Abstract
Background
The association of CD133 overexpression with clinicopathological significance and prognosis in patients with breast cancer remains controversial. We thus performed a meta-analysis to evaluate the role of CD133 expression in the development and prognosis of breast cancer.
Methods
The databases PubMed, Embase, and Cochrane Library (updated to August 1, 2016) were searched. Pooled odds ratios (ORs) or hazard ratios (HRs) with 95% confidence intervals (95% CI) were used to evaluate the impact of CD133 expression on clinicopathological features, overall survival, and disease-free survival.
Results
A total of 1,734 patients from 13 studies were subject to final analysis. The results showed a significant association between overexpression of CD133 and estrogen receptor status (OR 0.35, 95% CI 0.18–0.70), progesterone receptor status (OR 0.56, 95% CI 0.43–0.74), human epidermal growth factor-2 status (OR 1.81, 95% CI 1.33–2.45), lymph node metastasis (OR 1.98, 95% CI 1.34–2.92), and tumor histological grade (OR 1.79, 95% CI 1.26–2.54) in breast cancer. However, no significant correlation was found between upregulation of CD133 expression and onset age (OR 1.03, 95% CI 0.70–1.53) or tumor size (OR 1.29, 95% CI 0.80–2.09). Moreover, CD133-positive breast cancer patients had a higher risk of mortality (HR 1.91, 95% CI 1.21–3.03) and disease progression (HR 2.70, 95% CI 1.05–6.95).
Conclusion
This meta-analysis suggested that CD133 might be a predictor of clinical outcomes as well as prognosis and could be a potentially new gene therapy target for breast cancer patients.
Keywords: CD133, CSCs, breast cancer, prognosis, biomarker, meta-analysis
Introduction
Breast cancer is the most commonly occurring malignant tumor in women, with ~1.67 million new cases (25% of all cancers) diagnosed worldwide in 2012. It is the most frequent cause of cancer death (522,000 deaths, 14.7% of total) in females.1 From the time that distinct molecular subtypes were proposed by Perou et al in 2000,2 the combination of traditional pathological morphological classification and molecular subtyping has been applied to determine the optimal therapy for breast cancer patients. However, the prognosis of breast cancer patients remains unsatisfactory. Consequently, it is critical to predict prognosis through novel biomarkers that can serve as potential therapeutic targets in breast cancer patients.
There is a growing realization that a small subpopulation of cells with stem cell-like features resides in the tumor tissue and is known as cancer stem cells (CSCs).3 Their activity is achieved by self-renewal, unlimited proliferation and differentiation potential, and high tumorigenicity.4 Recently, it has been found that CSCs have similar specific cell surface molecular markers to stem cells such as CD44, CD24, ALDH1, and CD133. CD133, which is known as prominin-1, a pentaspan transmembrane cell surface glycoprotein with a molecular weight of 120 kDa, is located in plasma membrane protrusions. It was initially considered to be a marker of hematopoietic stem cells by Yin et al.5 Biological functions of CD133 include tumor initiation, cellular migration, vasculogenic mimicry, and drug resistance.6 Although CD133 has been studied intensely in various types of solid tumors, including lung cancer,7 renal cancer,8 esophageal carcinoma,9 and gastric cancer,10 the role of CD133 in breast cancer has not been verified.
In this meta-analysis, we aimed to evaluate the relationship between CD133 expression in breast cancer and clinicopathological features, including tumor size, lymph node metastasis, histological grade, onset age, receptor status (estrogen receptor [ER], progesterone receptor [PR], and human epidermal growth factor-2 [HER2]) as well as prognostic significance.
Methods
Literature search
The literature in the following electronic databases – PubMed, Embase, and the Cochrane Library (updated to August 1, 2016) – was systematically searched. We performed our search using the medical subject heading (MeSH) term “CD133” and its synonyms: “fudenine”, “prominin”, “PROML1”, and “AC141 antigen”. These keywords were then combined with “breast”, “mammary”, “cancer”, “neoplasm”, “carcinoma”, “prognosis”, and “survival” using the Boolean “OR” term or the Boolean “AND” term.
Study selection criteria
Study selection inclusion criteria were as follows: 1) patients diagnosed with breast cancer using pathological and histological examinations, 2) full text and published in English, 3) clinicopathological and survival (overall survival [OS] and disease-free survival [DFS]) outcomes were recorded, 4) CD133 expression was detected in primary breast tumors, and 5) outcomes were recorded using odds ratios (ORs) or hazard ratios (HRs) with 95% confidence intervals (CIs).
Exclusion criteria were as follows: 1) meeting abstracts, comments, case reports, reviews, and meta-analyses; 2) experiments on cell lines and animals; 3) metastatic or recurrent cancer; and 4) duplicate studies.
Quality assessment
The selected cohort studies were analyzed from three perspectives, selection, comparability, and outcomes, by two investigators independently, according to the Newcastle–Ottawa scale (NOS). The details of NOS table are shown in Table S1.
Data extraction
The following details were extracted using a predefined form: first author’s name, publication year, country, mean age, tumor stage, total number of included patients, median follow-up time, cutoff value, survival outcome, outcome method, and estimated HR.
Statistical analysis
This meta-analysis was performed using Stata Version 12.0 (Stata Corporation, College Station, TX, USA). For the pooled analysis of clinicopathological features, OR was evaluated. HR was applied as a measure of the prognostic value. Study heterogeneity was evaluated using the chi-square-based Q test and I2 statistic. Studies with an I2>50% or a P<0.05 was considered to have significant heterogeneity, and a random-effects model test was conducted. Otherwise, the fixed-effects model test was selected. Sensitivity analysis was performed to evaluate the stability of the pooled results. Publication bias was assessed using Begg’s funnel plots and Egger’s test. All P-values were two-sided and P<0.05 was considered statistically significant.
Results
Search results
A total of 424 citations were potentially identified for inclusion using the described search strategies. Through reviewing the title and abstracts, 381 papers were excluded. We then systematically read the full text of the remaining 43 articles and filtered out an additional 30 papers. Among the excluded papers, 13 studies were experimental studies, six studies were not correlated with target protein, three studies had overlapped data with other published trials, six studies had no sufficient survival data to analyze, and two studies are reviews. Ultimately, 13 studies11–23 were included (Figure 1).
