Abstract
PURPOSE
Keratoconus (KC) is a complex corneal dystrophy with multifactorial etiology. Previous studies have shown evidence of mitochondrial abnormalities in KC; however, the exact cause of these abnormalities remains unknown. The aim of this study was to identify if transforming growth factor-β (TGF-β) isoforms play a role in the regulation of mitochondrial proteins in human keratoconus cells (HKC).
MATERIALS AND METHODS
Human corneal fibroblasts (HCF) and HKC were isolated and cultured for four weeks in three different conditions: a) Control: MEM+10%FBS, b) MEM+10%FBS+TGF-β1, and c) MEM+10%FBS+TGF-β3. All samples were processed for mitochondrial damage analysis using real-time PCR.
RESULTS
We quantified and analyzed 84 mitochondrial and 5 housekeeping genes in HCFs and HKCs. Our data showed that when TGF-β1 and/or TGF-β3 were compared with control in HCFs, nine genes were significantly different; however, no genes were significantly regulated by the TGF-β isoforms in HKCs. Significant differences were also seen in seven genes when HFCs were compared to HKCs, in all three conditions.
CONCLUSIONS
Overall, our data supports the growing consensus that mitochondrial dysfunction is a key player in KC disease. These in vitro data show clear links between mitochondrial function and TGF-β isoforms, with TGF-β1 severely disrupting KC-mitochondrial function, while TGF-β3 maintained it, thus suggesting that TGF-β may play a role in KC-disease treatment.
Keywords: Keratoconus, Cornea, Mitochondria, Transforming Growth Factor-β, Mitochondrial dysfunction
INTRODUCTION
In the past three decades, the true role of mitochondria and mitochondrial regulation has been revealed in the human body and cells (1–7), and their role in cell signaling, differentiation, death, cycle, and growth has become known (8). Indeed, mitochondria are able to dictate the fate of a single cell, and ultimately, the fate and function of a whole tissue (9–13). Of all the organs in the human body, the human eye is one of the most affected by mitochondrial diseases—dominant optic atrophy, leber hereditary optic neuropathy, chronic progressive external ophthalmoplegia, and pigmentary retinopathy (14). These diseases are normally categorized as either primary or secondary. Primary mitochondrial diseases have a direct genetic impairment of mitochondrial function(s) in either the mitochondrial DNA (mtDNA) or nuclear DNA (nDNA); whereas, secondary mitochondrial diseases are the result of environmental factors and/or genetic disorders. Despite significant efforts to explore the mitochondrial role and regulation in ocular diseases, there is still a lot of work to be done.
In the cornea, one of the diseases that have been directly linked to mitochondrial dysfunction is keratoconus (KC). KC is a common (1:2000 people worldwide) non-inflammatory corneal dystrophy that typically begins at puberty and continues to progress until forty or fifty years of age (15–17). The results, depending on severity, are a cone-like bulging cornea with extreme thinning of the stromal layer. These characteristics cause severe visual impairments, and more than 20% of diagnosed cases opt for corneal transplantation to restore vision. While the etiology is still unknown, there is a long list of candidates, such as extreme eye rubbing, genetics, environmental factors, oxidative stress, and mitochondrial dysfunction (18–24).
To-date, the majority of studies in KC concentrate on abnormal antioxidant enzymes, and alterations in lipid peroxidation and nitric oxide pathways, which lead to increasing levels of oxidative stress and reactive nitrogen species (25–28). Almost all of these studies were performed using cadaver corneal tissue or tear fluid collected from individuals (29, 30) In a 3D in vitro stroma-like model, we characterized and quantified cellular and extracellular matrix (ECM) secreted by healthy human corneal fibroblasts (HCF) and human KC cells (HKC) (17, 29, 31). Also, over the last few years, we were able to establish the role and importance of transforming growth factor-β (TGF-β) and its isoforms in HKC regulation (17, 29, 31). TGF-β signaling is known to be required for normal tissue repair, including the corneal stroma; however, excessive TGF-β signaling can lead to severe tissue fibrosis, which is one of the main outcomes of KC disease.
Our studies aim to define any possible links between mitochondrial dysfunction in HKCs and the TGF-β isoforms when compared to HCFs. Here, we focus on two TGF-β isoforms, TGF-β1 and -β3, which have previously been reported to have opposite roles in the formation of myofibroblasts and fibrosis (32–39). Overall, our data supports the growing consensus that mitochondrial dysfunction is a key player in KC disease.
MATERIALS AND METHODS
Inclusion/exclusion criteria
The research followed the tenets of the Declaration of Helsinki, and has been approved by the institution’s ethics committee. Corneas collected for the control group had no ocular abnormalities or diseases. Any donor, for both control and KC groups, with a history of ocular surgery, diabetes, photocoagulation, trauma, inflammation, glaucoma, or ptosis were excluded. In addition, all samples underwent serology testing and were excluded if they tested positive for the folllowing: HIV, Hepatitis B, Hepatitis C, and Syphilis.
Isolation and primary cells
HCFs were isolated from healthy human corneas obtained from NDRI (National Disease Research Interchange; Philadelphia, PA) s. HKCs were isolated from human corneas with KC defects (17, 31), and were obtained from Dr. Jesper Hjortdal (Aarhus University Hospital, Aarhus, Denmark). All samples, for both HCFs and HKCs, underwent identical procedures in regards to isolation of corneal cells. Briefly, the corneal epithelium and endothelium were removed from the stroma by scraping with a razor blade. The stromal tissues were cut into small pieces (~2×2 mm) and placed in T25 culture flasks. Explants were allowed to adhere to the bottom of the flask at 37ºC for about 30 minutes and Eagle’s Minimum Essential Medium (EMEM: ATCC; Manassas, VA) containing 10% fetal bovine serum (FBS: Atlantic Biological’s; Lawrenceville, CA) and 1% antibiotic (Gibco® Antibiotic-Antimycotic: Life Technologies; Grand Island, NY) was added carefully without disturbing the explants. Upon 100% confluence (~1–2 weeks at 37ºC, 5%CO2), the cells were passaged into T75 culture flasks and cultivated for the following experiments.
