled semitargeted detection of multiply charged molecular species at enhanced signal-to-noise ratios (S/N), improved visualization of spatial distributions, and, most importantly, allowed detection of species which were unseen by ambient ionization MSI alone. Under static DESI-FAIMS conditions selected for transmission of doubly charged cardiolipins (CL), for example, detection of 71 different CL species was achieved in rat brain, 23 of which were not observed by DESI alone. Diagnostic CL were imaged in a human thyroid tumor sample with reduced interference of isobaric species. LMJ-SSP-FAIMS enabled detection of 84 multiply charged protein ions in rat brain tissue, 66 of which were exclusive to this approach. Spatial visualization of proteins in substructures of rat brain, and in human ovarian cancerous, necrotic, and normal tissues was achieved. Our results indicate that integration of FAIMS with ambient ionization MS allows improved detection and imaging of selected molecular species. We show that this methodology is valuable in biomedical applications of MSI for detection of multiply charged lipids and proteins from biological tissues.
Graphical abstract
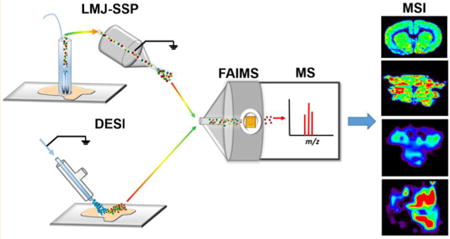
Mass spectrometry imaging (MSI) provides the outstanding ability of probing the spatial distribution of molecules in a sample surface with high specificity and sensitivity.1–3 In particular, ambient ionization MSI techniques have revolutionized the means by which spatial and molecular information is obtained from biological samples by enabling in situ, real time analysis of tissue samples with minimal pretreatment.4,5 Desorption electrospray ionization (DESI) is the most commonly used solvent-based ambient ionization MS technique to image and characterize lipids and metabolites in biological tissue samples.6,7 DESI has been increasingly applied for cancer diagnosis with the perspective of clinical use.8 In addition to DESI, other solvent-based ambient ionization techniques have been used to analyze biological tissue samples, including liquid microjunction surface sampling probe (LMJ-SSP)11 and nanoDESI.9,10
Despite providing a wealth of chemical and spatial information, inherent challenges of sample complexity using direct analysis by MSI have prevented comprehensive detection and characterization of molecular species.11,12 Naturally occurring lipids, for example, present enormous diversity of molecular structures and are observed over a relatively narrow mass-to-charge (m/z) range as molecular ions. Isobaric interferences in the mass spectrum complicate tandem MS analysis for structural characterization and obscures visualization of an ion's spatial distribution,13 an issue that has been previously addressed through MS/MS imaging.14 Lipids present in high concentration in biological tissues may also suppress the detection of other lipids at lower abundances.15 Protein imaging directly from biological samples is also an ongoing analytical challenge for ambient ionization techniques, although recent progress has been made.16 New approaches that integrate several analytical strategies are needed for improved imaging and characterization of molecules within biological tissue samples.17,18
Ion mobility has been increasingly applied to overcome issues in complex sample analysis by MS.19–21 In particular, high-field asymmetric waveform ion mobility spectrometry (FAIMS), or differential mobility separation, separates gas phase ions at atmospheric pressure on the basis of differences in their mobilities in electric fields prior to MS analysis.22 A high-frequency, asymmetric waveform is employed by alternating low and high electric fields perpendicular to the path through which the ions travel.23 This waveform, called the dispersion field (DF), causes the ions to be displaced from their initial trajectories, collide with the electrode walls, and be dissipated. However, a second, smaller DC voltage can be applied between the two electrode plates, creating a low-energy electric field that is superimposed on the DF. This compensation field (CF) counteracts the ion drift caused by the DF, allowing ions with specific mobilities to be transmitted for mass analysis. The highspeed gas phase separation provided by FAIMS has been shown to reduce chemical noise and to improve signal-to-noise ratios (S/N), sensitivity, and dynamic range.22 By optimizing the DF and CF, semiselective transmission of subsets of ions or classes of molecules can be achieved. The filtering capability of FAIMS allows only subsets of ions to enter and fill the mass analyzer, resulting in higher sensitivity in their detection. Several groups have explored the use of FAIMS, as well as other ion mobility techniques, for MSI applications.24–28 A lab-built DMS cell integrated with a DESI platform was used to image glycerophosphocholines (PC) in a mouse brain tissue, resulting in decreased chemical noise, as well as improved S/N and image contrast.29 More recently, liquid extraction surface analysis (LESA) was integrated with the FAIMS device we applied in our study to image proteins in mouse brain and liver tissue samples.30,31 Using the integrated LESA-FAIMS approach, 26 protein species in mouse brain and 29 protein species in mouse liver undetectable through LESA alone were observed.31 Here, we report an analytical approach coupling DESI or LMJ-SSP with FAIMS to image and characterize lipids and proteins from biological tissue sections.
In our study, we integrated and optimized a chip-based, highspeed ultraFAIMS device with DESI-MS/LMJ-SSP-MS and a high mass resolution mass spectrometer to image and characterize singly charged metabolites, singly- and doubly charged glycerophospholipids (GP) and glycosphingolipids, and multiply charged proteins in rat brain, human thyroid, and human ovarian cancer tissues. 2D-FAIMS sweep experiments were performed in tandem with DESI-MSI using a spot-by-spot approach to explore separation voltages for specific molecular classes of lipids and metabolites in a rat brain tissue section. Static, optimized FAIMS conditions were then used for doubly charged CL and gangliosides with no increase in analysis time. Multiply charged protein ions were imaged in biological tissues using LMJ-SSP-FAIMS with enhanced analytical performance. Using this approach, we show the first example of global protein imaging in human cancerous tissue by ambient ionization MSI. Our results indicate that integration of FAIMS with DESI or LMJ-SSP is valuable for imaging selected molecular classes in biological tissues.
Experimental Section
Chemicals
18:1 glycerophosphoinositol (PI), 18:1 glycerophosphoserine (PS), 18:1 glycerophosphoethanolamine (PE), 18:1 glycerophosphoglycerol (PG), 18:1 monoacylglycerophosphate (PA), 18:1 cardiolipin (CL), and a total ganglioside extract were purchased from Avanti Polar Lipids (Alabaster, AL). Fatty acids (FA) including oleic acid, lignoceric acid, and arachidonic acid, as well as metabolites n-acetylaspartic acid and 2-hydroxyglutarate were purchased from Sigma-Aldrich (St. Louis, MO). Standards were dissolved in chloroform/methanol (CHCl3/MeOH) 1:1 (v/v) at a concentration of 10 μg/mL. A ubiquitin standard (Sigma-Aldrich, St. Louis, MO) was dissolved in acetonitrile/water (ACN/H2O) 60:40 (v/v) with 0.1% formic acid at 1 μg/mL. Direct electrospray ionization (ESI) infusion of the standards was performed at a flow rate of 5 μL/min.