Figure 1.
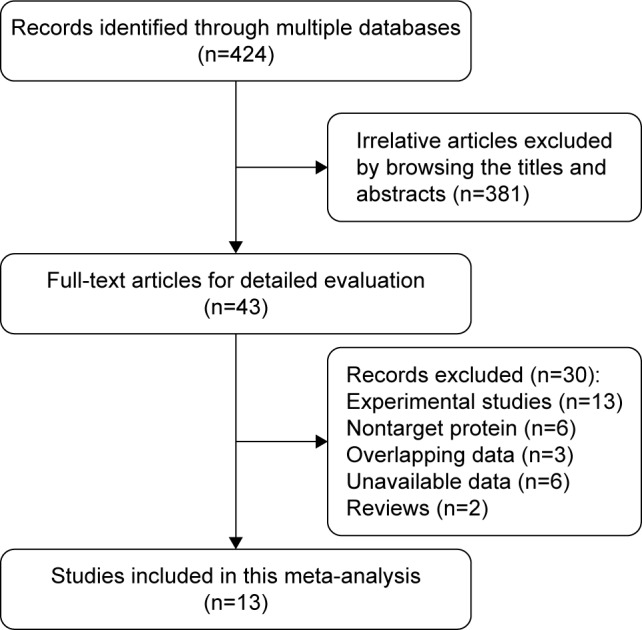
Flow diagram of the studies selection process.
Characteristics of included studies
The details of 13 included studies selected from the literature search are summarized in Table 1. In total, 13 eligible articles with 1,734 patients were analyzed for clinicopathological features, and five qualified studies with 879 patients were analyzed for survival outcomes. These cohort studies were conducted in eight regions (Italy, Taiwan, China, New Zealand, Japan, Turkey, Egypt, and Korea) and were published between 2009 and 2016 with a mean patient age ranging from 45.6 years to 61.8 years. Univariate analysis was applied for the survival data.
Table 1.
Characteristics of studies included in our meta-analysis
| Study | Year | Country | No of patients | Mean age (years) | Stage | Follow-up, months (range) | AB source | AB type | Scoring criteria | Cutoff value | Outcomes | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ieni et al11 | 2011 | Italy | 49 | 61.8 | I, II | 64.3 (6–136) | Rabbit | Polyclonal | ID score | NR | CF | 8 |
| Lin et al12 | 2015 | Taiwan | 49 | 52 | I–III | NR | NR | NR | Percentage | 10% | CF | 6 |
| Liu et al13 | 2009 | China | 74 | 49 | I–III | 41.6 (3–49.3) | NR | Polyclonal | Semiquantitative | 10% | CF | 6 |
| Currie et al14 | 2013 | New Zealand | 94 | NR | I–IV | 30.5 (1–60) | Mouse | Monoclonal | Semiquantitative | 10% | CF, OS, DFS | 8 |
| Di Bonito et al15 | 2012 | Italy | 204 | 56 | I–III | 42.5 (2–83) | NR | NR | Semiquantitative | 8% | CF | 6 |
| Aomatsu et al16 | 2012 | Japan | 102 | 55 | II, III | 48 (12–84) | NR | NR | Percentage | 10% | CF, OS, DFS | 8 |
| Kapucuoglu et al17 | 2015 | Turkey | 105 | 54 | I–IV | 45.05 (2–89) | NR | Polyclonal | Percentage | 10% | CF | 6 |
| Zhao et al18 | 2011 | China | 67 | 47 | I–III | 36 (1–64) | Rabbit | Polyclonal | Semiquantitative | 10% | OS, DFS | 8 |
| Mansour and Atwa19 | 2015 | Egypt | 120 | 49 | I–III | NR | Mouse | Monoclonal | Semiquantitative | 10% | CF | 6 |
| Kim et al20 | 2015 | Korea | 291 | 49 | I–III | 53.8 (4–97) | Rabbit | Polyclonal | Semiquantitative | 10% | CF, OS, DFS | 7 |
| Liu et al21 | 2013 | China | 134 | 47.5 | NR | NR | NR | NR | NR | NR | CF | 6 |
| Lv et al22 | 2016 | China | 120 | 48.7 | NR | NR | Rabbit | Monoclonal | Semiquantitative | 10% | CF | 7 |
| Han et al23 | 2015 | China | 325 | 45.6 | I–IV | 46.8 (15–108) | Mouse | Monoclonal | Semiquantitative | 10% | CF, OS | 8 |
Abbreviations: NR, not reported; AB, antibody; ID, intensity-distribution; CF, clinicopathological features; OS, overall survival; DFS, disease-free survival; NOS, Newcastle–Ottawa scale.