Cultures and TGF-βs
Both HCFs and HKCs were cultured on 6-well tissue culture plates and processed for RT2 Profiler™ PCR Array (Catalog #PAHS-087Z: Qiagen Inc.; Valencia, CA). Cells (1×106 cells/well) were seeded and cultured in EMEM + 10%FBS ± 0.1ng/mL of TGF-β1 (T1) or TGF-β3 (T3) (32–34). The cultures were grown for 4 weeks before further processing. Cultures without any growth factors served as controls. Fresh media was supplied to the cultures every other day for the duration of the experiment.
RT2 Profiler™ PCR Array
After 4 weeks, total RNA extraction (Ambion TRIzol® Plus RNA Purification Kit: Life Technologies) followed by cDNA synthesis (RT2 First Strand Kit [Catalog #33040]: Qiagen Inc.) using 500ng of RNA, was carried out according to the manufacturer’s protocol. Samples were then analyzed for a panel of 84 genes related to human mitochondria with the RT2 Profiler™ PCR Array Human Mitochondria plate (Catalog #PAHS-087Z: Qiagen Inc.). Each well was set up with a 25-μl reaction containing RT2 SYBR Green ROX qPCR Mastermix (Catalog #330520: Qiagen Inc.), 5ng of cDNA, and RNase free water (Catalog #10977-015: Life Technologies). StepOnePlus™ real-time PCR system (Life Technologies) was used for amplification of the samples according to standard manufacturer’s protocol. Qiagen Inc. web based data analysis software for RT2 Profiler™ PCR Array and MS-Excel were used for data analysis. All samples and probes were repeated at least three times.
Statistical Analysis
Data analysis for all samples was performed by t-test and one-way ANOVA using Prism 6 software (GraphPad Software, Inc.; La Jolla, CA), where p < 0.05 was considered statistically significant. Average values were calculated and plotted with standard error of the mean (SEM).
RESULTS
Mitochondrial gene panel
Information on mitochondrial function in KC derived corneal cells is very limited, if non-existent. Therefore, we quantified and analyzed the mitochondrial gene expressions in both healthy (HCFs) and diseased (HKCs) corneal cells, revealing significant differences between them. In addition, we investigate the role of T1 and T3 in mitochondrial gene expression. Table 1 shows the panel of mitochondrial genes that were significantly regulated, and Table 2 shows the gene name, description, and symbol of all the genes (84 plus 5 housekeeping) tested here.
Table 1.
Mitochondrial genes that were significantly regulated were analyzed and quantified: BNIP3, CPT1B, NEFL, PMAIP1, SLC25A2, SLC25A21, SLC25A31, SLC25A4, SOD2, UCP1, and UCP3. All experiments were repeated at least three times (n>=3).
| Symbol | Description | Gname |
|---|---|---|
| BNIP3 | BCL2/adenovirus E1B 19kDa interacting protein 3 | NIP3 |
| CPT1B | Carnitine palmitoyltransferase 1B (muscle) | CPT1-M/CPT1M/CPTI/CPTI-M/M-CPT1/MCCPT1/MCPT1 |
| NEFL | Neurofilament, light polypeptide | CMT1F/CMT2E/NF-L/NF68/NFL |
| PMAIP1 | Phorbol-12-myristate-13-acetate-induced protein 1 | APR/NOXA |
| SLC25A2 | Solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 2 | ORC2/ORNT2 |
| SLC25A21 | Solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21 | ODC/ODC1 |
| SLC25A31 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 | AAC4/ANT4/SFEC35kDa |
| SLC25A4 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | 1/AAC1/ANT/ANT 1/ANT1/MTDPS12/PEO2/PEO3/T1 |
| SOD2 | Superoxide dismutase 2, mitochondrial | IPOB/MNSOD/MVCD6 |
| UCP1 | Uncoupling protein 1 (mitochondrial, proton carrier) | SLC25A7/UCP |
| UCP2 | Uncoupling protein 2 (mitochondrial, proton carrier) | BMIQ4/SLC25A8/UCPH |
Table 2.
A list of 84 mitochondrial and 5 housekeeping genes tested in this study, as provided by the RT2 Profiler™ PCR Array. All experiments were repeated at least three times (n>=3).
