Abstract
Circulating tumor DNA (ctDNA) can be identified in the peripheral blood of patients and harbors the genomic alterations found in tumor tissues, which provides a noninvasive approach for detection of gene mutations. We conducted this meta-analysis to investigate whether ctDNA can be used for monitoring KRAS gene mutations in colorectal cancer (CRC) patients. Medline, Embase, Cochrane Library and Web of Science were searched for the included eligible studies in English, and data were extracted for statistical analysis according to the numbers of true-positive (TP), true-negative (TN), false-positive (FP) and false-negative (FN) cases. Sensitivity, specificity and diagnostic odds ratio (DOR) were calculated, and the area under the receiver operating characteristic curve (AUROC) was used to evaluate the diagnostic performance. After independent searching and reviewing, 21 studies involving 1,812 cancer patients were analyzed. The overall sensitivity, specificity and DOR were 0.67 (95% confidence interval [CI] =0.55–0.78), 0.96 (95% CI =0.93–0.98) and 53.95 (95% CI =26.24–110.92), respectively. The AUROC was 0.95 (95% CI =0.92–0.96), which indicated the high diagnostic accuracy of ctDNA. After stratified analysis, we found the higher diagnostic accuracy in subgroup of patients detected in blood sample of plasma. The ctDNA may be an ideal source for detection of KRAS gene mutations in CRC patients with high specificity and diagnostic value.
Keywords: cancer, KRAS, mutation, circulating tumor DNA
Introduction
Cancer is becoming the second most common cause for death in recent years.1 Surgery is the main treatment for early cancer patients; adjuvant and palliative treatments are mainly used in patients with advanced cancer, involving traditional chemotherapy and targeted therapy. Pathological test of tumor tissues is the gold standard method for histological diagnosing, and detection of gene mutations in tumor tissues is a directive factor for selection of targeted drugs.2 For non-small cell lung cancer (NSCLC), the status of EGFR gene mutations is a sensitive and reliable biomarker for EGFR-TKIs therapy (eg, gefitinib and erlotinib).3,4 For colorectal cancer (CRC), the anti-EGFR MoAb (eg, cetuximab) is now restricted to patients with wild-type KRAS gene, and the mutation status of BRAF gene is predictive of the drug resistance of anti-EGFR MoAb in wild-type KRAS patients.5,6 At the same time, detection of other gene mutations has also been used in patients with different cancers. For diagnosing genomic mutation, biopsy and surgery of primary and metastatic tumors are the main resources of tumor tissues, but they are invasive, uncomfortable and of high cost.7,8 Thus, new non-invasive methods for gene detection are increasingly focused on.
Circulating tumor DNA (ctDNA) is tumor DNA of cell-free DNA (cfDNA) circulating in plasma or serum of cancer patients,9 which is ~170 bp. ctDNA originates from tumor cells, which can be identified in blood samples, and rapidly disappears after surgery or chemotherapy. The level of ctDNA is significantly higher in cancer patients than in healthy controls, which can harbor the somatic genomic alterations found in tumor tissues.10,11 Unlike tissue biopsy, obtaining a sample of ctDNA is a noninvasive approach, with less risk and lower cost. Thus, detection of ctDNA is actively being explored and provides opportunities for minimally invasive cancer diagnosis, prognosis and tumor monitoring in cancer patients.12–15
Many studies have been carried out to validate the possibility of ctDNA as a new source for detection of gene mutations. On the basis of this background, we decided to conduct this meta-analysis to investigate the diagnostic value of ctDNA for detection of KRAS gene mutations in CRC patients. The results will help establishing whether ctDNA can be used for detection of gene mutations in cancer patients.
Materials and methods
Literature search strategy
Two investigators independently searched Medline, Embase, Cochrane library and Web of Science databases for potentially relevant articles between inception and January 31, 2016. The search heading terms and keywords were “carcinoma” or “cancer” or “neoplasm”, “serum” or “plasma” or “circulating”, “KRAS” and “mutation”, which were limited to English publications in human beings. Then, the results were manually searched for included studies and double-checked by a second investigator.
Selection criteria
We searched the full-text articles that investigated the comparison of detection of KRAS gene mutations between tumor tissue and the matched plasma or serum in CRC patients. The following publications were excluded: abstracts, news, letters, case reports and reviews. Publications retrieved from databases were first screened by titles and abstracts; full-text articles were reviewed and selected according to the inclusion criteria. The inclusion criteria were as follows: 1) CRC patients should be diagnosed by histopathological or cytological examination; 2) KRAS mutations should be detected in tumor tissue and the paired ctDNA; 3) sufficient information should be provided to conduct the statistical analysis; and 4) studies should be limited to human trials and published in English language. The study with most patients was included from those reported using the same technique by the same center.
Data extraction and quality assessment
Two investigators independently abstracted following information from each trial: name of the first author, year of publication, location where the study was performed, number of patients enrolled, stage of cancer (according to tumor–node–metastasis [TNM] criteria), storage method of tumor tissues, detection method of ctDNA, collection time of blood samples (before or after chemotherapy), format of blood samples (serum or plasma), true positive (TP), false negative (FN), false positive (FP) and true negative (TN). When the study included ≥2 methods of mutation detection, we used the data with the best sensitivity or specificity. The discrepancy between the 2 investigators was resolved by discussion among all the authors. Quality assessment of eligible studies was evaluated using QUADAS-2 (quality assessment of studies of diagnostic accuracy included in systematic reviews-2).
