Abstract
Purpose
How retinal bipolar cell interneurons are specified and assigned to specialized subtypes is only partially understood. In part, this is due to a lack of early pan- and subtype-specific bipolar cell markers. To discover these factors, we identified genes that were upregulated in Blimp1 (Prdm1) mutant retinas, which exhibit precocious bipolar cell development.
Methods
Postnatal day (P)2 retinas from Blimp1 conditional knock-out (CKO) mice and controls were processed for RNA sequencing. Genes that increased at least 45% and were statistically different between conditions were considered candidate bipolar-specific factors. Candidates were further evaluated by RT-PCR, in situ hybridization, and immunohistochemistry. Knock-in Tmem215-LacZ mice were used to better trace retinal expression.
Results
A comparison between Blimp1 CKO and control RNA-seq datasets revealed approximately 40 significantly upregulated genes. We characterized the expression of three genes that have no known function in the retina, Gsg1 (germ cell associated gene), Trnp1 (TMF-regulated nuclear protein), and Tmem215 (a predicted transmembrane protein). Germ cell associated gene appeared restricted to a small subset of cone bipolars while Trnp1 was seen in all ON type bipolar cells. Using Tmem215-LacZ heterozygous knock-in mice, we observed that β-galactosidase expression started early in bipolar cell development. In adults, Tmem215 was expressed by a subset of ON and OFF cone bipolar cells.
Conclusions
We have identified Gsg1, Tmem215, and Trnp1 as novel bipolar subtype-specific genes. The spatial and temporal pattern of their expression is consistent with a role in controlling bipolar subtype fate choice, differentiation, or physiology.
Keywords: retina, development, bipolar cell, amacrine cell, subtype, Blimp1 (Prdm1), Gsg1, Slitrk6, Trnp1, Tmem215, RNA-sequencing
The retina comprises seven major cell types, each of which is required for normal visual function. Photoreceptors detect light stimuli and transmit this information to bipolar cell interneurons. Bipolar cells relay signals to retinal ganglion cell (RGC) neurons that project to the brain. Owing to the complexity of visual information that must be processed, most retinal cell types are further specialized into subtypes with discrete functions, morphologies, and molecular characteristics. Bipolar cells have considerable subtype diversity. In mice, 14 bipolar cell subtypes are categorized based on how they synapse with photoreceptors, their physiology, and their dendritic and axonal morphologies.1–4 Rod bipolar cells synapse with rod photoreceptors and are activated by dim light signals. The remaining 13 bipolar subtypes preferentially synapse with cones. Five OFF subtypes (1, 2, 3a, 3b, 4) depolarize upon the loss of photic stimuli and the tips of their axons are localized to the outer-half of the inner plexiform layer (IPL). Eight ON subtypes (5i, 5o, 5t, XBC, 6, 7, 8, and 9) typically depolarize at the onset of light stimuli and their axons ramify in the inner-half of the IPL.1–4 Together, these bipolar subtypes make up approximately 7% of the cells in the mouse retina.5 The fundamental question of how bipolar cell diversity is programmed during development remains only partially answered.
Bipolar cells permanently exit the cell cycle late in mouse retinal development, starting around the time of birth and continuing to roughly postnatal day (P)7.6–9 Loss-of-function experiments have shown that the transcription factors Otx2 and Vsx2 (Chx10) are necessary for bipolar cell development.10–13 Otx2 is required for the formation of multiple cell types during retinogenesis and marks bipolar cells and photoreceptors in the mature mouse retina.12–14 Vsx2 is expressed by all proliferative retinal progenitors, but it is not robustly expressed in postmitotic cells until P4.15,16 Indeed, definitive bipolar markers are not appreciably seen until P4 or later,16 demonstrating a temporal gap between cell cycle exit and differentiation. Vsx2 has been shown to repress photoreceptor gene expression, arguing that it is permissive for bipolar cell development.17,18 The transcription factors Ascl1 and Neurod4 (Math3) are made in proliferative progenitors and/or postmitotic precursor cells. Ascl1 or Neurod4 overexpression along with Vsx2 can generate excess bipolar cells.19 The combined loss of Ascl1 and Neurod4 strongly reduces bipolar cell formation.20,21 Due to limited marker availability, whether Ascl1 and Neurod4 combine to regulate bipolar cell fate choice as an entire group or whether they control specific subtype genesis is unclear. Other transcription factors involved in bipolar development, including Bhlhb4, Bhlhb5 (Bhlhe22), Isl1, Lhx3, Lhx4, Prdm8, and Vsx1, appear to be expressed in postmitotic committed bipolar cells.16,22–34 Loss-of-function analysis suggests that these genes control bipolar cell subtype choice or differentiation. For example, Bhlhb4 deletion causes the progressive loss of rod bipolar cells while Bhlhb5 mutants do not form type 2 cone bipolar cells.23,24,27 Together, these data provide only a partial explanation for the mechanisms that control bipolar cell commitment and subtype choice.
A major barrier to uncovering the mechanisms of bipolar cell development is a lack of early pan and subtype-specific markers. Mice lacking the transcription factor Blimp1 (Prdm1) generate excess bipolar cells at the expense of photoreceptors.15,35,36 Bipolar cells are formed precociously in Blimp1 conditional knock-out (CKO) retinas. We compared gene expression in Blimp1 CKO retinas to controls at P2, which precedes normal bipolar-specific gene expression onset. This provided a sensitive assay for the unbiased detection of early bipolar-specific factors by RNA sequencing (RNA-seq). This profiling technique was sensitive and robust; we identified several known genes and approximately two dozen novel candidate bipolar-specific factors. We characterized the expression of three of these candidates in more detail. Candidate genes Gsg1, Trnp1, and Tmem215 were expressed in discrete subsets of bipolar cells, broadening the portfolio of markers that describe developing bipolar cells. The characterization of the remaining candidate genes is likely to increase this portfolio even further. The specificity and timing of Gsg1, Trnp1, and Tmem215 suggests that they regulate different aspects of bipolar subtype choice and differentiation.
Materials and Methods
Animals
Heterozygous Blimp1 (αPax6-Cre-IRES-GFP::Blimp1Flox/+) control and conditional knockout (CKO) (αPax6-Cre-IRES-GFP::Blimp1Flox/Flox) mice were generated and genotyped as previously described.15 These mice were used at P2, P5, and P7 for RNA-seq, RT-PCR, and histologic techniques (below). Wild-type C57BL/6J mice (strain #664, Jackson Laboratories, Bar Harbor, ME, USA) were used for histology at multiple ages. To generate Tmem215-LacZ gene trap mice, cryopreserved (Tmem215tm1(KOMP)Vlcg) heterozygous embryos from the Knockout Mouse Project (KOMP)37 repository were acquired from the Mutant Mouse Regional Resource Center (University of California Davis, Davis, CA, USA). The embryos were rederived by the University of Colorado Denver Bioengineering Core and the resulting animals were crossed to C57BL/6J mice. The allele Tmem215-LacZ was detected by PCR with the following primers at 60°C annealing: 5′-GTCTGTCCTAGCTTCCTCACTG and 5′-GTCAGAGATAGCAAGAAAGAG, yielding a 279-bp product. Tmem215-LacZ heterozygous mice were used for histology or crossed to Blimp1 CKO mice to generate Tmem215-LacZ::αPax6-Cre-IRES-GFP::Blimp1Flox/Flox animals. All animals were used in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and with the approval of the University of Colorado Denver IACUC.
RNA Sequencing
We collected eyes from five P2 Blimp1 CKO and heterozygous control mice. From each animal, one retina was dissected in PBS and homogenized in 0.5 mL TRIzol (Thermo Fisher Scientific, Waltham, MA, USA). The other eye was processed for immunohistochemistry (below) and the presence or absence of Blimp1 confirmed by immunostaining. Total RNA was purified from TRIzol according to the manufacturer's instructions. We further purified the RNA using a commercial kit (RNeasy; Qiagen, Valencia, CA, USA) according the manufacturer's protocol. Total RNA was submitted to the University of Colorado Genomics and Microarray Core Facility for quality control and labeling. The 10 samples were labeled with unique barcodes for RNA-seq using a commercial kit (Illumina TruSeq mRNA Library Preparation Kit; Illumina, San Diego, CA, USA). Samples were sequenced in 1 × 100 mode on a sequencing instrument (Illumina HiSeq 2000; Illumina) to generate approximately 20 million informative fragments per sample. Sequencing of RNA was analyzed by applying a custom computational pipeline consisting of the open-source gSNAP, Cufflinks, and R for sequence alignment and ascertainment of differential gene expression.38–41 Reads of RNA were aligned to the mouse genome (MM9) by gSNAP; expression (fragments per kilobase exon per million mapped reads [FPKM]) derived by Cufflinks; and differential expression analyzed with ANOVA in R. We used the following criteria to define bipolar-specific gene candidates: upregulated >1.45-fold versus heterozygous controls, expression >1 FPKM in Blimp1 CKO samples, false discovery rate (FDR) <0.45, and P < 0.05.
Reverse Transcription PCR
We dissected the retinas from three P2 Blimp1 CKO and heterozygous control mice in PBS and homogenized each pair of retinas separately in 0.5 mL TRIzol. Total RNA was purified as above and treated with commercial endonuclease (RNase-free DNase; Promega, Madison, WI, USA) for 1 hour; the six samples were further purified with a RNeasy kit, as described above. Reverse transcription was done using a complementary DNA synthesis kit according to the manufacturer's protocol (iScript; Bio-Rad Laboratories, Inc., Temecula, CA, USA). We conducted RT-PCR with primers to candidate and house-keeping genes (for primers, see Supplementary Table S2) using Sso Fast Supermix (Bio-Rad Laboratories, Inc.) and a CFX Connect thermocycler (Bio-Rad Laboratories, Inc.) according to the manufacturer's instructions. Expression differences were calculated using the ΔΔCt method.42 Differences were compared using unpaired Student's t-tests. We considered P < 0.05 to be statistically significant.