| Symbol | Description | Gname |
|---|---|---|
| AIFM2 | Apoptosis-inducing factor, mitochondrion-associated, 2 | AMID/PRG3/RP11-367H5.2 |
| AIP | Aryl hydrocarbon receptor interacting protein | ARA9/FKBP16/FKBP37/SMTPHN/XAP-2/XAP2 |
| BAK1 | BCL2-antagonist/killer 1 | BAK/BAK-LIKE/BCL2L7/CDN1 |
| BBC3 | BCL2 binding component 3 | JFY-1/JFY1/PUMA |
| BCL2 | B-cell CLL/lymphoma 2 | Bcl-2/PPP1R50 |
| BCL2L1 | BCL2-like 1 | BCL-XL/S/BCL2L/BCLX/BCLXL/BCLXS/Bcl-X/PPP1R52/bcl-xL/bcl-xS |
| BID | BH3 interacting domain death agonist | FP497 |
| BNIP3 | BCL2/adenovirus E1B 19kDa interacting protein 3 | NIP3 |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | ARF/CDK4I/CDKN2/CMM2/INK4/INK4A/MLM/MTS-1/MTS1/P14/P14ARF/P16/P16-INK4A/P16INK4/P16INK4A/P19/P19ARF/TP16 |
| COX10 | COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast) | - |
| COX18 | COX18 cytochrome c oxidase assembly homolog (S. cerevisiae) | COX18HS |
| CPT1B | Carnitine palmitoyltransferase 1B (muscle) | CPT1-M/CPT1M/CPTI/CPTI-M/M-CPT1/MCCPT1/MCPT1 |
| CPT2 | Carnitine palmitoyltransferase 2 | CPT1/CPTASE/IIAE4 |
| DNM1L | Dynamin 1-like | DLP1/DRP1/DVLP/DYMPLE/EMPF/HDYNIV |
| FIS1 | Fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) | TTC11 |
| TIMM10B | Fracture callus 1 homolog (rat) | FXC1/TIM10B/Tim9b |
| GRPEL1 | GrpE-like 1, mitochondrial (E. coli) | HMGE |
| HSP90AA1 | Heat shock protein 90kDa alpha (cytosolic), class A member 1 | EL52/HSP86/HSP89A/HSP90A/HSP90N/HSPC1/HSPCA/HSPCAL1/HSPCAL4/HSPN/Hsp89/Hsp90/LAP2 |
| HSPD1 | Heat shock 60kDa protein 1 (chaperonin) | CPN60/GROEL/HLD4/HSP-60/HSP60/HSP65/HuCHA60/SPG13 |
| IMMP1L | IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) | IMP1/IMP1-LIKE |
| IMMP2L | IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) | IMMP2L-IT1/IMP2/IMP2-LIKE |
| LRPPRC | Leucine-rich PPR-motif containing | CLONE-23970/GP130/LRP130/LSFC |
| MFN1 | Mitofusin 1 | hfzo1/hfzo2 |
| MFN2 | Mitofusin 2 | CMT2A/CMT2A2/CPRP1/HSG/MARF |
| MIPEP | Mitochondrial intermediate peptidase | HMIP/MIP |
| MPV17 | MpV17 mitochondrial inner membrane protein | MTDPS6/SYM1 |
| MSTO1 | Misato homolog 1 (Drosophila) | MST |
| MTX2 | Metaxin 2 | - |
| NEFL | Neurofilament, light polypeptide | CMT1F/CMT2E/NF-L/NF68/NFL |
| OPA1 | Optic atrophy 1 (autosomal dominant) | MGM1/NPG/NTG/largeG |
| PMAIP1 | Phorbol-12-myristate-13-acetate-induced protein 1 | APR/NOXA |
| RHOT1 | Ras homolog gene family, member T1 | ARHT1/MIRO-1/MIRO1 |
| RHOT2 | Ras homolog gene family, member T2 | ARHT2/C16orf39/MIRO-2/MIRO2/RASL |
| SFN | Stratifin | YWHAS |
| SH3GLB1 | SH3-domain GRB2-like endophilin B1 | Bif-1/PPP1R70/dJ612B15.2 |
| SLC25A1 | Solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 | CTP/D2L2AD/SEA/SLC20A3 |
| SLC25A10 | Solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 | DIC |
| SLC25A12 | Solute carrier family 25 (mitochondrial carrier, Aralar), member 12 | AGC1/ARALAR |
| SLC25A13 | Solute carrier family 25, member 13 (citrin) | ARALAR2/CITRIN/CTLN2 |
| SLC25A14 | Solute carrier family 25 (mitochondrial carrier, brain), member 14 | BMCP1/UCP5 |
| SLC25A15 | Solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 | D13S327/HHH/ORC1/ORNT1 |
| SLC25A16 | Solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 | D10S105E/GDA/GDC/HGT.1/ML7/hML7 |
| SLC25A17 | Solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 | PMP34 |
| SLC25A19 | Solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 | DNC/MCPHA/MUP1/THMD3/THMD4/TPC |
| SLC25A2 | Solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 2 | ORC2/ORNT2 |
| SLC25A20 | Solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 | CAC/CACT |
| SLC25A21 | Solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21 | ODC/ODC1 |
| SLC25A22 | Solute carrier family 25 (mitochondrial carrier: glutamate), member 22 | EIEE3/GC1/NET44 |
| SLC25A23 | Solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 | APC2/MCSC2/SCaMC-3 |
| SLC25A24 | Solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 | APC1/SCAMC-1 |
| SLC25A25 | Solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 | MCSC/PCSCL/RP11-395P17.4/SCAMC-2 |
| SLC25A27 | Solute carrier family 25, member 27 | UCP4 |
| SLC25A3 | Solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 | PHC/PTP |
| SLC25A30 | Solute carrier family 25, member 30 | KMCP1 |
| SLC25A31 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 | AAC4/ANT4/SFEC35kDa |
| SLC25A37 | Solute carrier family 25, member 37 | MFRN/MFRN1/MSC/MSCP/PRO1278/PRO1584/PRO2217 |
| SLC25A4 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | 1/AAC1/ANT/ANT 1/ANT1/MTDPS12/PEO2/PEO3/T1 |
| SLC25A5 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | 2F1/AAC2/ANT2/T2/T3 |
| SOD1 | Superoxide dismutase 1, soluble | ALS/ALS1/HEL-S-44/IPOA/SOD/hSod1/homodimer |
