Abstract
Protein kinases are frequently regulated by intramolecular autoinhibitory interactions between protein modules that are reversed when these modules bind other ‘activating’ protein or membrane-bound targets. One group of kinases, the MAP/microtubule affinity-regulating kinases (MARKs) contain a poorly understood regulatory module, the KA1 (kinase associated-1) domain, at their C-terminus. KA1 domains from MARK1 and several related kinases from yeast to humans have been shown to bind membranes containing anionic phospholipids, and peptide ligands have also been reported. Deleting or mutating the C-terminal KA1 domain has been reported to activate the kinase in which it is found — also suggesting an intramolecular autoinhibitory role. Here, we show that the KA1 domain of human MARK1 interacts with, and inhibits, the MARK1 kinase domain. Using site-directed mutagenesis, we identify residues in the KA1 domain required for this auto-inhibitory activity, and find that residues involved in autoinhibition and in anionic phospholipid binding are the same. We also demonstrate that a ‘mini’ MARK1 becomes activated upon association with vesicles containing anionic phospholipids, but only if the protein is targeted to these vesicles by a second signal. These studies provide a mechanistic basis for understanding how MARK1 and its relatives may require more than one signal at the membrane surface to control their activation at the correct location and time. MARK family kinases have been implicated in a plethora of disease states including Alzheimer’s, cancer, and autism, so advancing our understanding of their regulatory mechanisms may ultimately have therapeutic value.
Introduction
Precise control of protein kinase activity is crucial for normal cell signaling. In many cases this is achieved by noncatalytic domain(s) within modular proteins that interact directly with the kinase domain to exert either a positive or negative (autoinhibitory) influence [1]. Src and protein kinase C (PKC) are particularly well-studied illustrations [1, 2]. The Src kinase domain is autoinhibited by intramolecular interactions with SH2 and SH3 domains within the same protein, disrupted when these domains become engaged by other protein ligands. PKC and its relatives also assume an autoinhibited conformation, stabilized by intramolecular domain/domain interactions that are disrupted when the lipid-binding C1 and C2 domains both bind targets in the cell membrane [2]. It is now clear that many modular protein kinases share this basic regulatory principle [3]. In addition to C1 and C2 domains, several other lipid-binding modules are frequently found alongside kinase domains, including pleckstrin homology (PH), phox homology, FERM, and kinase associated-1 (KA1) domains [3–5]. Structural studies have provided valuable insights into the precise mechanisms of intramolecular regulation for Src and PKC family members [6–8], as well as FAK [9]. Details are also now emerging for how PH domains exert their lipid-modulated autoinhibitory effects [10, 11], but relatively little is known for other lipid-binding domains.
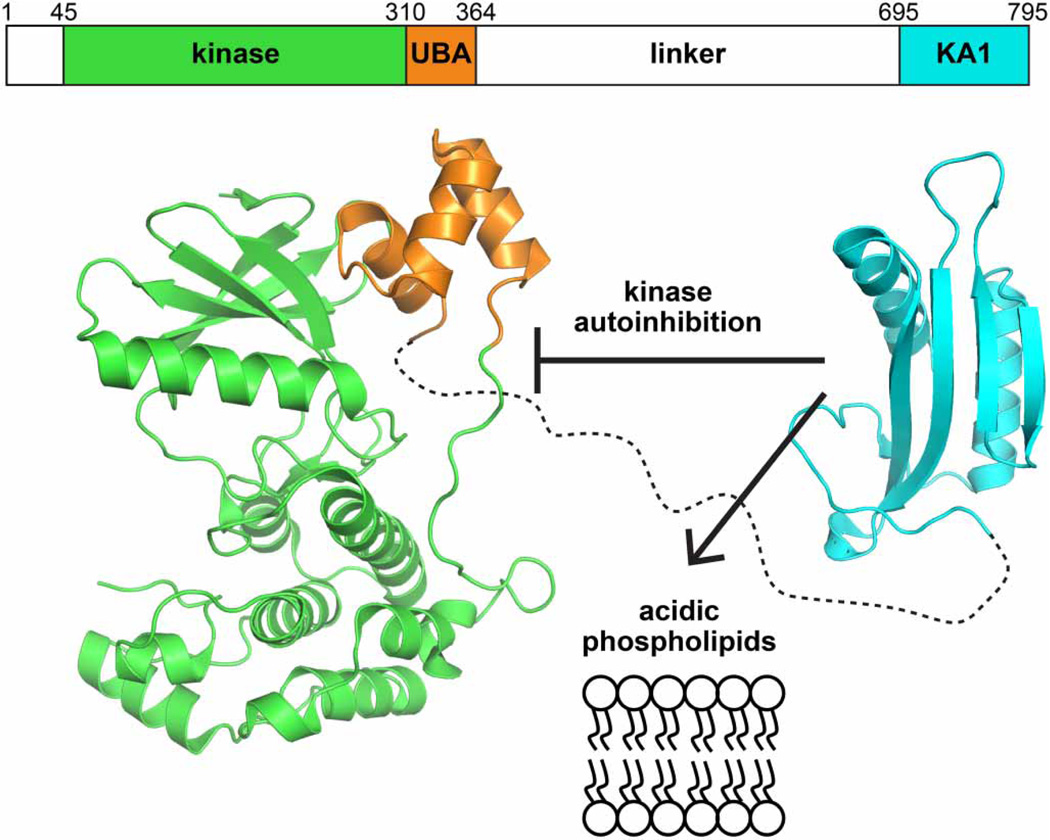
We previously reported that the KA1 domains of MARK/PAR1 family kinases bind acidic phospholipids [12], and numerous studies have suggested an autoregulatory role for the (typically C-terminal) KA1 domain in these (and related) kinases [13]. The MARKs (for MAP/microtubule affinity-regulating kinases) form a subfamily of the calcium/calmodulin-dependent protein kinase group [14] and are closely related to the AMP-activated protein kinases. Their domain composition is depicted in Figure 1, with an N-terminal Ser/Thr kinase domain followed successively by an ubiquitin-associated (UBA) domain, a linker (or ‘spacer’) of ~300 amino acids that is predicted to be unstructured [13, 15], and the C-terminal KA1 domain. Crystal structures of kinase–UBA domain fragments of MARK kinases [16–19] and other relatives [20, 21] have shown that the UBA domain abuts (and regulates) the kinase domain [13] as shown in Figure 1. Whether the KA1 domain also interacts directly with the kinase domain in cis to autoinhibit it is not yet clear. Deletion studies have suggested such an autoinhibitory role for the KA1 domain in MELK [22] and in the Saccharomyces cerevisiae Kin1/2 orthologs [23]. Similarly, mutations in the deduced KA1 domain in Chk1 (in the same branch of the kinome) were shown to promote constitutive activation of this kinase in the absence of DNA damage [24]. On the other hand, a structure of a related kinase called SAD (for synapses of amphids defective) indicated that the KA1 domain is only loosely involved in intramolecular interactions [21] — with an autoinhibitory domain adjacent to the KA1 domain (and unique to SAD kinases) that directly interacts with the kinase domain.
Figure 1. Proposed roles of the MARK1 KA1 domain.
The C-terminal KA1 domain (cyan) of MARK1 (PDB: 3OSE) is separated from the kinase (green) and UBA (orange) domains (PDB: 2HAK) by a linker of ~300 amino acids that is predicted to be unstructured [13, 15]. The KA1 domain has been separately implicated in autoinhibition of kinase activity [13], binding to anionic phospholipid membranes [12], and binding to Gab1 (not shown) [49]. Numbering is shown for human MARK1.
Motivated by these observations, we initiated efforts to understand how KA1 domains in MARK family kinases regulate kinase activity. In this report, we show that the recombinant MARK1 KA1 domain can bind and inhibit the human MARK1 kinase–UBA fragment in trans, and we use mutagenesis to identify the face of the KA1 domain that mediates this binding. Furthermore, we show that a ‘mini’ MARK1 construct (lacking most of the linker region between the kinase and KA1 domains) is autoinhibited and can be activated by KA1 domain mutations. Moreover, ‘mini’ MARK1 can be activated in a KA1-dependent manner by enforced recruitment to vesicles containing anionic phospholipids. Our findings provide new insights into the role of the KA1 domain in MARK1 regulation. MARK1 and its relatives play diverse roles in cell physiology, most notably in establishing cell polarity through phosphorylation of Tau protein [25, 26]. MARK family kinases have been implicated in multiple diseases, including Alzheimer’s disease, autism, and cancer [27–30]. Thus, advancing our understanding of how kinases in this family are regulated should help illuminate new approaches for therapeutic targeting that will complement those that target the kinase domain itself.