Tissue Samples
Rat brain samples were obtained from BioreclamationIVT (Hicksville, NY). Banked frozen human tissue samples including human thyroid oncocytic tumor, ovarian cancer, and normal ovarian tissues were obtained from Cooperative Human Tissue Network (CHTN) under approved IRB protocol. Samples were stored in a −80 °C freezer until sectioned. Tissue samples were sectioned at a thickness of 16 μm using a CryoStar NX50 cryostat (Thermo Scientific, San Jose, CA). After sectioning, the glass slides were stored in a −80 °C freezer. Prior to MSI, the glass slides were dried for ∼15 min. For protein analysis, sections were washed in ethanol for 10 s, followed by a wash in chloroform for 10 s to remove excess lipids.
FAIMS
An ultraFAIMS device (Owlstone Ltd., Cambridge, UK) using an ND chip (Owlstone), was used for all experiments performed. The ND chip has a trench length of 97.0 μm, a gap width of 101.51 μm, and a chip thickness of 700 μm. The chip-region temperature was set to 90 °C. At this temperature, the DF can be set to any value from 0 to 280.98 Townsends (Td) and the CF from −14.84 to 14.84 Td.
DESI-MSI
A 2D Omni Spray (Prosolia Inc., Indianapolis, IN) coupled to a Q Exactive mass spectrometer (Thermo Scientific, San Jose, CA) was used for tissue profiling and imaging. An unheated extended transfer tube of 5 in. with an inner diameter of 0.067 in. was lab-built to fit the FAIMS platform. DESI-MSI was performed in the negative ion mode from m/z 100–1500, using the Q Exactive mass spectrometer which allows for high mass accuracy (<5 ppm mass error) and high mass resolution (70 000 resolving power at m/z 200) measurements. The spatial resolution of the imaging experiments was 200 μm. The histologically compatible solvent system dimethylformamide/acetonitrile (DMF/ACN) 1:1 (v/v) was used for analysis at a flow rate of 1.2 μL/min with 5 kV applied to the solvent.32 The N2 pressure was set to 180 psi. The capillary temperature of the mass spectrometer was set to 300 °C.
LMJ-SSP MSI
A LMJ-SSP system, the Flowprobe (Prosolia Inc. Indianapolis, IN), coupled to a Q Exactive mass spectrometer (Thermo Scientific, San Jose, CA) was used for tissue profiling and imaging. LMJ-SSP-MSI was performed in the positive ion mode from m/z 250–2000, using the hybrid Quadrupole-Orbitrap mass spectrometer. The spatial resolution of the imaging experiments was ∼630 μm. The solvent system of acetonitrile/water (ACN/H2O) 60:40 (v/v) with 0.1% formic acid was used for analysis at a flow rate of 40 μL/min, as suggested by the manufacturer (10–100 μL/min). The N2 pressure was between 30 and 70 psi and was adjusted manually when necessary to maintain the liquid microjunction throughout experiments. Leaking of the solvent from the probe is avoided through alterations in pressure applied to the ESI source but can occur during operation. The voltage applied to the solvent within the ESI nozzle was 4 kV, and the capillary temperature of the mass spectrometer was 300 °C.
Tissue Staining
The same tissue sections analyzed by DESI-MSI and adjacent sections analyzed by LMJ-SSP-MSI were stained using standard H&E staining protocol. Pathologic evaluation was performed using light microscopy.
Lipid and Protein Identification
Lipid species were identified using high mass accuracy measurements and higher-energy collision induced dissociation (HCD) tandem MS analysis, performed on the Q Exactive at 70 000 resolving power (m/z 200). Lipid fragmentation patterns were compared to literature reports and used in conjunction with data from Lipidmaps database (www.lipidmaps.org) for identification. For protein identification, mass spectra from sequential profiling experiments with and without the FAIMS separation were obtained. The data was deconvoluted using the Xtract function within Xcalibur software using a S/N threshold of 3 to obtain monoisotopic masses of protein species detected during experiments. Protein species were analyzed by top-down collision induced dissociation (CID) tandem MS, performed on a hybrid LTQ-Orbitrap Elite mass spectrometer (Thermo Scientific, San Jose, CA) at 120 000 resolving power at m/z 200. Protein fragmentation patterns were input into ProSight Lite software (http://prosightlite.northwestern.edu/) and compared with protein amino acid sequences that have been previously observed within the tissue types analyzed. Protein amino acid sequences were obtained from the UniProt database.
2D Imaging Data Analysis
Xcalibur RAW files were converted into images using FireFly data conversion software (Prosolia, Inc. Indianapolis, IN) and then uploaded into the open source imaging software packages BioMap (Novartis) or MSiReader33 for visualization. The interpolate function within BioMap was used to smooth the pixels within the images. All images are normalized to the maximum ion intensity within all the spectra (all pixels) used to create the image.
Results and Discussion
Optimization of FAIMS for Separation of Lipids and Metabolites
To evaluate FAIMS separation voltages for lipids and metabolites, a synthetic mixture of standards was prepared and analyzed using ESI and DESI in the negative ion mode. The mixture of lipids contained species from the main classes of GP commonly detected from biological tissues in the negative ion mode including singly charged PI, PS, PE, PG, and PA, doubly charged CL and gangliosides, three common FA (oleic, lignoceric, and arachidonic), and two representative metabolites, n-acetylaspartic acid (NAA) and 2-hydroxyglutarate. For ESI experiments, the synthetic mixture was directly infused while a 2D FAIMS field sweep was performed in which the DF was stepped in 10 Td increments from 150 to 280 Td with a CV sweep from −1 to +4 Td occurring at every DF value. Increasing DF causes an increase in separation efficiency while decreasing total ion current. Thus, an optimal DF was chosen as the field at which lipids and metabolites were most clearly separated in the ion chromatogram without detrimental loss of total ion current. At this selected DF value, an optimal CF for each species was chosen as the voltage at which the specific ion had a maximum total intensity.