| SOD2 | Superoxide dismutase 2, mitochondrial | IPOB/MNSOD/MVCD6 |
| STARD3 | StAR-related lipid transfer (START) domain containing 3 | CAB1/MLN64/es64 |
| TAZ | Tafazzin | BTHS/CMD3A/EFE/EFE2/G4.5/LVNCX/Taz1 |
| TIMM10 | Translocase of inner mitochondrial membrane 10 homolog (yeast) | TIM10/TIM10A |
| TIMM17A | Translocase of inner mitochondrial membrane 17 homolog A (yeast) | TIM17/TIM17A |
| TIMM17B | Translocase of inner mitochondrial membrane 17 homolog B (yeast) | DXS9822/TIM17B |
| TIMM22 | Translocase of inner mitochondrial membrane 22 homolog (yeast) | TEX4/TIM22 |
| TIMM23 | Translocase of inner mitochondrial membrane 23 homolog (yeast) | TIM23 |
| TIMM44 | Translocase of inner mitochondrial membrane 44 homolog (yeast) | TIM44 |
| TIMM50 | Translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) | TIM50/TIM50L |
| TIMM8A | Translocase of inner mitochondrial membrane 8 homolog A (yeast) | DDP/DDP1/DFN1/MTS/TIM8 |
| TIMM8B | Translocase of inner mitochondrial membrane 8 homolog B (yeast) | DDP2/TIM8B |
| TIMM9 | Translocase of inner mitochondrial membrane 9 homolog (yeast) | TIM9/TIM9A |
| TOMM20 | Translocase of outer mitochondrial membrane 20 homolog (yeast) | MAS20/MOM19/TOM20 |
| TOMM22 | Translocase of outer mitochondrial membrane 22 homolog (yeast) | 1C9-2/MST065/MSTP065/TOM22 |
| TOMM34 | Translocase of outer mitochondrial membrane 34 | HTOM34P/TOM34/URCC3 |
| TOMM40 | Translocase of outer mitochondrial membrane 40 homolog (yeast) | C19orf1/D19S1177E/PER-EC1/PEREC1/TOM40 |
| TOMM40L | Translocase of outer mitochondrial membrane 40 homolog (yeast)-like | RP11-297K8.10/TOMM40B |
| TOMM70A | Translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) | - |
| TP53 | Tumor protein p53 | BCC7/LFS1/P53/TRP53 |
| TSPO | Translocator protein (18kDa) | BPBS/BZRP/DBI/IBP/MBR/PBR/PBS/PKBS/PTBR/mDRC/pk18 |
| UCP1 | Uncoupling protein 1 (mitochondrial, proton carrier) | SLC25A7/UCP |
| UCP2 | Uncoupling protein 2 (mitochondrial, proton carrier) | BMIQ4/SLC25A8/UCPH |
| UCP3 | Uncoupling protein 3 (mitochondrial, proton carrier) | SLC25A9 |
| UXT | Ubiquitously-expressed transcript | ART-27/STAP1 |
| ACTB | Actin, beta | BRWS1/PS1TP5BP1 |
| B2M | Beta-2-microglobulin | - |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | G3PD/GAPD |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | HGPRT/HPRT |
| RPLP0 | Ribosomal protein, large, P0 | L10E/LP0/P0/PRLP0/RPP0 |
Role of TGF-β isoforms in mitochondrial gene regulation
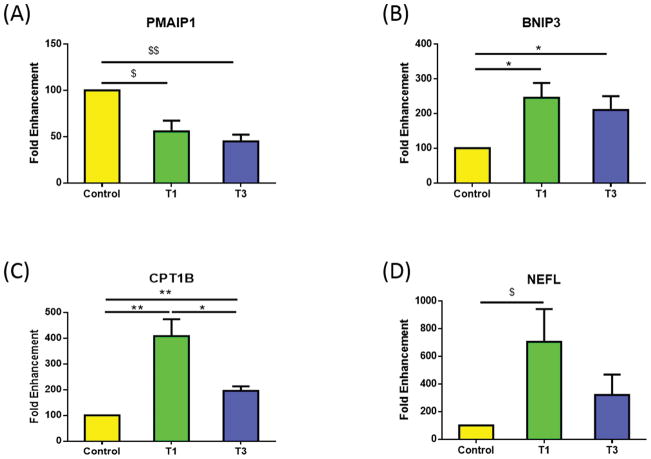
We identified nine total mitochondrial genes (9/84 genes) that were significantly regulated by T1 and/or T3 in HCFs when compared to controls. Figure 1A and B show a pro-apoptotic member of the BCL-2 protein family, PMAIP1 (Phorbol-12-myristate-13-acetate-induced protein 1), also known as Noxa (40, 41). PMAIP1 was significantly down-regulated following both T1 and T3 treatment (Fig. 1A: p<0.01 and p<0.001, respectively). Interestingly, BNIP3 (BCL2/adenovirus E1B 19kDa interacting protein 3), which contains a BH3 domain (42, 43), was significantly up-regulated in both T1 and T3 when compared to control (Fig. 1B: p<0.05).
Figure 1.
Mitochondrial genes that were significantly regulated by T1 and/or T3, in HCFs, when compared to controls: A) PMAIP1 was significantly down-regulated with T1 and T3; B) BNIP3 was significantly up-regulated with T1 and T3; C) CPT1B was significantly higher with T1 when compared to control and T3. T3 was significantly higher compared to control; D) NEFL was significantly higher with T1 compared to controls All samples were repeated at least three times, with p<0.05 considered to be statistically significant ($$$p<0.0001, $$p<0.001, $p<0.01, ***p<0.0005, **p<0.005, *p<0.05).
Carnitine palmitoyltransferase 1B (CPT1B) was significantly up-regulated by T1 and T3 (p<0.005 and p<0.005, respectively), as shown in Figure 1C. T3 was significantly lower than T1 (p<0.05), indicating a trend to maintain CPT1B expression to control levels (Fig. 1C). Neurofilament, light polypeptide (NEFL) was significantly up-regulated by T1 (p<0.01) compared to control, as shown in Figure 1D; whereas, T3 down-regulated NEFL expression compare to T1, however it did not reach significance (p=0.075).
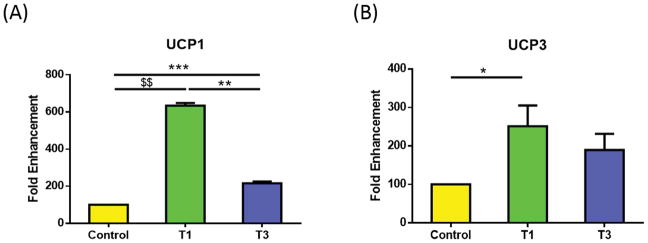
Two of the five known uncoupling proteins (UCPs) were significantly regulated in our study—UCP1 and UCP3. Figure 2A shows UCP1 significantly up-regulated in T1 and T3 compared to controls (p<0.0001 and p<0.0005, respectively), with T1 significantly higher than T3 (Fig. 2A; p<0.005). On the other hand, UCP3 expression was only significantly up-regulated in T1 (Fig. 2B; p<0.05) as compared with control. UCP1 and UCP3 are known to play a key role in regulating ATP synthesis and metabolic homeostasis (44).
Figure 2.