Immunohistochemistry
Retinal tissue was fixed in 2% paraformaldehyde (PFA) for 1 to 2 hours at room temperature, cryopreserved through 30% sucrose, frozen in OCT (Sakura, Torrance, CA, USA), and cryosectioned at 10 to 12 μm thickness. For immunostaining, sections were blocked in a previously described solution containing 5% milk and 0.5% Triton X-10015 for 2 to 4 hours at room temperature. The sections were incubated with primary antibodies (below) diluted in milk block solution for 12 to 18 hours at room temperature. Fluorescently conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA) diluted in milk block solution were added to the sections for one hour at room temperature. Primary antibodies used were: mouse anti-Ap2α (1:250, clone 5E4; Developmental Studies Hybridoma Bank, Iowa City, IA, USA); chicken anti–β-galactosidase (β-gal; 1:2000, AB9361; Abcam, Cambridge, MA, USA); goat anti-Bhlhb5 (1:1000, sc-6045; Santa Cruz Biotechnology, Inc., Dallas, TX, USA); mouse anti-Cabp5 (1:10, a gift from F. Haeseleer, University of Washington)43; mouse anti-Calretinin (1:750) (MAB1568, Milipore, Billerica, MA, USA); mouse anti-Calsenilin (1:2000, 05-756; Milipore); rabbit anti-GAD65/67 (1:500, AB1511; Milipore); goat anti-GlyT1 (1:2000, AB1770; Milipore); rabbit anti-HCN4 (1:500, APC-052; Alomone Labs Ltd., Jerusalem, Israel); mouse anti-Isl1/2 (1:250, clone 39.4D5; Developmental Studies Hybridoma Bank); goat anti-Otx2 (1:200, BAF1979; R&D Systems, Minneapolis, MN, USA); rabbit anti-Pax6 (1:500, 901301; BioLegend, Inc., San Diego, CA, USA); mouse anti-PKARIIβ (1:3000, 610625; BD Biosciences, San Jose, CA, USA); mouse anti-PKCα (1:250, P5704; Sigma-Aldrich Corp., St. Louis, MO, USA); rabbit anti-Scgn (1:5000, RD181120100; Biovendor LLC, Ashville, NC, USA); goat anti-Sox2 (1:100, sc17320; Santa Cruz Biotechnology); guinea pig anti-Trnp1 (1:200, a gift from M. Götz, Helmholtz Zentrum Muenchen)44; and rabbit anti-Vsx1 (1:250, a gift from E. Levine, Vanderbilt University).31 We used a laser scanning confocal microscope (C2+; Nikon Instruments, Inc., Melville, NY, USA) to acquire 1024 × 1024 pixel photographs of retinal sections one laser line at a time. Three to five z-stacks (1–1.5 μm per slice) were acquired and minimally processed with ImageJ (http://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD, USA)45 and a raster graphics editor (Photoshop; Adobe Systems, Inc., San Jose, CA, USA) to generate maximum intensity z-stack projections. Marker overlaps were secondarily verified by examining the XZ and YZ orthogonal projections of the images. For quantification of Trnp1 immunostaining, 3108 Trnp1+ cells were counted from 107 adult C57BL/6J sections representing six eyes costained with amacrine- and bipolar-specific markers. For quantification of Tmem215-LacZ, 4680 β-gal+ cells were counted from 141 sections of four adult Tme215-LacZ heterozygous mice. For adult Gsg1 in situ experiments (below), 209 Scgn+ cells (five sections, two mice) and 138 Cabp5+ cells (four sections, two mice) were scored for the presence or absence of Gsg1 fluorescent puncta.
In Situ Hybridization
Wild-type C57BL/6 and Blimp1 CKO retinas from 3 to 10 animals were used for in situ hybridization at multiple ages. We used a tissue assay kit (ViewRNA ISH; Affymetrix, Santa Clara, CA, USA) with a signal amplification kit (ViewRNA Chromogenic; Affymetrix) for single-plex fluorescent detection of mRNA. Custom probes to genes of interest were acquired from Affymetrix (ViewRNA Type I). In situ hybridization was conducted on 12-μm cryosections (see above) according to the manufacturer's instructions, but with some modifications as conditions needed to be optimized for each probe. For Tmem215 (VB1-15511-01; Affymetrix), sections were fixed for 16.5 hours in 4% PFA at 4°C, incubated in protease solution for 10 minutes, hybridized with probe for 3 hours, and developed with Fast-Red substrate for 40 minutes. For Trnp1 (VB1-16821-01; Affymetrix), sections were fixed for 1.5 hours in 4% PFA at room temperature, protease treated for 10 minutes, hybridized for 2.5 hours, and developed with Fast-Red for 40 minutes. For Slitrk6 (VB1-15510-01; Affymetrix), sections were fixed for 16.5 hours in 4% PFA at 4°C, treated with protease for 15 minutes, hybridized for 3 hours, and developed with Fast-Red for 30 minutes. For Gsg1 (VB1-17855-01; Affymetrix), sections were fixed for 30 minutes with 4% PFA at room temperature, protease treated for 10 minutes, incubated with probe for 3 hours, and developed for 40 minutes with Fast-Red. After the in situ hybridization, sections were counterstained with DAPI (4′,6-diamidino-2-phenylindole) or immunostained as described above. The number, intensity, and size of Fast-Red positive puncta varied between retinal sections and animals, but the overall spatial pattern was reproducible. Puncta tended to be localized within and near the cell nucleus, making quantification with nuclear markers problematic. Immunostaining sections first was incompatible with the in situ protocol. Sections of Blimp1 CKO adjacent to those used for in situ were separately immunostained for Blimp1 (see above) to establish the rough extent of the knock-out domain of the peripheral retina. This typically amounted to approximately one half of the total retinal length in a given section.
Results
Identification of Genes Differentially Regulated in Blimp1 Mutant Retinas
The genes that regulate bipolar cell genesis and subtype formation are poorly understood. In mice that lack Blimp1 (Prdm1), roughly three times more bipolar cell interneurons are generated at the expense of photoreceptors during development.15,36 Starting around birth, bipolar-specific genes become precociously expressed in these mutants.15,36 Since definitive bipolar markers are not observed until approximately P4 in wild-type mice,16 we reasoned that genes upregulated in Blimp1 mutant retinas prior to P4 would specifically mark and/or promote bipolar cell development. We therefore decided to examine gene expression at P2, roughly 2 days after precocious bipolar gene expression onset in Blimp1 mutants, but still well before their onset in wild-type retinas.
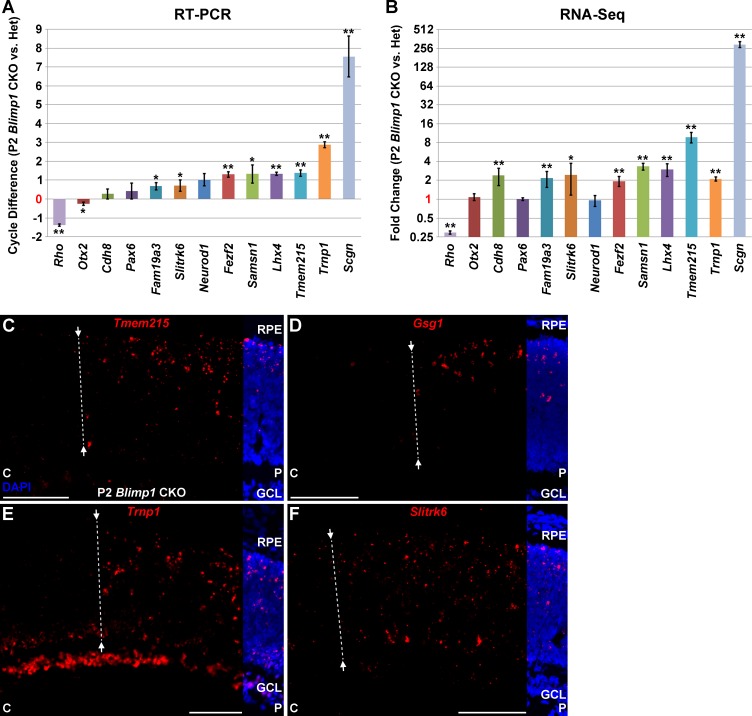
To identify the full repertoire of precocious genes, we compared the transcriptome of αPax6-Cre-IRES-GFP;Blimp1Flox/+ (heterozygous, control) and αPax6-Cre-IRES-GFP;Blimp1Flox/Flox (CKO, experimental) retinas at P2 by RNA sequencing (Fig. 1). In these mice, the peripheral part of the retina expresses Cre recombinase. Thus, peripheral retinas lack either one or both Blimp1 alleles, while the central retina remains essentially wild-type (Fig. 1). We collected RNA from the entire retinas from five eyes of each genotype for RNA-seq analysis. After mapping sequencing reads to genes and normalizing for sequencing depth and transcript length (FPKM), the average expression values for each gene were calculated. Overall, the variability in expression values between replicates was modest (Supplementary Table S1). Next, the ratio between mutant and heterozygous retinas was calculated and converted to fold change (Supplementary Table S1). We reasoned that fold-change values that are significantly greater than 1 represent bipolar-specific genes while those significantly less than −1 represent photoreceptor-specific genes (Fig. 1). We anticipated difficulty detecting downregulated genes because the wild-type areas of the retina in each sample robustly express several photoreceptor markers at P2, diluting the changes seen by Blimp1 loss-of-function. Nonetheless, we identified 40 genes that were significantly reduced more than 1.5-fold in mutants compared to heterozygous controls (P < 0.05, FDR < 0.45; Table 1). Despite a lower power to detect downregulated genes in this experiment, we observed a clear decrease in Blimp1 (Prdm1) expression in mutants (Table 1). As expected, the majority of these downregulated factors were rod and cone photoreceptor-specific genes like Arr3, Pde6b, Rho, Thrb, and Cnga1 (bold text, Table 1). We did not anticipate difficulty identifying upregulated genes in this experiment because few bipolar genes are expressed in wild-type retinas at this age. We observed 40 genes that were significantly upregulated more than 1.45-fold in mutants compared to heterozygous controls (P < 0.05, FDR < 0.45; Table 2). As predicted, several of these genes are known markers of bipolar cells such as Vsx1, Vsx2, Scgn, Lhx3, Lhx4, Fezf2, Grm6, Isl1, and Bhlhb5 (Bhlhe22; bold text, Table 2). Several of the other upregulated genes in this list have not been characterized in the retina and in some cases, any other tissues. We next validated a subset of these upregulated genes by quantitative RT-PCR on a different set of P2 heterozygous control and Blimp1 CKO retinas (Figs. 2A, 2B). We examined one gene that decreased (Rho); three genes that were not significantly changed (Otx2, Pax6, and Neurod1); and nine genes that increased (Cdh8, Fam19a3, Slitrk6, Fezf2, Samsn1, Lhx4, Tmem215, Trnp1, and Scgn) by RNA-seq (Fig. 2B). Most of these genes showed similar changes by RT-PCR except for Cdh8 (unchanged) and Otx2 (slight decrease; Fig. 2A). The small statistically significant decrease in Otx2 was likely due to the unusually small variance from the RT-PCR replicates (Fig. 2A), while the discordance in Cdh8 (Figs. 2A, 2B) could represent a false positive from the RNA-seq analysis. However, since Cdh8 has been shown to be expressed in type 2 cone OFF bipolar cells,46 it is more likely that the RT-PCR result represents a false negative result. On the whole, these data indicate that our RNA-seq assay can robustly detect precociously expressed bipolar cell-specific genes.