Meta-analysis of clinicopathological parameters
CD133 expression and ER, PR, and HER2 status
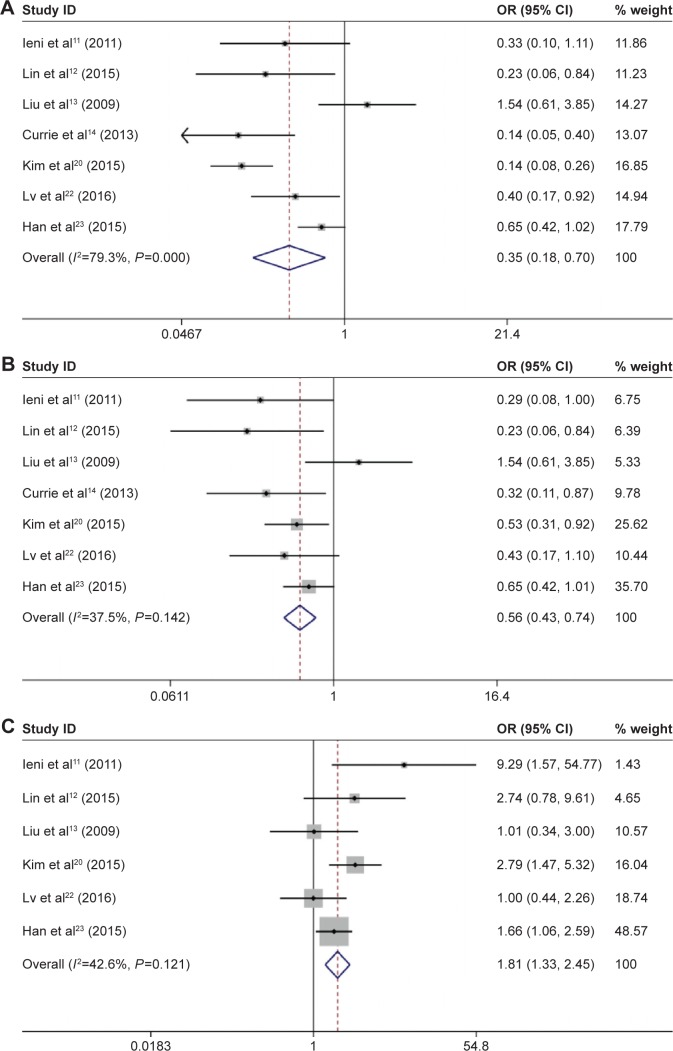
The pooled ORs indicated that overexpression of CD133 was significantly associated with ER status (positive vs negative: OR 0.35, 95% CI 0.18–0.70; Figure 2A), PR status (positive vs negative: OR 0.56, 95% CI 0.43–0.74; Figure 2B), and HER2 status (≥2+ vs 1+, OR 1.81, 95% CI 1.33–2.45; Figure 2C). In the subgroup analysis of ER status, there were no significant heterogeneity in the group of non-Asian (I 2=11.1%, P=0.289) and group of mean age was >50 years (I 2=0.0%, P=0.689). The details are shown in Table 2.
Figure 2.
Forest plots of ORs for the correlation between CD133 overexpression and ER, PR, and HER2.
Notes: (A) OR for the relation between CD133 overexpression and ER; (B) OR for the relation between CD133 overexpression and PR; and (C) OR for the relation between CD133 overexpression and HER2. Weights are from random-effects analysis.
Abbreviations: OR, odds ratio; CI, confidence interval; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor-2.
Table 2.
Stratified analysis of pooled hazard ratios or odds ratios of breast cancer patients with CD133 expression on survival data and clinicopathological features
| Categories | Region
|
Sample sizes
|
Mean age
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Asian
|
non-Asian
|
<150
|
>150
|
<50
|
>50
|
|||||||||||||
| HR/OR | I2H | PH | HR/OR | I2H | PH | HR/OR | I2H | PH | HR/OR | I2H | PH | HR/OR | I2H | PH | HR/OR | I2H | PH | |
| OS | 2.22 (1.38, 3.57) | 61.8% | 0.049 | 0.78 (0.32, 1.93) | – | – | 1.98 (0.84, 4.68) | 71.2% | 0.031 | 1.70(1.05, 2.75) | 43.8% | 0.182 | 1.70 (1.05, 2.75) | 43.8% | 0.182 | 1.68 (0.40, 6.96) | 85.1% | 0.01 |
| DFS | 4.01 (1.83, 8.79) | 72.7% | 0.026 | 0.80 (0.37, 1.73) | – | – | 2.68 (0.68, 10.58) | 90.2% | 0.000 | 2.72 (1.39, 5.33) | – | – | 4.89 (1.54, 15.52) | 82.3% | 0.017 | 0.80 (0.37, 1.73) | – | – |
| ER | 0.42 (0.19, 0.96) | 83.9% | 0.000 | 0.20 (0.08, 0.47) | 11.1% | 0.289 | 0.38 (0.16, 0.88) | 69.2% | 0.011 | 0.31 (0.07, 1.36) | 93.7% | 0.000 | 0.48 (0.19, 1.21) | 87.4% | 0.000 | 0.28 (0.11,0.68) | 0% | 0.689 |
| PR | 0.61 (0.46, 0.82) | 42.3% | 0.139 | 0.30 (0.14, 0.67) | 0% | 0.904 | 0.49 (0.31,0.78) | 54.5% | 0.067 | 0.60 (0.43, 0.85) | 0% | 0.569 | 0.64 (0.48, 0.87) | 35.1% | 0.202 | 0.26 (0.10, 0.64) | 0% | 0.805 |
| HER2 | 1.70(1.25, 2.32) | 25.4% | 0.252 | 9.29 (1.57, 54.77) | – | – | 1.57 (0.92, 2.67) | 53.3% | 0.093 | 1.94(1.34, 2.81) | 41.4% | 0.191 | 1.65 (1.20, 2.27) | 37.5% | 0.187 | 4.28 (1.58, 11.57) | 17.7% | 0.270 |
| Age | 1.01 (0.70, 1.47) | 22.6% | 0.264 | 0.94 (0.32, 2.70) | 80.5% | 0.006 | 0.98 (0.52, 1.82) | 67.3% | 0.009 | 1.00 (0.70, 1.44) | 0% | 0.460 | 1.31 (0.90, 1.91) | 24.6% | 0.250 | 0.77 (0.38, 1.58) | 44.7% | 0.179 |
| T | 1.50 (0.86, 2.61) | 64.1% | 0.010 | 0.83 (0.44, 1.57) | 12.4% | 0.285 | 1.04 (0.48, 2.27) | 66.7% | 0.010 | 1.70(1.04, 2.79) | 52.7% | 0.121 | 1.98 (1.27, 3.07) | 39.9% | 0.155 | 0.68 (0.27, 1.75) | 54.6% | 0.111 |
| N | 2.42(1.67, 3.51) | 49.2% | 0.046 | 1.11 (0.45, 2.75) | 72.1% | 0.028 | 2.14 (1.33, 3.44) | 57.7% | 0.015 | 1.65 (0.73, 3.71) | 83.4% | 0.002 | 2.74 (1.89, 3.97) | 45.4% | 0.089 | 1.23 (0.62, 2.45) | 52.4% | 0.098 |
| G | 1.85 (1.40, 2.46) | 33.1% | 0.175 | 1.50 (0.99, 2.28) | 58.7% | 0.046 | 2.30 (1.66, 3.20) | 40.2% | 0.100 | 1.29 (0.92, 1.81) | 0% | 0.413 | 1.70 (1.29, 2.22) | 41.5% | 0.129 | 1.32 (0.79, 2.21) | 13.6% | 0.328 |
Abbreviations: OS, overall survival; DFS, disease-free survival; HR, hazard ratio; OR, odds ratio; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor-2; T, tumor size; N, lymph node; G, tumor histological grade; H, heterogeneity.