Experimental procedures
Expression constructs and protein purification
Constructs encoding various human MARK1 fragments with a noncleavable N-terminal hexahistidine tag were generated in pET21a (EMD Millipore) for expression in Escherichia coli. MARK1 fragments included the kinase–UBA fragment (residues 45–371: ΔN-KINWT-UBA; WT, wild type), a T215E-mutated form of the kinase–UBA fragment (ΔN-KINTE-UBA), the MARK1 KA1 domain (residues 683–795), and variants of ‘mini’ MARK1 T215E (from which residues 383–681 have been deleted from the linker). Other mutations and truncations are described in the text. A version of ΔN-KINWT-UBA was also generated with a tobacco etch virus (TEV) protease cleavage site inserted between the hexahistidine tag and the kinase domain. KA1 domain point mutations were introduced using ‘round-the-horn’ site-directed mutagenesis [31]. cDNA encoding human Gab1 was a gift of Dr Benjamin Neel (NYU Langone Medical Center, New York, NY). The region encoding residues 152–251 of Gab1 was amplified by PCR to introduce a noncleavable N-terminal hexahistidine tag and was subcloned into pET21a. Expression constructs for the hexahistidine-tagged KA1 domains from S. cerevisiae Chk1 (residues 412–527) and Kcc4p (residues 917–1037) were generated in pET21a as described previously [12].
All constructs were transformed into OverExpress C41(DE3) E. coli and induced for expression using 1 mM IPTG [32]. Cultures harboring the Gab1 fragment were induced for 4 h at 37°C, whereas others were induced at 25°C for 16 h. Hexahistidine-tagged proteins were purified at 4°C essentially as described previously [12]. Harvested cell pellets were resuspended in lysis buffer containing 25 mM Tris (pH 8), 150 mM NaCl, 5% (v/v) glycerol, with 1 mM phenylmethylsulfonyl fluoride, and 5 mM β-mercaptoethanol. The suspension was sonicated, and protein was purified from clarified cell lysates using Ni-NTA affinity (step elution with 0.3 M imidazole), cation exchange [Resource S run in 25 mM MES (pH 6), 5% (v/v) glycerol, 5 mM β-mercaptoethanol, with a NaCl gradient of 150–1000 mM], and size exclusion chromatography (SEC) on a Superose 12 column (GE Healthcare) in 20 mM HEPES (pH 7.5), 150 mM NaCl, and 1 mM TCEP (SEC buffer, which was also used for protein storage at 4°C). The Gab1 fragment was purified using Ni-NTA affinity and SEC without an ion exchange step. Purification of the MARK1 kinase–UBA fragment with a TEV-cleavable N-terminal hexahistidine tag involved an additional dialysis step concurrent with TEV cleavage [against 25 mM Tris (pH 8), 300 mM NaCl, 5% (v/v) glycerol, and 10 mM β-mercaptoethanol] followed by removal of the cleaved tag by Ni-NTA chromatography prior to cation exchange. Removal of the hexahistidine tag was crucial for concentrating the kinase–UBA fragment to beyond 5 mg/ml. Tag-cleaved ΔN-KINWT-UBA was used for all experiments except surface plasmon resonance (SPR).
Surface plasmon resonance
SPR experiments were performed on a BIAcore 3000 instrument (GE Healthcare) essentially as described previously [12, 33, 34]. The MARK1 kinase–UBA domain fragment (with hexahistidine tag intact) was immobilized on a CM5 biosensor chip by amine coupling in 10 mM Na acetate (pH 7.0). The running buffer for all experiments was 20 mM HEPES (pH 7.5), 3 mM EDTA, 0.005% (v/v) Surfactant P20, containing 0.15 M NaCl. An extended wash with running buffer was performed following each injection to remove bound protein and to restore the signal to baseline. Preparation of lipid vesicles and immobilization onto L1 biosensor chips for SPR analysis were performed as described previously [12, 34]. 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC), l-α-phosphatidylserine (PS), l-α-phosphatidic acid (PA), and 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] nickel salt (DOGS-Ni-NTA) were obtained from Avanti Polar Lipids, Inc. (Alabaster, AL).
Analytical ultracentrifugation
Sedimentation velocity analytical ultracentrifugation (SV-AUC) was performed using an Optima XL-A instrument (Beckman Coulter, Fullerton, CA) with an Ti-60 rotor held at 4°C and 40 000 rpm. Samples were diluted in SEC buffer, loaded into two-channel charcoal Epon centerpieces between quartz window pieces, and monitored at 280 nm. Buffer and sample-dependent variables were calculated using chemical composition with the program SEDNTERP [35], and size-distribution analysis c(s) was carried out using the Lamm equation within the program SEDFIT [36]. Results were displayed using the GraphPad Prism (GraphPad Software, Inc., San Diego, CA).
Small-angle X-ray scattering
All data were collected in-house on a custom Rigaku PSAXS S-Max3000 system with Osmic mirror optics (Osmic, Inc., Troy, MI), a three-pinhole enclosed flight path, a vacuum chamber with custom 4°C cooled sample mount, a gas-filled multi-wire CCD detector, and a Rigaku MicroMax-007 HF microfocus rotating anode X-ray generator (Rigaku America, Woodland, TX). Protein samples were exposed for 30–60 min, and forward scatter was subjected to circular averaging using SAXSgui ( JJ X-ray Systems Aps, Lyngby, Denmark) to create a one-dimensional intensity profile. Buffer blanks were subtracted, and PRIMUS [37] was used to calculate Guinier region estimates of extrapolated forward scatter at the zero angle (I0) and radius of gyration (Rg). The Guinier region was defined such that q*Rg was held between 0.4 and 1.8. All samples were monodispersed as judged by linearity within the Guinier region (Supplementary Figure S1A) and had good agreement when these parameters were recalculated with GNOM [38]. The maximum dimension (Dmax) was also obtained using GNOM [38]. I0 values were normalized by mass concentration (I0/c) for direct comparison along with a lysozyme standard (14.6 kDa). I0/c for the sample containing 5.3 mg/ml tag-cleaved MARK1 kinase–UBA domain fragment was chosen as the reference value of 1.0.
Thermal shift assays
A real-time PCR instrument was used to read increased fluorescence of SYPRO Orange dye (Invitrogen, Carlsbad, CA). A 15 µl aliquot of 2 µM KA1 domain was mixed with 5 µl of 300-fold diluted dye in 384-well white microplates. Absorbance was read as the temperature of samples was increased from 20°C to 95°C over 90 min. Traces were normalized based on minimum and maximum signal, and plotted using GraphPad Prism.
Kinase assays
MARK1 kinase activity was assayed essentially as previously reported [26]. The assay conditions were 25 mM HEPES (pH 7.5), 1 mM DTT, 5 mM MgCl2, 100 µM ATP, and 100 µM Tau-derived peptide substrate (NVKSKIGSTENLK, Genscript). Enzyme (in SEC buffer) was diluted 5-fold into the assay and reactions were performed at 25°C. Reaction progress was monitored using trace amounts of γ-32P-labeled ATP (~20 µCi per experiment). Phosphorylated peptides were captured by spotting an aliquot of the reaction mixture at each time point onto phosphocellulose paper and immediately quenching with a 0.5% phosphate solution. After three washes in this solution plus one in acetone, incorporated radioactivity was measured by scintillation counting with appropriate background correction. Reaction velocity was calculated as peptide substrate phosphorylated per enzyme molecule per minute. For assays in which vesicles were also included, vesicle preparations were not preincubated with enzyme but added to the master mix. Final protein concentration in each assay was 5 µM, except for the T215E kinase–UBA fragment, which was assayed at 0.5 µM because of its increased activity. The anaplastic lymphoma kinase (ALK) tyrosine kinase was used as a positive control (with its own specific peptide) as described previously [39].