Mitochondrial genes that were significantly regulated by T1 and/or T3, in HCFs, when compared to controls: A) UCP1 was significantly higher with T1 when compared to control and T3. T3 was significantly higher compared to control; B) UCP 3 was significantly higher with T1 compared to control. All samples were repeated at least three times, with p<0.05 considered to be statistically significant ($$$p<0.0001, $$p<0.001, $p<0.01, ***p<0.0005, **p<0.005, *p<0.05).
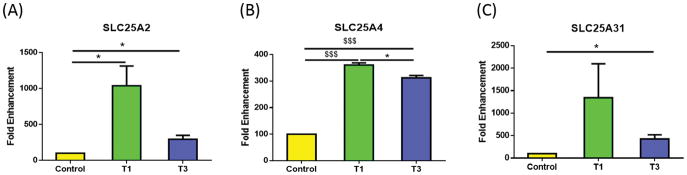
In addition, we identified significant regulation of three membrane transport proteins known as solute carriers, or SLCs—SLC25A2, SLC25A4, and SLC25A31. Figure 3A shows SLC25A2 significantly up-regulated by both T1 and T3 as compared with control (p<0.05), with T3 seemingly down-regulated compared to T1; however, the difference did not reach significance (p=0.0566). As with SLC25A2, SLC25A4 was significantly up-regulated in both T1 and T3 as compared to control (Fig. 3B; p<0.0001). Mutations in SLC25A4 gene have been shown to result in autosomal dominant progressive external opthalmoplegia and familial hypertrophic cardiomyopathy (45). The third SLC to be regulated was SLC25A31, which was only significantly up-regulated with the T3 treatment as compared with control (Fig. 3C; p<0.05). T1 treatment was not significantly regulated compared to the control (p=0.17). Interestingly, in HKCs, we found no mitochondrial genes that were significantly regulated by the TGF-β isoforms.
Figure 3.
Mitochondrial genes that were significantly regulated by T1 and/or T3, in HCFs, when compared to controls: A) SLC25A2 was significantly up-regulated with T1 and T3; B) SLC25A4 was significantly higher with T1 when compared to control and T3. T3 was significantly higher compared to control; C) SLC25A31 was significantly higher with T3 compared to control. All samples were repeated at least three times, with p<0.05 considered to be statistically significant ($$$p<0.0001, $$p<0.001, $p<0.01, ***p<0.0005, **p<0.005, *p<0.05).
Differences in mitochondrial gene expressions between HCFs and HKCs
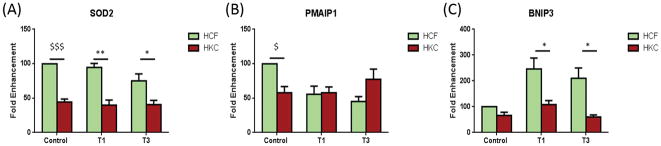
We identified seven mitochondrial genes (7/84 genes) that were significantly different between HCFs and HKCs, either in control, T1, or T3. Figure 4A shows significant down-regulation of SOD2 in HKCs, when compared to HCFs, under all three conditions (Controls, p<0.0001; T1, p<0.005; and T3, p<0.05). Mutations in this gene have been associated with idiopathic cardiomyopathy, sporadic motor neuron disease, and cancer (46, 47).
Figure 4.
Mitochondrial genes that were significantly different between HFCs and HKCs, either in controls, T1, or T3. A) SOD2 was significantly up-regulated in HCFs when compared to HKCs, independent of treatment; B) PMAIP1 was significantly down-regulated in control HKCs, compared to HCFs; C) BNIP3 was significantly down-regulated in HKCs when compared with HCFs under T1 and T3 treatments. All samples were repeated at least three times, with p<0.05 considered to be statistically significant ($$$p<0.0001, $$p<0.001, $p<0.01, ***p<0.0005, **p<0.005, *p<0.05).
PMAIP1 was significantly up-regulated in HCF controls compared to HKC controls (p<0.01) as shown in Figure 4B; however, as previously shown in Figure 1A, PMAIP1 was down-regulated by the TGF-β isoforms in HCFs, therefore, no significant difference was observed between the HCFs and HKCs with these samples (Fig. 4B). Interestingly, the opposite was true with BNIP3, which was significantly up-regulated in HCFs under T1 and T3 treatment when compared to HKCs (Fig. 4C; p<0.05), but not with control samples.
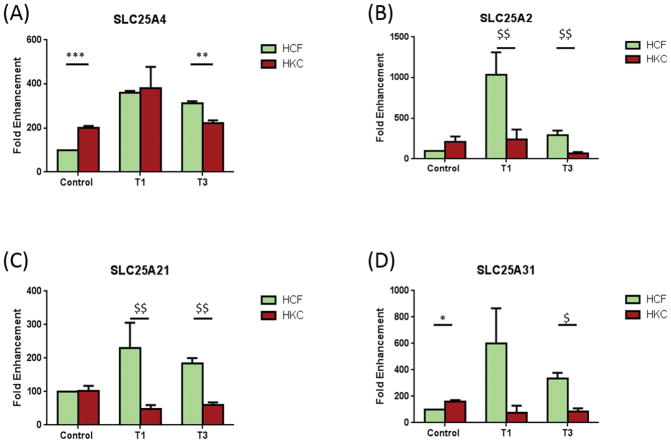
SLC25A4 was significantly up-regulated in HKCs when compared to HCF controls (Fig. 5A; p<0.0005). However, upon T3 treatment, HKCs were significantly lower than HCFs (Fig. 5A; p<0.005). No difference was seen between HCFs and HKCs under T1 treatment. SLC25A2 (Fig. 5B) and SLC25A21 (Fig. 5C) were both up-regulated in HCFs, compared to HKCs, upon T1 and T3 treatment (p<0.001). SLC25A31 (Fig. 5D) was up-regulated in control HKCs when compared to HCFs (p<0.01); however, SLC25A31 expression was down-regulated following T3 treatment in HKCs compared to HCFs (Fig. 5D; p<0.01).
Figure 5.