ESI experiments yielded an optimal DF of 220 Td for the highest transmittance and separation of lipids and small metabolites by FAIMS. At this DF, optimal CFs were determined for small metabolites (CF = −0.10 Td), singly charged GP (CF = 0.99 Td), singly charged FA (CF = 1.17 Td), doubly charged gangliosides (CF = 1.80 Td), doubly charged CL (CF = 2.20 Td), and triply charged gangliosides (CF = 2.71 Td; Figure S1). A trend in optimized CF values was observed, in which higher CF values were ideal for transmission of larger, multiply charged ions and lower CF values were more suitable for the transmission of smaller, singly charged ions. Note that while different optimized CF values allowed separation of doubly charged CL and gangliosides from singly charged GP species, separation of different subclasses of singly charged GP was not clearly observed. DESI-FAIMS optimization experiments performed on the synthetic lipid and metabolite mixture yielded similar optimized CF values for the species of interest at DF of 220 Td. For both ESI and DESI, a drop in total ion current of 1 order of magnitude or more was observed with FAIMS separation for all experiments performed. Planar FAIMS devices are known to cause an exponential decrease in ion transmission with increasing residence time within the device in order to achieve higher separation resolving power.34 Thus, despite the short residence time associated with this chip-based device, this method is not immune to this decrease in total ion transmission. Nevertheless, further decrease of noise leads to an overall increase in the S/N for selected molecular ions.34
Optimization of the DESI-FAIMS system was also performed for imaging lipids and metabolites from a tissue sample in the negative ion mode. A spot-by-spot DESI analysis of a rat brain tissue section was performed using a methodology similar to the one employed for ESI. At a spatial resolution (step size) of 200 μm, the DF was kept constant at the optimized 220 Td while a 1D FAIMS sweep (CF= −1 to 3 Td) was performed (Figure 1A). A total of eight mass spectra, each corresponding to a 0.5 Td CF increment, were acquired at each spot. Using this approach, a total of 6 s of analysis time was required for each spot (pixel). The spectra for each field were extracted and compiled into 2D ion images, yielding eight sets of 2D DESI-MS images at specific DF and CF values that highlight different molecular classes transmitted at the optimized voltages (Figure 1B). Although time-consuming, this DESI-FAIMS-MSI experiment allowed optimization of separation voltages for metabolites, FA, singly charged GP, doubly charged CL, and gangliosides, directly from a tissue sample. For example, preferential transmission and highest relative abundances of metabolites were observed at CF = 0–0.5 Td. At these parameters, clear spatial distribution of NAA, recently described as a glioma oncometabolite,35 was observed mainly in the brain cortex. In contrast, doubly charged species (CL and gangliosides), which are difficult to image within complex biological tissue samples due to their low abundances, are nearly exclusively observed at CF= 2–2.5 Td. We note that a slight drop in signal intensity was observed with analysis time in each spot (Figure S2), which could account for lower image quality for the species observed at higher CF values. At CF range 2.5–3 Td, little to no ion transmission through the FAIMS device was observed. The same optimal ion transmission voltages were obtained in replicate profiling experiments for the lipid classes investigated.
Figure 1.
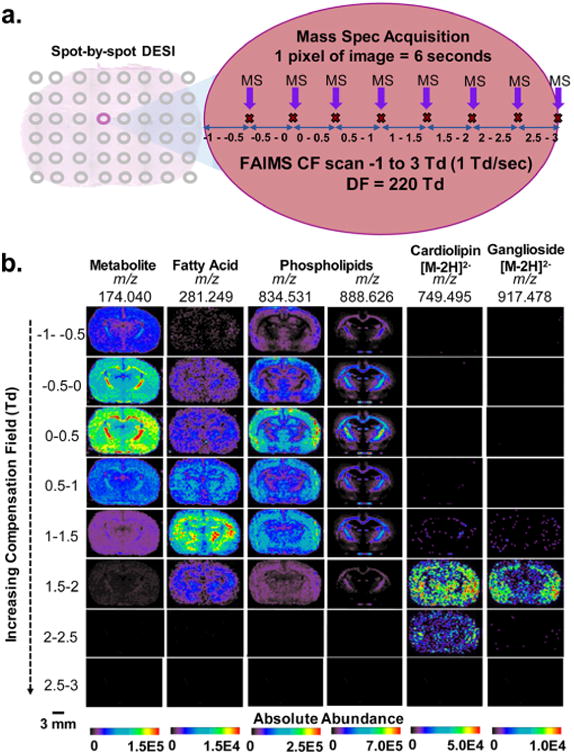
2D-FAIMS, spot-by-spot DESI-MS imaging of a rat brain tissue section. (a) Schematic of the 2D FAIMS sweep experiment used to create multiple sets of DESI-MS ion images at varied CF values in one tissue section. Each 200 μm spot on the tissue was analyzed for 6 s for a total of 8 MS acquisitions, in which a FAIMS sweep occurred, allowing MS acquisition correlated to different CF ranges. (b) DESI-FAIMS-MS ion images for a rat brain tissue section for 6 representative lipid and metabolite species, each acquired at DF = 220 Td and a different CF range, thus highlighting the increased transmission of different molecular species at each CF value. Small metabolites transmit at the lowest CF (–1 to +0.5 Td), followed by FA and singly charged complex lipids at midrange CF (0.5–1.5 Td), and doubly charged CL and gangliosides at the high-range CF (1.5– 2.5 Td). The images for each ion (vertical columns) are normalized to the highest ion intensity of that ion, and not across all ions. Scale bar = 3 mm.
DESI-FAIMS Semiselective Imaging of Cardiolipins and Gangliosides in Rat Brain Sections
We further explored the DESI-FAIMS methodology for optimized imaging of doubly charged CL and gangliosides from biological tissue samples. CL are a complex class of GP that exist almost exclusively within the inner mitochondrial membrane of cells. These unique lipids are important structural and functional components of both normal and diseased tissues, and have been increasingly explored in a variety of pathologies.36–38 We first performed DESI-FAIMS-MSI of doubly charged CL within rat brain tissue sections at the optimized FAIMS parameters in the negative ion mode. In this static FAIMS approach, both the DF and CF remain constant throughout the entire tissue imaging experiment at the optimized settings (DF = 220 Td, CF = 2.20 Td). Thus, DESI-MSI was operated in the usual rastering mode (0.5 s/pixel), without increase of analysis time, and with a spatial resolution of 200 μm. An evident increase in the S/N of CL species in comparison to other GP species was observed in the mass spectra obtained using DESI-FAIMS-MSI when compared to DESI-MSI alone (Figure 2). For example, using DESI-FAIMS-MSI, an increase in the S/N from 116 to 203 for an average of 50 spectra was observed for the most abundant CL species (m/z 749.495, CL (20:4/20:4/30:4/18:1), mass error of –0.93 ppm), with minimal change in signal intensity (from 1.78 × 104 to 1.98 × 104) resulting in images that highlight the CL species over other GP within the tissue sample (Figures 2c and S4). Overall, about 50% increase in the S/N was observed for all CL species detected with the DESI-FAIMS-MSI approach. Most importantly, an increase in the number of detectable CL species was observed with the addition of FAIMS. For example, m/z 797.494, identified as CL(84:18) with a mass error of 1.00 ppm, was only observed by DESI-FAIMS-MSI and undetected with DESI-MSI alone. In total, 71 CL species were detected and identified using DESI-FAIMS-MSI, compared to 48 CL species detected when using DESI-MSI alone, as summarized in Table S1. Acyl chain composition, but not their positions, was tentatively assigned based on tandem MS patterns. When tandem MS fragmentation patterns for low abundance CL were unclear, tentative identification was based on accurate mass measurements alone.