Results
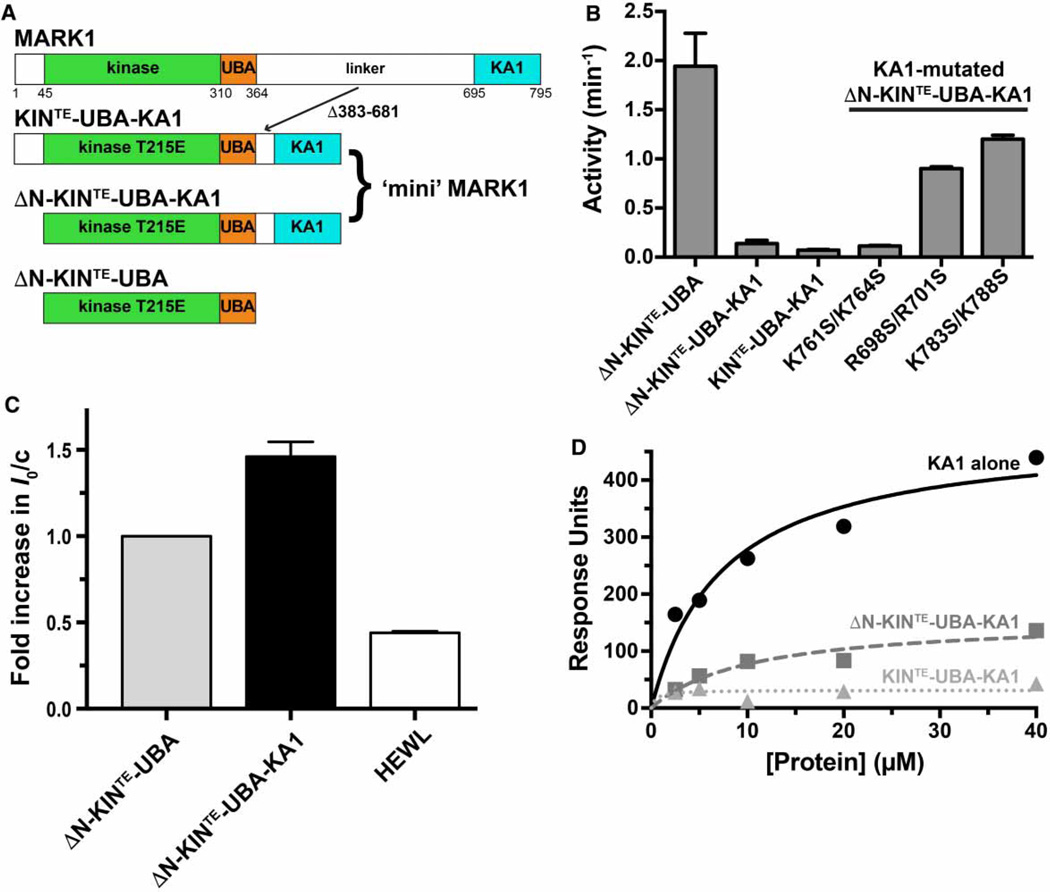
The MARK1 KA1 domain binds the kinase–UBA fragment in trans
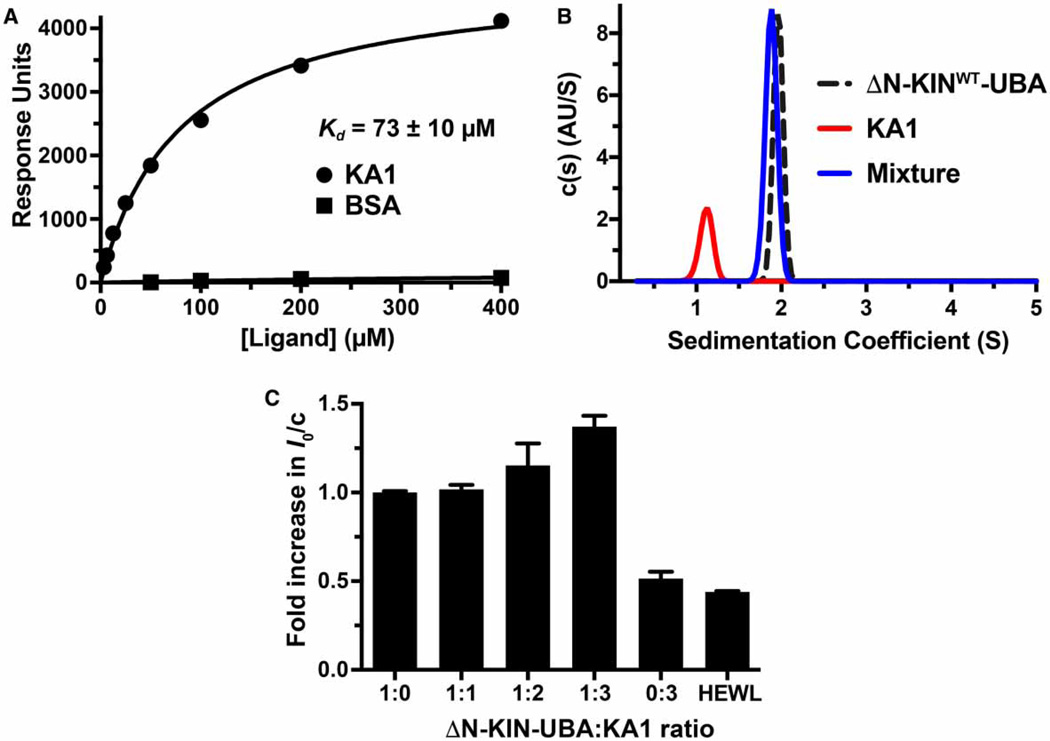
We began by testing the hypothesis that the MARK1 kinase–UBA fragment and the KA1 domain interact with one another when present in separate polypeptides. We immobilized the hexahistidine-tagged kinase–UBA fragment on a CM5 BIAcore sensor chip and used SPR to assess the ability of the KA1 domain to bind when it flowed over the resulting surface. As shown in Figure 2A, the KA1 bound robustly — albeit weakly — to the immobilized kinase–UBA fragment, with an apparent dissociation constant (Kd) of 73±10 µM. Increasing ionic strength reduced both affinity and the maximum extent of this binding (data not shown), indicating that it is driven substantially by electrostatic interactions. The interaction between the kinase–UBA fragment and the KA1 domain could also be detected directly in solution using SV-AUC. When studied alone by SV-AUC, the KA1 domain and the kinase–UBA fragment sediment with sedimentation coefficients of ~1 and ~2 Svedbergs (S), respectively (Figure 2B). When the two proteins are mixed, however (at a protein concentration of 35 µM), the KA1 domain appears to be largely ‘chased’ into the faster sedimenting peak, which is likely to represent a KA1/kinase–UBA heterodimer (Figure 2B). To investigate the absolute molecular mass of the complex formed between these two proteins, we used small-angle X-ray scattering (SAXS). The extrapolated forward scatter at the zero angle (I0) in an SAXS experiment, when normalized for mass concentration, reflects the weight-average molecular weight (MW) of the particles in solution. Alone (at 140 µM), the kinase–UBA fragment gives a normalized I0/c value (arbitrarily set to 1) that is 2.4 times that measured for hen egg white lysozyme (Supplementary Table S1 and Figure 2C), consistent with the ratio of their molecular masses (37.7 and 14.3 kDa, respectively: ratio is 2.6), and arguing that the kinase–UBA fragment is a monomer. When excess KA1 domain (MW = 14.2 kDa) is added, the normalized I0/c for the complex saturates at 1.4 times that for the kinase–UBA fragment, consistent with formation of a 1:1 complex (which would have a molecular mass of 51.9 kDa, i.e. 1.4 times that of the kinase–UBA fragment) — although it should be noted that scattering by the free excess KA1 domain will reduce the normalized I0/c value (and weight-average molecular weight). Values for the radius of gyration (Rg) and maximum particle diameter (Dmax) also increase upon the addition of KA1 domains, providing additional evidence that the KA1 domain forms a complex with the kinase–UBA fragment (Supplementary Table S1). Thus, using three separate biophysical methods we can robustly detect a likely 1:1 complex between the KA1 domain of MARK1 and the kinase–UBA fragment. The interaction is quite weak when studied in trans, as expected since these domains are covalently linked to one another in the natural protein. Indeed, for this interaction to be modulated in regulating MARK1, it is important that the affinity in cis is not too high so that trans interactions with the KA1 domain (membrane binding for example) are able to compete effectively.
Figure 2. Evidence for direct binding of the MARK1 KA1 domain to the kinase–UBA domain fragment.
(A) SPR analysis of the MARK1 KA1 domain binding to the MARK1 kinase–UBA domain fragment (residues 45–371: ΔN-KINWT-UBA) immobilized on a CM5 sensor chip. The binding curve shown is representative of three independent repeats. Fitting to each individual binding curve yields a mean Kd value (±standard deviation) of 73 ± 10 mM for the KA1 domain. No binding was observed for the BSA control. Note that ΔN-KINWT-UBA from which the hexahistidine tag had not been proteolytically removed was used for these studies. (B) Analytical ultracentrifugation sedimentation velocity c(s) distribution analysis of tag-cleaved kinase–UBA domain fragment at 35 mM (black dashed trace), 35 mM KA1 domain alone (red trace), and a mixture of both (blue trace). The fact that there is no detectable free KA1 domain in this mixture suggests that the apparent affinity in this assay is stronger than that measured in the SPR experiment, which could result from restrictions on KA1 binding imposed by covalent immobilization of the kinase–UBA fragment on the SPR sensor chip. Note also that the sedimentation coefficient for the mixture is slightly smaller than that for the kinase–UBA alone, suggesting that the complex may be more compact. Similar c(s) distributions were obtained for two independent protein preparations. (C) Normalized I0 SAXS analysis of the tag-cleaved kinase–UBA domain fragment held at 140 µM (5.3 mg/ml) as KA1 domain was added in trans at increasing concentrations from 140 µM (1:1 ratio) to 420 µM (1:3 ratio). The KA1 domain-only control contained 420 µM protein, and the hen egg white lysozyme (HEWL) control was at 700 µM (10.5 mg/ml). The mean values from three independent experiments (±standard deviation) are shown, with the exception of the 1:3 ratio sample, for which data for two protein preparations are shown (each measured in triplicate). Rg and Dmax values for all samples are listed in Supplementary Table S1.