Statistical analysis
The publication bias was detected using Deeks funnel plot, and P<0.05 suggested the significant publication bias. The detection of KRAS gene mutation in tumor tissues acted as the gold standard for diagnosing gene mutation in this analysis. We calculated the sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), corresponding 95% confidence intervals (95% CIs) and area under the ROC curve (AUROC), according to the numbers of TP, TN, FP and FN. Subgroup analyses were also conducted according to different clinical data. Statistical heterogeneity was evaluated by Q test and inconsistency index (I2), with significant heterogeneity set at P≤0.05 and I2>50%. All statistical analyses were performed using STATA software (version 12.1, StataCorp LP) with the Midas module.
Results
Eligible studies
After independent searching, 6,033 records were retrieved from these databases. After reviewing the titles and abstracts, 5,977 records were excluded. After full-text articles were reviewed, 20 publications were considered to be eligible for inclusion in our analysis (Figure 1).15–34 In the article reported by Morgan et al,25 the blood samples including serum and plasma were analyzed. Subsequently, 21 eligible studies were included for analyzing the effectiveness of ctDNA for detection of KRAS gene mutations in CRC patients.
Figure 1.
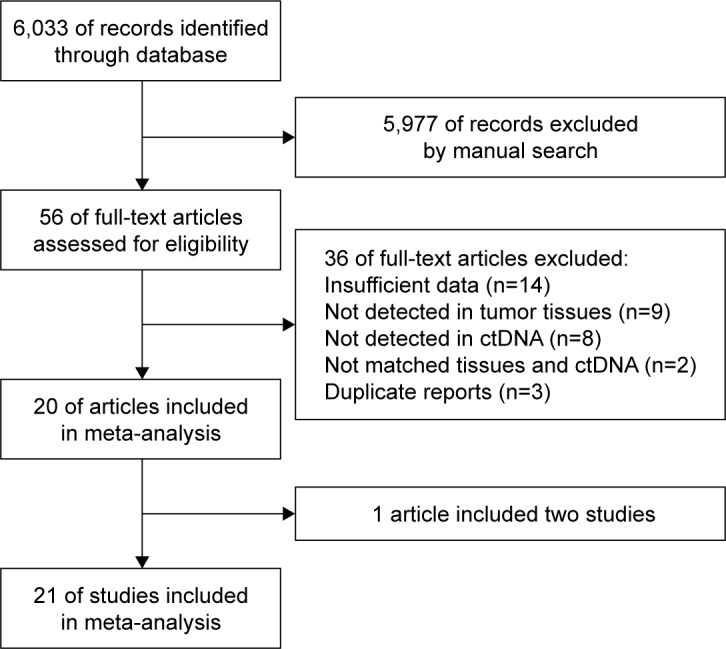
A flow chart showing the studies included in this meta-analysis.
Study characteristics
Our analysis included 21 eligible studies, which are shown in Table 1. In total, 5 studies were from China, 4 from France, 3 from the USA, 2 from Italy, 2 from Japan, 1 from Switzerland, 1 from multi-country, 1 from Denmark, 1 from Korea and 1 from the Netherlands. A total of 1,812 CRC patients were included. In all, 18 studies detected both codon 12 and codon 13 of KRAS gene. A total of 3 studies detected only codon 12 of KRAS gene. In all, 14 studies reported the exact collection times of both tumor tissue samples and blood samples, which were collected before chemotherapy. A total of 16 circulating blood samples were plasma and 5 samples were serum. In all, 12 formalin-fixed and paraffin-embedded (FFPE) tumor tissue samples were used for detection of KRAS gene mutations; 4 tumor tissues were frozen before detection. Various detection methods were used, in which the allele refractory mutation system – quantitative polymerase chain reaction (ARMS-qPCR) was the most frequently reported.
Table 1.
Characteristics of eligible studies
| First author | Year | Country | Patients | TNM | Tissue | Blood | Collection | Method | Codon | Sensitivity | Specificity | TP | FN | FP | TN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bettegowda et al16 | 2014 | Multi | 206 | Advanced | NA | Plasma | NA | dPCR | 12, 13 | 0.872 | 0.992 | 68 | 10 | 1 | 127 |
| Danese et al17 | 2015 | Italy | 85 | I–IV | Frozen | Plasma | Before | ARMS-qPCR | 12, 13 | 0.815 | 0.931 | 22 | 5 | 4 | 54 |
| Kim et al18 | 2015 | Korea | 65 | Advanced | FFPE | Serum | NA | RFLP-PCR | 12, 13 | 0.581 | 0.765 | 18 | 13 | 8 | 26 |
| Kopreski et al19 | 2000 | USA | 135 | NA | FFPE | Plasma | Before | RFLP-PCR | 12 | 0.829 | 0.930 | 29 | 6 | 7 | 93 |
| Kuo et al20 | 2014 | China | 52 | I–IV | NA | Plasma | NA | PNA-PCR | 12, 13 | 1.000 | 0.703 | 15 | 0 | 11 | 26 |
| Lefebure et al21 | 2010 | France | 24 | IV | NA | Serum | NA | Real-time PCR | 12, 13 | 0.500 | 1.000 | 7 | 7 | 0 | 10 |
| Lin et al22 | 2014 | China | 133 | I–IV | Frozen | Plasma | Before | MassARRAY | 12, 13 | 0.456 | 1.000 | 41 | 49 | 0 | 43 |
| Liu et al23 | 2012 | China | 62 | NA | Frozen | Plasma | Before | Nested cold-PCR | 12 | 0.750 | 0.920 | 9 | 3 | 4 | 46 |
| Miyano et al24 | 2012 | Japan | 42 | I–IV | FFPE | Plasma | Before | PNA-PCR | 12, 13 | 0.615 | 0.931 | 8 | 5 | 2 | 27 |