Figure 2.
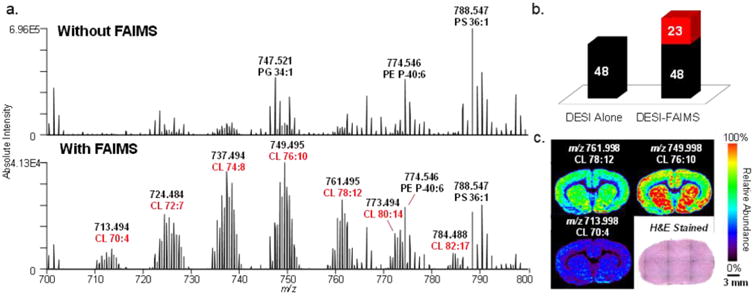
Static DESI-FAIMS-MSI of CL in rat brain tissue. (a) Representative negative ion mode DESI mass spectra acquired without and with FAIMS in the m/z 700–800 range at the optimized doubly charged CL parameters (DF = 220 Td, CF = 2.20 Td), showing a clear increase in the relative abundance of CL species. Without FAIMS and with FAIMS spectra are averages of one DESI line scan, consisting of 68 and 66 scans, respectively. (b) Chart of CL species detected with DESI alone and the DESI-FAIMS integrated approach. (c) 2D DESI-FAIMS ion images for selected CL species (spatial resolution of 200 μm). Voxel versions of the same ion images are shown in Figure S15.
Imaging of doubly charged gangliosides was performed using serial rat brain tissue sections at optimized FAIMS parameters (DF = 220 Td, CF = 1.8 Td) in the negative ion mode. Gangliosides are a subclass of glycosphingolipids which are most abundant within the nervous system. Gangliosides have important cellular functions, such as cell signaling and calcium homeostasis, and have been associated with multiple diseases including Alzheimer's and Tay-Sachs diseases.39,40 DESI-FAIMS-MSI yielded an increased relative abundance and S/N of gangliosides compared to DESI-MSI alone. For example, the S/N of the most abundant ganglioside species detected, identified as trisialotetrahexosylganglioside at m/z 1063.530 (GT1 d36:1), doubled using FAIMS. Out of the 21 doubly charged ganglioside species detected by DESI-FAIMS-MSI, 7 were undetectable by traditional DESI-MSI (Table S2). Gangliosides present at low relative abundances were tentatively identified by accurate mass alone. Note that while FAIMS allowed improved detection of triply charged gangliosides at CF = 2.71 Td using a standard lipid mixture, this charge state was not observed in biological tissues by DESI-FAIMS or DESI alone due to low abundance of gangliosides at this charge state under our experimental conditions.
DESI-FAIMS-MSI of Cardiolipins in Human Oncocytic Thyroid Tumors
We have recently described a diverse group of CL as molecular markers of oncocytic thyroid tumors, including oxidized CL, adducts of CL and PC, and monolysocardiolipins.41 However, many of the diagnostic CL are detected at low relative abundances and within close m/z values to other interfering GP, which complicates ion isolation for tandem MS experiments, obscures visualization of their 2D distribution, and adds complexity to data analysis. Thus, we applied the integrated DESI-FAIMS-MSI approach to improve imaging of CL in oncocytic thyroid tumors. Two serial sections from an oncocytic thyroid tumor were subjected to DESI-MSI under identical conditions in the negative ion mode, one with static mode FAIMS at the optimized CL voltages, and one without FAIMS separation. As shown in Figure 3a, DESI-FAIMS-MSI allowed enhanced detection of CLs within the oncocytic thyroid tumor tissue section with reduced interference from other GP in the m/z 700–800 range. For example, using DESI-MSI alone, PE(O-38:5) at m/z 750.545 had a signal intensity 4 times that of the doubly charged CL(76:9) at m/z 750.502. When FAIMS is applied, CL(76:9) had a signal intensity double that of PE(O-38:5), which facilitates ion isolation, fragmentation, identification, and visualization of its 2D distribution. DESI-FAIMS-MSI revealed high relative abundance of CL within the entire thyroid tissue section, as shown in Figure 3b for m/z 723.479, m/z 737.494, and m/z 747.473 identified as CL(18:2/18:2/18:2/18:2 and/or 20:4/18:2/18:2/16:0), CL(20:4/18:2/18:1/18:1, 20:3/18:2/18:2/18:1, 20:2/18:2/18:2/18:2) and CL(22:6/20:4/18:2/16:0), respectively. These results indicate the presence of oncocytic thyroid tumor in the entire tissue section, which was confirmed by histopathologic evaluation of the same H&E stained tissue section.
Figure 3.
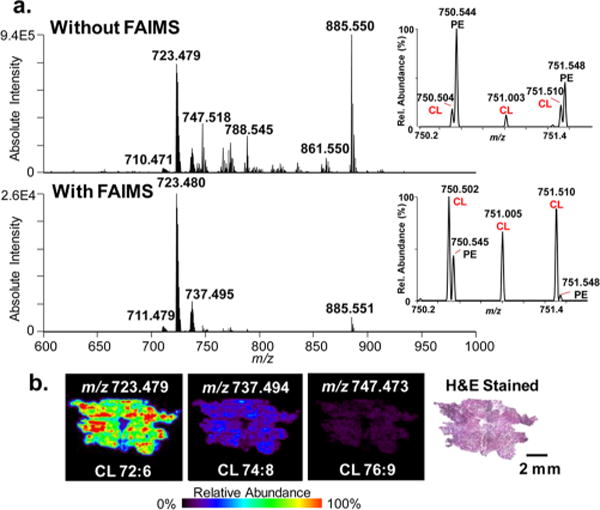
Static DESI-FAIMS-MSI of CLs in an oncocytic thyroid tumor tissue. (a) Representative negative ion mode DESI mass spectra acquired without and with FAIMS in the complex lipid region (m/z 700–900) at the optimized doubly charged CL parameters (DF = 220 Td, CF = 2.20 Td). Zoom in show spectra from m/z 750–752. Without FAIMS and with FAIMS spectra are averages of a DESI line scan, consisting of 57 and 54 scans, respectively. (b) DESI-FAIMS-MS ion images and optical images of the H&E stained oncocytic thyroid tumor sections for three CL species, m/z 723.479, m/z 737.494, and m/z 747.473, showing high relative intensities of CL, a feature of oncocytic tumors (spatial resolution of 200 μm). Scale bar = 2 mm.