Trans-inhibition of MARK1 kinase activity by the KA1 domain
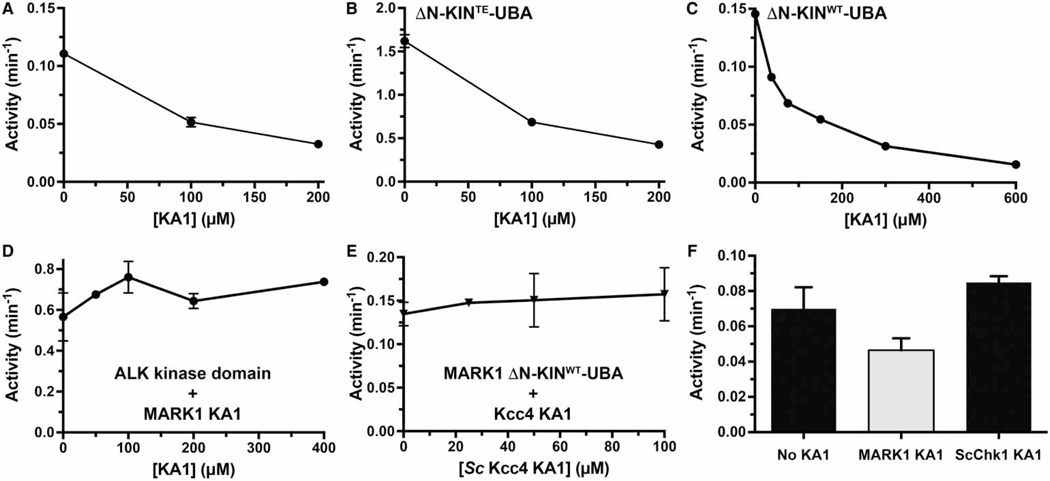
To determine whether KA1 domain binding to the kinase–UBA fragment can inhibit kinase activity in trans (a requirement for cis-autoinhibition), we implemented a previously described kinase assay for MARK1 using a Tau-derived peptide substrate [26]. We assayed both WT and T215E-mutated kinase–UBA fragments as increasing amounts of KA1 domain were added to the reaction. The T215E mutation is designed to mimic (in part) activation loop phosphorylation of MARK1 by LKB1 [40], increasing its catalytic activity. As shown in Figure 3A, the activity of the kinase–UBA fragment is weak (similar to the basal activity of a receptor tyrosine kinase [39]), but is elevated ~15-fold by the T215E phosphomimetic mutation (Figure 3B). Bona fide phosphorylation at this site and others, as well as other influences in the intact protein, are required for full activity [13]. Nonetheless, the addition of KA1 domain had a clear inhibitory effect on both WT (Figure 3A) and T215E (Figure 3B) MARK1 kinase–UBA fragments, and 90% inhibition of the kinase activity could be achieved when KA1 domain was added at concentrations >200 µM (Figure 3C). An approximate IC50 value in the range of 100 µM for kinase–UBA inhibition by the KA1 domain is suggested by the data in Figure 3C. This in turn suggests that Kd for the inhibitory interaction between the KA1 and kinase domains is ~100 µM (similar to that measured in Figure 2A), ~10-fold weaker than KA1 domain binding to membranes containing acidic phospholipids (~10 µM, [12]). For intact MARK1, KA1/membrane interactions in trans would have to compete with cis KA1/kinase interactions to activate MARK1, and hence would need to be significantly stronger — as observed.
Figure 3. Inhibition of MARK1 kinase activity in trans by the KA1 domain.
(A and B) The purified MARK1 KA1 domain was added at increasing concentrations to the MARK1 kinase–UBA domain fragment with the WT kinase domain (ΔN-KINWT-UBA) at 5 µM (A) or harboring the activating T215E activation loop mutation (ΔN-KINTE-UBA) at 0.5 µM (B). Kinase activities of the mixtures were assayed in triplicate using a Tau-derived peptide as a substrate. Values plotted are means ± standard deviation. (C) Titration of up to 600 µM MARK1 KA1 domain into the assay with ΔN-KINWT-UBA resulted in up to ~90% inhibition compared with the kinase–UBA domain fragment alone. By contrast, as shown in (D), the MARK1 KA1 domain (added at up to 400 µM) had no inhibitory effect on activity of the ALK tyrosine kinase domain at 2 µM, and (E) the KA1 domain from S. cerevisiae Kcc4 [12] did not inhibit the MARK1 kinase–UBA domain fragment (present at 5 µM). Also confirming KA1/kinase domain specificity (F), adding the S. cerevisiae Chk1 KA1 domain (at 75 µM) had no effect on activity of the MARK1 kinase–UBA domain fragment (at 5 µM) under the same conditions that the MARK1 KA1 domain (at 75 µM) achieved ~40% inhibition. Mean values ± standard deviation for at least three replicates are reported.
To establish specificity of this effect, we showed that the MARK1 KA1 domain at concentrations >200 µM does not inhibit the ALK tyrosine kinase domain (Figure 3D). Moreover, neither the KA1 domain from the S. cerevisiae MARK/PAR kinase family member Kcc4 [12] nor that from S. cerevisiae Chk1 [24] had a significant effect on MARK1 kinase activity (Figure 3E,F). These data suggest that the MARK1 KA1 domain has a specific regulatory effect on the kinase domain in the same molecule through intramolecular versions of the domain/ domain interactions recapitulated here in trans.
A basic patch on the KA1 domain surface implicated in membrane binding also mediates kinase inhibition
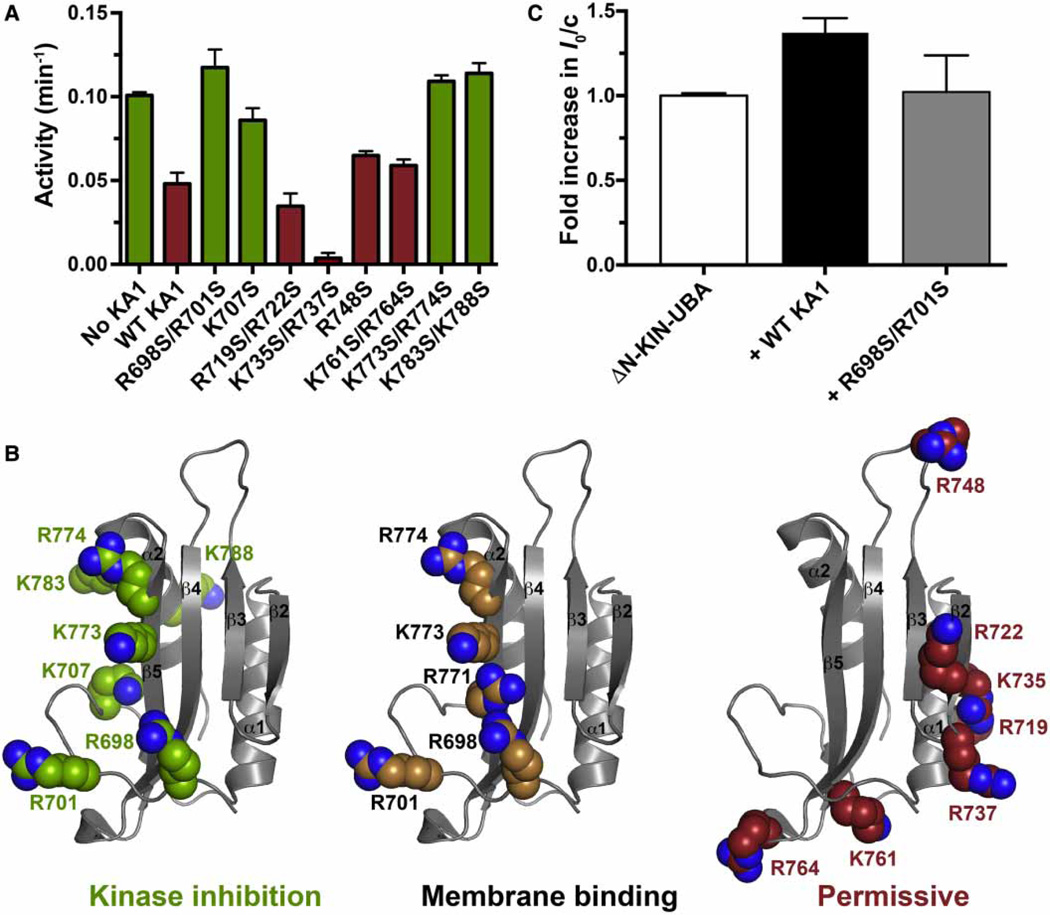
In our previous structural studies of the MARK1 KA1 domain [12] we noted several basic patches on the surface, and found that mutations in at least one of these disrupt binding to anionic membrane surfaces. We evaluated the same set of KA1 domain variants for their ability to inhibit the MARK1 kinase–UBA fragment in trans, to test the hypothesis that the same surface of the KA1 domain might be involved in both cis-autoinhibitory interactions and membrane interactions (which trans activate MARK1). As shown in Figure 4A, several — but not all — KA1 domain variants in which basic residues at the surface had been replaced by serine failed to suppress activity of the kinase–UBA fragment. In particular, the R698S/R701S, K707S, K773S/ R774S, and K783S/K788S variants all lost inhibitory activity. These residues lie around strands β1 and β5 of the KA1 domain as well as helix α2, in a region that clearly overlaps with — and extends beyond — the region that we previously implicated in membrane binding (Figure 4B). As also shown in Figure 4B (right-most panel), the basic residues at which mutations failed to prevent trans kinase inhibition or membrane binding (i.e. the ‘permissive’ residues) lie in a distinct region of the KA1 domain surface. To exclude the possibility that the loss of trans-inhibitory activity in Figure 4A resulted simply from structural destabilization of the KA1 domain, melting temperatures were measured for each mutated variant, monitoring with SYPRO Orange fluorescence. Although a range of melting temperatures was observed (42–62°C), no mutated KA1 domain displayed a melting temperature of <40°C (Supplementary Figure S2A), and there was no correlation between melting temperature and inhibitory activity (Supplementary Figure S2B). Furthermore, SDS–PAGE showed similar expression levels and purity of all mutated KA1 domain variants (Supplementary Figure S2C). The most stable of the KA1 domain variants with impaired trans kinase inhibition ability (R698S/R701S) was also analyzed for its capacity to bind to the kinase–UBA fragment using SAXS. As shown in Figure 4C, the increase in I0/c seen (at 140 µM protein) when the WT KA1 domain was added to the kinase–UBA fragment was not as pronounced with the R698S/ R701S variant, suggesting that this KA1 domain variant binds more weakly to the kinase–UBA fragment. Adding the mutated KA1 domain also did not increase Rg or Dmax much beyond the values (Rg = 28.1 ± 0.4 Å, Dmax = 75 Å; Supplementary Table S1) measured for the ΔN-KIN-UBA fragment alone, reaching only 27.2 ± 0.2 Å (Rg) and 85 Å (Dmax).