Mitochondrial genes that were significantly different between HFCs and HKCs, either in controls, T1, or T3. A) SLC25A4 was significantly up-regulated in control HKCs compared to control HCFs. T3 treatment led to a significant down-regulation of SLC25A4 in HKCs compared to HCFs; B) SLC25A2 was significantly down-regulated in HKCs, compared to HCFs, in both T1 and T3; C) SLC25A21 was significantly down-regulated in HKCs, compared to HCFs, in both T1 and T3; D) SLC25A31 was significantly up-regulated in control HKCs compared to control HCFs. T3 treatment led to a significant down-regulation of SLC25A31 in HKCs compared to HCFs. All samples were repeated at least three times, with p<0.05 considered to be statistically significant ($$$p<0.0001, $$p<0.001, $p<0.01, ***p<0.0005, **p<0.005, *p<0.05).
DISCUSSION
Keratoconus is a multifactorial disease involving complex interaction of both genetic and environmental factors (15, 16, 18–24, 48). The majority of studies narrowly focus on the identification of a single gene as the possible cause of KC; however, a large number of these studies have not been confirmed and the association of the disease with a single gene has not been established (17, 31, 48). More recently, studies have focused on the role of mitochondria as regulators of oxidative damage to tissue and/or cells. Indeed, KC has been linked to oxidative damage since the 1990s, where KC corneas were found to be oxidatively damaged, and to have various abnormalities in stress-related enzymes (13, 14, 26–28) and metabolites (17). However, to the authors’ knowledge, no group has linked mitochondrial dysfunction or regulation of mitochondrial genes to KC derived cells in vitro.
Our study examined cells isolated from human corneal stroma from healthy individuals and those with KC. The aim of our study was to identify and report any differences in mitochondrial genes between these cell types. Furthermore, we investigated the role of two TGF-β isoforms, T1 and T3, which are known to play a role in KC and corneal fibrosis in general (17, 31–34).
Our most intriguing data from these experiments was that the HCFs’ mitochondrial gene profile was more affected by the TGF-β isoforms than the HKC, 9/84 compared with 0/84 genes affected. This supports our previous work where we reported that HKCs were terminally differentiated into the myofibroblast phenotype, and HCFs were not (17, 31). Our data here suggests that HKCs’ mitochondrion were probably damaged to a point that the cells simply could not respond to any further TGF-β stimulation. Interestingly, with HCFs, T1 and T3 did not always have the same result, which agrees with previous data on the effects of T1 and T3 on fibrosis (17, 31–34). In the present study, T3 acted opposite to T1 and “maintained” the expression of the following mitochondrial genes at control levels: NEFL, CPT1B, UCP1, SLC25A2, and SLC25A31. NEFL (Fig. 1D) has been reported to be regulated in HCFs in response to dexamethasone, a therapeutic agent (49). CPT1B (Fig. 1C), and more specifically L-carnitine, was found to protect against corneal stress activation (50). SLC25A2 and A31 (Fig. 3A and C), are both mitochondrial carriers and any dysregulation will almost certainly alter cell and/or tissue mechanisms (51). Although A2 and A31 act through different substrate carrier proteins (ornithine and adenine nucleotide, respectively), they are both vital for normal cellular/tissue function (52).
When HCFs and HKCs were directly compared, several genes were different between the two cell types, indicating inherent differences—BNIP3, PMAIP1, SOD2, SLC25A2, SLC25A4, SLC25A21, and SLC25A31. Both SLC25A2 and A31 were significantly lower in HKC-T3 when compared to HCF-T3; however, their expression was higher in HKC-C compared to HCF-C (Fig. 5B and D). This indicates a potential regulatory role of T3 for these two mitochondrial carriers and significant alterations in KC diseased cells. BNIP3 and PMAIP1 (Fig. 4C and B, respectively) are both members of the BCL-2 family, and they act as anti- or pro-apoptotic regulators that are involved in a wide variety of cellular functions. Their role in keratoconus is unknown, however, based on our data, it is rather safe to assume they are key players on regulating cellular response (53). SOD2 was a very intriguing gene during our analysis. SOD2 is a member of the iron/manganese superoxide dismutase family. Mutations in this gene have been associated with idiopathic cardiomyopathy (IDC), sporadic motor neuron disease, and cancer, and mice lacking SOD2 die shortly after birth, indicating that unchecked levels of superoxide are incompatible with mammalian life (46). In ocular health, SOD2 down-regulation is known to cause elevated oxidative stress and ROS production, with potential mitochondrial membrane loss and early cell apoptosis (54). In our study, SOD2 expression was consistently lower in HKCs when compared to HCFs, independent of treatment (Fig. 4A). This agrees with KC corneal cell loss as the disease progresses, and it would be interesting to further investigate.
Overall, our study reports a panel of mitochondrial genes that are differentially regulated in corneal KC derived cells. Future studies are necessary in order to validate those genes and understand their mechanism. Mitochondrial function is key for the health of human tissues and the optimum cellular activity. We, therefore, believe that we can dissect specific mitochondrial dysfunctions in HKCs in vitro in order to develop future therapeutic agents.
CONCLUSIONS
Overall, our data supports the growing consensus that mitochondrial dysfunction is a key player in KC disease. Our data shows clear links between mitochondrial function and TGF-β isoforms. Clearly further studies are necessary in order to unravel mitochondrial mechanisms in KC.
Acknowledgments
FUNDING
This work was supported by the National Institutes of Health/National Eye Institute grants EY023568 and EY020886 (DK), and, in part, by an unrestricted grant (DMEI) from Research to Prevent Blindness (New York, NY USA).
Supported, in part, by an unrestricted grant from Research to Prevent Blindness, (New York, NY USA).
Footnotes
DECLARATION OF INTERESTS
The authors declare that there is no conflict of interest.