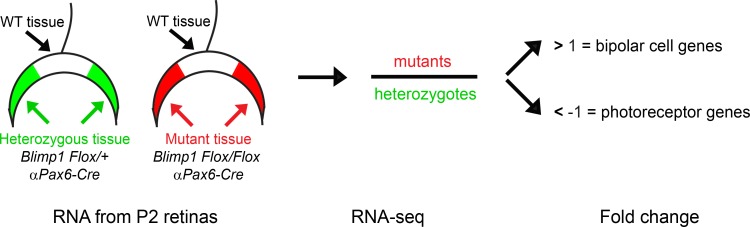
Figure 1.
Experimental design. Mice lacking Blimp1 precociously form bipolar cells at the expense of photoreceptors starting around birth. We collected retinas from αPax6-Cre::Blimp1Flox/+ heterozygous control (green) and αPax6-Cre::Blimp1Flox/Flox conditional knockout (red) mice at P2 for gene expression profiling. The transgene αPax6-Cre drives recombination in roughly half of the retina (arrows), leaving the central region un-recombined (wild-type). We collected RNA from whole retinas and subjected to RNA-seq analysis. By comparing expression values in mutants to heterozygous controls, we predict that upregulated genes will mark precocious developing bipolar cells and downregulated genes to mark photoreceptors. Since there are few bipolar genes expressed at P2 in wild-type retinas, this assay is especially sensitive to detect upregulated genes (i.e., early bipolar-specific factors). In contrast, the presence of photoreceptors in the P2 retina diminishes the power to detect downregulated genes.
Table 1.
Genes Downregulated in Blimp1 CKO Retinas
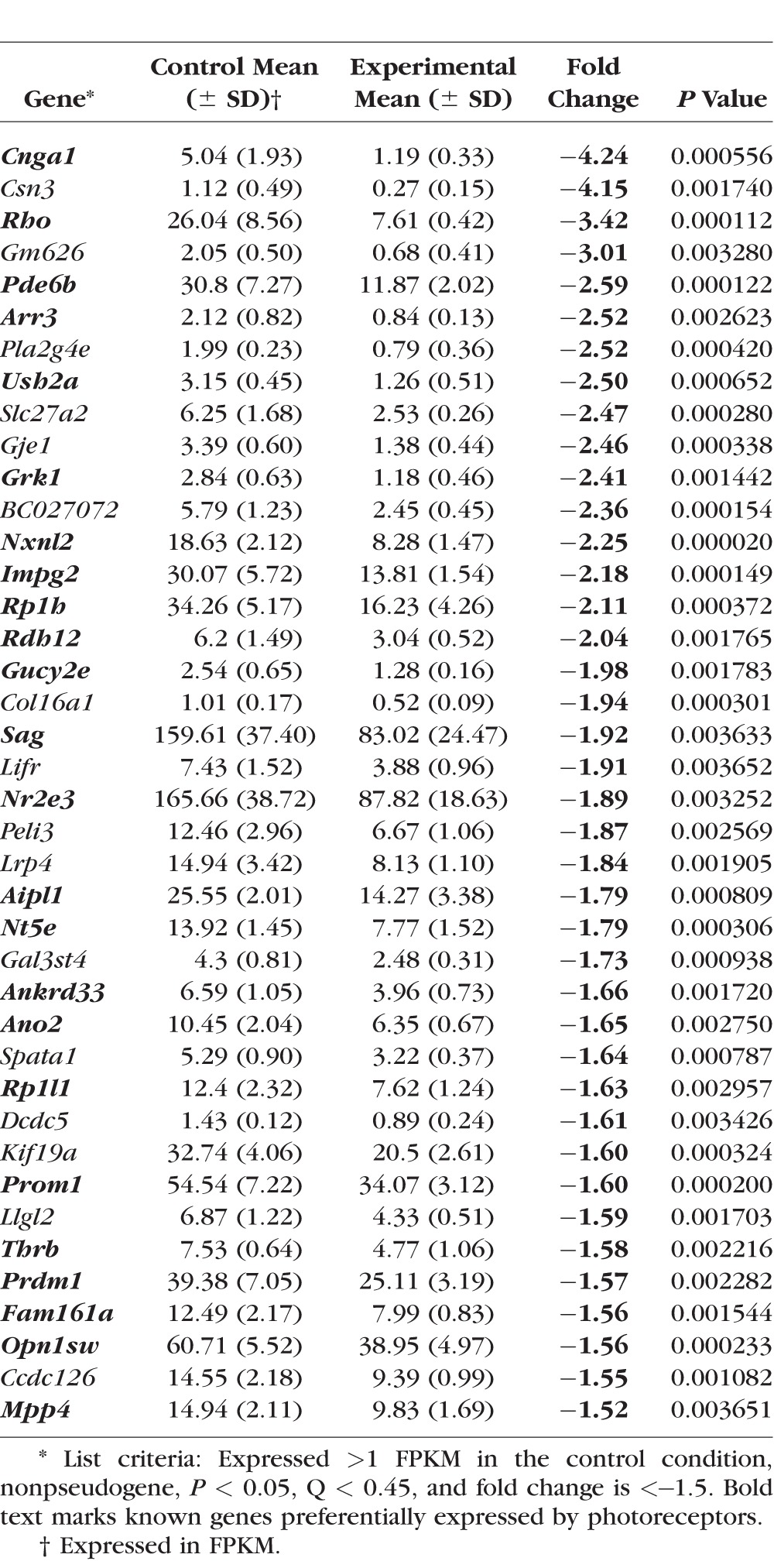
Table 2.
Genes Upregulated in Blimp1 CKO Retinas
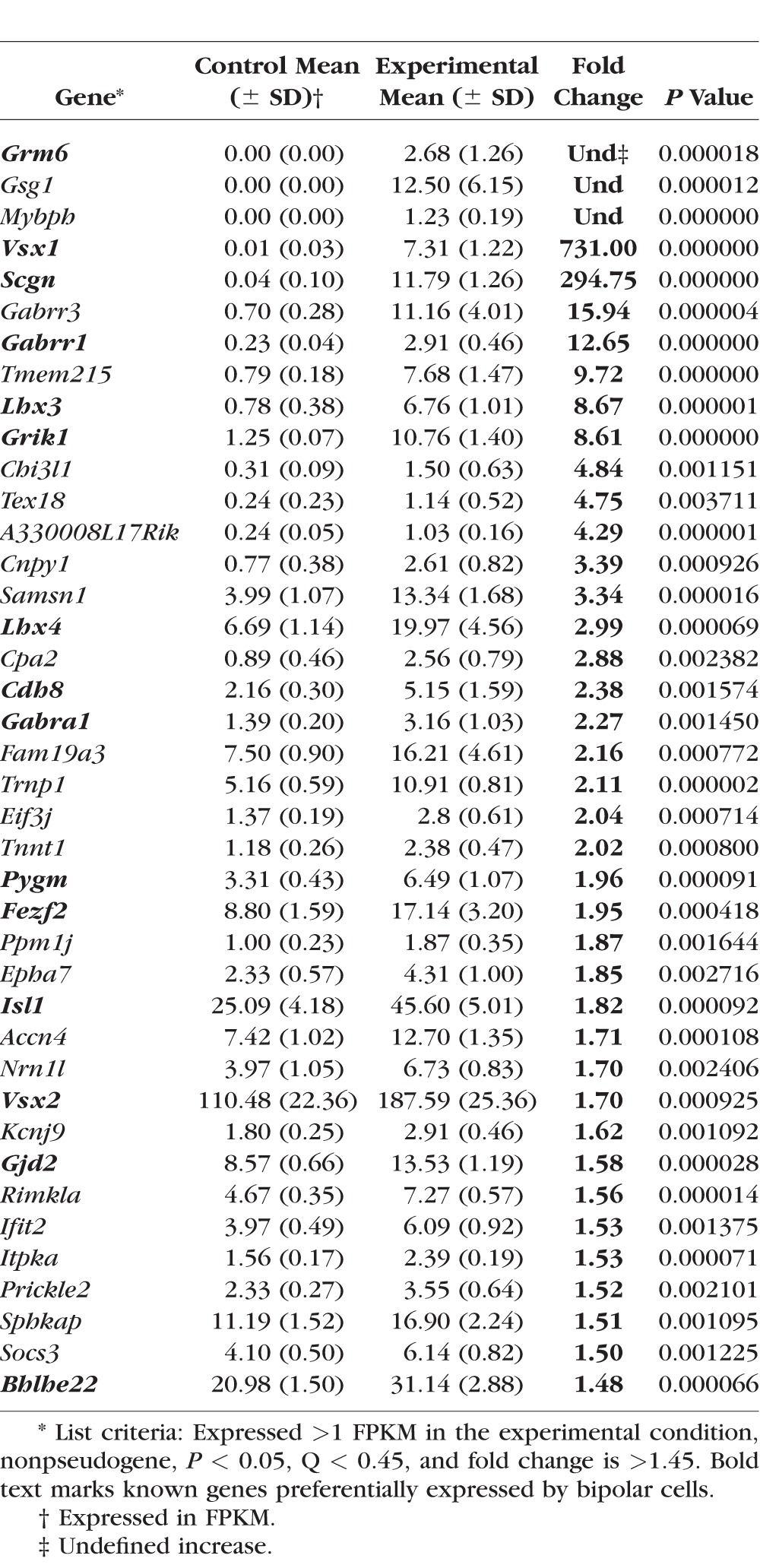
Figure 2.
Precociously upregulated gene expression in P2 Blimp1 CKO retinas. (A, B) Gene expression quantification. Expression of several known and novel bipolar-specific candidate genes measured by RT-PCR (A) and RNA-seq (B). There is good agreement between these methods, with the exception of Cdh8, which was increased by RNA-seq, but not RT-PCR. Otx2 is unchanged by RNA-seq, but is modestly lower by RT-PCR. As progenitor, ganglion, and amacrine cell numbers are grossly equivalent between Blimp1 heterozygous and CKO animals, we did not expect to see changes in Pax6 or Neurod4 expression. The rod specific gene Rho is decreased in mutants as expected, while known bipolar genes Lhx4, Fezf2, and Scgn are increased. * P < 0.05. ** P < 0.01. (C–F) In situ hybridization (red) of four candidate bipolar-specific genes in P2 Blimp1 CKO retinas. DAPI (blue) staining is cropped to show retinal structure. The central (C) retina is left and the peripheral (P) retina is toward the right. The dotted line marks the approximate edge of Blimp1 deletion. (C) Tmem215 signal is extensive in the peripheral Blimp1 CKO retina and is absent from the ganglion cell layer (GCL). The signal in the central retina is sparse. (D) Expression of Gsg1 is modest in the peripheral retina and is essentially absent centrally and within the GCL. (E) Trnp1 is widely expressed throughout the peripheral retina. It is expressed in the GCL in both peripheral and central retina. (F) Expression of Slitrk6 is robust in the peripheral retina, while little signal is seen centrally. Scale bar: 100 μm for each panel.