Figure 4. Mapping the autoinhibitory interface of the MARK1 KA1 domain.
(A) The noted purified KA1 domain variants (at 100 µM) were added to 5 µM kinase–UBA domain fragment (ΔN-KINWT-UBA, with hexahistidine tag removed) and kinase activity assessed in triplicate. Mean values ± standard deviation are plotted. (B) The MARK1 KA1 domain (PDB: 3OSE) is shown in gray cartoon representation with selected basic side chains shown as spheres. Mutation of those colored green in the left-hand panel prevented the KA1 domain from inhibiting kinase activity in trans in (A). Mutation of those colored tan in the middle panel impaired KA1 domain binding to membranes containing acidic phospholipids in our previous studies [12], whereas mutation of the side chains colored red in the right-most panel did not affect trans kinase inhibition or membrane binding, so we have termed them ‘permissive’. Our data suggest that the binding sites for kinase inhibition and membrane association are co-located on the KA1 domain surface. (C) Analysis of increases in weight-average molecular weight (MW) by monitoring the intensity of forward scatter (I0) in SAXS experiments shows that the WT KA1 domain forms a complex with tag-cleaved ΔN-KINWT-UBA (at 140 µM) — showing the same data as in (C) — but the R698S/R701S variant (which does not inhibit kinase activity) does so less robustly, suggesting that this KA1 domain variant binds more weakly to the kinase–UBA fragment. KA1 domains were added at 280 µM. Rg and Dmax for ΔN-KINWT-UBA plus mutated KA1 domain were 27.2 ± 0.2 Å and 85 Å respectively, compared with 28.1 ± 0.4 Å and 75 Å respectively, for ΔN-KINWT-UBA alone and 30 ± 1.0 Å and 95 Å for ΔN-KINWT-UBA plus (2×) WT KA1 domain (Supplementary Table S1). Mean and standard deviations for at least three measurements from two independent protein preparations are shown.
Taken together, these results indicate that the same face of the MARK1 KA1 domain is involved in both membrane binding and autoinhibitory interactions with the kinase domain, in turn suggesting that membrane binding by the KA1 domain could activate MARK1 by disengaging such cis-autoinhibitory interactions. Importantly, all of the KA1 domain variants tested in Figure 4A that retained the ability to inhibit the MARK1 kinase–UBA fragment also retained WT membrane association in our earlier studies [12]. Although the basic character of the lipid-/kinase-binding surface appears to be conserved across KA1 domains (allowing relatively nonspecific membrane binding), the structural details of this surface differ significantly, which may allow distinct specific protein-binding (kinase domain) partners.
Linker-truncated ‘mini’ MARK1 variants are autoinhibited
As mentioned in the Introduction section, several studies have indicated that deleting the KA1 domain from MARK/PAR family kinases promotes their activation [13]. In an effort to do the reverse, we effectively appended the KA1 domain to the kinase–UBA fragment to ask whether this domain can exert a cis-autoinhibitory influence when included in the same molecule. This amounts to deleting 299 amino acids (residues 383–681) from the linker (predicted to be unstructured, [13, 15]) between the UBA domain and the KA1 domain (Figure 5A). In parallel, we were also interested in determining whether the ~45 amino acid predicted unstructured region at the amino-terminus has any influence on the catalytic activity of this construct.
Figure 5. Intramolecular autoinhibition of MARK1 by the KA1 domain.
(A) Diagram of MARK1 constructs. Full-length MARK1 is shown at the top, and ‘mini’ MARK1 was generated by deleting residues 383–681 from the linker between the UBA and KA1 domains, to yield KIN-UBA-KA1. The T215E activation loop phosphomimetic mutation was also introduced to yield KINTE-UBA-KA1. A second ‘mini’ MARK1 construct is identical except that it lacks the N-terminal 44 residues (ΔN-KINTE-UBA-KA1), and a T215E version of the kinase–UBA domain fragment (ΔN-KINTE-UBA) was also generated. All constructs contain an N-terminal hexahistidine tag. (B) A comparison of the kinase activities of KINTE-UBA-KA1 and ΔN-KINTE-UBA-KA1 (assayed at 5 µM) with that of ΔN-KINTE-UBA (assayed at 0.5 µM) shows that KA1 domain deletion from ‘mini’ MARK1 increases activity by ~25-fold or more. Enhanced activity of variants of ΔN-KINTE-UBA-KA1 with a mutated KA1 domain (assayed at 1 µM) further shows that alterations that disrupt kinase inhibition in trans by the KA1 domain (R698S/R701S or K783S/K788S; Figure 4) increase the activity of ‘mini’ MARK1 by ~12–17-fold. By contrast, permissive mutations (K761S/K764S) have no effect. Activities were assayed in triplicate, and mean values ± standard deviation plotted. (C) SAXS analysis shows that the weight-average molecular weight (MW) of tag-cleaved ΔN-KINTE-UBA-KA1 (at 140 µM) plotted as a relative I0 value is 1.4-fold greater than that for ΔN-KINTE-UBA (at 140 µM). Comparison with Figure 2C shows that the addition of the KA1 domain in trans has the same effect on I0 as appending it in cis, increasing I0/c by the same factor (~1.4), and increasing Rg from 28.1 ± 0.4 Å (for ΔN-KINTE-UBA) to 31.7 ± 1.7 Å for ΔN-KINTE-UBA-KA1 and Dmax from 75 to 95 Å — similar to the increases seen when KA1 domain is added in trans (Supplementary Table S1). A HEWL control is also shown. The mean and standard deviations for at least three measurements from two independent protein preparations are shown. (D) SPR analysis of binding to membranes containing acidic phospholipids for the isolated MARK1 KA1 domain, ΔN-KINTE-UBA-KA1 ‘mini’ MARK1, and KINTE-UBA-KA1 ‘mini’ MARK1. DOPC membranes containing 20% (mole/mole) PS were immobilized on L1 sensor chips as described previously [12]. Binding curves are representative of at least three independent repeats and were fit separately. The Kd value (±standard deviation) estimated for the MARK1 KA1 domain binding to these vesicles was 7.4 ± 2.6 µM. Neither ΔN-KINTE-UBA-KA1 nor KINTE-UBA-KA1 ‘mini’ MARK1 gave robust signals, and binding was too weak to quantitate.