CD133 expression and age, tumor, node, and grade
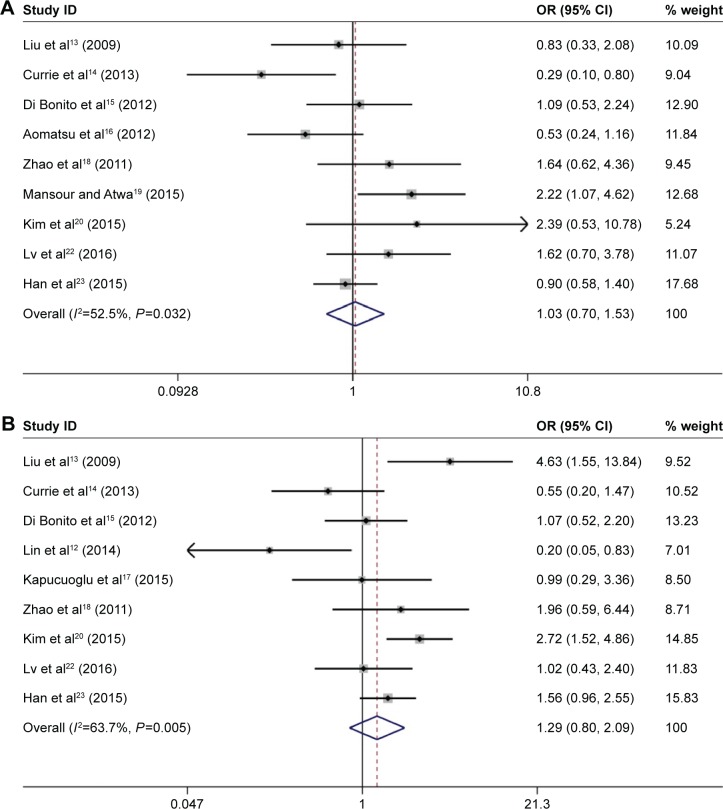
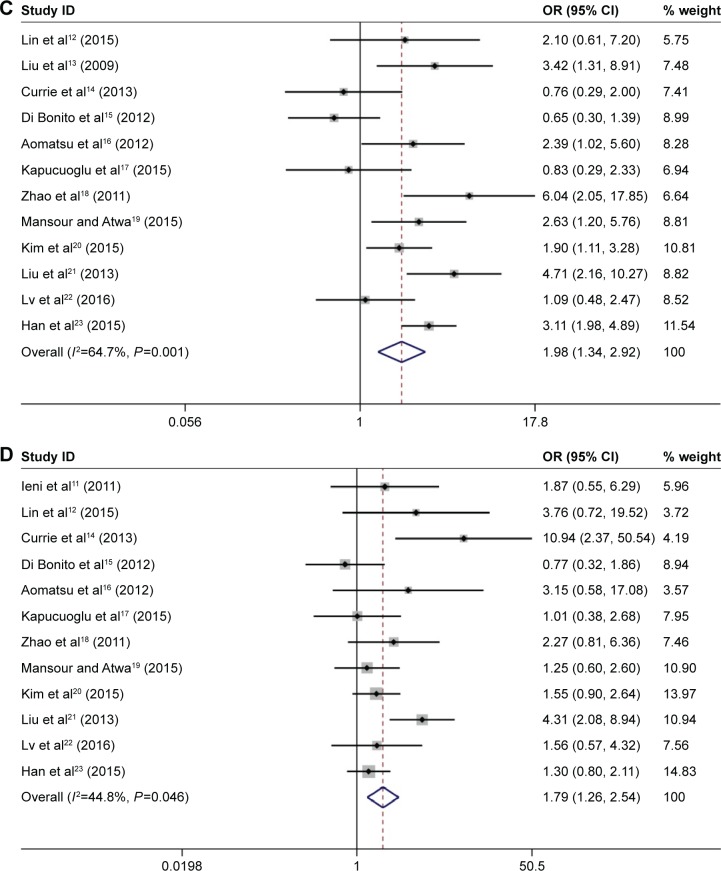
Our results showed that there was no significant association between CD133 high expression and onset age (≥50 vs <50 OR 1.03, 95% CI 0.70–1.53; Figure 3A) and tumor size (≥2 cm vs <2 cm, OR 1.29, 95% CI 0.80–2.09; Figure 3B). However, breast cancer with CD133 expression was associated with lymph node metastasis (positive vs negative: OR 1.98, 95% CI 1.34–2.92; Figure 3C) and tumor histological grade (III vs I–II: OR 1.79, 95% CI 1.26–2.54; Figure 3D). We further performed subgroup analysis from three aspects: region, sample size, and mean age. The results showed that in the group of sample size >150, there was a significant correlation between CD133 expression and tumor size (pooled OR 1.70, 95% CI 1.04–2.79). The details of subgroup analysis are shown in Table 2.
Figure 3.
Forest plots of ORs for the correlation between CD133 overexpression and age, tumor size, lymph node metastasis, and tumor histological grade.
Notes: (A) OR for the relation between CD133 overexpression and age; (B) OR for the relation between CD133 overexpression and tumor size; (C) OR for the relation between CD133 overexpression and lymph node metastasis; and (D) OR for the relation between CD133 overexpression and tumor histological grade. Weights are from random-effects analysis.
Abbreviations: OR, odds ratio; CI, confidence interval.