| Morgan et al25 | 2012 | USA | 71 | IV | FFPE | Plasma | Before | ARMS-qPCR | 12, 13 | 0.313 | 0.949 | 10 | 22 | 2 | 37 |
| Morgan et al25 | 2012 | USA | 71 | IV | FFPE | Serum | Before | ARMS-qPCR | 12, 13 | 0.281 | 1.000 | 9 | 23 | 0 | 39 |
| Mulcahy et al26 | 2000 | Switzerland | 14 | NA | NA | Plasma | Before | MASA-PCR | 12 | 0.857 | 1.000 | 6 | 1 | 0 | 7 |
| Perrone et al27 | 2014 | Italy | 12 | NA | FFPE | Plasma | Before | ME-PCR | 12, 13 | 0.000 | 1.000 | 0 | 5 | 0 | 7 |
| Pu et al28 | 2013 | China | 115 | I–IV | FFPE | Serum | Before | Nested PCR | 12, 13 | 0.243 | 0.949 | 9 | 28 | 4 | 74 |
| Ryan et al29 | 2003 | the Netherlands | 78 | I–IV | FFPE | Serum | Before | ME-PCR | 12, 13 | 0.756 | 0.973 | 31 | 10 | 1 | 36 |
| Sakai et al30 | 2015 | Japan | 15 | NA | FFPE | Plasma | NA | NGS | 12, 13 | 0.714 | 1.000 | 5 | 2 | 0 | 8 |
| Sefrioui et al31 | 2015 | France | 34 | IV | FFPE | Plasma | Before | dPCR | 12, 13 | 0.688 | 1.000 | 11 | 5 | 0 | 18 |
| Spindler et al32 | 2015 | Denmark | 211 | IV | FFPE | Plasma | Before | ARMS-qPCR | 12, 13 | 0.800 | 0.958 | 112 | 28 | 3 | 68 |
| Taly et al33 | 2013 | France | 50 | IV | Frozen | Plasma | NA | qPCR | 12, 13 | 0.789 | 0.935 | 15 | 4 | 2 | 29 |
| Thierry et al15 | 2014 | France | 95 | IV | NA | Plasma | NA | qPCR | 12, 13 | 0.923 | 0.982 | 36 | 3 | 1 | 55 |
| Xu et al34 | 2014 | China | 242 | IV | FFPE | Plasma | Before | PNA-PCR | 12, 13 | 0.547 | 0.904 | 64 | 53 | 12 | 113 |
Abbreviations: TNM, tumor–node–metastasis; TP, true positive; FN, false negative; FP, false positive; TN, true negative; NA, not applicable; dPCR, digital polymerase chain reaction; ARMS-qPCR, allele refractory mutation system – quantitative polymerase chain reaction; FFPE, formalin-fixed and paraffin-embedded; RFLP-PCR, restriction fragment length polymorphism – polymerase chain reaction; PNA-PCR, peptide nucleic acid-mediated polymerase chain reaction; PCR, polymerase chain reaction; MASA-PCR, mutant specific alleles – polymerase chain reaction; ME-PCR, mutant-enriched polymerase chain reaction, NGS, next-generation sequencing; qPCR, quantitative polymerase chain reaction.
Study quality
We assessed the quality of eligible studies by QUADAS-2 and found that the quality of all studies was good (Table 2). The Deeks regression test was performed, and no significant publication bias was detected (P=0.060; Figure 2).
Table 2.
QUADAS-2 score of eligible studies
| First author | Risk of bias
|
Application concerns
|
|||||
|---|---|---|---|---|---|---|---|
| Patient selection | Index test | Reference standard | Flow and timing | Patient selection | Index test | Reference standard | |
| Bettegowda et al16 | L | U | U | L | L | L | L |
| Danese et al17 | L | U | U | L | L | L | L |
| Kim et al18 | L | U | U | L | L | L | L |
| Kopreski et al19 | L | U | U | L | L | L | L |
| Kuo et al20 | L | U | U | L | L | L | L |
| Lefebure et al21 | L | U | U | L | L | L | L |
| Lin et al22 | L | L | L | L | L | L | L |
| Liu et al23 | L | U | U | L | L | L | L |
| Miyano et al24 | L | U | U | L | L | L | L |
| Morgan et al25 | L | L | L | L | L | L | L |
| Morgan et al25 | L | L | L | L | L | L | L |
| Mulcahy et al26 | L | U | U | L | L | L | L |
| Perrone et al27 | L | U | U | L | L | L | L |
| Pu et al28 | L | U | U | L | L | L | L |
| Ryan et al29 | L | U | U | L | L | L | L |
| Sakai et al30 | L | U | U | L | L | L | L |
| Sefrioui et al31 | L | H | L | L | L | L | L |
| Spindler et al32 | L | L | L | L | L | L | L |
| Taly et al33 | L | U | U | L | L | L | L |
| Thierry et al15 | L | U | U | L | L | L | L |
| Xu et al34 | L | L | L | L | L | L | L |
Abbreviations: L, low; U, unclear; H, high; QUADAS-2, quality assessment of studies of diagnostic accuracy included in systematic reviews-2.
Figure 2.