References
- 1.Dianov GL, Souza-Pinto N, Nyaga SG, Thybo T, Stevnsner T, Bohr VA. Base excision repair in nuclear and mitochondrial DNA. Prog nucleic acid res mol biol. 2001;68:285–97. doi: 10.1016/s0079-6603(01)68107-8. [DOI] [PubMed] [Google Scholar]
- 2.de Souza-Pinto NC, Mason PA, Hashiguchi K, Weissman L, Tian J, Guay D, et al. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair. 2009;8(6):704–19. doi: 10.1016/j.dnarep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ingman M, Kaessmann H, PaÈaÈbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408(6813):708–13. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 4.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331(6158):717–9. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 5.Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza A, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242(4884):1427–30. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 6.McFarland R, Taylor R, Turnbull D. Mitochondrial disease—its impact, etiology, and pathology. Curr top dev biol. 2007;77:113–55. doi: 10.1016/S0070-2153(06)77005-3. [DOI] [PubMed] [Google Scholar]
- 7.Schaefer AM, McFarland R, Blakely EL, He L, Whittaker RG, Taylor RW, et al. Prevalence of mitochondrial DNA disease in adults. Ann neurol. 2008;63(1):35–9. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- 8.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Current Biology. 2006;16(14):R551–R60. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 9.Lesnefsky EJ, Moghaddas S, Tandler B, Kerner J, Hoppel CL. Mitochondrial dysfunction in cardiac disease: ischemia–reperfusion, aging, and heart failure. J Mol Cell Cardiol. 2001;33(6):1065–89. doi: 10.1006/jmcc.2001.1378. [DOI] [PubMed] [Google Scholar]
- 10.Zeviani M, Di Donato S. Mitochondrial disorders. Brain. 2004;127(10):2153–72. doi: 10.1093/brain/awh259. [DOI] [PubMed] [Google Scholar]
- 11.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6(5):389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherer TB, Betarbet R, Greenamyre JT. Environment, mitochondria, and Parkinson’s disease. Neuroscientist. 2002;8(3):192–7. doi: 10.1177/1073858402008003004. [DOI] [PubMed] [Google Scholar]
- 13.Pieczenik SR, Neustadt J. Mitochondrial dysfunction and molecular pathways of disease. Exp mol pathol. 2007;83(1):84–92. doi: 10.1016/j.yexmp.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Schrier SA, Falk MJ. Mitochondrial disorders and the eye. Curr opin ophthalmol. 2011;22(5):325. doi: 10.1097/ICU.0b013e328349419d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordan LT. Keratoconus: diagnosis and treatment. Int ophthalmol clin. 1997;37(1):51–63. doi: 10.1097/00004397-199703710-00005. [DOI] [PubMed] [Google Scholar]
- 16.Edrington T, Zadnik K, Barr J. Keratoconus. Optom clin. 1994;4(3):65–73. [PubMed] [Google Scholar]
- 17.Karamichos D, Hutcheon A, Rich C, Trinkaus-Randall V, Asara J, Zieske J. In vitro model suggests oxidative stress involved in keratoconus disease. Sci rep. 2014;4 doi: 10.1038/srep04608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahi A, Davies P, Ruben M, Lobascher D, Menon J. Keratoconus and coexisting atopic disease. Br J Ophthalmol. 1977;61(12):761–4. doi: 10.1136/bjo.61.12.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasset A, Hinson W, Frias J. Keratoconus and atopic diseases. Ann Ophthalmol. 1978;10(8):991–4. [PubMed] [Google Scholar]
- 20.Bawazeer AM, Hodge WG, Lorimer B. Atopy and keratoconus: a multivariate analysis. Br J ophthalmol. 2000;84(8):834–6. doi: 10.1136/bjo.84.8.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cullen J, Butler H. Mongolism (Down’s syndrome) and keratoconus. Br J ophthalmol. 1963;47(6):321. doi: 10.1136/bjo.47.6.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shapiro MB, France TD. The ocular features of Down’s syndrome. Am J ophthalmol. 1985;99(6):659–63. doi: 10.1016/s0002-9394(14)76031-3. [DOI] [PubMed] [Google Scholar]
- 23.Robertson I. Keratoconus and the Ehlers-Danlos syndrome: a new aspect of keratoconus. The Med J Aust. 1975;1(18):571–3. doi: 10.5694/j.1326-5377.1975.tb111590.x. [DOI] [PubMed] [Google Scholar]
- 24.Woodward E, Morris M. Joint hypermobility in keratoconus. Ophthal Physl Opt. 1990;10(4):360–2. doi: 10.1111/j.1475-1313.1990.tb00882.x. [DOI] [PubMed] [Google Scholar]
- 25.Pathak D, Nayak B, Singh M, Sharma N, Tandon R, Sinha R, et al. Mitochondrial complex 1 gene analysis in keratoconus. Mol vis. 2011;17:1514. [PMC free article] [PubMed] [Google Scholar]
- 26.Abu-Amero KK, Azad TA, Kalantan H, Sultan T, Al-Muammar AM. Mitochondrial sequence changes in keratoconus patients. IOVS. 2014;55(3):1706–10. doi: 10.1167/iovs.14-13938. [DOI] [PubMed] [Google Scholar]
- 27.Wojcik KA, Kaminska A, Blasiak J, Szaflik J, Szaflik JP. Oxidative stress in the pathogenesis of keratoconus and Fuchs endothelial corneal dystrophy. Int J mol sci. 2013;14(9):19294–308. doi: 10.3390/ijms140919294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atilano SR, Coskun P, Chwa M, Jordan N, Reddy V, Le K, et al. Accumulation of mitochondrial DNA damage in keratoconus corneas. IOVS. 2005;46(4):1256–63. doi: 10.1167/iovs.04-1395. [DOI] [PubMed] [Google Scholar]
- 29.Karamichos D, Zieske J, Sejersen H, Sarker-Nag A, Asara JM, Hjortdal J. Tear metabolite changes in keratoconus. Exp Eye Res. 2015 doi: 10.1016/j.exer.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balasubramanian SA, Wasinger VC, Pye DC, Willcox MD. Preliminary identification of differentially expressed tear proteins in keratoconus. Mol Vis. 2013;19:2124–34. [PMC free article] [PubMed] [Google Scholar]