Meta-analysis of OS and DFS
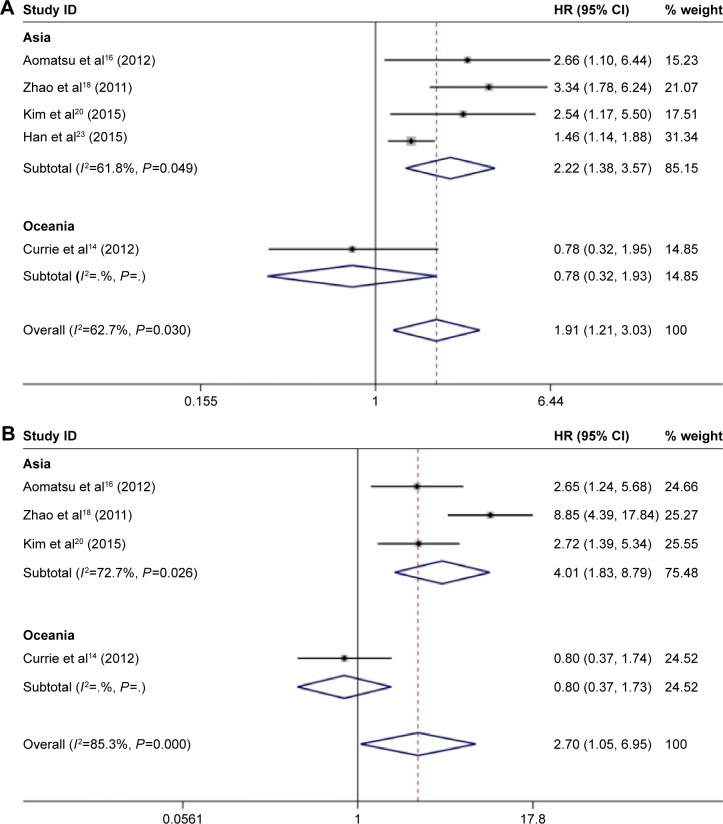
The overall analysis of five studies revealed that CD133-positive breast cancer patients had a higher risk of mortality (pooled HR 1.91, 95% CI 1.21–0.03; Figure 4A) with heterogeneity (I2=62.7%, P=0.03). Meanwhile, the pooled results of four studies showed that increased CD133 expression in breast cancer patients had poorer DFS (pooled HR 2.70, 95% CI 1.05–6.95, Figure 4B) with significant heterogeneity (I2=85.3%, P=0.04). We further performed subgroup analysis according to region and sample size. In subgroup analysis of region, we found that there was a different trend between the Asian and non-Asian groups. Patients in the Asian group with tumors that showed high expression of CD133 tended to have a poorer OS (HR 2.22, 95% CI 1.38–3.57; Figure 4A) and DFS (HR 4.01, 95% CI 1.83–8.79; Figure 4B), while there was no significant association between high-level CD133 expression and OS (HR 0.78, 95% CI 0.32–1.93; Figure 4A) or DFS (HR 0.80, 95% CI 0.37–1.73; Figure 4B) in the non-Asian group. The details of subgroup analysis are shown in Table 2.
Figure 4.
Forest plots of HRs for the association between CD133 overexpression and survival.
Notes: (A) HR for the relation between CD133 overexpression and OS and (B) HR for the relation between CD133 overexpression and DFS. Weights are from random-effects analysis.
Abbreviations: HR, hazard ratio; CI, confidence interval; OS, overall survival; DFS, disease-free survival.
Sensitivity analysis and publication bias
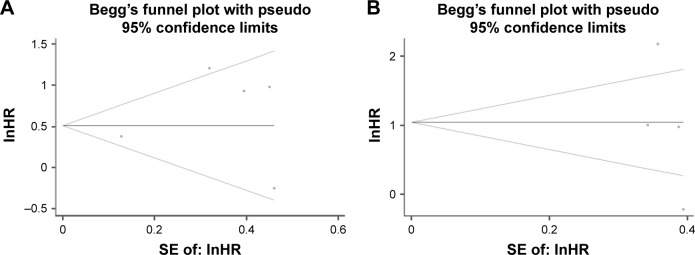
We further performed sensitivity analysis to gauge the stability of our results with respect to clinicopathological characteristics as well as OS and DFS. The plots illustrated the robustness of our results because excluding any single study did not significantly influence pooled ORs or HRs (Figure 5). Egger’s test and Begg’s funnel plots were used to assess publication bias in this meta-analysis. Both the tests indicated that there was no publication bias for pooled ER (PEgger=0.654), PR (PEgger=0.310), HER2 (PEgger=0.560), age (PEgger=0.784), tumor size (PEgger=0.263), lymph node metastasis, (PEgger=0.523), or histological grade (PEgger=0.166) as well as OS (PEgger=0.806) or DFS (PEgger=0.308).
Figure 5.
(A) Begg’s funnel plot for the assessment of publication bias in analysis of OS; (B) Begg’s funnel plot for the assessment of publication bias in analysis of DFS.
Abbreviations: In, napierian logarithm; HR, hazard ratio; SE, standard error; OS, overall survival; DFS, disease-free survival.
Discussion
CSCs have been a hot topic of debate in the field of malignant tumor biology since its fundamental theory was put forward. It has been considered that the tumor is composed of tumor cells and CSCs, which are a rare subpopulation of cells in solid tumors with the capability of self-renewal, differentiation potential, and initiating tumors.4,24 CSCs are at the root of tumor formation that can lead to various degrees of differentiation and are the source that enables the tumor to keep growing and spreading.25 This recognition of the importance of CSCs in tumors has not only led to new directions and perspectives that resulted in a reexamination of the causes of tumor initiation, development, and therapeutic resistance but also provided new ideas for early diagnosis and treatment.
CSCs can be distinguished from tumor cells through identification of specific molecular surface markers such as CD24, CD44, ALDH1, and ESA (epithelial specific antigen). Several meta-analysis studies have been performed to evaluate the association between biomarkers of CSCs and prognosis in various malignancies. A meta-analysis performed by Wang et al26 revealed that CD24 overexpression was significantly correlated with shortened OS in breast cancer patients. Wei et al27 conducted a pooled analysis of ALDH1 expression in lung cancer; the results showed that higher ALDH1 levels were associated with decreased DFS and OS.