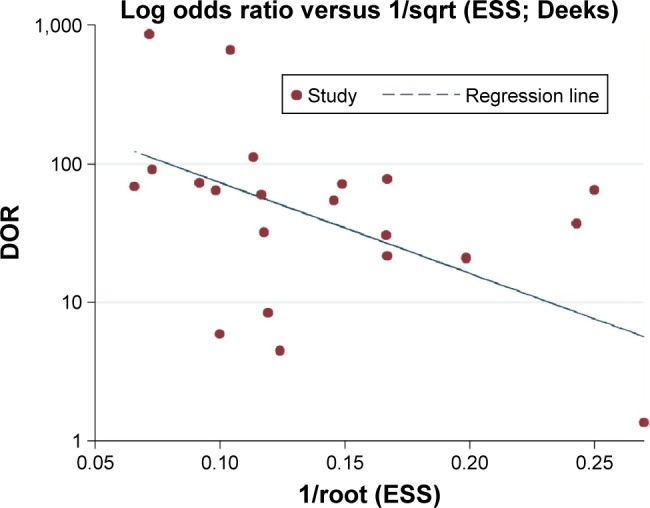
The Deeks regression line showing the publication bias of studies.
Abbreviations: ESS, effective sample size; DOR, diagnostic odds ratio.
Meta-analysis of specificity and sensitivity in overall population
In our analysis, the overall pooled specificity was 0.96 (95% CI =0.93–0.98), the pooled sensitivity was 0.67 (95% CI =0.55–0.78), the DOR was 53.95 (95% CI =26.24–110.92) and the AUROC was 0.95 (95% CI =0.92–0.96) (Figure 3 and Table 3). Specificity can quantify the ability of avoiding FPs, so the high specificity indicated high accuracy of ctDNA for detection of KRAS gene mutations. Sensitivity refers to the ability of correctly detecting patients who do have the condition. Considering the trade-off between sensitivity and specificity, AUROC is used to measure the diagnostic performance of ctDNA. When AUROC is >0.9, the diagnostic accuracy is high enough for a detection method to act as an effective maker. The PLR was 18.33 (>10), suggesting that it could be used for confirming KRAS gene mutations. The NLR was 0.34 (>0.1), showing that a negative result was 34% likely to be an FN (Figure 4 and Table 3). The between-study heterogeneity was evaluated by chi-square test (Q =53.58, P=0.000<0.05) and I-square test (I2=96.27, 95% CI =93.48–99.06), which suggested the significantly high heterogeneity. From the likelihood ratio (LR) scattergram, we found that ctDNA was located in the right upper quadrant, indicating ctDNA could be used for confirming KRAS gene mutations (Figure 5).
Figure 3.
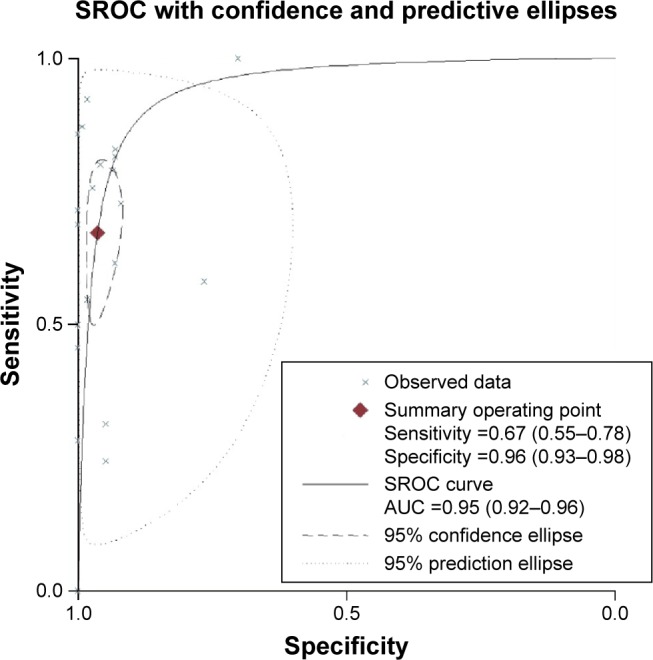
The SROC curve of ctDNA for detection of KRAS gene mutations.
Abbreviations: SROC, summary receiver operating characteristic; AUC, area under the curve.
Table 3.
Meta-analysis of subgroup
| Subgroup analyses | n | Sensitivity | Specificity | AUROC | DOR | PLR | NLR |
|---|---|---|---|---|---|---|---|
| All | 21 | 0.67 (0.55–0.78) | 0.96 (0.93–0.98) | 0.95 (0.92–0.96) | 53.95 (26.24–110.92) | 18.33 (10.14–33.16) | 0.34 (0.24–0.49) |
| TNM stage | |||||||
| Advanced | 10 | 0.66 (0.50–0.79) | 0.97 (0.93–0.99) | 0.95 (0.93–0.97) | 71.52 (21.18–241.57) | 24.88 (9.36–66.17) | 0.35 (0.22–0.54) |
| I–IV | 6 | 0.73 (0.40–0.92) | 0.93 (0.86–0.97) | 0.94 (0.92–0.96) | 37.80 (14.18–100.79) | 11.02 (6.18–19.66) | 0.29 (0.11–0.77) |
| Storage of tissue | |||||||
| FFPE | 12 | 0.55 (0.40–0.69) | 0.96 (0.92–0.98) | 0.93 (0.90–0.95) | 29.19 (12.47–68.33) | 13.63 (6.88–27.01) | 0.47 (0.34–0.65) |
| Frozen | 4 | 0.70 (0.51–0.84) | 0.95 (0.88–0.98) | 0.94 (0.91–0.96) | 48.17 (18.44–125.87) | 15.07 (6.29–36.09) | 0.31 (0.18–0.54) |
| Format of blood | |||||||
| Plasma | 16 | 0.74 (0.60–0.84) | 0.97 (0.93–0.98) | 0.96 (0.94–0.97) | 77.45 (39.03–153.72) | 20.93 (11.58–37.82) | 0.27 (0.17–0.43) |
| Serum | 5 | 0.47 (0.29–0.66) | 0.96 (0.84–0.99) | 0.83 (0.79–0.86) | 22.16 (4.30–114.30) | 12.25 (2.69–55.84) | 0.55 (0.39–0.79) |
| Detection method | |||||||
| ARMS-qPCR | 4 | 0.58 (0.30–0.81) | 0.96 (0.91–0.98) | 0.96 (0.94–0.97) | 34.90 (11.17–109.07) | 15.38 (6.96–34.01) | 0.44 (0.23–0.85) |
| Detection site | |||||||
| Codon 12 | 21 | 0.67 (0.55–0.78) | 0.96 (0.93–0.98) | 0.95 (0.92–0.96) | 53.95 (26.24–110.92) | 18.33 (10.14–33.16) | 0.34 (0.24–0.49) |
| Codon 12, 13 | 18 | 0.65 (0.50–0.77) | 0.97 (0.93–0.99) | 0.95 (0.92–0.96) | 55.56 (23.97–128.81) | 20.26 (9.97–41.18) | 0.37 (0.25–0.54) |
Note: Data presented as n (95% confidence interval).