- 31.Karamichos D, Zareian R, Guo X, Hutcheon AE, Ruberti JW, Zieske JD. Novel in vitro model for keratoconus disease. J Funct Biomater. 2012;3(4):760–75. doi: 10.3390/jfb3040760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karamichos D, Hutcheon A, Zieske J. Reversal of fibrosis by TGF-β3 in a 3D in vitro model. Exp Eye Res. 2014;124:31–6. doi: 10.1016/j.exer.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karamichos D, Rich CB, Zareian R, Hutcheon AE, Ruberti JW, Trinkaus-Randall V, et al. TGF-β3 stimulates stromal matrix assembly by human corneal keratocyte-like cells. IOVS. 2013;54(10):6612–9. doi: 10.1167/iovs.13-12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karamichos D, Hutcheon A, Zieske J. Transforming growth factor-β3 regulates assembly of a non-fibrotic matrix in a 3D corneal model. J Tissue Eng Regen Med. 2011;5(8):e228–e38. doi: 10.1002/term.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gatherer D, Ten Dijke P, Baird D, Akhurst R. Expression of TGF-beta isoforms during first trimester human embryogenesis. Development. 1990;110(2):445–60. doi: 10.1242/dev.110.2.445. [DOI] [PubMed] [Google Scholar]
- 36.Millan FA, Denhez F, Kondaiah P, Akhurst RJ. Embryonic gene expression patterns of TGF beta 1, beta 2 and beta 3 suggest different developmental functions in vivo. Development. 1991;111(1):131–43. doi: 10.1242/dev.111.1.131. [DOI] [PubMed] [Google Scholar]
- 37.Pelton R, Dickinson M, Moses H, Hogan B. In situ hybridization analysis of TGF beta 3 RNA expression during mouse development: comparative studies with TGF beta 1 and beta 2. Development. 1990;110(2):609–20. doi: 10.1242/dev.110.2.609. [DOI] [PubMed] [Google Scholar]
- 38.Pelton RW, Saxena B, Jones M, Moses HL, Gold LI. Immunohistochemical localization of TGF beta 1, TGF beta 2, and TGF beta 3 in the mouse embryo: expression patterns suggest multiple roles during embryonic development. J Cell Biol. 1991;115(4):1091–105. doi: 10.1083/jcb.115.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmid P, Cox D, Bilbe G, Maier R, McMaster GK. Differential expression of TGF beta 1, beta 2 and beta 3 genes during mouse embryogenesis. Development. 1991;111(1):117–30. doi: 10.1242/dev.111.1.117. [DOI] [PubMed] [Google Scholar]
- 40.Hijikata M, Kato N, Sato T, Kagami Y, Shimotohno K. Molecular cloning and characterization of a cDNA for a novel phorbol-12-myristate-13-acetate-responsive gene that is highly expressed in an adult T-cell leukemia cell line. J Virol. 1990;64(10):4632–9. doi: 10.1128/jvi.64.10.4632-4639.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jansson AK, Emterling AM, Arbman G, Sun X-F. Noxa in colorectal cancer: a study on DNA, mRNA and protein expression. Oncogene. 2003;22(30):4675–8. doi: 10.1038/sj.onc.1206655. [DOI] [PubMed] [Google Scholar]
- 42.Dejean LM, Martinez-Caballero S, Manon S, Kinnally KW. Regulation of the mitochondrial apoptosis-induced channel, MAC, by BCL-2 family proteins. Biochim Biophys Acta. 2006;1762(2):191–201. doi: 10.1016/j.bbadis.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Reed JC, Zha H, Aime-Sempe C, Takayama S, Wang HG. Structure-function analysis of Bcl-2 family proteins. Regulators of programmed cell death. Adv Exp Med Biol. 1996;406:99–112. [PubMed] [Google Scholar]
- 44.Liu J, Li J, Li W-J, Wang C-M. The role of uncoupling proteins in diabetes mellitus. J Diabetes Res. 2013;2013 doi: 10.1155/2013/585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park K-P, Kim H-S, Kim E-S, Park Y-E, Lee C-H, Kim D-S. SLC25A4 and C10ORF2 mutations in autosomal dominant progressive external ophthalmoplegia. J Clin Neurol. 2011;7(1):25–30. doi: 10.3988/jcn.2011.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Huang T-T, Carlson EJ, Melov S, Ursell PC, Olson JL, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nature genetics. 1996;(11):376–81. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 47.Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, et al. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol genomics. 2003;16(1):29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- 48.Barr JT, Wilson BS, Gordon MO, Rah MJ, Riley C, Kollbaum PS, et al. Estimation of the incidence and factors predictive of corneal scarring in the Collaborative Longitudinal Evaluation of Keratoconus (CLEK) Study. Cornea. 2006;25(1):16–25. doi: 10.1097/01.ico.0000164831.87593.08. [DOI] [PubMed] [Google Scholar]
- 49.Liu L, Walker EA, Kissane S, Khan I, Murray PI, Rauz S, et al. Gene expression and miR profiles of human corneal fibroblasts in response to dexamethasone. IOVS. 2011;52(10):7282–8. doi: 10.1167/iovs.11-7463. [DOI] [PubMed] [Google Scholar]
- 50.Flanagan JL, Simmons PA, Vehige J, Willcox MD, Garrett Q. Role of carnitine in disease. Nutr Metab. 2010;7:30. doi: 10.1186/1743-7075-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker JE. The mitochondrial transporter family. Curr opin struct biol. 1992;2(4):519–26. [Google Scholar]
- 52.Porter RK. Mammalian mitochondrial inner membrane cationic and neutral amino acid carriers. Biochim Biophys Acta. 2000;1459(2):356–62. doi: 10.1016/s0005-2728(00)00172-9. [DOI] [PubMed] [Google Scholar]
- 53.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288(5468):1053–8. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 54.Liu C, Ogando D, Bonanno JA. SOD2 contributes to anti-oxidative capacity in rabbit corneal endothelial cells. Mol Vis. 2011;17:2473. [PMC free article] [PubMed] [Google Scholar]