We next examined upregulated candidate genes from the RNA-seq screen (Table 2) by immunohistochemistry and in situ hybridization in P2 Blimp1 CKO retinas (Figs. 2C–F and data not shown). We limited our analysis to four of these candidate bipolar-specific genes (Tmem215, Gsg1, Trnp1, and Slitrk6). Expression of Tmem215 was sparsely observed in the central (wild-type) regions of P2 Blimp1 CKO retinas (Fig. 2C). As predicted from the RNA-seq data, there was considerably more signal in the peripheral (mutant) areas of these retinas (Fig. 2C). Signal was absent from the RPE and the ganglion cell layer (GCL), consistent with expression in precocious nascent bipolar cells (Fig. 2C). Gsg1 was essentially absent from the central retina, but robustly expressed in the mutant peripheral retina (Fig. 2D). Expression was more modest than Tmem215 and absent from the RPE and GCL, consistent with expression in bipolar cells (Fig. 2D). Expression of Trnp1 was seen in the central and peripheral retina (Fig. 2E). Expression levels in the GCL were roughly equal across the retina, while expression in the rest of the developing retina was conspicuously higher in the peripheral retina (Fig. 2E). This pattern is most consistent with Trnp1 expression in multiple cell types, including bipolar cells. Expression of Slitrk6 was observed sparsely in the central retina and more robustly in the peripheral areas of P2 Blimp1 CKO retinas (Fig. 2F). There was a modest signal in the GCL, but most of the expression in the peripheral retina was consistent with precocious bipolar cell localization (Fig. 2F). The increased expression of all four of these genes corresponded to the Blimp1 mutant area of the retina, where it appeared to label a greater number of cells. These data suggest that Slitrk6, Gsg1, Trnp1, and Tmem215 are expressed by bipolar cells. Further characterization of each of these genes is detailed below.
Slitrk6 and Gsg1 Are Expressed by a Subpopulation of Bipolar Cells
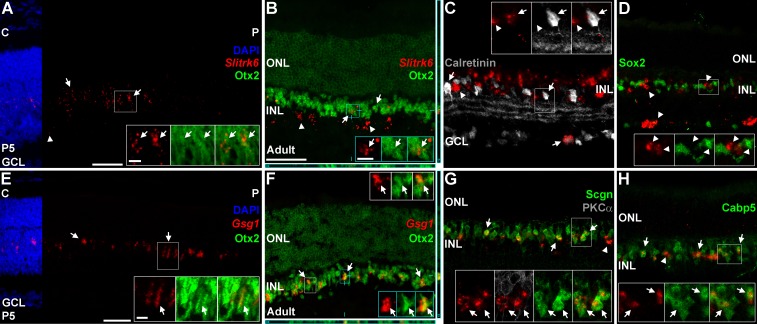
We examined Slitrk6 and Gsg1 expression both at P5 and in the mature retina where bipolar cells and their subtypes can be readily distinguished. After the in situ process, we immunostained sections with cell type–specific antibodies (Fig. 3). At P5, we observed Slitrk6 expression only in the central area of the wild-type retina (Fig. 3A). This overlapped extensively, but not completely, with the bipolar marker Otx2 (Table 3; Fig. 3A). No appreciable signal was seen in wild-type retinas at younger ages, suggesting that Slitrk6 expression in bipolar cells starts at P5. Expression of Slitrk6 in the adult retina has been previously examined by in situ hybridization, but the signal was diffuse in the GCL and inner nuclear layer (INL), and not precisely localized to any given cell type.47 Consistent with these observations, we observed relatively sparse Slitrk6 expression within the INL and GCL (Figs. 3B–D). Colabeling showed that some of the Slitrk6 signal in the INL overlapped with Otx2, consistent with bipolar cell identity (Fig. 3B). Slitrk6 signal in the INL and GCL partially overlapped with calretinin, a marker of multiple amacrine and RGC subpopulations48 (Fig. 3C). We did not observe Slitrk6 overlap with Sox2, a marker of Müller glia and cholinergic amacrine cells49–51 (Fig. 3D). These data indicate that Slitrk6 is made by a subset of bipolar and amacrine cells, and perhaps by a subset of RGCs. We were unable to further investigate these subtypes because the in situ protocol for Slitrk6 (see methods) was incompatible with nearly all of our cell type–specific antibodies.
Figure 3.
Bipolar cells labeled with Slitrk6 and Gsg1. (A–D) Postnatal day 5 (A) and adult (B–D) wild-type retinas labeled for Slitrk6 mRNA (red) and cell-type specific markers (gray/green). (A) At P5, Slitrk6 signal (red) is seen in the central (C), but not peripheral (P) retina. Most of the Slitrk6 signal overlaps with Otx2 (green, arrows, insets) while the remaining signal in the inner retina does not (arrowheads). Nuclear counterstaining with DAPI is cropped for clarity. (B) Slitrk6 signal in the adult INL overlaps highly with the bipolar cell marker Otx2 (green, arrows, insets). Some Slitrk6 signal in the INL does not overlap with Otx2 (arrowheads), consistent with amacrine cell identity. Slitrk6 is also seen in the GCL, where it likely indicates amacrine and/or ganglion cell identity. Orthogonal XZ and YZ views are shown below and to the right with blue lines highlighting the X and Y positions. (C) Expression of Slitrk6 partially overlaps with the amacrine and ganglion cell marker calretinin (gray; arrows, insets). Arrowheads mark Slitrk6+ cells that are located in the inner aspect of the INL that do not express calretinin, which are likely amacrine cells. (D) Slitrk6 (arrowheads) does not overlap with Sox2, which marks Müller glia and starburst amacrines. Together, these results suggest that Slitrk6 marks a subset of bipolars, amacrines, and possibly ganglion cells. (E–H) Postnatal day 5 (E) and adult (F–H) wild-type retinas labeled for Gsg1 mRNA (red) and cell-type specific markers (gray/green). (E) At P5, Gsg1 signal (red) shows a central to peripheral gradient. The Gsg1 signal overlaps extensively with Otx2 (green, arrows, insets). Nuclear staining with DAPI is cropped for clarity. (F) Gsg1 is sparse in the adult retina and is limited to the INL where it overlaps with Otx2 (green, arrows, insets). This suggests that Gsg1 marks a subpopulation of bipolar cells. The orthogonal views are as above. (G) Gsg1 overlaps highly with Scgn+ cone bipolar cells (green, arrows, insets), but not PKCα+ rod bipolar cells (gray, insets only). Approximately 30% of the Scgn+ bipolars contain Gsg1 signal. (H) Gsg1 overlaps highly with Cabp5 (green, arrows, insets), but only 21% of the Cabp5+ cells have an overlapping Gsg1 signal. Arrowheads mark the occasional cell that appears Cabp5 negative. These results suggest that Gsg1 marks types 3 and/or type 5 cone bipolar cells. Scale bars: 50 μm for (A–E) and (F–H); 10 μm for insets in (A–E) and (F–H).
Table 3.
Bipolar Cell Markers Used in This Study

We did not observe Gsg1 signal in the wild-type retina until P5, where it showed a central to peripheral gradient of expression (Fig. 3E). Gsg1 signal was sparse and colocalized with Otx2 (Fig. 3E). In the adult retina, Gsg1 expression was tightly limited to a small number of cells in the outer half of the INL, where bipolar cells reside (Figs. 3F–H). Gsg1 signal appeared to overlap with Otx2 expression, suggesting that Gsg1 is bipolar-specific (Fig. 3F). We next costained Gsg1 hybridized sections with a panel of bipolar subtype-specific markers (Table 3). As with Slitrk6, most of our antibodies were incompatible with Gsg1 in situ hybridization. Nonetheless, we were able to colabel sections with the rod bipolar marker PKCα52 and with Scgn, which labels several subtypes of cone bipolar cells53 (Table 3; Fig. 3G). We did not observe Gsg1 signal in PKCα+ rod bipolars, but rather it appeared that the Gsg1 signal overlapped extensively with Scgn (Fig. 3G). This suggests that Gsg1 labels types 2 to 6 cone bipolar cells (Table 3). Since only a subset of the Scgn+ bipolar cells were labeled (29.7% ± 4.8% SD), it is likely that Gsg1 is made by one or two bipolar subtypes. To further narrow down which subtypes are labeled by Gsg1, we costained with Cabp5, a marker of rod bipolars and types 3 and 5 cone bipolars54 (Table 3; Fig. 3H). It appeared that the Gsg1 signal overlapped highly with Cabp5 (Fig. 3H), but only a subset (20.9% ± 2.3% S.D) of the Cabp5+ cells contained Gsg1 signal. Together, these data argue that Gsg1 preferentially labels type 3 and/or type 5 cone bipolar cells (Table 4). We were unable to further discriminate between these subtypes due to incompatibility between the in situ protocol and our other antibodies.
Table 4.
Bipolar Cell Subsets Marked by Gsg1, Trnp1, and Tmem215

Trnp1 Marks ON Type Bipolar Cells in the Mouse Retina
Owing to the limits of immunostaining following in situ hybridization, we used a well-characterized antibody44 to better track Trnp1 expression in the retina. We started by immunostaining wild-type retinas from several embryonic time-points, but none showed any detectable Trnp1 staining (data not shown). We next examined wild-type retinas at postnatal time-points. From P0 to P5, we did not observe any Trnp1 expression in the retina (Figs. 4A, 4B, and data not shown). This contrasts with the in situ data from P2 Blimp1 CKO retinas, which showed Trnp1 signal in the GCL. This raises the possibility that Trnp1 expression in the GCL is below the detection threshold of this antibody, the in situ signal is spurious, or that Trnp1 mRNA is not translated at these stages. Starting around P7, nuclear-localized Trnp1 signal was abundant in the INL of the retina (Fig. 4C). We costained with Otx2 to mark bipolar cells and with Pax6 to mark other cell types in the INL12,14,55 at P7, P10, and in adult retinas (Figs. 4C–E). At all stages, Trnp1 nuclear staining overlapped with Otx2, but not Pax6 (Figs. 4C–E, 4J). Every Trnp1 cell expressed Otx2, but only approximately 53% of Otx2+ bipolar cells made Trnp1 (Fig. 4J). Next, we looked at bipolar subtype-specific markers (Table 3). All of the Trnp1+ cells coexpressed Isl1/2, a marker of cone ON and rod bipolar cells25,26 (Fig. 4F). We then stained for PKCα and Scgn to mark rod bipolars and types 2 through 6 cone bipolars, respectively. Consistent with the Isl1/2 results, we observed that only a subset of Scgn+ cone bipolars coexpressed Trnp1 (28.1% ± 8.7% SD) (Figs. 4G, 4J). Nearly all of the PKCα+ rod bipolars expressed Trnp1 (89.4% ± 9.6% SD) (Figs. 4G, 4J). We found that Trnp1 did not appreciably overlap with the cone OFF bipolar markers Bhlhb5, PKARIIβ,56 and Calsenilin (Csen)57 (Figs. 4H–J and data not shown). Taken together, these data suggest that Trnp1 is expressed early in bipolar cell maturation by all ON type bipolar cells in the retina (Table 4).