Abbreviations: AUROC, area under the ROC curve; DOR, diagnostic odds ratio; PLR, positive likelihood ratio; NLR, negative likelihood ratio; TNM, tumor–node–metastasis; FFPE, formalin-fixed and paraffin-embedded; ARMS-qPCR, allele refractory mutation system – quantitative polymerase chain reaction.
Figure 4.
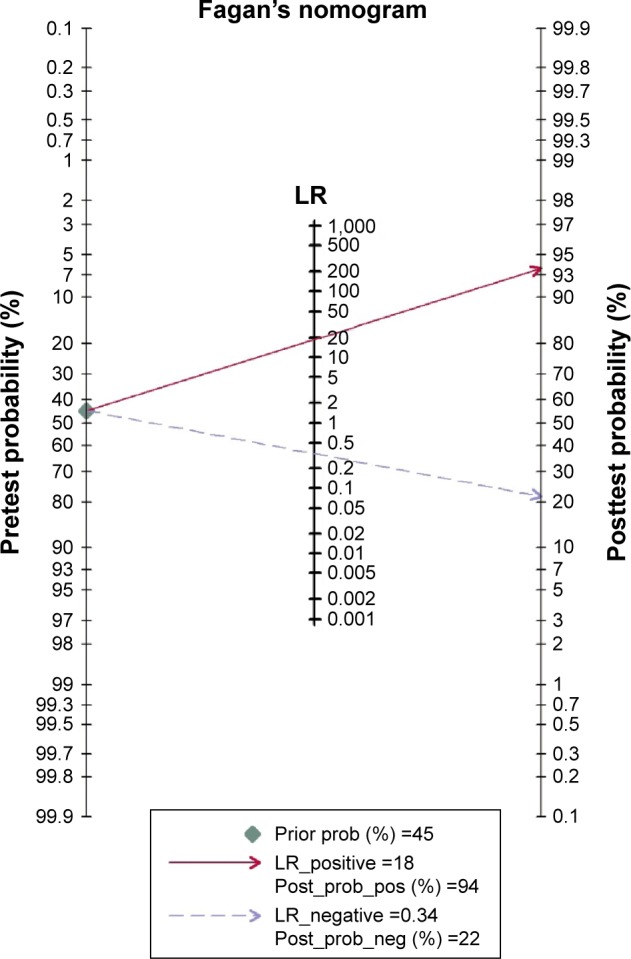
Fagan’s nomogram of ctDNA for detection of KRAS gene mutations.
Abbreviation: LR, likelihood ratio.
Figure 5.
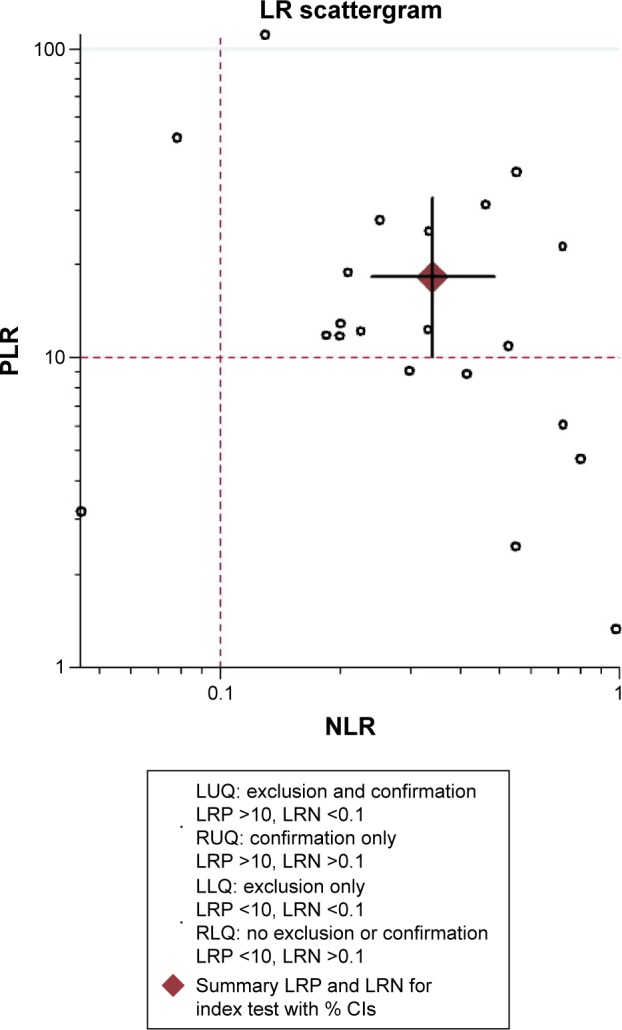
LR scattergram of ctDNA for detection of KRAS gene mutations.