As shown in Figure 5B, the most active form of MARK1 that we were able to generate was the kinase–UBA fragment lacking the N-terminus (ΔN) and harboring the T215E activation loop mutation (ΔN-KINTE-UBA). Simply appending the KA1 domain to this protein (via a 32 amino acid linker) to give ΔN-KINTE-UBA-KA1 (or ‘mini’ MARK1) reduced activity by 15–20-fold. Restoring the N-terminus in this protein (in KINTE-UBA-KA1) had little effect. These data therefore argue that the appended KA1 domain exerts an intramolecular (or cis) auto-inhibitory effect on kinase activity in ΔN-KINTE-UBA-KA1 and KINTE-UBA-KA1. This conclusion is further supported by the finding that KA1 domain mutations that abrogate trans-inhibition of the kinase–UBA fragment by the isolated KA1 domain (R698S/R701S and K783S/K788S) enhanced ‘mini’ MARK1 activity, suggesting that these mutations disrupt cis-autoinhibitory interactions (Figure 5B). By contrast, the K761S/K764S mutation pair, which did not prevent trans-inhibition by the KA1 domain, also did not activate ‘mini’ MARK1. Another piece of data suggesting that interactions between the KA1 and kinase domains are similar whether in cis or in trans came from SAXS experiments. I0/c, Rg, and Dmax values measured for ΔN-KINTE-UBA-KA1 (‘mini’ MARK1) are similar to those measured when the KA1 domain was added in trans to KINTE-UBA (see Figure 5C). Moreover, despite containing the complete KA1 domain, SPR studies showed that the ΔN-KINTE-UBA-KA1 and KINTE-UBA-KA1 fragments bind acidic membrane much less effectively than the isolated KA1 domain (Figure 5D). This finding is consistent with the idea that the surface of the KA1 domain responsible for membrane association is occluded by interaction with the kinase domain in these multidomain constructs. In other words, just as the KA1 domain appears to autoinhibit the kinase domain, so does the kinase domain ‘auto’ inhibit the ability of the KA1 domain in the same protein to bind membranes.
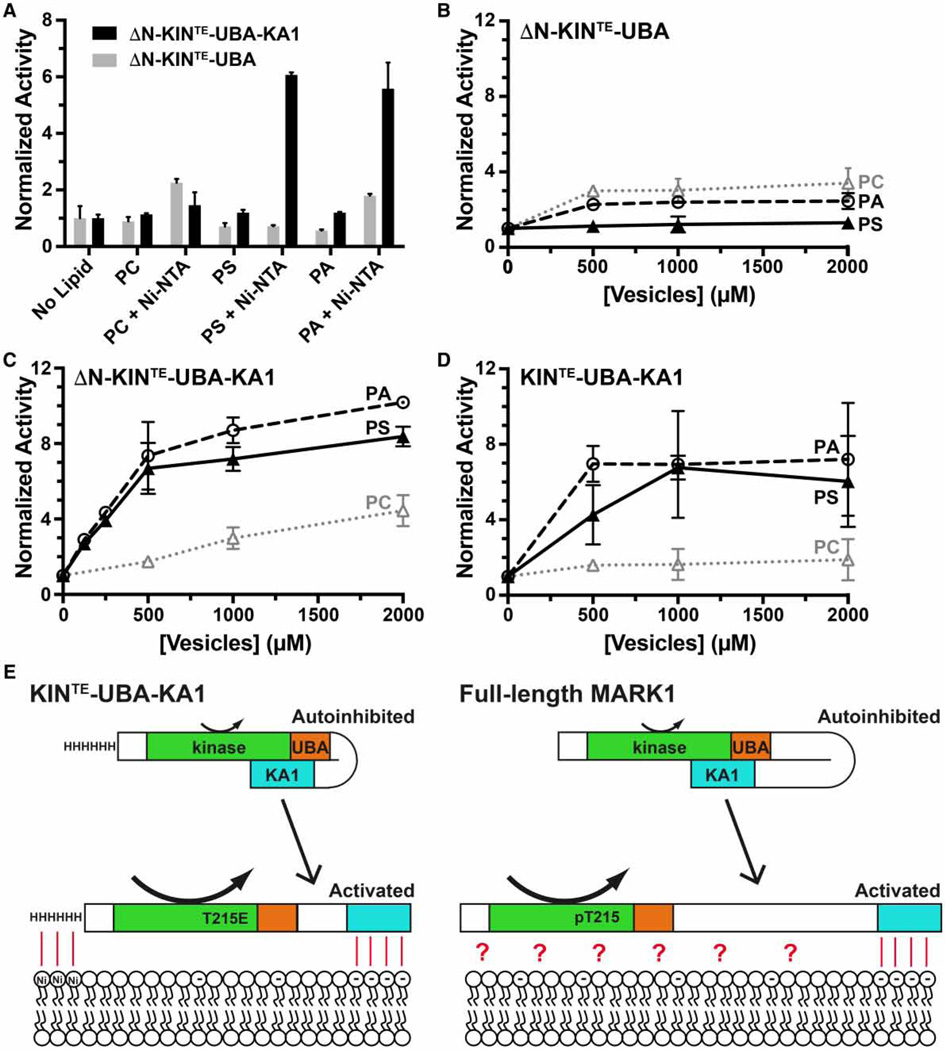
Anionic phospholipids can activate autoinhibited MARK1
If the KA1 domain uses its membrane-binding surface to inhibit the kinase domain in ‘mini’ MARK1, we reasoned that binding to acidic membrane surfaces might reverse the autoinhibition if membrane binding has sufficient affinity. Efforts to activate ΔN-KINTE-UBA-KA1 or ‘mini’ MARK1 with vesicles containing 20% (mole/ mole) PS or PA on their own did not show significant activation, however, even at the highest lipid concentrations employed (Supplementary Figure S3A). In addition, adding these vesicles to the MARK1 kinase–UBA fragment did not reverse the ability of the KA1 domain to inhibit kinase activity in trans (Supplementary Figure S3B). We reasoned that the weak membrane binding of the KA1 domain (Kd > 5 µM) might not be sufficient to allow it to compete on its own with the intramolecular kinase/KA1 interactions. We therefore included DOGS-Ni-NTA lipids (5% mole/mole), so that the N-terminal hexahistidine tag of the proteins (retained in these experiments) would tether them to the vesicle surface. A similar strategy was previously employed for assessing autoinhibition of Ste5, a scaffold protein within the yeast MAPK pathway, and for its activation by membrane phosphoinositides [41]. Just as with Ste5, recruiting ‘mini’ MARK1 variants KINTE-UBA-KA1 and ΔN-KINTE-UBA-KA1 to vesicles through their hexahistidine tags allowed acidic phospholipids to promote MARK1 kinase activity by up to ~6-fold (Figure 6A) or ~10-fold (Figure 6C,D) when lipid concentration reached 2 mM. PS and PA both had similar effects, consistent with their similar binding affinities for the KA1 domain [12]. This activating effect was only seen for the ‘mini’ MARK1 proteins, and not for the kinase–UBA fragment (Figure 6B–D), showing that it depends on the presence of the KA1 domain. These data therefore argue that MARK1 localized to the membrane can be activated by anionic phospholipids present in that membrane, probably through their interaction with the KA1 domain, leading to relief of autoinhibition as suggested in the cartoon in Figure 6E. As we reported in our studies of the S. cerevisiae MARK/PAR kinases, two membrane-associated components appear to be required for activation — septins and acidic phospholipids in the case of Kcc4 [12]. In the activation of ‘mini’ MARK1 observed in Figure 6, binding of the hexahistidine tag to Ni-NTA lipids appears to be capable of substituting for septin binding.
Figure 6. Lipid activation of ‘mini’ MARK1.