Figure 4.
Trnp1 marks ON type bipolar cells. Immunostaining of developing and adult retinas with Trnp1 (green) and cell type-specific markers. (A–E) Trnp1 costaining with Otx2 (red) and Pax6 (gray) at multiple ages. Otx2 is cropped and Pax6 shown only in the insets for clarity. At P0 (A) and P5 (B), no Trnp1 immunostaining is detected in the retina. (C) Starting at P7, Trnp1 nuclear staining is seen in the INL, where it overlaps completely with Otx2 (arrows, insets). The same pattern of Trnp1 expression is seen at P10 (D) and in adult (E) sections. Pax6+ amacrine cells in the ONL (arrowheads, insets) do not coexpress Trnp1 at any age. (F–K) Adult sections stained with Trnp1 and bipolar subtype specific markers (red/gray). (F) Cells that are Trnp1+ coexpress Isl1/2 (red, arrows, insets), which marks ON type bipolar cells in the retina. Starburst amacrines labeled by Isl1/2 (arrowheads) do not express Trnp1. (G–G'') A section showing Trnp1, Scgn (gray) and PKCα (red) costaining. A subset of Trnp1+ cells coexpresses Scgn (arrowheads, insets) or PKCα (arrows, insets). Nearly all of the PKCα+ rod bipolar cells express Trnp1 (G''), but only a fraction of Scgn+ cone bipolars are Trnp1+ (G'). (H) Type 2 cone OFF bipolar cells marked by Bhlhb5 staining (arrowheads, insets) do not coexpress Trnp1. Bhlhb5+ amacrine cells are marked with an “a”. (I) Calsenilin-positive type 4 cone OFF bipolar cells (arrowheads, insets) do not coexpress Trnp1. Scale bars: (A–E, G–H) 25 μm for panels and 10 μm for insets; (F) 100 μm and 10 μm for insets; (I) 50 μm and 10 μm for the insets. (J) Quantification of Trnp1 staining in the adult wild-type retina. The left panel shows the fraction of Trnp1+ cells that coexpress a cell-type specific marker. The right panel shows what percentage of a given population of cells expresses Trnp1. Error bars represent SD.
Tmem215 Is Expressed by a Complex Subset of Bipolar and Amacrine Cells
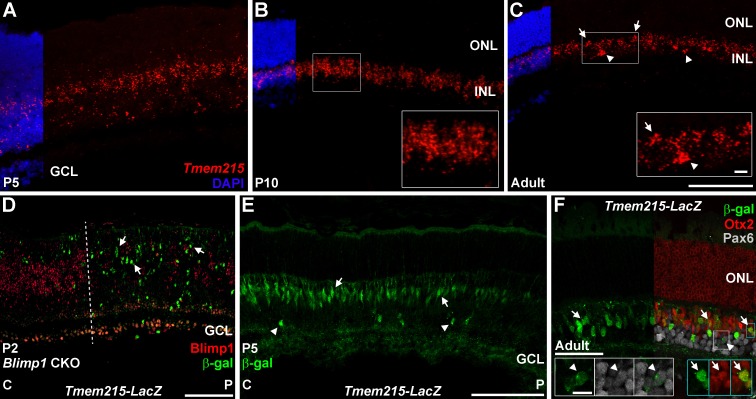
We conducted Tmem215 in situ hybridization on P5, P10, and adult wild-type retinas. At P5, the Tmem215 signal was limited to the nascent INL, with higher intensity in the outer aspect where bipolar cells reside (Fig. 5A). By P10, a strong band of Tmem215 signal was seen in the outer portion of the INL (Fig. 5B). In adult mice, the Tmem215 signal labeled a subset of cells in the outer and inner portions of the INL, consistent with bipolar and amacrine cell labeling (Fig. 5C). Due to incompatibilities with antibodies, we sought a different approach to label Tmem215. We acquired a Tmem215-LacZ gene trap knock-in line from the KOMP Repository.37 These mice are designed to express β-gal in the Tmem215 pattern. To test this, we first crossed Tmem215-LacZ animals to Blimp1 CKO mice and immunostained for β-gal and Blimp1. At P2, we observed a strong upregulation of β-gal expression in the peripheral (mutant) retina (Fig. 5D). We did not observe β-gal+ cells in the central-most (wild-type) retina or that coexpressed Blimp1 (Fig. 5D). We observed rare, weakly β-gal+ cells in the central retina of P4 Tmem215-LacZ mice (data not shown); but by P5, robust β-gal expression was seen in a central to peripheral gradient, mimicking the progression of bipolar cell development (Fig. 5E). Staining with β-gal was equivalent to the Tmem215 in situ pattern, with most of the β-gal expression in the bipolar cell area and fewer instances of amacrine cells (Fig. 5E). In adult Tmem215-LacZ mice, β-gal+ cells were localized to the INL (Fig. 5F). The majority (∼90%) of β-gal+ cells coexpressed Otx2 and a smaller fraction (∼9%) coexpressed the amacrine marker Pax6 (Figs. 5F, 6J). These β-gal+ cells only represented approximately 39% of the Otx2+ bipolar and 8% of the amacrine cell populations (Figs. 5F, 6J).
Figure 5.
Tmem215-LacZ mimics Tmem215 expression in the retina. (A–C) Tmem215 in situ hybridization (red) in wild-type retinas. Nuclear counterstaining with DAPI (blue) is cropped for clarity. (A) Tmem215 signal in the central P5 retina is localized to the area of the retina containing bipolar cells and amacrines. (B) By P10, Tmem215 signal is strongly localized to the INL (inset). (C) In the adult retina, Tmem215 clusters signal in the outer INL (arrows, inset) suggest bipolar cell staining while less frequent clusters in the inner INL indicate amacrine identity (arrowheads, inset). (D–F) Tmem215-LacZ knock-in mice stained for β-galactosidase (green). (D) Postnatal day 2 Blimp1 CKO::Tmem215-LacZ transgenic retina. Central (C) is to the left while peripheral (P) is right. Blimp1 immunostaining (red) shows substantial deletion right of the dotted line. There are more β-gal+ cells (arrows) in the Blimp1 deleted peripheral region, mimicking the Tmem215 in situ data (Fig. 2). None of the β-gal+ cells in any region coexpresses Blimp1. (E) In P5 Tmem215-LacZ heterozygous mice, β-gal+ cells are localized in the future INL and primarily show bipolar cell morphology (arrows). A few cells with amacrine morphology are seen (arrowheads) and there is a central-to-peripheral gradient of β-gal expression, mimicking the normal developmental progression of bipolar cell genesis. (F) In adults, β-gal+ cells are localized to the INL. Most of the β-gal+ cells coexpress Otx2 (red) (arrows, blue insets) while a smaller population coexpresses Pax6 (gray, arrowheads, white insets). The Tmem215-LacZ transgene closely matches the Tmem215 pattern. Scale bars: (A–E) 100 μm for panels and 10 μm for insets; (F) 50 μm for the panel and 10 μm for insets.
Figure 6.
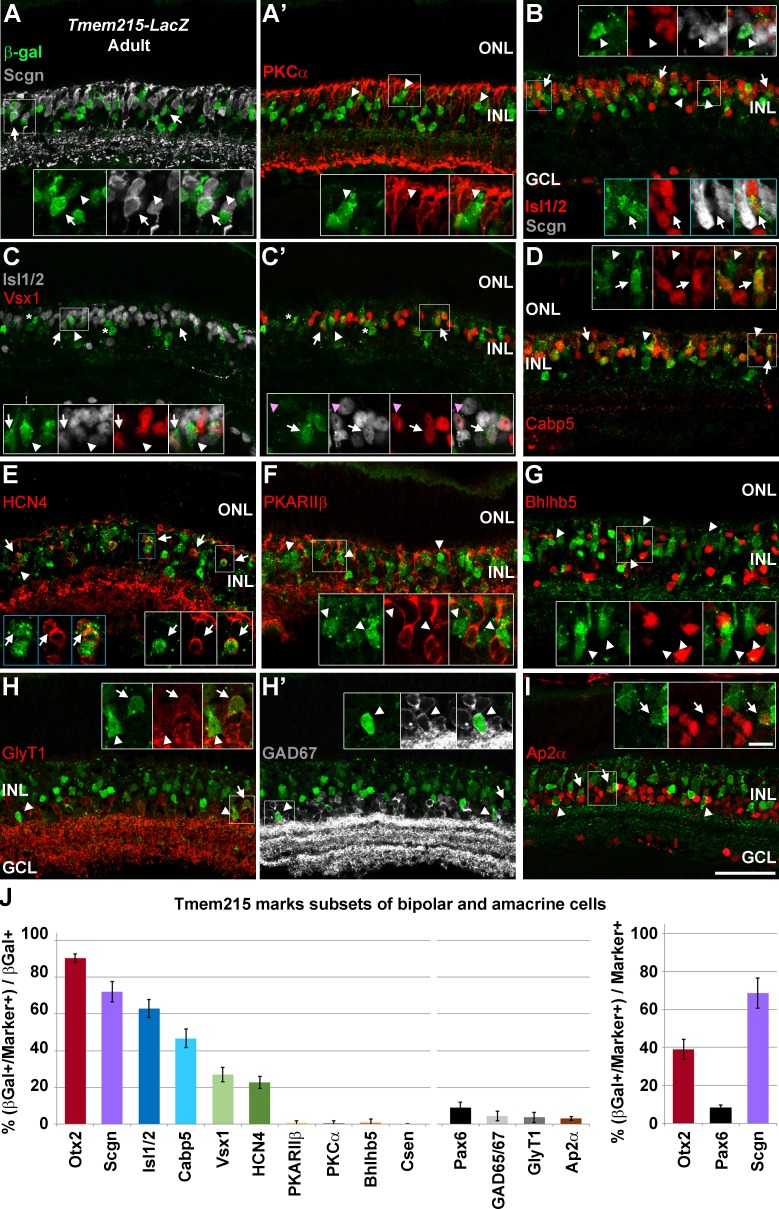
Tmem215 marks subsets of bipolar and amacrine cells. Adult Tmem215-LacZ heterozygous mice stained for β-gal (green) and cell-type specific markers (red/gray). (A–A') Section stained with Scgn (gray) and PKCα (red). A large fraction of β-gal+ cells coexpress Scgn (arrows, insets), but none overlap with PKCα (arrowheads, insets). (B) Costaining with Scgn (gray) and Isl1/2 (red) to mark ON bipolar cells. A subset of β-gal+ cells coexpress Isl1/2 (arrows, blue insets). Other β-gal+ cells coexpress only Scgn (arrowheads), marking them as cone OFF bipolars. Thus, Tmem215-LacZ marks both ON and OFF cone bipolar cells. (C–C') Costaining with Isl1/2 (gray) and Vsx1 (red), which mark types 1, 2, and 7 cone bipolars. A subset of β-gal+ cells coexpress Vsx1 and Isl1/2 (arrows, insets), marking them as type 7 cone ON bipolars. However, not all type 7 cone bipolars were β-gal+ (magenta arrowheads, insets). Some β-gal+ cells expressed Isl1/2, but not Vsx1 (arrowheads, insets). Of the β-gal+ cells that did not express Isl1/2, none coexpressed Vsx1 (asterisks). This argues that types 1 and 2 cone bipolars are not Tmem215+. Isl1/2+ amacrine cells do not coexpress β-gal. (D) A subset of β-gal+ cells coexpress Cabp5 (red, arrows, insets), which marks types 3 and 5 cone bipolars. (E) β-gal costaining with HCN4 to mark type 3a cone OFF bipolars. Most HCN4+ bipolar cells coexpress β-gal (arrows, insets), though some HCN4+ cells in the inner INL lack β-gal staining (arrowheads). (F) β-gal expression (arrowheads, insets) does not overlap with PKARIIβ (red), a marker of type 3b cone bipolars. (G) Type 2 cone OFF bipolars marked with Bhlhb5 did not express β-gal (arrowheads, insets). We did not see β-gal overlap with the type 4 cone OFF bipolar marker Csen (data not shown). (H–H') Retinas stained with the amacrine markers GlyT1 (red, glycinergic) and GAD65/67 (gray, GABAergic). Roughly equal fractions coexpress GlyT1 (arrows, insets) and GAD65/67 (arrowheads, insets). There are no β-gal+ displaced amacrine cells seen. (I) A subset of the β-gal+ amacrine cells (arrowheads, insets) coexpress Ap2α (red) (arrows, insets). Scale bars: 50 μm for panels and 10 μm for insets. (J) Quantification of β-gal+ cells. The left panel shows the percentage of β-gal+ cells that coexpress a cell type–specific marker. There are approximately 9 β-gal+ bipolar cells (Otx2+) for every amacrine cell (Pax6+). The right panel shows the fraction of a cell type–specific marker population that coexpresses β-gal+. Error bars show SD.