Abbreviations: LR, likelihood ratio; NLR, negative likelihood ratio; PLR, positive likelihood ratio; CI, confidence interval; LUQ, left upper quadrant; LRP, likelihood ratio for positive results; RUQ, right upper quadrant; LLQ, left lower quadrant; RLQ, right lower quadrant; LRN, likelihood ratio for negative results.
Subgroup analysis
To determine the effect of potential confounding factors, we performed subgroup analyses measured by AUROC and stratified according to TNM stage, storage method of tumor tissues, blood sample format, ctDNA detection method and detection site of KRAS gene mutation (Table 3). For blood samples, the AUROC was higher in plasma (0.96, 95% CI =0.94–0.97) than in serum (0.83, 95% CI =0.79–0.86), indicating the higher diagnostic accuracy (P=0.000<0.05). With regard to detection methods, ARMS-qPCR was the most frequently used (0.96, 95% CI =0.94–0.97). However, there was no statistically significant difference in diagnostic accuracy between different cancer stages, storage methods of tumor tissues, ctDNA detection method and detection site of KRAS gene mutation.
Discussion
Cancer is the main cause of death, and effective treatment can significantly reduce the mortality of patients with carcinoma. Besides the traditional chemotherapy based on pathological diagnosis, targeted therapy has been widely used according to gene mutation status. Gene evaluation of tumor tissue is the gold standard for assessing mutation status, but it is usually carried out only once because of the invasiveness and costliness. However, genomic alterations may vary in primary and metastatic tumor tissues with the progression of cancer, which need the repetitive genotyping.
As we know, cancer generation and progression are associated with numerous genetic and epigenetic factors, some of which can be detected in gene alternations of tumor tissues. ctDNA, also known as part of cfDNA, can be released into blood and other body fluids from tumor tissues, carrying tumor-related genetic and epigenetic alterations, and can be more informative, specific and accurate than protein biomarkers. Several observations not only suggested the role of DNA in heredity but also provided the evidence for the occurrence of DNA beyond the confinement of cells. In 1977, Leon et al9 reported that the level of cfDNA in the peripheral blood of cancer patients was notably higher than that in healthy controls, especially in those patients with metastatic sites, and significantly reduced after radiation. In 1994, Sorenson et al35 and Vasioukhin et al36 found that RAS gene mutations can be detected in blood cfDNA. As newly improved methods, the detection rate of ctDNA for KRAS and BRAF gene mutations in blood of CRC patients has also been reported in different studies. In 2014, Thierry et al15 reported that ctDNA showed 98% specificity and 92% sensitivity of KRAS gene mutations and 100% specificity and sensitivity of BRAF gene mutations. Kuo et al20 pointed out that the detection rate of cfDNA for KRAS gene mutations in plasma was higher (50%) than in primary tumor tissues (28.8%). However, Perrone et al27 revealed that the KRAS gene mutation rate of cfDNA in plasma was very low (3%) compared to the matched adenocarcinoma tissues (45%). Inconsistent results made the use of ctDNA screening to be unclear. In 2015, Qiu et al14 compared the diagnostic value of ctDNA for detection of EGFR gene mutations with tumor tissues by a meta-analysis and suggested ctDNA as a highly specific but relatively low-sensitive biomarker in NSCLC patients. In spite of these findings, the clinical utilization of ctDNA for therapeutic monitoring is not yet widespread because of the different results in different studies. Now, we conducted this meta-analysis to suppose the consistency of KRAS gene mutations between ctDNA and the matched tumor tissues in CRC patients, for assessing the possibility of ctDNA used for gene monitoring.
As we know, anti-EGFR MoAb is now restricted to the patients with wild-type KRAS gene.5 Thus, detection of KRAS gene mutations in CRC patients has become a routine clinical test. Now, we analyzed the diagnostic performance of ctDNA for detection of KRAS gene mutations in CRC patients through meta-analysis in order to assess the clinical value of ctDNA detection. In our meta-analysis, publication bias was assessed by a regression of diagnostic log odds ratio against the inverse of the square root of the effective sample size,37 and P=0.060 for the slope coefficient indicated the absence of publication bias and the accuracy of meta-analysis. Our analysis showed that the overall pooled specificity was 96% and the pooled sensitivity was 67%, which were almost as same as what had been reported for ctDNA for detection of EGFR gene mutations (specificity 96%, sensitivity 62%).14 According to the guideline of AUROC and DOR,38 ctDNA detection of KRAS gene mutations also had high diagnostic accuracy and discriminatory value in CRC patients. LR is also used for evaluating diagnostic accuracy and clinical utility of diagnostic test.37 On basis of the results of PLR and NLR in our analysis, ctDNA was located in the right upper quadrant, indicating that it could be served as a test for confirmation of KRAS gene mutations in CRC patients.