LMJ-SSP-FAIMS-MSI of Proteins in Rat Brain Tissue Section
We next pursued the integration of LMJ-SSP and FAIMS for biological tissue imaging. LMJ-SSP provides similar molecular information as DESI from biological tissues with higher sensitivity yet lower spatial resolution and image quality, as limited by the probe tip diameter (∼630 μm).42 Metabolite and lipid optimization experiments performed using LMJ-SSP-FAIMS in the negative ion mode yielded similar results to those obtained by DESI-FAIMS. Yet, we found that positive ion mode LMJ-SSP-FAIMS allowed obvious improvements for imaging multiply charged protein species from biological tissue samples. Thus, we focused our efforts on evaluating and optimizing LMJ-SSP-FAIMS for protein imaging. Voltage optimization was performed for midmass range proteins (4–12 kDa) through LMJ-SSP-FAIMS profiling of a rat brain tissue section while conducting a 2D-FAIMS sweep from DF 230–280 Td and CF 0–4 Td. Ubiquitin, a well-known protein with a molecular weight of 8.5 kDa, was used for optimization. The highest abundance of its 10+ charge state at m/z 857.467 was found at DF = 220 Td and CF = 2.55 Td, and thus all static FAIMS experiments for protein imaging were performed at these settings.
LMJ-SSP profiling experiments of rat brain tissue sections were performed with and without FAIMS separation in the positive ion mode under identical experimental conditions. A clear increase in the relative abundances of multiply charged protein peaks was observed in the mass spectra when FAIMS was applied at optimized voltages for protein transmission (Figure 4a, from m/z 600–1300), despite a 1 order magnitude drop in total ion current. In total, 84 species attributed to proteins and/or proteoforms were detected using LMJ-SSP-FAIMS, 66 of which were not detected by LMJ-SSP alone. For example, the protein isotope pattern for the 9+ ion at m/z 821.218 was clearly resolved with a S/N of 28 using LMJ-SSP-FAIMS, while undetectable above a S/N threshold of 3 with LMJ-SSP alone (Figure 4b). Using LMJ-SSP alone, 67 proteins were detected, 49 of which were not detected by LMJ-SSP-FAIMS. This effect is attributed to lower transmission of proteins with molecular weight outside of the optimized FAIMS parameters (4–12 kDa), as well as to the loss of total signal intensity associated with FAIMS.34 Out of the six protein species detected by both approaches, four were observed at a slightly higher absolute signal intensity using FAIMS, while enhancement of S/N was observed for all six of the proteins. For example, a 7-fold increase in S/N (from 23 to 180) was observed for the 10+ ion cluster at m/z 857.468 when FAIMS was applied (Figure 4c). A list of the deconvoluted molecular weights of all protein species detected using LMJ-SSP-FAIMS and LMJ-SSP alone is shown in Figure S6.
Figure 4.
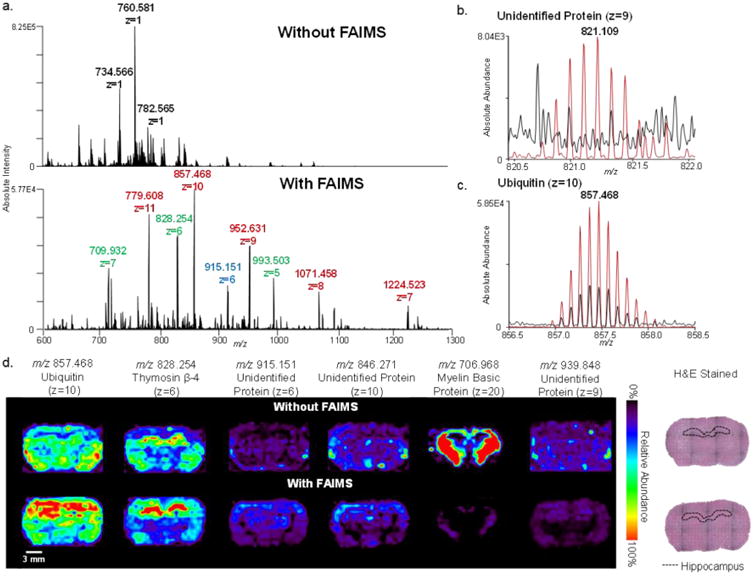
Static LMJ-SSP-FAIMS-MS profiling and imaging of rat brain tissue for protein detection. (a) LMJ-SSP-MS spectra of a rat brain tissue section, analyzed without FAIMS and with FAIMS optimized for transmission of midmass range proteins (4–12 kDa). Different colored labels represent different charge states of same proteins. (b) Isotope patterns of ubiquitin (z = 10), and (c) an unidentified protein (z = 9), with (red) and without (black) FAIMS applied, showing absolute signal intensity and S/N enhancements with the addition of FAIMS. Without FAIMS and with FAIMS mass spectra are averages of a line scan, consisting of 75 scans each. (d) LMJ-SSP-MS and LMJ-SSP-FAIMS-MS ion images of rat brain tissue for six representative protein species including ubiquitin, thymosin β-4, MBP, and three unidentified proteins (spatial resolution ∼630 μm). Voxel versions of the same ion images are shown in Figure S15. Scale bar = 3 mm.
In an effort to identify detected proteins, top-down CID tandem MS was performed on protein ions during rat brain tissue LMJ-SSP analysis. The 9+ charge state at m/z 857.370 (MW = 8559.61 Da) was identified as ubiquitin with a P-score of 8.9 × 10−53 (32% sequence coverage, Figure S7, Table S3). The 6+ ions at m/z 828.256 (MW = 4960.48 Da) were identified as thymosin β-4 with a P-score of 2.5 × 10−11 (33% sequence coverage, Figure S8, Table S4). Myelin basic protein (MBP) was tentatively attributed to the 20+ ion cluster at m/z 706.968 (MW = 14113.23 Da) with a P-score of 3.1 × 10−6 (21% sequence coverage). The cluster of 18+ ions at m/z 845.271 (MW = 15187.75 Da) was identified as hemoglobin α-1/2 with a P-score of 9.5 × 10−18 (21% sequence coverage, Figure S9, Table S5). High mass accuracy measurements were used to confirm protein identity, yielding mass errors below 10 ppm for all identified protein ions.