CD133, a transmembrane cell surface glycoprotein, was initially found in hematopoietic stem cells and is considered to be a specific molecular biomarker of hematopoietic stem cells.28 In recent years, CD133 as a stem cell marker was demonstrated to be expressed in many types of solid tumors, such as liver, colorectal, and ovarian cancers.29–31 However, the prognostic role of CD133 expression in breast cancer is still controversial. Kim et al20 suggested that CD133 high-expression patients had shorter OS and DFS than CD133 low-expression cases. Conversely, Currie et al14 found no significant difference between CD133 high expression and CD133 low expression in breast cancer patients regarding survival time. In view of the inconsistent conclusions on the impact of CD133 expression in breast cancer patients, it was necessary to conduct a meta-analysis to evaluate the prognostic value of CD133 in breast cancer.
Based on our comprehensive analysis of published studies, we found that overexpression of CD133 was significantly associated with ER-positive status, PR-positive status, HER2-positive status, lymph node metastasis, and high histological grade. However, there was no significant association between CD133 high expression and large tumor size or late onset age. Furthermore, the overall analysis of prognosis revealed that CD133-positive breast cancer patients had a higher risk of mortality and a poorer DFS. In subgroup analysis by region, there was a difference between the Asian and non-Asian groups. In the Asian group, there was a significant association between CD133-positive breast cancer and poorer OS and DFS.
It was gradually discovered that several signaling pathways such as Hedgehog, Wnt, Notch, and NF-κB were involved in the CSC development, progression, differentiation, and metastasis.32 Based on the blockade of these signaling pathways, targeted therapy provides a new method to attack surface molecules of CSCs. Before 2010, gemtuzumab/ozogamicin, an antibody–drug conjugate of a recombinant humanized anti-CD33 monoclonal antibody, had been used in targeted therapy for clinical applications in acute myeloid leukemia patients, but has been pulled out of market due to high toxicity.33,34 Moreover, a recently published report in Nature Communications revealed that self-renewal of CD133 cells by IL6/Notch3 signaling regulates therapeutic resistance in metastatic breast cancer.35 Similarly, the results of our meta-analysis demonstrated that patients with high CD133-expressing tumors tended to have poorer survival. Consequently, targeted drugs that act on CD133 have the potential to be applied in the clinic, and breast cancer patients with high CD133 expression levels may benefit from them.
Previously, there was a similar meta-analysis published in 2010, which evaluated the association of CSCs with clinical outcome.36 They presented statistics on two indicators, CD44+/CD24−/low and ALDH1. However, the key point of our study focused on CD133 expression and its association with clinicopathological features and prognosis. Furthermore, statistical analysis of earlier studies on survival data was calculated using risk ratios (RRs). It is believed that the statistics of HRs take into consideration differences in end events, and also take into account the time to reach the end point and censored data. Consequently, the survival data statistic of HRs is calculated in our meta-analysis.
There were limitations in our meta-analysis. First, eligible studies were incorporated with diverse TNM stage and histological grade that may have potentially influenced the results. Second, although we collected all eligible studies for evaluating the association between CD133 expression and survival data, the sample size was not large enough, which in turn weakened the statistical power of the results. Finally, in this present analysis, the influence of bias could not be completely excluded.
Conclusion
The present results provide some evidence on the clinical outcome and prognostic value of CD133 in breast cancer patients. High CD133 expression predicted a worse OS and DFS. CD133 markers may potentially serve as prognostic markers and novel potential therapeutic targets in breast cancer. Large-scale and standard cohort studies are required for further confirmation.
Supplementary material
Table S1.
Newcastle–Ottawa quality assessment scale (cohort studies)
| Study | Selection
|
Comparability
|
Outcome
|
Score | |||||
|---|---|---|---|---|---|---|---|---|---|
| Representativeness of the exposed cohort | Selection of the nonexposed cohort | Ascertainment of exposure | Demonstration that outcome of interest was not present at start of study | Comparability of cohorts on the basis of the design or analysis | Assessment of outcome | Was follow-up long enough for outcomes to occur? | Adequacy of follow-up of cohorts | ||
|
|
|||||||||
| a. Truly representative of the average (described) in the community ★ b. Somewhat representative of the average in the community ★ c. Selected group of users, for example, nurses, volunteers d. No description of the derivation of the cohort |
a. Community controls ★ b. Drawn from a different source c. No description of the derivation of the nonexposed cohort |
a. Secure record (eg, surgical records) ★ b. Structured interview c. Written self- report d. No description |
a. Yes (end point) ★ b. No |
a. Study controls for the most important factor ★ b. Study controls for any additional factor. (This criteria could be modified to indicate specific control for a second important factor) ★ |
a. Independent blind assessment ★ b. Record linkage ★ c. Self-report d. No description |
a. Yes (select an adequate follow-up period for outcome of interest) ★ b. No |
a. Complete follow-up – all subjects accounted for ★ b. Subjects lost to follow-up unlikely to introduce bias – small number lost >80% (select an adequate %) follow-up, or description provided of those lost ★ c. Follow-up rate <80% (select an adequate %) and no description of those lost d. No statement |
||
|
| |||||||||
| Ieni et al11 | b★ | ★ | ★ | ★ | a★ | a★ | ★ | b★ | 8 |
| Lin et al12 | b★ | ★ | ★ | ★ | a★ | a★ | 6 | ||
| Liu et al13 | b★ | ★ | ★ | ★ | a★ | a★ | 6 | ||
| Currie et al14 | b★ | ★ | ★ | ★ | a★ | a★ | ★ | a★ | 8 |
| Di Bonito et al15 | b★ | ★ | ★ | ★ | a★ | ★ | 6 | ||
| Aomatsu et al16 | b★ | ★ | ★ | ★ | a★ | a★ | ★ | a★ | 8 |
| Kapucuoglu et al17 | b★ | ★ | ★ | ★ | a★ | a★ | 6 | ||
| Zhao et al18 | b★ | ★ | ★ | ★ | a,b★★ | a★ | ★ | 8 | |
| Mansour and Atwa19 | b★ | ★ | ★ | ★ | a★ | a★ | 6 | ||
| Kim et al20 | b★ | ★ | ★ | ★ | a★ | a★ | ★ | 7 | |
| Liu et al21 | b★ | ★ | ★ | ★ | a★ | a★ | 6 | ||
| Lv et al22 | b★ | ★ | ★ | ★ | a,b★★ | a★ | 7 | ||
| Han et al23 | b★ | ★ | ★ | ★ | a,b★★ | a★ | ★ | 8 | |
Notes: A study can be awarded a maximum of one star for each numbered item within the “selection” and “outcome” categories. A maximum of two stars can be given for “comparability”.