To define which bipolar and amacrine subtypes expressed Tmem215, we costained adult Tmem215-LacZ heterozygous mice for β-gal and several subtype-specific markers (Table 3; Fig. 6). We observed that a large subset of β-gal + cells coexpressed Scgn (72.2% ± 5.5% SD), though not all Scgn+ cells were β-gal+ (Figs. 6A, 6J). This suggests that Tmem215 marks mostly types 2 through 6 cone bipolar cells in addition to other bipolar subtypes. Immunostaining for PKCα did not reveal any appreciable overlap with β-gal, arguing that rod bipolars do not express Tmem215 (Fig. 6A'). Next, we immunostained sections with Isl1/2 to label ON type bipolars and with either Scgn or Vsx1 to mark subsets containing both ON and OFF cone bipolar subtypes (Table 3; Figs. 6B–C). We observed that the majority (63.0% ± 4.9% SD) of β-gal+ cells coexpressed Isl1/2, indicating that they are mostly cone ON bipolar cells (Figs. 6B, 6C, 6J). We observed that β-gal+ bipolars coexpressed Isl1/2 and Scgn or just Isl1/2, indicating that Tmem215 marks cone ON bipolar subtypes in addition to just types 5 and 6 (that are marked by Scgn; Fig. 6B). This also indicates that Tmem215 labels at least some of the types 2 through 4 cone OFF bipolar cells. A fraction of the β-gal+ bipolar cells coexpressed Vsx1, which marks types 1, 2, and 7 cone bipolars28 (Figs. 6C, 6J). However, these β-gal+/Vsx1+ cells always coexpressed Isl1/2, indicating that they are type 7 cone ON bipolar cells and not types 1 or 2 OFF bipolars (Fig. 6C). Not all type 7 cone ON bipolars expressed β-gal (Fig. 6C'). We next examined Cabp5 expression, which marks rod bipolars and types 3 and 5 cone bipolars. Since rod bipolars were not found to express β-gal, we expected that only a subset of the Cabp5+ cells would coexpress β-gal (Fig. 6D). Indeed, only 33% of the Cabp5+ cells made β-gal, whereas 46% of the β-gal+ cells made Cabp5, suggesting that types 3 and 5 cone bipolar cells express Tmem215 (Figs. 6D, 6J). We next used antibodies to HCN4 to mark type 3a cone OFF bipolar cells and small subset of amacrines56,58 (Table 3; Fig. 6E). We observed that approximately 22% of the β-gal+ cells coexpressed HCN4 (Figs. 6E, 6J). As was the case with type 7 cells, not all of the HCN4+ cells in the INL were colabeled with β-gal (63.3% ± 10.4% +SD; Fig. 6E). We then costained β-gal+ cells with markers of the remaining three cone OFF bipolar populations (Table 3); PKARIIβ (type 3b); Bhlhb5 (type 2); and Csen (type 4) (Figs. 6F, 6G). There was no appreciable overlap of β-gal with PKARIIβ (Fig. 6F), Bhlhb5 (Fig. 6G), or Csen (not shown and Fig. 6J) indicating that Tmem215 does not mark types 2, 3b, or 4 cone OFF bipolar cells. Taken together, these combinations of marker overlap suggest that Tmem215 marks type 3a cone OFF bipolar cells and types 5, 6, and 7 cone ON bipolars (Table 4). We were unable to distinguish between the three type 5 bipolar cell subtypes. Nor were we able to specifically mark type 6, 8, 9, or XBC bipolar cells. Since there are Isl1/2+ cone ON bipolars that make β-gal, but not Scgn, it is possible that type 8, 9, or XBCs also express Tmem215 (Table 4). We observed some variability in the number of β-gal+ cells between animals, suggesting that β-gal expression does not mark all Tmem215+ cells. This may explain why some type 3a cone OFF bipolars (HCN4+) and type 7 cone ON bipolars (Vsx1+/Isl1/2+) did not coexpress β-gal. Alternatively, only subsets of these bipolar subtypes might express Tmem215.
Lastly, we looked at amacrine cell subtypes that express Tmem215. We costained adult Tmem215-LacZ heterozygous retinas with β-gal and three broad amacrine subtype markers (Figs. 6H–J). We observed that approximately half of the β-gal+ amacrine population expressed GlyT1, a marker of glycinergic amacrine cells48,59 (Figs. 6H, 6J). Similarly, approximately half of the β-gal+ cells coexpressed GAD65/67, which marks GABAergic amacrines48 (Figs. 6H', 6J). Lastly, approximately a third of the β-gal+ amacrines expressed Ap2α, a marker of a large complex set of amacrine subtypes in the retina60 (Figs. 6I, 6J). Together, these data indicate that Tmem215 marks multiple subtypes of amacrine cells in the retina. We found Tmem215 does not mark any displaced amacrine subtypes, since all of the β-gal+ cells have their somas in the INL.
Discussion
The mechanisms that govern bipolar interneuron fate choice and subtype identity are only partially understood. The transcription factor Blimp1 represses bipolar cell fate choice during retinal development.15,35,36 We took advantage of precocious bipolar cell development in Blimp1 CKO mice to screen for novel pan- and subtype-specific developmental markers of bipolar cells. From this, we identified several genes that have not been characterized during bipolar cell genesis. Of the genes we examined in more detail (Slitrk6, Gsg1, Trnp1, and Tmem215), each marked a different subpopulation of bipolar cells. The expression patterns of several other precociously upregulated genes from our study, such as Cnpy1 and Samsn1, remain to be characterized. Based on the spatial specificity of the genes we have tested to date, it is likely that many of the remaining candidates will show bipolar subtype-specific expression. Thus, precociously expressed bipolar-specific genes may function as novel regulators of class and subtype fate choice. Whether Slitrk6, Gsg1, Trnp1, and Tmem215 control bipolar cell development remains to be tested.
Slitrk6 and Gsg1 Proteins
Slitrk6 belongs to a family of single-pass transmembrane proteins characterized by their ability to affect neurite outgrowth.61 Messenger RNA expression of Slitrk6 was previously detected in the mature INL and GCL of the mouse retina,47 similar to what we observed. Other data suggest that Slitrk6 is made earlier in development,62,63 though we saw little signal in the retina at P2 or younger stages. This may be due to the use of different in situ probes and hybridization techniques. Humans with homozygous SLITRK6 nonsense mutations display high (i.e., severe) myopia and sensorineural deafness.47 Mice with a Slitrk6 mutation have similar phenotypes, but also show delayed maturation of ribbon synapses in the outer plexiform layer. This implies that Slitrk6 expression in bipolar cells is important for synapse formation and/or maturation.47 The apparent expression of Slitrk6 by only a subset of bipolar cells may explain why the ribbon synapse phenotype in Slitrk6 knockout mice is relatively mild. Better tools are needed to precisely characterize which subtypes of bipolars, amacrines, and possibly ganglion cells express Slitrk6. This information will be important for fully understanding how Slitrk6 regulates retinal development and physiology.
The expression pattern of Gsg1 has not been characterized previously in the mouse retina. Prior work showed that the amount of Gsg1 transcription was decreased in retinas that lacked Otx2 (loss of bipolar cells).64 Conversely, Blimp1 mutant mice showed increased Gsg1 (and several other genes we observed) expression by microarray analysis at P6,36 a time when wild-type bipolar cells are normally differentiating. Together, these data are consistent with our findings that Gsg1 is bipolar cell–specific. Starting around P5, we observed Gsg1 mRNA in a small subset of bipolar cells. The in situ pattern of Gsg1 was most consistent with expression in types 3 and/or 5 cone bipolars. While we cannot rule out that other cone bipolar subtypes make Gsg1, the sparse in situ pattern suggests that only one or two subtypes are labeled. Little is known about Gsg1 expression and function. It has been best characterized in spermatids, where it appears to be an endoplasmic reticulum (ER)-localized membrane protein.65 Gsg1 interacts with testis-specific poly A polymerase (TPAP), sequestering it to the ER. This localization of TPAP may be required for spermatid maturation.65,66 In the retina, Gsg1 may affect mRNA polyadenylation within specific bipolar cell subtypes to uniquely control their physiology or development. The precise expression pattern and function of Gsg1 in the retina remains to be tested.
TMF-Regulated Nuclear Protein
Initially, Trnp1 was characterized in immortalized cell lines. This 223 amino acid nuclear-localized protein promotes cell cycle progression of cultured cells.67 More recently, Trnp1 expression has been described in the developing cerebral cortex.44,68,69 Expression of Trnp1 was seen in neural stem cells.44,68 Gain- and loss-of-function experiments in the mouse cerebral cortex showed that high levels of Trnp1 promotes neural stem cell self-renewal and tangential expansion. In contrast, lower levels of Trnp1 promote radial expansion, with increased intermediate progenitors and basal radial glial cells leading to the folding of the otherwise smooth murine cerebral cortex.44,68 Thus, Trnp1 acts like a master regulator of neural stem cell fate. In addition to neural stem cells, Trnp1 is also transiently expressed in postmitotic cortical neurons.