In consideration of the influences of confounding factors on diagnostic accuracy, subgroup analyses were conducted on basis of some common covariates. Many studies have reported that the level of ctDNA was associated with tumor burden and stage.22,39,40 ctDNA has also been reported with a higher diagnostic value for detection of EGFR gene mutations in patients with advanced stage of NSCLC.14 However, we did not find any significant diagnostic difference for detection of KRAS gene mutation in different stages of CRC patients, which may be due to insufficient data of I–IV stages. The mechanism of release of ctDNAg from tumor cells is not clear, so the level of ctDNA in peripheral blood may not be in accordance with the TNM stage of cancer. That is to say, ctDNA may be useful for CRC patients in different stages. We also did not find the diagnostic accuracy difference of ctDNA between different detection sites of KRAS gene mutation, while may be for the relatively few studies and low mutation rate of codon 13 in CRC patients. In general, ctDNA is obtained from plasma or serum; FFPE or frozen tumor tissue is usually used. Our further stratified analysis indicated that the detection of ctDNA extracted from plasma was more accurate than that from serum, which was consistent with what had been reported in other studies.14 In 2015, Qiu et al pointed out that ctDNA extracted from plasma had higher diagnostic accuracy than that from serum for detection of EGFR gene mutations in NSCLC patients.14 Though FFPE may influence the nucleic acids, we still did not find any significant diagnostic accuracy difference of AUROC between FFPE and frozen tumor tissues. What is more, ARMS-qPCR was the most frequently used and had also been proved with high diagnostic performance. The limitations of our meta-analysis were also discussed to reveal the misinterpretation of present results. First, the relatively small size and potentially important differences in subgroup studies may result in statistical biases. Second, our analysis showed high heterogeneity, but none of the present characteristics could declare the heterogeneity. Because of the insufficient reporting of included studies, we could not analyze the probable influence of some potential factors, eg, ethnicity, pathological diagnosis of carcinoma, collection time of blood, chemotherapy situation and the detailed tumor stage, which might be the main sources of heterogeneity. Third, we did not find any significant difference in diagnostic accuracy for KRAS gene mutations after stratified analyses according to cancer stage and the storage method of tumor tissues. These results were in consonance with those studies of ctDNA for detection of other gene mutations, which could be by reason of the relatively few studies and the unclear records in details. Further studies are still needed to account for these issues.
This meta-analysis revealed that ctDNA detection might be useful for repetitive and noninvasive genotyping of KRAS gene mutations in patients with carcinoma, especially for those patients without the opportunity of biopsy at the decision-making points of treatment. This would be an ideal method of gene detection, particularly for cancer patients who are resistant or unable to catch tumor tissues.
Conclusion
ctDNA may help providing information of diagnostic screen, personalized medicine selection and real-time monitoring for cancer patients.
Acknowledgments
This study was supported by the funding from First Affiliated Hospital of Chinese People’s Liberation Army General Hospital. We would like to thank Doctor Jianhua Zhu, Doctor Guanghui Li and Doctor He Zhang for their help and support in study work.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Centers of Diseases Control and Prevention Health Status and Determinants. Life Expectancy and Mortality. 2015. [Accessed May 1, 2016]. Available from: http://www.cdc.gov/nchs/data/hus/hus15.pdf#019.
- 2.National Comprehensive Cancer Network [webpage on the Internet] Guidelines for Detection, Prevention, & Risk Reduction. 2017. [Accessed December 24, 2016]. Available from: http://www.nccn.org/professionals/physician_gls/f_guidelines.asp.
- 3.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Molecular predictors of outcome with gefitinib in a phase III placebo-controlled study in advanced non-small-cell lung cancer. J Clin Oncol. 2006;24(31):5034–5042. doi: 10.1200/JCO.2006.06.3958. [DOI] [PubMed] [Google Scholar]
- 4.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 5.Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to anti-EGFR treatment in colorectal cancer: a cohort study. Lancet Oncology. 2005;6(5):279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 6.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359(17):1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 7.Hol L, de Jonge V, van Leerdam ME, et al. Screening for colorectal cancer: comparison of perceived test burden of guaiac-based faecal occult blood test, faecal immunochemical test and flexible sigmoidoscopy. Eur J Cancer. 2010;46(11):2059–2066. doi: 10.1016/j.ejca.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 8.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646–650. [PubMed] [Google Scholar]
- 10.Anker P, Mulcahy H, Chen XQ, Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev. 1999;18(1):65–73. doi: 10.1023/a:1006260319913. [DOI] [PubMed] [Google Scholar]
- 11.Gormally E, Caboux E, Vineis P, Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat Res. 2007;635(2–3):105–117. doi: 10.1016/j.mrrev.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Koyanagi K, Mori T, O’Day SJ, Martinez SR, Wang HJ, Hoon DS. Association of circulating tumor cells with serum tumor-related methylated DNA in peripheral blood of melanoma patients. Cancer Res. 2006;66(12):6111–6117. doi: 10.1158/0008-5472.CAN-05-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori T, O’Day SJ, Umatani N, et al. Predictive utility of circulating methylated DNA in serum of melanoma patients receiving biochemotherapy. J Clin Oncol. 2005;23(36):9351–9358. doi: 10.1200/JCO.2005.02.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu M, Wang J, Xu Y, et al. Circulating tumor DNA is effective for the detection of EGFR mutation in non-small cell lung cancer: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2015;24(1):206–212. doi: 10.1158/1055-9965.EPI-14-0895. [DOI] [PubMed] [Google Scholar]