Acknowledgments
This study was supported by the grants from the National Natural Science Foundation of China (no 81372811) and Science and Technology Agency of Liaoning Province (no 2013225049).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Alison MR, Islam S, Wright NA. Stem cells in cancer: instigators and propagators? J Cell Sci. 2010;123(pt 14):2357–2368. doi: 10.1242/jcs.054296. [DOI] [PubMed] [Google Scholar]
- 4.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 5.Miraglia S, Godfrey W, Yin AH, et al. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood. 1997;90(12):5013–5021. [PubMed] [Google Scholar]
- 6.Nadal R, Ortega FG, Salido M, et al. CD133 expression in circulating tumor cells from breast cancer patients: potential role in resistance to chemotherapy. Int J Cancer. 2013;133(10):2398–2407. doi: 10.1002/ijc.28263. [DOI] [PubMed] [Google Scholar]
- 7.Wang W, Chen Y, Deng J, et al. The prognostic value of CD133 expression in non-small cell lung cancer: a meta-analysis. Tumour Biol. 2014;35(10):9769–9775. doi: 10.1007/s13277-014-2270-9. [DOI] [PubMed] [Google Scholar]
- 8.Cheng B, Yang G, Jiang R, et al. Cancer stem cell markers predict a poor prognosis in renal cell carcinoma: a meta-analysis. Oncotarget. 2016;7(40):65862–65875. doi: 10.18632/oncotarget.11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sui YP, Jian XP, Ma LI, et al. Prognostic value of cancer stem cell marker CD133 expression in esophageal carcinoma: a meta-analysis. Mol Clin Oncol. 2016;4(1):77–82. doi: 10.3892/mco.2015.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wen L, Chen XZ, Yang K, et al. Prognostic value of cancer stem cell marker CD133 expression in gastric cancer: a systematic review. PLoS One. 2013;8(3):e59154. doi: 10.1371/journal.pone.0059154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ieni A, Giuffre G, Adamo V, Tuccari G. Prognostic impact of CD133 immunoexpression in node-negative invasive breast carcinomas. Anticancer Res. 2011;31(4):1315–1320. [PubMed] [Google Scholar]
- 12.Lin CH, Liu CH, Wen CH, Ko PL, Chai CY. Differential CD133 expression distinguishes malignant from benign papillary lesions of the breast. Virchows Arch. 2015;466(2):177–184. doi: 10.1007/s00428-014-1695-2. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q, Li JG, Zheng XY, Jin F, Dong HT. Expression of CD133, PAX2, ESA, and GPR30 in invasive ductal breast carcinomas. Chin Med J (Engl) 2009;122(22):2763–2769. [PubMed] [Google Scholar]
- 14.Currie MJ, Beardsley BE, Harris GC, et al. Immunohistochemical analysis of cancer stem cell markers in invasive breast carcinoma and associated ductal carcinoma in situ: relationships with markers of tumor hypoxia and microvascularity. Hum Pathol. 2013;44(3):402–411. doi: 10.1016/j.humpath.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Di Bonito M, Cantile M, Collina F, et al. Overexpression of cell cycle progression inhibitor Geminin is associated with tumor stem-like phenotype of triple-negative breast cancer. J Breast Cancer. 2012;15(2):162–171. doi: 10.4048/jbc.2012.15.2.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aomatsu N, Yashiro M, Kashiwagi S, et al. CD133 is a useful surrogate marker for predicting chemosensitivity to neoadjuvant chemotherapy in breast cancer. PLoS One. 2012;7(9):e45865. doi: 10.1371/journal.pone.0045865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapucuoglu N, Bozkurt KK, Baspinar S, et al. The clinicopathological and prognostic significance of CD24, CD44, CD133, ALDH1 expressions in invasive ductal carcinoma of the breast: CD44/CD24 expression in breast cancer. Pathol Res Pract. 2015;211(10):740–747. doi: 10.1016/j.prp.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Zhao P, Lu Y, Jiang X, Li X. Clinicopathological significance and prognostic value of CD133 expression in triple-negative breast carcinoma. Cancer Sci. 2011;102(5):1107–1111. doi: 10.1111/j.1349-7006.2011.01894.x. [DOI] [PubMed] [Google Scholar]
- 19.Mansour SF, Atwa MM. Clinicopathological significance of CD133 and ALDH1 cancer stem cell marker expression in invasive ductal breast carcinoma. Asian Pac J Cancer Prev. 2015;16(17):7491–7496. doi: 10.7314/apjcp.2015.16.17.7491. [DOI] [PubMed] [Google Scholar]
- 20.Kim SJ, Kim YS, Jang ED, Seo KJ, Kim JS. Prognostic impact and clinicopathological correlation of CD133 and ALDH1 expression in invasive breast cancer. J Breast Cancer. 2015;18(4):347–355. doi: 10.4048/jbc.2015.18.4.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu TJ, Sun BC, Zhao XL, et al. CD133+ cells with cancer stem cell characteristics associates with vasculogenic mimicry in triple-negative breast cancer. Oncogene. 2013;32(5):544–553. doi: 10.1038/onc.2012.85. [DOI] [PubMed] [Google Scholar]
- 22.Lv X, Wang Y, Song Y, Pang X, Li H. Association between ALDH1+/CD133+ stem-like cells and tumor angiogenesis in invasive ductal breast carcinoma. Oncol Lett. 2016;11(3):1750–1756. doi: 10.3892/ol.2016.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han Z, Chen Z, Zheng R, Cheng Z, Gong X, Wang D. Clinicopathological significance of CD133 and CD44 expression in infiltrating ductal carcinoma and their relationship to angiogenesis. World J Surg Oncol. 2015;13:56. doi: 10.1186/s12957-015-0486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison SJ, Wandycz AM, Hemmati HD, Wright DE, Weissman IL. Identification of a lineage of multipotent hematopoietic progenitors. Development. 1997;124(10):1929–1939. doi: 10.1242/dev.124.10.1929. [DOI] [PubMed] [Google Scholar]