LMJ-SSP with and without FAIMS separation was performed on serial rat brain tissue sections in the positive ion mode. Figure 4 shows 2D ion images of selected protein species obtained using LMJ-SSP with a spatial resolution of ∼630 μm. The increase in S/N achieved with LMJ-SSP-FAIMS approach allowed for clear visualization of the spatial distribution of the proteins within the optimized transmission range (Figure 4d). For example, the spatial distribution of the 9+ charge state of an unidentified protein (m/z 939.848, MW = 5449.60 Da) by LMJ-SSP alone was unclear, with signal due to protein detection and background noise observed throughout the tissue section and the surrounding glass slide. However, the LMJ-SSP-FAIMS image clearly shows protein distribution at higher relative abundances within the white matter of the rat brain. The same trend was seen for the protein detected at m/z 915.151. LMJ-SSP-FAIMS-MSI also allowed visualization of the spatial distribution of the 6+ charge state of thymosin β-4 (m/z 828.256) within the hippocampus of the rat brain (delineated over optical image of H&E stained tissues), while the 10+ charge state of ubiquitin (m/z 857.370) was distributed homogeneously throughout the tissue section. Protein distributions obtained by LMJ-SSP-MSI were reproducible (n = 4 for each approach, Figure S13), although small alterations in experimental parameters including pressure and probe distance to the sample slightly affected ion intensity, which most noticeably impacts ion images obtained for ions that are not localized in a specific region of the tissue. Similar spatial distributions for these proteins described have been recently reported using other ambient ionization MSI techniques.16,31
Proteins with molecular weights outside the optimized FAIMS voltages are observed at lower absolute intensities when compared to LMJ-SSP alone. For example, MBP has a molecular weight of 15 kDa which is outside the 4-12 kDa range. LMJ-SSP-FAIMS-MSI of its 20+ charge state at m/z 706.968 still allows for visualization of its spatial distribution despite a 2-fold reduction in intensity. Note that lipid ions could still be detected and imaged using the FAIMS conditions optimized from proteins, although at lower relative and absolute abundances. For example, Figure S14 shows the ion images obtained from an unwashed rat brain tissue section for m/z 760.584, PC (34:1), m/z 810.597, PC (38:4), and m/z 857.370, ubiquitin, which were detected during the same LMJ-SSP-FAIMS-MSI experiment.
LMJ-SSP-FAIMS-MSI of Proteins in Ovarian Cancer Tissue Sections
The integrated LMJ-SSP-FAIMS approach was applied to image proteins in human normal ovarian and high grade serous ovarian cancer tissue samples. 2D ion images revealed distinct spatial distribution of various protein ions within the tissue sections, as shown in Figure 5b for four selected protein ions: 10+ ion cluster at m/z 857.963 (MW = 8559.54 Da), identified as ubiquitin (P-score = 1.6 × 10−20, 13% sequence coverage, mass error of 9.35 ppm, Figure S10, Table S6); 5+ ion cluster at m/z 993.501 (MW = 4960.46 Da), identified as thymosin β-4 (P-score = 1.6 × 10−5, 12% sequence coverage, mass error of 6.15 ppm, Figure S11, Table S7); 16+ ion cluster at m/z 946.311 (MW = 15116.76 Da), identified as hemoglobin α-subunit (P-score = 5.3 × 10−11, 26% sequence coverage, mass error of −7.94 ppm, Figure S12, Table S8); and the 9+ ion cluster at m/z 1122.038 (MW = 10084.27 Da), proposed as calcyclin (P-score = 5.4 × 10−12, 7% sequence coverage, mass error of 4.46 ppm). Histopathologic evaluation of H&E stained serial sections revealed the presence of necrotic tissue adjacent to the tumor cells in the tumor tissue, while the normal tissue was homogeneously composed of normal ovarian tissue. Comparison of the 2D ion images and the annotated optical images of the H&E stained samples revealed that ubiquitin, thymosin β-4, and calcyclin showed higher relative abundances within the tumor region of the ovarian tumor sample compared with both the necrosis region and the normal ovarian tissue. Hemoglobin α-subunit, however, showed higher relative abundances within the necrosis region compared to the adjacent tumor region and the normal ovarian tissue. The mass spectra obtained showed distinct protein distribution for high grade serous ovarian cancer, necrotic ovarian tissue, and normal ovarian tissue under optimized LMJ-SSP-FAIMS conditions (Figure 5a).
Figure 5.
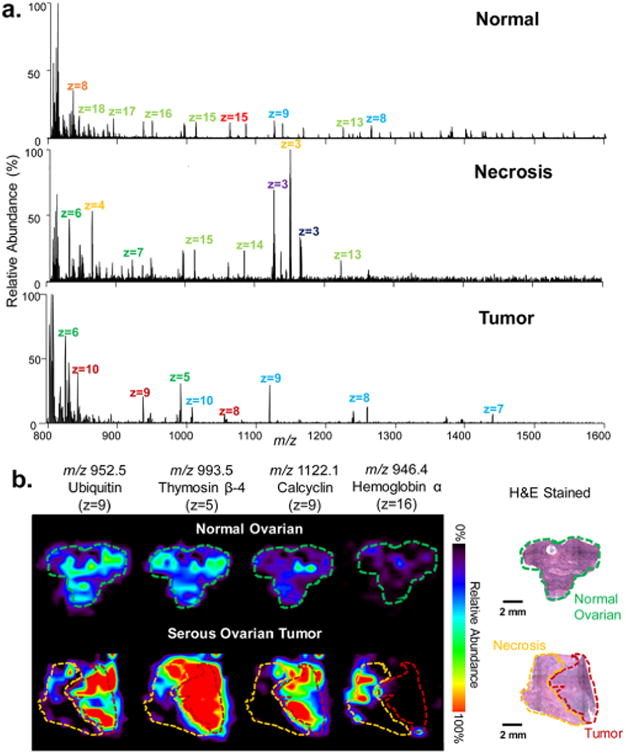
Static LMJ-SSP-FAIMS-MS profiling and imaging of human normal and cancerous ovarian tissues. (a) LMJ-SSP-FAIMS-MS spectra of normal ovarian, necrotic, and serous ovarian cancer samples in which different colored labels represent different charge states of same protein species. (b) LMJ-SSP-FAIMS-MS ion images of ubiquitin, thymosin β-4, calcyclin, and hemoglobin α-subunit for a normal ovarian tissue sample compared with the high grade serous ovarian tumor sample, containing both necrotic and tumor regions (spatial resolution is ∼630 μm). Optical images of H&E stained sections show regions of normal ovarian, necrotic, and high grade serous ovarian tumor. Voxel versions of the same ion images are shown in Figure S15. Scale bar = 2 mm.