- 15.Thierry AR, Mouliere F, El Messaoudi S, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20(4):430–435. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 16.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danese E, Minicozzi AM, Benati M, et al. Comparison of genetic and epigenetic alterations of primary tumors and matched plasma samples in patients with colorectal cancer. PLoS One. 2015;10(5):e0126417. doi: 10.1371/journal.pone.0126417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim ST, Chang WJ, Jin L, Sung JS, Choi YJ, Kim YH. Can serum be used for analyzing the KRAS mutation status in patients with advanced colorectal cancer? Cancer Res Treat. 2015;47(4):796–803. doi: 10.4143/crt.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopreski MS, Benko FA, Borys DJ, Khan A, McGarrity TJ, Gocke CD. Somatic mutation screening: identification of individuals harboring K-ras mutations with the use of plasma DNA. J Natl Cancer Inst. 2000;92(11):918–923. doi: 10.1093/jnci/92.11.918. [DOI] [PubMed] [Google Scholar]
- 20.Kuo YB, Chen JS, Fan CW, Li YS, Chan EC. Comparison of KRAS mutation analysis of primary tumors and matched circulating cell-free DNA in plasmas of patients with colorectal cancer. Clin Chim Acta. 2014;433:284–289. doi: 10.1016/j.cca.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 21.Lefebure B, Charbonnier F, Di Fiore F, et al. Prognostic value of circulating mutant DNA in unresectable metastatic colorectal cancer. Ann Surg. 2010;251(2):275–280. doi: 10.1097/SLA.0b013e3181c35c87. [DOI] [PubMed] [Google Scholar]
- 22.Lin JK, Lin PC, Lin CH, et al. Clinical relevance of alterations in quantity and quality of plasma DNA in colorectal cancer patients: based on the mutation spectra detected in primary tumors. Ann Surg Oncol. 2014;21(suppl 4):S680–S686. doi: 10.1245/s10434-014-3804-5. [DOI] [PubMed] [Google Scholar]
- 23.Liu P, Liang H, Xue L, et al. Potential clinical significance of plasma-based KRAS mutation analysis using the COLD-PCR/Taq-Man(®) -MGB probe genotyping method. Exp Ther Med. 2012;4(1):109–112. doi: 10.3892/etm.2012.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyano S, Hanazawa K, Kitabatake T, Fujisawa M, Kojima K. Detecting KRAS mutations in peripheral blood of colorectal cancer patients by peptide nucleic acid clamp PCR. Exp Ther Med. 2012;4(5):790–794. doi: 10.3892/etm.2012.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan SR, Whiteley J, Donald E, et al. Comparison of KRAS mutation assessment in tumor DNA and circulating free DNA in plasma and serum samples. Clin Med Insights Pathol. 2012;5:15–22. doi: 10.4137/CPath.S8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulcahy HE, Lyautey J, Lederrey C, et al. Plasma DNA K-ras mutations in patients with gastrointestinal malignancies. Ann N Y Acad Sci. 2000;906:25–28. doi: 10.1111/j.1749-6632.2000.tb06585.x. [DOI] [PubMed] [Google Scholar]
- 27.Perrone F, Lampis A, Bertan C, et al. Circulating free DNA in a screening program for early colorectal cancer detection. Tumori. 2014;100(2):115–121. doi: 10.1177/030089161410000201. [DOI] [PubMed] [Google Scholar]
- 28.Pu X, Pan Z, Huang Y, et al. Comparison of KRAS/BRAF mutations between primary tumors and serum in colorectal cancer: biological and clinical implications. Oncol Lett. 2013;5(1):249–254. doi: 10.3892/ol.2012.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryan BM, Lefort F, McManus R, et al. A prospective study of circulating mutant KRAS2 in the serum of patients with colorectal neoplasia: strong prognostic indicator in postoperative follow up. Gut. 2003;52(1):101–108. doi: 10.1136/gut.52.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakai K, Tsurutani J, Yamanaka T, et al. Extended RAS and BRAF mutation analysis using next-generation sequencing. PLoS One. 2015;10(5):e0121891. doi: 10.1371/journal.pone.0121891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sefrioui D, Sarafan-Vasseur N, Beaussire L, et al. Clinical value of chip-based digital-PCR platform for the detection of circulating DNA in metastatic colorectal cancer. Dig Liver Dis. 2015;47(10):884–890. doi: 10.1016/j.dld.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 32.Spindler KL, Pallisgaard N, Andersen RF, Brandslund I, Jakobsen A. Circulating free DNA as biomarker and source for mutation detection in metastatic colorectal cancer. PLoS One. 2015;10(4):e0108247. doi: 10.1371/journal.pone.0108247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taly V, Pekin D, Benhaim L, et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem. 2013;59(12):1722–1731. doi: 10.1373/clinchem.2013.206359. [DOI] [PubMed] [Google Scholar]
- 34.Xu JM, Liu XJ, Ge FJ, et al. KRAS mutations in tumor tissue and plasma by different assays predict survival of patients with metastatic colorectal cancer. J Exp Clin Cancer Res. 2014;33:104. doi: 10.1186/s13046-014-0104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev. 1994;3(1):67–71. [PubMed] [Google Scholar]
- 36.Vasioukhin V, Anker P, Maurice P, Lyautery J, Lederrey C, Stroun M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br J Haematol. 1994;86(4):774–779. doi: 10.1111/j.1365-2141.1994.tb04828.x. [DOI] [PubMed] [Google Scholar]
- 37.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58(9):882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240(4857):1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- 39.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nygaard AD, Spindler KG, Pallisgaard N, Anderson RF, Jakobsen A. Levels of cell-free DNA and plasma KRAS during treatment of advanced NSCLC. Oncol Rep. 2014;31:969–974. doi: 10.3892/or.2013.2906. [DOI] [PubMed] [Google Scholar]