- 25.Kummermehr JC. Tumour stem cells – the evidence and the ambiguity. Acta Oncol. 2001;40(8):981–988. doi: 10.1080/02841860152708279. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Wang Q, Wang Q, Wang Y, Chen J. Prognostic significance of CD24 and CD44 in breast cancer: a meta-analysis. Int J Biol Markers. 2016 Jul 27; doi: 10.5301/jbm.5000224. Epub. [DOI] [PubMed] [Google Scholar]
- 27.Wei D, Peng JJ, Gao H, Zhang T, Tan Y, Hu YH. ALDH1 expression and the prognosis of lung cancer: a systematic review and meta-analysis. Heart Lung Circ. 2015;24(8):780–788. doi: 10.1016/j.hlc.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 28.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241(4861):58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, Ge C, Zhao F, et al. NRBP2 overexpression increases the chemosensitivity of hepatocellular carcinoma cells via Akt signaling. Cancer Res. 2016;76(23):7059–7071. doi: 10.1158/0008-5472.CAN-16-0937. [DOI] [PubMed] [Google Scholar]
- 30.Stanisavljevic L, Myklebust MP, Leh S, Dahl O. LGR5 and CD133 as prognostic and predictive markers for fluoropyrimidine-based adjuvant chemotherapy in colorectal cancer. Acta Oncol. 2016;55(12):1425–1433. doi: 10.1080/0284186X.2016.1201215. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Q, Chen A, Song H, Tao J, Yang H, Zuo M. Prognostic value of cancer stem cell marker CD133 in ovarian cancer: a meta-analysis. Int J Clin Exp Med. 2015;8(3):3080–3088. [PMC free article] [PubMed] [Google Scholar]
- 32.Matsui WH. Cancer stem cell signaling pathways. Medicine (Baltimore) 2016;95(1 suppl 1):S8–S19. doi: 10.1097/MD.0000000000004765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterlin P, Guillaume T, Delaunay J, et al. Similarity of fractionated versus single dose(s) of gemtuzumab ozogamicin as part of the MIDAM salvage regimen in relapsed/refractory acute myeloid leukemia patients. Semin Hematol. 2016;53(3):216–217. doi: 10.1053/j.seminhematol.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Kell J. The addition of gemtuzumab ozogamicin to chemotherapy in adult patients with acute myeloid leukemia. Expert Rev Anticancer Ther. 2016;16(4):377–382. doi: 10.1586/14737140.2016.1162099. [DOI] [PubMed] [Google Scholar]
- 35.Sansone P, Ceccarelli C, Berishaj M, et al. Self-renewal of CD133(hi) cells by IL6/Notch3 signalling regulates endocrine resistance in metastatic breast cancer. Nat Commun. 2016;7:10442. doi: 10.1038/ncomms10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou L, Jiang Y, Yan T, et al. The prognostic role of cancer stem cells in breast cancer: a meta-analysis of published literatures. Breast Cancer Res Treat. 2010;122(3):795–801. doi: 10.1007/s10549-010-0999-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Newcastle–Ottawa quality assessment scale (cohort studies)
| Study | Selection
|
Comparability
|
Outcome
|
Score | |||||
|---|---|---|---|---|---|---|---|---|---|
| Representativeness of the exposed cohort | Selection of the nonexposed cohort | Ascertainment of exposure | Demonstration that outcome of interest was not present at start of study | Comparability of cohorts on the basis of the design or analysis | Assessment of outcome | Was follow-up long enough for outcomes to occur? | Adequacy of follow-up of cohorts | ||
|
|
|||||||||
| a. Truly representative of the average (described) in the community ★ b. Somewhat representative of the average in the community ★ c. Selected group of users, for example, nurses, volunteers d. No description of the derivation of the cohort |
a. Community controls ★ b. Drawn from a different source c. No description of the derivation of the nonexposed cohort |
a. Secure record (eg, surgical records) ★ b. Structured interview c. Written self- report d. No description |
a. Yes (end point) ★ b. No |
a. Study controls for the most important factor ★ b. Study controls for any additional factor. (This criteria could be modified to indicate specific control for a second important factor) ★ |
a. Independent blind assessment ★ b. Record linkage ★ c. Self-report d. No description |
a. Yes (select an adequate follow-up period for outcome of interest) ★ b. No |
a. Complete follow-up – all subjects accounted for ★ b. Subjects lost to follow-up unlikely to introduce bias – small number lost >80% (select an adequate %) follow-up, or description provided of those lost ★ c. Follow-up rate <80% (select an adequate %) and no description of those lost d. No statement |
||
|
| |||||||||
| Ieni et al11 | b★ | ★ | ★ | ★ | a★ | a★ | ★ | b★ | 8 |
| Lin et al12 | b★ | ★ | ★ | ★ | a★ | a★ | 6 | ||
| Liu et al13 | b★ | ★ | ★ | ★ | a★ | a★ | 6 | ||
| Currie et al14 | b★ | ★ | ★ | ★ | a★ | a★ | ★ | a★ | 8 |
| Di Bonito et al15 | b★ | ★ | ★ | ★ | a★ | ★ | 6 | ||
| Aomatsu et al16 | b★ | ★ | ★ | ★ | a★ | a★ | ★ | a★ | 8 |
| Kapucuoglu et al17 | b★ | ★ | ★ | ★ | a★ | a★ | 6 | ||
| Zhao et al18 | b★ | ★ | ★ | ★ | a,b★★ | a★ | ★ | 8 | |
| Mansour and Atwa19 | b★ | ★ | ★ | ★ | a★ | a★ | 6 | ||
| Kim et al20 | b★ | ★ | ★ | ★ | a★ | a★ | ★ | 7 | |
| Liu et al21 | b★ | ★ | ★ | ★ | a★ | a★ | 6 | ||
| Lv et al22 | b★ | ★ | ★ | ★ | a,b★★ | a★ | 7 | ||
| Han et al23 | b★ | ★ | ★ | ★ | a,b★★ | a★ | ★ | 8 | |
Notes: A study can be awarded a maximum of one star for each numbered item within the “selection” and “outcome” categories. A maximum of two stars can be given for “comparability”.