Based on our RNA-seq experiments and the findings in cortex, we expected to see Trnp1 in bipolar cells and progenitors. We observed modest Trnp1 mRNA expression by in situ hybridization in the developing retina, but it was primarily localized to the ganglion cell layer that is devoid of any progenitor cells. We did not observe any Trnp1 immunostaining in wild-type retinas until P7, where it specifically and permanently marked bipolar cells. This discordance between mRNA and protein labeling could be due to nonspecific signal detection from the in situ hybridization protocol. However, since there was a modest number of Trnp1 RNA-seq reads in control retinas at P2, Trnp1 protein may be below the threshold of antibody detection or the mRNA may not be translated until later time-points. While Trnp1 immunostaining labeled only ON type bipolar cells in the adult retina, a small seemingly random population of these bipolars was Trnp1 negative. This further suggests that Trnp1 immunostaining is not completely sensitive.
Trnp1 regulates neural stem cell fate choice in the developing cortex; however, the onset of Trnp1 protein expression in the retina at P7 suggests that it influences bipolar cell maturation or physiology instead of subtype fate choice. This may also be the case in the cortex, where Trnp1 is expressed by newly formed neurons. While the exact cellular functions of Trnp1 are unknown, it has been localized to euchromatin in the nucleus, suggesting that it promotes gene expression.44,67 This raises the possibility that Trnp1 functions downstream of subtype fate choices in the retina by activating the expression of genes important for ON bipolar function, such as Grm6 (mGluR6) and Gnao (Goα). Trnp1 may synergize with or act downstream of Isl1, a transcription factor that is also expressed by ON type bipolar cells.25,26 Loss-of-function studies are ongoing to test whether Trnp1 controls subtype choice in the retina or ON bipolar physiology. These studies will also reveal whether Trnp1 plays an earlier role in retinal development.
Tmem215 Expression
Using RNA-seq, in situ hybridization and a LacZ knock-in mouse line, we characterized Tmem215 expression in the developing retina. However, these techniques yielded slightly different results. Sequencing of RNA on control P2 retinas suggested that Tmem215 was expressed at very low levels or in only a few cells. Few if any β-gal+ cells were seen in Tmem215-LacZ mice at P2. In contrast, Tmem215 in situ hybridization signal occasionally yielded a broader expression pattern at P2. Starting at P5, the Tmem215 in situ hybridization signal and the β-gal+ immunostaining patterns were equivalent. Thus, there may be nonspecific in situ probe binding at P2 when there are few Tmem215 transcripts available. In the mature retina, Tmem215-LacZ marked a complex subset of cone bipolar cells and amacrines. We observed that types 3a, 5, and 7 cone bipolars expressed β-gal and that types 1, 2, 3b, and 4 lacked β-gal staining. We were unable to determine definitively whether the remaining cone bipolar subtypes were β-gal positive or negative. Not every type 3a, 5, or 7 cone bipolar made β-gal. This suggests that there is either additional subtype diversity in these populations or that β-gal expression was below the threshold of detection. In support of the latter, we observed modest variability in the number of β-gal+ cells between Tmem215-LacZ animals. The types of bipolars that coexpressed β-gal did not change between animals suggesting that the Tmem215-LacZ gene trap allele incompletely labels specific subsets of cone bipolar cells. Further supporting our findings, transcriptome profiling of sorted bipolar cells showed that Tmem215 expression is enriched in bipolar cells compared to other retinal cell types.70
Since Tmem215 expression starts early in bipolar cell development, it could play a role in bipolar cell subtype fate choice, maturation, and/or physiology. Little is known about how Tmem215 functions. It is a predicted transmembrane protein that may interact with MAGI1, a protein localized to cell junctions.71–74 MAGI1 can influence cell signaling events, thus Tmem215 may mediate transduction of cell-cell signals during bipolar genesis to regulate subtype fate choice. It has also been shown that MAGI1 is important for synapse formation in Caenorhabditis elegans,75 where it helps shuttle ionotropic glutamate receptors to synapses. As these receptors are seen in OFF type bipolar cells, Tmem215 may regulate synaptic maturation and/or function. Since Tmem215 is expressed by both ON and OFF bipolar cells, it may regulate common physiologic features of these subtypes. Alternatively, Tmem215 may regulate different functions in each subtype. This is analogous to Vsx1, which controls gene expression differently in ON versus OFF cone bipolar cells.28 Comparing bipolar cell development between homozygous Tmem215-LacZ gene trap mice and heterozygous controls will distinguish between these possibilities.
Supplementary Material
Acknowledgments
The authors thank David Beebe, Tatiana Eliseeva, Miriam Esgleas, Noah Goodson, Magdalena Götz, Françoise Haeseleer, Ed Levine, Taylor Mills, and Jhenya Nahreini for technical advice and antibodies; Katrina Diener, Bifeng Gao, and Ted Shade for assistance with RNA sequence collection; and Abby Zamora and the University of Colorado Denver Biotechnology Core for assistance with Tmem215-LacZ mice. The mouse strain Tmem215-LacZ was created from ES cell clone 10442a-F5, obtained from the KOMP Repository (www.komp.org) and generated by Regeneron Pharmaceuticals, Inc. J. A. Brzezinski IV is a Boettcher Investigator.
Supported in part by NIH R01-EY024272 and a Challenge Grant to the University of Colorado Denver Department of Ophthalmology from Research to Prevent Blindness, Inc.
Disclosure: K.U. Park, None; G. Randazzo, None; K.L. Jones, None; J.A. Brzezinski IV, None
References
- 1. Greene MJ, Kim JS, Seung HS; The EyeWirers. . Analogous convergence of sustained and transient inputs in parallel on and off pathways for retinal motion computation. Cell Rep. 2016; 14: 1892–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Helmstaedter M,, Briggman KL,, Turaga SC,, Jain V,, Seung HS,, Denk W. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature. 2013; 500: 168–174. [DOI] [PubMed] [Google Scholar]
- 3. Euler T,, Haverkamp S,, Schubert T,, Baden T. Retinal bipolar cells: elementary building blocks of vision. Nat Rev Neurosci. 2014; 15: 507–519. [DOI] [PubMed] [Google Scholar]
- 4. Ichinose T,, Fyk-Kolodziej B,, Cohn J. Roles of ON cone bipolar cell subtypes in temporal coding in the mouse retina. J Neurosci. 2014; 34: 8761–8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jeon CJ,, Strettoi E,, Masland RH. The major cell populations of the mouse retina. J Neurosci. 1998; 18: 8936–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sidman RL. Histogenesis of mouse retina studied with thymidine H3. : Smelser GK, Structure of the Eye. New York, NY: Academic Press; 1961: 487–506. [Google Scholar]
- 7. Rapaport DH,, Wong LL,, Wood ED,, Yasumura D,, LaVail MM. Timing and topography of cell genesis in the rat retina. J Comp Neurol. 2004; 474: 304–324. [DOI] [PubMed] [Google Scholar]
- 8. Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985; 212: 199–205. [DOI] [PubMed] [Google Scholar]
- 9. Morrow EM,, Chen CM,, Cepko CL. Temporal order of bipolar cell genesis in the neural retina. Neural Dev. 2008; 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burmeister M,, Novak J,, Liang MY,, et al. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996; 12: 376–384. [DOI] [PubMed] [Google Scholar]
- 11. Green ES,, Stubbs JL,, Levine EM. Genetic rescue of cell number in a mouse model of microphthalmia: interactions between Chx10 and G1-phase cell cycle regulators. Development. 2003; 130: 539–552. [DOI] [PubMed] [Google Scholar]
- 12. Nishida A,, Furukawa A,, Koike C,, et al. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003; 6: 1255–1263. [DOI] [PubMed] [Google Scholar]
- 13. Sato S,, Inoue T,, Terada K,, et al. Dkk3-Cre BAC transgenic mouse line: a tool for highly efficient gene deletion in retinal progenitor cells. Genesis. 2007; 45: 502–507. [DOI] [PubMed] [Google Scholar]
- 14. Fossat N,, Le Greneur C,, Beby F,, et al. A new GFP-tagged line reveals unexpected Otx2 protein localization in retinal photoreceptors. BMC Dev Biol. 2007; 7: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brzezinski JA,, Lamba DA,, Reh TA. Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development. 2010; 137: 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim DS,, Ross SE,, Trimarchi JM,, Aach J,, Greenberg ME,, Cepko CL. Identification of molecular markers of bipolar cells in the murine retina. J Comp Neurol. 2008; 507: 1795–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dorval KM,, Bobechko BP,, Fujieda H,, Chen S,, Zack DJ,, Bremner R. CHX10 targets a subset of photoreceptor genes. J Biol Chem. 2006; 281: 744–751. [DOI] [PubMed] [Google Scholar]
- 18. Livne-Bar I,, Pacal M,, Cheung MC,, et al. Chx10 is required to block photoreceptor differentiation but is dispensable for progenitor proliferation in the postnatal retina. Proc Natl Acad Sci U S A. 2006; 103: 4988–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hatakeyama J,, Tomita K,, Inoue T,, Kageyama R. Roles of homeobox and bHLH genes in specification of a retinal cell type. Development. 2001; 128: 1313–1322. [DOI] [PubMed] [Google Scholar]
- 20. Akagi T,, Inoue T,, Miyoshi G,, et al. Requirement of multiple bHLH genes for retinal neuronal subtype specification. J Biol Chem. 2004; 279: 28492–28498. [DOI] [PubMed] [Google Scholar]
- 21. Tomita K,, Moriyoshi K,, Nakanishi S,, Guillemot F,, Kageyama R. Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 2000; 19: 5460–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Balasubramanian R,, Bui A,, Ding Q,, Gan L. Expression of LIM-homeodomain transcription factors in the developing and mature mouse retina. Gene Expr Patterns. 2014; 14: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feng L,, Xie X,, Joshi PS,, et al. Requirement for Bhlhb5 in the specification of amacrine and cone bipolar subtypes in mouse retina. Development. 2006; 133: 4815–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang L,, Hu F,, Feng L,, et al. Bhlhb5 is required for the subtype development of retinal amacrine and bipolar cells in mice. Dev Dyn. 2014; 243: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Elshatory Y,, Everhart D,, Deng M,, Xie X,, Barlow RB,, Gan L. Islet-1 controls the differentiation of retinal bipolar and cholinergic amacrine cells. J Neurosci. 2007; 27: 12707–12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Elshatory Y,, Deng M,, Xie X,, Gan L. Expression of the LIM-homeodomain protein Isl1 in the developing and mature mouse retina. J Comp Neurol. 2007; 503: 182–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bramblett DE,, Pennesi ME,, Wu SM,, Tsai MJ. The transcription factor Bhlhb4 is required for rod bipolar cell maturation. Neuron. 2004; 43: 779–793. [DOI] [PubMed] [Google Scholar]