Conclusions
We developed and optimized an analytical approach integrating DESI and LMJ-SSP with a chip-based FAIMS device for imaging biological tissue samples. This method allowed for partial separation of analytes after desorption or extraction from biological samples, resulting in decreased chemical noise and increased detection of selected molecular species. Reducing interferences through FAIMS separation improved the S/N for the species of interest, aiding in spectra interpretation and improving ion image quality. Optimization of DF and CF values for semitargeted transmission of different molecular classes can be performed through 2D-FAIMS sweeping experiments during a profiling or a more time-consuming imaging assay, directly from tissue samples.
We show through several examples that this approach is particularly useful for semiselective imaging of subsets of multiply charged lipids and proteins at optimized FAIMS conditions, with no increase in analysis time. For example, improvement in the S/N, ion abundances, and number of doubly charged CL and gangliosides species at optimized FAIMS conditions were observed in rat brain tissue sections. DESI-FAIMS-MSI allowed enhanced detection and imaging of diagnostic CL in human oncocytic thyroid tumor with reduced interference from other GP species, enabling improved ion isolation and identification by tandem MS. LMJ-SSP-FAIMS-MSI was particularly powerful for imaging multiply charged protein ions from biological tissue samples. Optimized FAIMS parameters for proteins in the 4–12 kDa range allowed increase in the S/N, number detected, and visualization of the 2D spatial distribution of 84 protein species within rat brain, even with an overall decrease in total ion current. LMJ-SSP-FAIMS-MSI of human ovarian tissue samples enabled detection, identification, and correlation of spatial distribution of several proteins within the heterogeneous regions of the tissue samples, including regions of tumor, necrotic, and normal ovarian tissues. This is the first example of global protein imaging in human cancer tissue by ambient ionization MSI.
Our results show that FAIMS can be successfully integrated with DESI and LMJ-SSP for improved detection and imaging of subsets of molecular species in biological tissues. While separation using the planar FAIMS device caused a decrease in total ion abundance of 1 order of magnitude or more, an increase in S/N ratio due to concomitant reduction of noise level contributed to improved data quality for MS imaging experiments.34 Thus, addition of FAIMS to a DESI or LMJ-SSP MSI workflow at optimized conditions is valuable for the detection and spatial visualization of otherwise undetectable lipids and proteins of diagnostic importance in biological tissues. Yet, while FAIMS provides a separation capability to increase data quality in ambient ionization MS imaging, ion suppression and matrix effects during ionization are still pertinent and should be considered as these may hinder ionization of molecules of interest.43
Further optimization of the integrated system is being pursued to increase sensitivity and ion transmission. As with other MSI techniques, the extent of molecular information obtained is significantly less than that achieved with standard HPLC-MS approaches. For example, HPLC-MS proteomic assays of selected tissue regions obtained by laser capture microdissection can be performed at similar spatial resolution as LMJ-SSP, although more time-consuming and labor intensive.44 Evaluation of the molecular coverage achieved when compared to standard chromatographic separation techniques will be sought to better quantify the utility of our approach. However, as ambient ionization MSI provides molecular and spatial information at a fraction of the time, without extensive sample preparation, we expect the integrated approach described here to be valuable in biomedical applications targeted at imaging specific diagnostic lipids and proteins which are otherwise difficult to detect in biological tissues.
Supplementary Material
Tables of identified compounds, MS/MS data, ESI-FAIMS optimization data, time-dependent DESI data, CL images for rat brain, summary of detected proteins, LMJ-SSP image replicates, and images illustrating ability to detect both lipids and proteins. (PDF)
Acknowledgments
This work was supported by The Welch Foundation (Grant F-1895), the National Institutes of Health (grant 4R00CA190783-02), and the National Science Foundation (Grant CHE-1559839). We thank Dr. Wendong Yu (Department of Pathology and Immunology, Baylor College of Medicine) and Dr. Jinsong Liu (Department of Pathology, The University of Texas MD Anderson Cancer Center) for pathologic analysis. We are grateful to Jialing Zhang, Marta Sans Escofet, and John Lin for assistance with experiments and data analysis. Human tissue samples were provided by the CHTN which is funded by the National Cancer Institute.
Footnotes
Supporting Information: The Supporting Information is available free of charge via the Internet at The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.6b02798.
Notes: The authors declare no competing financial interest.