(A) Kinase activities of the ΔN-KINTE-UBA-KA1 ‘mini’ MARK1 and a variant lacking the KA1 domain (ΔN-KINTE-UBA) were assayed in triplicate following the addition of 200 µM DOPC, in vesicles into which various combinations of other phospholipids had been incorporated. Values are plotted as mean ± standard deviation, normalized to activity in the absence of added lipid. The data show that vesicles containing PS or PA robustly activated ‘mini’ MARK1, but only when they also contained 5% (mole/mole) DOGS-Ni-NTA, which binds the hexahistidine tag on both constructs — left intact for these experiments. PS or PA was included at 20% (mole/mole) when present as the only addition, or 15% when added alongside DOGS-Ni-NTA. (B) The kinase activity of ΔN-KINTE-UBA (lacking the KA1 domain) was unaffected by the addition of vesicles containing 5% (mole/mole) DOGS-Ni-NTA plus 95% PC alone (dotted gray line), 80% PC plus 15% PA (dashed black line), or 80% PC plus 15% PS (solid black line). Vesicles were titrated up to 2 mM total added lipid. (C) By contrast, histidine-tagged ΔN-KINTE-UBA-KA1 ‘mini’ MARK1 was robustly activated by the vesicles containing DOGS-Ni-NTA plus PA or PS, as was KINTE-UBA-KA1 (D). We present a hypothetical model in (E) where MARK1 can be activated by the presence of anionic phospholipids (and their binding to the KA1 domain), but only when another entity contributes to membrane recruitment. This role is assumed by the hexahistidine tag binding to DOGS-Ni-NTA in our in vitro studies, but might be provided by septins in yeast orthologs [12, 43], and/or other components (denoted by the red question mark) in MARK1 — as summarized in the text. Curved arrows indicate low kinase activity for the autoinhibited state, and elevated kinase activity in the membrane-bound state.
Discussion
We previously showed that the KA1 domain of several kinases in the MARK/PAR family of kinases can bind acidic phospholipids and promote recruitment of their host proteins to the cytoplasmic face of the plasma membrane [12]. A number of other studies have indicated that the KA1 domain — typically found at the C-terminus of these kinases — plays an intramolecular autoinhibitory role [13, 22–24]. By analogy with the other membrane-associated domains in protein kinases [2], notably the C1 and C2 domains of PKC and the PH domain of Akt, for example, it seemed reasonable to suspect that the KA1 domain might interact directly with the kinase domain in MARK/PAR kinases and play an autoinhibitory role. Our data suggest that this is the case for MARK1, and further that the kinase-binding site overlaps with the membrane-binding site so that association with the two targets is mutually exclusive. As with PKC and Akt, this allows a regulatory mode in which membrane binding can simultaneously relocalize the kinase and enhance its catalytic activity.
Although the regulatory role of the KA1 domain suggested by our data and by previous studies appears relatively straightforward, it is clear that it is only one of several components involved in both regulation and localization of MARK/PAR family kinases [13]. Several phosphorylation events play key roles, and binding partners for the membrane-proximal UBA domain could play important positive or negative roles [13]. Additional auto-inhibitory interactions may also occur, as seen for the unique autoinhibitory sequence adjacent to the KA1 domain in SAD kinases [21], which has been seen crystallographically to interact with helix αC in the kinase domain. The linker between the UBA and KA1 domain has also been suggested to contain binding sites for several targets, including membrane lipids and proteins [42–47]. Indeed, a recent study of Hsl1 [43] argues that co-ordinated septin binding to elements in the linker region of Hsl1 with membrane binding by the C-terminal KA1 domain is responsible for its recruitment to septin collars at the bud neck. Our data suggest that such co-operation of multiple domains can reverse kinase autoinhibition (at least in MARK1), and hence will have the capacity to activate the kinase at the correct location through ‘coincidence detection’ [5]. In the case of our ‘mini’ MARK1 proteins, we find that a hexahistidine tag can mimic the septin association elements in vitro if we include Ni-NTA lipids in our vesicles. It is not clear for MARK1 and its close relatives what might be the second target analogous to septins. One intriguing possibility is death-associated protein kinase, which has been reported to relieve autoinhibition of MARK1 and MARK2 by binding to the ‘linker’ region [48] to modulate microtubule assembly and neuronal differentiation, and may do so in concert with KA1 domain engagement by anionic membrane surfaces.
Our data argue that the KA1 domain has regulatory properties that resemble lipid-binding domains in other protein kinases [2, 3, 5]. Moreover, like several of these domains, notably C2 and PH domains, KA1 domains may bind both membrane and protein targets [49] — just as protein or peptide targets have been suggested for PH domains [50] and C2 domains [51]; for example. Yang et al. [49] reported that the KA1 domain of PAR1b/MARK2 and other human MARK family kinases interacts with Gab1 by binding to an ~100 amino acid stretch (aa 152–250) in its unstructured region — potentially linking receptor tyrosine kinase signaling to control of cell polarity. Indeed, this study suggests that Gab1 acts as a scaffold to facilitate phosphorylation of PAR3 and possibly other polarity-regulating proteins by MARK2 [49]. Just as acidic vesicles can activate our ‘mini’ MARK1 in vitro, overexpression of the KA1-binding region of Gab1 appears to increase MARK2 activation in cells. It is not clear whether Gab1 must bind the KA1 domain in addition to membranes to achieve this effect, or whether it binds to the same binding site on the KA1 domain as do acidic phospholipids. We were not able to detect activation of purified ‘mini’ MARK1 with the Gab1152–251 segment in vitro, and the addition of excess Gab1152–251 did not reverse the trans-inhibitory effect of the KA1 domain on MARK1 kinase activity (Supplementary Figure S3C). Gab1 may bind a separate surface on the KA1 domain from that involved in kinase inhibition, or may require additional cellular components for its binding to — and activation of — MARK1 in cells. Studies of kinases in this family indicate that numerous activating signals must co-operate in order to activate them at the right place and at the correct time. Our data argue that the KA1 domain plays an important role in this — at least for MARK1 — and a role that is analogous to that seen for other protein modules with defined binding targets in other kinases. As with these other examples, the additional mechanistic understanding of the MARK/PAR family kinases should ultimately be useful in devising ways to inhibit their aberrant activation in pathological situations, such as tauopathies [13], by targeting allosteric activation rather than the kinase domain itself.
Supplementary Material
Acknowledgments
We thank members of the Ferguson and Lemmon laboratories for valuable discussion and reagents. We also thank Dr Benjamin Neel for providing Gab1 cDNA, and Drs Kushol Gupta and Steve Stayrook for assisting with SAXS and AUC data collection and analysis.
Funding
This work was funded in part by National institutes of Health grant [R01-CA112552 to K.M.F.] and postdoctoral fellowship [F32-GM115098 to R.P.E.].
Abbreviations
- DOGS-Ni-NTA
1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1- carboxypentyl)iminodiacetic acid)succinyl] nickel salt
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- FAK
focal adhesion kinase
- FERM
band four-point-one ezrin radixin moesin
- IPTG
isopropyl b-d-1-thiogalactopyranoside
- KA1
kinase associated-1
- MAP
microtubule-associated protein
- MARKs
microtubule affinity-regulating kinases
- MELK
maternal embryonic leucine zipper kinase
- MES
2-(N-morpholino)ethanesulfonic acid
- MW
molecular weight
- PA
l-α-phosphatidic acid
- PAR1
partitioning-defective 1
- PH
pleckstrin homology
- PKC
protein kinase C
- PS
l-α-phosphatidylserine
- SAD
synapses of amphids defective
- SAXS
small-angle X-ray scattering
- SAD
synapses of amphids defective
- SEC
size exclusion chromatography
- SH
Src homology
- SPR
surface plasmon resonance
- SV-AUC
sedimentation velocity analytical ultracentrifugation
- TCEP
tris(2-carboxyethyl)phosphine
- TEV
tobacco etch virus
- UBA
ubiquitin-associated domain
- WT
wild type
Footnotes
Author Contribution
R.P.E. performed and analyzed all experiments. R.P.E., M.A.L., and K.M.F. designed the experiments and wrote the manuscript together.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Pawson T, Kofler M. Kinome signaling through regulated protein-protein interactions in normal and cancer cells. Curr. Opin. Cell Biol. 2009;21:147–153. doi: 10.1016/j.ceb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Newton AC. Lipid activation of protein kinases. J. Lipid Res. 2009;50(Suppl):S266–S271. doi: 10.1194/jlr.R800064-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leonard TA, Hurley JH. Regulation of protein kinases by lipids. Curr. Opin. Struct. Biol. 2011;21:785–791. doi: 10.1016/j.sbi.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell. Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 6.Antal CE, Callender JA, Kornev AP, Taylor SS, Newton AC. Intramolecular C2 domain-mediated autoinhibition of protein kinase C βII. Cell Rep. 2015;12:1252–1260. doi: 10.1016/j.celrep.2015.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonard TA, Różycki B, Saidi LF, Hummer G, Hurley JH. Crystal structure and allosteric activation of protein kinase C βII. Cell. 2011;144:55–66. doi: 10.1016/j.cell.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sicheri F, Kuriyan J. Structures of Src-family tyrosine kinases. Curr. Opin. Struct. Biol. 1997;7:777–785. doi: 10.1016/s0959-440x(97)80146-7. [DOI] [PubMed] [Google Scholar]