- 28. Shi Z,, Trenholm S,, Zhu M,, et al. Vsx1 regulates terminal differentiation of type 7 ON bipolar cells. J Neurosci. 2011; 31: 13118–13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kerschensteiner D,, Liu H,, Cheng CW,, et al. Genetic control of circuit function: Vsx1 and Irx5 transcription factors regulate contrast adaptation in the mouse retina. J Neurosci. 2008; 28: 2342–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ohtoshi A,, Wang SW,, Maeda H,, et al. Regulation of retinal cone bipolar cell differentiation and photopic vision by the CVC homeobox gene Vsx1. Curr Biol. 2004; 14: 530–536. [DOI] [PubMed] [Google Scholar]
- 31. Clark AM,, Yun S,, Veien ES,, et al. Negative regulation of Vsx1 by its paralog Chx10/Vsx2 is conserved in the vertebrate retina. Brain Research. 2008; 1192: 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blackshaw S,, Harpavat S,, Trimarchi J,, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004; 2: E247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jung CC,, Atan D,, Ng D,, et al. Transcription factor PRDM8 is required for rod bipolar and type 2 OFF-cone bipolar cell survival and amacrine subtype identity. Proc Natl Acad Sci U S A. 2015; 112: E3010–E3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ross SE,, McCord AE,, Jung C,, et al. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron. 2012; 73: 292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brzezinski JA,, Uoon Park K,, Reh TA. Blimp1 (Prdm1) prevents re-specification of photoreceptors into retinal bipolar cells by restricting competence. Dev Biol. 2013; 384: 194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Katoh K,, Omori Y,, Onishi A,, Sato S,, Kondo M,, Furukawa T. Blimp1 suppresses Chx10 expression in differentiating retinal photoreceptor precursors to ensure proper photoreceptor development. J Neurosci. 2010; 30: 6515–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Valenzuela DM,, Murphy AJ,, Frendewey D,, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003; 21: 652–659. [DOI] [PubMed] [Google Scholar]
- 38. Baird NL,, Bowlin JL,, Cohrs RJ,, Gilden D,, Jones KL. Comparison of varicella-zoster virus RNA sequences in human neurons and fibroblasts. J Virol. 2014; 88: 5877–5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bradford AP,, Jones K,, Kechris K,, et al. Joint MiRNA/mRNA expression profiling reveals changes consistent with development of dysfunctional corpus luteum after weight gain. PLoS One. 2015; 10: e0135163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Henderson HH,, Timberlake KB,, Austin ZA,, et al. Occupancy of RNA polymerase II phosphorylated on serine 5 (RNAP S5P) and RNAP S2P on varicella-zoster virus genes 9, 51, and 66 is independent of transcript abundance and polymerase location within the gene. J Virol. 2015; 90: 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Maycotte P,, Jones KL,, Goodall ML,, Thorburn J,, Thorburn A. Autophagy supports breast cancer stem cell maintenance by regulating IL6 secretion. Mol Cancer Res. 2015; 13: 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Livak KJ,, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 43. Rieke F,, Lee A,, Haeseleer F. Characterization of Ca2+-binding protein 5 knockout mouse retina. Invest Ophthalmol Vis Sci. 2008; 49: 5126–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stahl R,, Walcher T,, De JuanRomero C,, et al. Trnp1 regulates expansion and folding of the mammalian cerebral cortex by control of radial glial fate. Cell. 2013; 153: 535–549. [DOI] [PubMed] [Google Scholar]
- 45. Schneider CA,, Rasband WS,, Eliceiri KW. NIH. Image to ImageJ: 25 years of image analysis. Nature Methods. 2012; 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Duan X,, Krishnaswamy A,, De la Huerta I,, Sanes JR. Type II cadherins guide assembly of a direction-selective retinal circuit. Cell. 2014; 158: 793–807. [DOI] [PubMed] [Google Scholar]
- 47. Tekin M,, Chioza BA,, Matsumoto Y,, et al. SLITRK6 mutations cause myopia and deafness in humans and mice. J Clin Invest. 2013; 123: 2094–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Haverkamp S,, Wassle H. Immunocytochemical analysis of the mouse retina. J Comp Neurol. 2000; 424: 1–23. [PubMed] [Google Scholar]
- 49. Taranova OV,, Magness ST,, Fagan BM,, et al. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006; 20: 1187–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Surzenko N,, Crowl T,, Bachleda A,, Langer L,, Pevny L. SOX2 maintains the quiescent progenitor cell state of postnatal retinal Müller glia. Development. 2013; 140: 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cherry TJ,, Trimarchi JM,, Stadler MB,, Cepko CL. Development and diversification of retinal amacrine interneurons at single cell resolution. Proc Natl Acad Sci U S A. 2009; 106: 9495–9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Greferath U,, Grunert U,, Wassle H. Rod bipolar cells in the mammalian retina show protein kinase C-like immunoreactivity. J Comp Neurol. 1990; 301: 433–442. [DOI] [PubMed] [Google Scholar]
- 53. Puthussery T,, Gayet-Primo J,, Taylor WR. Localization of the calcium-binding protein secretagogin in cone bipolar cells of the mammalian retina. J Comp Neurol. 2010; 518: 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ghosh KK,, Bujan S,, Haverkamp S,, Feigenspan A,, Wassle H. Types of bipolar cells in the mouse retina. J Comp Neurol. 2004; 469: 70–82. [DOI] [PubMed] [Google Scholar]
- 55. de Melo J,, Qiu X,, Du G,, Cristante L,, Eisenstat DD. Dlx1, Dlx2, Pax6, Brn3b, and Chx10 homeobox gene expression defines the retinal ganglion and inner nuclear layers of the developing and adult mouse retina. J Comp Neurol. 2003; 461: 187–204. [DOI] [PubMed] [Google Scholar]
- 56. Mataruga A,, Kremmer E,, Muller F. Type 3a and type 3b OFF cone bipolar cells provide for the alternative rod pathway in the mouse retina. J Comp Neurol. 2007; 502: 1123–1137. [DOI] [PubMed] [Google Scholar]
- 57. Haverkamp S,, Specht D,, Majumdar S,, et al. Type 4 OFF cone bipolar cells of the mouse retina express calsenilin and contact cones as well as rods. J Comp Neurol. 2008; 507: 1087–1101. [DOI] [PubMed] [Google Scholar]
- 58. Wassle H,, Puller C,, Muller F,, Haverkamp S. Cone contacts, mosaics, and territories of bipolar cells in the mouse retina. J Neurosci. 2009; 29: 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wassle H,, Koulen P,, Brandstatter JH,, Fletcher EL,, Becker CM. Glycine and GABA receptors in the mammalian retina. Vision Res. 1998; 38: 1411–1430. [DOI] [PubMed] [Google Scholar]
- 60. Bassett EA,, Pontoriero GF,, Feng W,, et al. Conditional deletion of activating protein 2alpha (AP-2alpha) in the developing retina demonstrates non-cell-autonomous roles for AP-2alpha in optic cup development. Mol Cell Biol. 2007; 27: 7497–7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Aruga J,, Mikoshiba K. Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol Cell Neurosci. 2003; 24: 117–129. [DOI] [PubMed] [Google Scholar]
- 62. Aruga J. Slitrk6 expression profile in the mouse embryo and its relationship to that of Nlrr3. Gene Expr Patterns. 2003; 3: 727–733. [DOI] [PubMed] [Google Scholar]
- 63. Beaubien F,, Cloutier JF. Differential expression of Slitrk family members in the mouse nervous system. Dev Dyn. 2009; 238: 3285–3296. [DOI] [PubMed] [Google Scholar]
- 64. Omori A,, Akasaka K,, Kurokawa D,, Amemiya S. Gene expression analysis of Six3, Pax6, and Otx in the early development of the stalked crinoid Metacrinus rotundus. Gene Expr Patterns. 2011; 11: 48–56. [DOI] [PubMed] [Google Scholar]
- 65. Choi HS,, Lee SH,, Kim H,, Lee Y. Germ cell-specific gene 1 targets testis-specific poly(A) polymerase to the endoplasmic reticulum through protein-protein interactions. FEBS Lett. 2008; 582: 1203–1209. [DOI] [PubMed] [Google Scholar]
- 66. Kashiwabara S,, Noguchi J,, Zhuang T,, et al. Regulation of spermatogenesis by testis-specific, cytoplasmic poly(A) polymerase TPAP. Science. 2002; 298: 1999–2002. [DOI] [PubMed] [Google Scholar]
- 67. Volpe M,, Shpungin S,, Barbi C,, et al. Trnp: a conserved mammalian gene encoding a nuclear protein that accelerates cell-cycle progression. DNA Cell Biol. 2006; 25: 331–339. [DOI] [PubMed] [Google Scholar]
- 68. Pilz GA,, Shitamukai A,, Reillo I,, et al. Amplification of progenitors in the mammalian telencephalon includes a new radial glial cell type. Nat Commun. 2013; 4: 2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Martinez-Martinez MA,, Romero CD,, Fernandez V,, Cardenas A,, Gotz M,, Borrell V. A restricted period for formation of outer subventricular zone defined by Cdh1 and Trnp1 levels. Nature Communications. 2016; 7: 11812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Siegert S,, Cabuy E,, Scherf BG,, et al. Transcriptional code and disease map for adult retinal cell types. Nat Neurosci. 2012; 15: 487–495, S481–S482. [DOI] [PubMed] [Google Scholar]
- 71. Stetak A,, Hajnal A. The C. elegans MAGI-1 protein is a novel component of cell junctions that is required for junctional compartmentalization. Dev Biol. 2011; 350: 24–31. [DOI] [PubMed] [Google Scholar]
- 72. Luck K,, Fournane S,, Kieffer B,, Masson M,, Nomine Y,, Trave G. Putting into practice domain-linear motif interaction predictions for exploration of protein networks. PLoS One. 2011; 6: e25376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ide N,, Hata Y,, Nishioka H,, et al. Localization of membrane-associated guanylate kinase (MAGI)-1/BAI-associated protein (BAP) 1 at tight junctions of epithelial cells. Oncogene. 1999; 18: 7810–7815. [DOI] [PubMed] [Google Scholar]
- 74. Wegmann F,, Ebnet K,, Du Pasquier L,, Vestweber D,, Butz S. Endothelial adhesion molecule ESAM binds directly to the multidomain adaptor MAGI-1 and recruits it to cell contacts. Exp Cell Res. 2004; 300: 121–133. [DOI] [PubMed] [Google Scholar]
- 75. Stetak A,, Horndli F,, Maricq AV,, van den Heuvel S,, Hajnal A. Neuron-specific regulation of associative learning and memory by MAGI-1 in C. elegans. PLoS One. 2009; 4: e6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.