References
- 1.Dill AL, Eberlin LS, Ifa DR, Cooks RG. Chem Commun. 2011;47:2741–2746. doi: 10.1039/c0cc03518a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fletcher JS, Vickerman JC, Winograd N. Curr Opin Chem Biol. 2011;15:733–740. doi: 10.1016/j.cbpa.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seeley EH, Schwamborn K, Caprioli RM. J Biol Chem. 2011;286:25459–25466. doi: 10.1074/jbc.R111.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu CP, Dill AL, Eberlin LS, Cooks RG, Ifa DR. Mass Spectrom Rev. 2013;32:218–243. doi: 10.1002/mas.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu CC, Dorrestein PC. Curr Opin Biotechnol. 2015;31:24–34. doi: 10.1016/j.copbio.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiseman JM, Ifa DR, Song QY, Cooks RG. Angew Chem, Int Ed. 2006;45:7188–7192. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- 7.Takats Z, Wiseman JM, Gologan B, Cooks RG. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 8.Ifa DR, Eberlin LS. Clin Chem. 2016;62:111–123. doi: 10.1373/clinchem.2014.237172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ. Anal Chem. 2012;84:141–148. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laskin J, Lanekoff I. Anal Chem. 2016;88:52–73. doi: 10.1021/acs.analchem.5b04188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen HW, Gamez G, Zenobi R. J Am Soc Mass Spectrom. 2009;20:1947–1963. doi: 10.1016/j.jasms.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 12.Hankin JA, Murphy RC. Anal Chem. 2010;82:8476–8484. doi: 10.1021/ac101079v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yetukuri L, Ekroos K, Vidal-Puig A, Oresic M. Mol BioSyst. 2008;4:121–127. doi: 10.1039/b715468b. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh Y, Casale R, Fukuda E, Chen JW, Knemeyer I, Wingate J, Morrison R, Korfmacher W. Rapid Commun Mass Spectrom. 2006;20:965–972. doi: 10.1002/rcm.2397. [DOI] [PubMed] [Google Scholar]
- 15.Lou XW, van Dongen JLJ, Vekemans J, Meijer EW. Rapid Commun Mass Spectrom. 2009;23:3077–3082. doi: 10.1002/rcm.4224. [DOI] [PubMed] [Google Scholar]
- 16.Hsu CC, Chou PT, Zare RN. Anal Chem. 2015;87:11171–11175. doi: 10.1021/acs.analchem.5b03389. [DOI] [PubMed] [Google Scholar]
- 17.Kertesz V, Weiskittel TM, Van Berkel GJ. Anal Bioanal Chem. 2015;407:2117–2125. doi: 10.1007/s00216-014-8287-5. [DOI] [PubMed] [Google Scholar]
- 18.Lombard-Banek C, Moody SA, Nemes P. Angew Chem, Int Ed. 2016;55:2454–2458. doi: 10.1002/anie.201510411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins DC, Lee ML. Anal Bioanal Chem. 2002;372:66–73. doi: 10.1007/s00216-001-1195-5. [DOI] [PubMed] [Google Scholar]
- 20.Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH. J Mass Spectrom. 2008;43:1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- 21.Ray JA, Kushnir MM, Yost RA, Rockwood AL, Meikle AW. Clin Chim Acta. 2015;438:330–336. doi: 10.1016/j.cca.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 22.Guevremont R. Journal of Chromatography A. 2004;1058:3–19. [PubMed] [Google Scholar]
- 23.Kolakowski BM, Mester Z. Analyst. 2007;132:842–864. doi: 10.1039/b706039d. [DOI] [PubMed] [Google Scholar]
- 24.Da Costa C, Turner M, Reynolds JC, Whitmarsh S, Lynch T, Creaser CS. Anal Chem. 2016;88:2453–2458. doi: 10.1021/acs.analchem.5b04595. [DOI] [PubMed] [Google Scholar]
- 25.Manicke NE, Belford M. J Am Soc Mass Spectrom. 2015;26:701–705. doi: 10.1007/s13361-015-1096-z. [DOI] [PubMed] [Google Scholar]
- 26.Griffiths RL, Dexter A, Creese AJ, Cooper H. J Analyst. 2015;140:6879–6885. doi: 10.1039/c5an00933b. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Smith BK, Mark L, Nemes P, Nazarian J, Vertes A. Int J Mass Spectrom. 2015;377:681–689. [Google Scholar]
- 28.Roscioli KM, Tufariello JA, Zhang X, Li SX, Goetz GH, Cheng GL, Siems WF, Hill HH. Analyst. 2014;139:1740–1750. doi: 10.1039/c3an02113k. [DOI] [PubMed] [Google Scholar]
- 29.Galhena AS, Harris GA, Kwasnik M, Fernandez FM. Anal Chem. 2010;82:9159–9163. doi: 10.1021/ac102340h. [DOI] [PubMed] [Google Scholar]
- 30.Sarsby J, Griffiths RL, Race AM, Bunch J, Randall EC, Creese AJ, Cooper HJ. Anal Chem. 2015;87:6794–6800. doi: 10.1021/acs.analchem.5b01151. [DOI] [PubMed] [Google Scholar]
- 31.Griffiths RL, Creese AJ, Race AM, Bunch J, Cooper HJ. Anal Chem. 2016;88:6758–6766. doi: 10.1021/acs.analchem.6b01060. [DOI] [PubMed] [Google Scholar]
- 32.Eberlin LS, Ferreira CR, Dill AL, Ifa DR, Cheng L, Cooks RG. ChemBioChem. 2011;12:2129–2132. doi: 10.1002/cbic.201100411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robichaud G, Garrard KP, Barry JA, Muddiman DC. J Am Soc Mass Spectrom. 2013;24:718–721. doi: 10.1007/s13361-013-0607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swearingen KE, Moritz RL. Expert Rev Proteomics. 2012;9:505–517. doi: 10.1586/epr.12.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarmusch AK, Pirro V, Baird Z, Hattab EM, Cohen-Gadol AA, Cooks RG. Proc Natl Acad Sci U S A. 2016;113:1486–1491. doi: 10.1073/pnas.1523306113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji J, Kline AE, Amoscato A, Samhan-Arias AK, Sparvero LJ, Tyurin VA, Tyurina YY, Fink B, Manole MD, Puccio AM, Okonkwo DO, Cheng JP, Alexander H, Clark RSB, Kochanek PM, Wipf P, Kagan VE, Bayir H. Nat Neurosci. 2012;15:1407–1413. doi: 10.1038/nn.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sapandowski A, Stope M, Evert K, Evert M, Zimmermann U, Peter D, Paege I, Burchardt M, Schild L. Mol Cell Biochem. 2015;410:175–185. doi: 10.1007/s11010-015-2549-1. [DOI] [PubMed] [Google Scholar]
- 38.Corcelli A, Angelini R, Lobasso S, Bowron A, Steward C. FASEB J. 2015;29 doi: 10.1194/jlr.D059824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandhoff K, Harzer K. J Neurosci. 2013;33:10195–10208. doi: 10.1523/JNEUROSCI.0822-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu RK, Tsai YT, Ariga T. Neurochem Res. 2012;37:1230–1244. doi: 10.1007/s11064-012-0744-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Yu W, Ryu S, Lin J, Buentello G, Tibshirani R, Suliburk JW, Eberlin LS. Cancer Res. 2016 doi: 10.1158/0008-5472.CAN-16-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prideaux B, El Naggar MS, Zimmerman M, Wiseman JM, Li X, Dartois V. Int J Mass Spectrom. 2015;377:699–708. doi: 10.1016/j.ijms.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen H, Gamez G, Zenobi R. J Am Soc Mass Spectrom. 2009;20:1947–1963. doi: 10.1016/j.jasms.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 44.Zang L, Toy DP, Hancock WS, Sgroi DC, Karger BL. J Proteome Res. 2004;3:604–612. doi: 10.1021/pr034131l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables of identified compounds, MS/MS data, ESI-FAIMS optimization data, time-dependent DESI data, CL images for rat brain, summary of detected proteins, LMJ-SSP image replicates, and images illustrating ability to detect both lipids and proteins. (PDF)