- 9.Lietha D, Cai X, Ceccarelli DFJ, Li Y, Schaller MD, Eck MJ. Structural basis for the autoinhibition of focal adhesion kinase. Cell. 2007;129:1177–1187. doi: 10.1016/j.cell.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, Vogan EM, Nocka LM, Rosen CE, Zorn JA, Harrison SC, et al. Autoinhibition of Bruton’s tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate. eLife. 2015;20:4. doi: 10.7554/eLife.06074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu WI, Voegtli WC, Sturgis HL, Dizon FP, Vigers GPA, Brandhuber BJ. Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PLoS ONE. 2010;5:e12913. doi: 10.1371/journal.pone.0012913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moravcevic K, Mendrola JM, Schmitz KR, Wang Y-H, Slochower D, Janmey PA, et al. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell. 2010;143:966–977. doi: 10.1016/j.cell.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marx A, Nugoor C, Panneerselvam S, Mandelkow E. Structure and function of polarity-inducing kinase family MARK/Par-1 within the branch of AMPK/Snf1-related kinases. FASEB J. 2010;24:1637–1648. doi: 10.1096/fj.09-148064. [DOI] [PubMed] [Google Scholar]
- 14.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 15.Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucl. Acids Res. 2015;43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marx A, Nugoor C, Müller J, Panneerselvam S, Timm T, Bilang M, et al. Structural variations in the catalytic and ubiquitin-associated domains of microtubule-associated protein/microtubule affinity regulating kinase (MARK) 1 and MARK2. J. Biol. Chem. 2006;281:27586–27599. doi: 10.1074/jbc.M604865200. [DOI] [PubMed] [Google Scholar]
- 17.Cao L-S, Wang J, Chen Y, Deng H, Wang Z-X, Wu J-W. Structural basis for the regulation of maternal embryonic leucine zipper kinase. PLoS ONE. 2013;8:e70031. doi: 10.1371/journal.pone.0070031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panneerselvam S, Marx A, Mandelkow E-M, Mandelkow E. Structure of the catalytic and ubiquitin-associated domains of the protein kinase MARK/Par-1. Structure. 2006;14:173–183. doi: 10.1016/j.str.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 19.Murphy JM, Korzhnev DM, Ceccarelli DF, Briant DJ, Zarrine-Afsar A, Sicheri F, et al. Conformational instability of the MARK3 UBA domain compromises ubiquitin recognition and promotes interaction with the adjacent kinase domain. Proc. Natl Acad. SciU.SA. 2007;104:14336–14341. doi: 10.1073/pnas.0703012104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Jiao Z-H, Zheng L-S, Zhang Y-Y, Xie S-T, Wang Z-X, et al. Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature. 2009;459:1146–1149. doi: 10.1038/nature08075. [DOI] [PubMed] [Google Scholar]
- 21.Wu J-X, Cheng Y-S, Wang J, Chen L, Ding M, Wu J-W. Structural insight into the mechanism of synergistic autoinhibition of SAD kinases. Nat. Commun. 2015;6:8953. doi: 10.1038/ncomms9953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beullens M, Vancauwenbergh S, Morrice N, Derua R, Ceulemans H, Waelkens E, et al. Substrate specificity and activity regulation of protein kinase MELK. J. Biol. Chem. 2005;280:40003–40011. doi: 10.1074/jbc.M507274200. [DOI] [PubMed] [Google Scholar]
- 23.Elbert M, Rossi G, Brennwald P. The yeast par-1 homologs Kin1 and Kin2 show genetic and physical interactions with components of the exocytic machinery. Mol. Biol. Cell. 2004;16:532–549. doi: 10.1091/mbc.E04-07-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong E-Y, Smits VAJ, Fumagallo F, Piscitello D, Morrice N, Freire R, et al. KA1-targeted regulatory domain mutations activate Chk1 in the absence of DNA damage. Sci. Rep. 2015;5:10856. doi: 10.1038/srep10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. PMID:7758115. [DOI] [PubMed] [Google Scholar]
- 26.Drewes G, Ebneth A, Preuss U, Mandelkow E-M, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 27.Chin JY, Knowles RB, Schneider A, Drewes G, Mandelkow E-M, Hyman BT. Microtubule-affinity regulating kinase (MARK) is tightly associated with neurofibrillary tangles in Alzheimer brain: a fluorescence resonance energy transfer study. J. Neuropathol. Exp. Neurol. 2000;59:966–971. doi: 10.1093/jnen/59.11.966. [DOI] [PubMed] [Google Scholar]
- 28.Timm T, Marx A, Panneerselvam S, Mandelkow E, Mandelkow E-M. Structure and regulation of MARK, a kinase involved in abnormal phosphorylation of Tau protein. BMC Neurosci. 2008;9(Suppl 2):S9. doi: 10.1186/1471-2202-9-S2-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maussion G, Carayol J, Lepagnol-Bestel A-M, Tores F, Loe-Mie Y, Milbreta U, et al. Convergent evidence identifying MAP/microtubule affinity-regulating kinase 1 (MARK1) as a susceptibility gene for autism. Hum. Mol. Genet. 2008;17:2541–2551. doi: 10.1093/hmg/ddn154. [DOI] [PubMed] [Google Scholar]
- 30.Goodwin JM, Svensson RU, Lou HJ, Winslow MM, Turk BE, Shaw RJ. An AMPK-Independent signaling pathway downstream of the LKB1 tumor suppressor controls Snail1 and metastatic potential. Mol. Cell. 2014;55:436–450. doi: 10.1016/j.molcel.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemsley A, Arnheim N, Toney MD, Cortopassi G, Galas DJ. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 1989;17:6545–6551. doi: 10.1093/nar/17.16.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 33.Ferguson KM, Darling PJ, Mohan MJ, Macatee TL, Lemmon MA. Extracellular domains drive homo- but not hetero-dimerization of erbB receptors. EMBO J. 2000;19:4632–4643. doi: 10.1093/emboj/19.17.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, et al. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 35.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe A, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge, UK: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 36.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 38.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 39.Bresler SC, Wood AC, Haglund EA, Courtright J, Belcastro LT, Plegaria JS, et al. Differential inhibitor sensitivity of anaplastic lymphoma kinase variants found in neuroblastoma. Science Transl. Med. 2011;3:108ra114. doi: 10.1126/scitranslmed.3002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zalatan JG, Coyle SM, Rajan S, Sidhu SS, Lim WA. Conformational control of the Ste5 scaffold protein insulates against MAP kinase misactivation. Science. 2012;337:1218–1222. doi: 10.1126/science.1220683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan SM, Nie WC, He F, Jia ZW, Gao XD. Kin2, the budding yeast ortholog of animal MARK/PAR-1 kinases, localizes to the sites of polarized growth and may regulate septin organization and the cell wall. PLoS ONE. 2016;11:e0153992. doi: 10.1371/journal.pone.0153992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finnigan GC, Sterling SM, Duvalyan A, Liao EN, Sargsyan A, Garcia G, III, et al. Coordinate action of distinct sequence elements localizes checkpoint kinase Hsl1 to the septin collar at the bud neck in Saccharomyces cerevisiae. Mol. Biol. Cell. 2016;27:2213–2233. doi: 10.1091/mbc.E16-03-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crutchley J, King KM, Keaton MA, Szkotnicki L, Orlando DA, Zyla TR, et al. Molecular dissection of the checkpoint kinase Hsl1p. Mol. Biol. Cell. 2009;20:1926–1936. doi: 10.1091/mbc.E08-08-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Longtine MS, Fares H, Pringle JR. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 1998;143:719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okuzaki D, Nojima H. Kcc4 associates with septin proteins of Saccharomyces cerevisiae. FEBS Lett. 2001;489:197–201. doi: 10.1016/s0014-5793(01)02104-4. [DOI] [PubMed] [Google Scholar]
- 47.Hanrahan J, Snyder M. Cytoskeletal activation of a checkpoint kinase. Mol. Cell. 2003;12:663–673. doi: 10.1016/j.molcel.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 48.Wu P-R, Tsai P-I, Chen G-C, Chou H-J, Huang Y-P, Chen Y-H, et al. DAPK activates MARK1/2 to regulate microtubule assembly, neuronal differentiation, and tau toxicity. Cell Death Differ. 2011;18:1507–1520. doi: 10.1038/cdd.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z, Xue B, Umitsu M, Ikura M, Muthuswamy SK, Neel BG. The signaling adaptor GAB1 regulates cell polarity by acting as a PAR protein scaffold. Mol. Cell. 2012;47:469–483. doi: 10.1016/j.molcel.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lemmon MA. Pleckstrin homology domains: not just for phosphoinositides. Biochem. Soc. Trans. 2004;32:707–711. doi: 10.1042/BST0320707. [DOI] [PubMed] [Google Scholar]
- 51.Benes CH, Wu N, Elia AEH, Dharia T, Cantley LC, Soltoff SP. The C2 domain of PKCδ is a phosphotyrosine binding domain. Cell. 2005;121:271–280. doi: 10.1016/j.cell.2005.02.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.