Abstract
Introduction
African Americans’ (AAs) late-onset Alzheimer’s disease (LOAD) genetic risk profile is incompletely understood. Including clinical covariates in genetic analyses using informed conditioning might improve study power.
Methods
We conducted a genome-wide association study (GWAS) in AAs employing informed conditioning in 1825 LOAD cases and 3784 cognitively normal controls. We derived a posterior liability conditioned on age, sex, diabetes status, current smoking status, educational attainment, and affection status, with parameters informed by external prevalence information. We assessed association between the posterior liability and a genome-wide set of single-nucleotide polymorphisms (SNPs), controlling for APOE and ABCA7, identified previously in a LOAD GWAS of AAs.
Results
Two SNPs at novel loci, rs112404845 (P = 3.8 × 10−8), upstream of COBL, and rs16961023 (P = 4.6 × 10−8), downstream of SLC10A2, obtained genome-wide significant evidence of association with the posterior liability.
Discussion
An informed conditioning approach can detect LOAD genetic associations in AAs not identified by traditional GWAS.
Keywords: Alzheimer’s disease, Genome-wide association study (GWAS), African Americans, Informed conditioning on clinical covariates, COBL, SLC10A2, APOE, ABCA7, Age, Sex differences, Diabetes, Smoking, Education, Resveratrol
1. Introduction
Late-onset Alzheimer’s disease (LOAD) in African Americans (AAs) is influenced by multiple genetic, clinical, and environmental factors [1–3]. AAs are at increased risk of LOAD compared with non-Hispanic whites [4–6]. Nonetheless, knowledge about the genetic architecture of LOAD comes disproportionately from studies of non-Hispanic whites. The relative lack of data presents a substantial barrier to understanding LOAD mechanisms in AAs [3]. The APOE ε4 allele is a well-established genetic risk factor for LOAD in AAs [7]. Whereas >20 LOAD risk genes have been identified from genome-wide association studies (GWASs) for non-Hispanic whites, only two loci have been identified from GWAS for AAs [1,8]. A GWAS by Reitz et al. [1] found that, in addition to the APOE ε4 allele, a variant in the ABCA7 gene (rs115550680) was significantly associated with LOAD in AAs.
Most genetic association studies in LOAD, including those of AAs, adjust for age, sex, and population substructure (PC) only. For quantitative traits analyzed by linear regression, introducing other nonconfounding covariates into a genetic study could enhance detection of additional loci by accounting for some of the variance in the outcome. However, for case-control association studies, including nonconfounding covariates in a logistic regression model can actually reduce power to detect an association because case-control ascertainment can create an artificial correlation between the genetic variant and a covariate, and each additional covariate reduces the precision of estimates [9,10]. Zaitlen et al. [11] recently showed that using an informed conditioning approach, nonconfounding covariates could be included in a case-control study with an increase in power compared with models that do not include covariates. Informed conditioning is based on the liability threshold model with parameters informed by external prevalence information. In this approach, first a liability model is constructed using covariates’ independent effect estimates, in the form of trait prevalences at different covariate levels. Next, an association is tested between a genetic variant and the residuals from the liability model [11]. Informed conditioning has been applied with success to other phenotypes including stroke, type-2 diabetes, prostate cancer, lung cancer, breast cancer, reheumatoid arthritis, age-related macular degeneration, and end-stage kidney disease [11,12]. In the present study, we conducted a GWAS in the AA cohort of Reitz et al. [1], employing informed conditioning on LOAD status and several well-established LOAD risk factors, obtaining genome-wide significant evidence of association at two novel loci.
2. Methods
2.1. Study population
Study population included 1825 well-characterized AA LOAD cases and 3784 cognitively normal AA controls from 9 Alzheimer’s Disease Genetic Consortium (ADGC) datasets: Adult Changes in Thought, Alzheimer’s Disease Centers 1&2 (ADC1&2), ADC3, ADGC, Chicago Health and Aging Project (CHAP), Indianapolis, Genetic and Environmental Risk Factors for Alzheimer Disease Among AAs (GenerAAtions), Multi-Institutional Research on Alzheimer Genetic Epidemiology (MIRAGE) 300k, and MIRAGE 660k. The ADGC dataset contained participants from several studies including the AA AD Genetics Study, the ADCs, CHAP, Mayo Clinic, Mount Sinai School of Medicine, Religious Orders Study/Memory and Aging Project/Minority Aging Research Study (MARS)/Rush Clinical Core (CORE), University of Miami (UM)/Vanderbilt University (VU), University of Pittsburgh, Washington Heights Columbia Aging Project, and Washington University. A detailed description of subject recruitment and phenotyping has been described previously [1]. Two of these studies were family based (i.e., contained related participants: MIRAGE and GenerAAtions), whereas the other studies included only unrelated participants. Age of symptom onset was available for most cases. For the remaining cases, surrogate age information was available (age at ascertainment for Indianapolis, age at diagnosis for CHAP and MARS/CORE, and age at death for autopsy cases from UM/VU). Age of last examination or age of death was available for controls. We excluded cases younger than age 60. Ascertainment of additional risk factor data (educational attainment, diabetes status, and current smoking status) in each parent study has been described previously [13–20]. For the present study, we defined low educational attainment as ≤8 years of education. Because only a subset of the subjects from the ADGC dataset had additional risk factor data available, we divided the dataset into those with new risk factor data (ADGC1) and those without new risk factor data (ADGC2). Individuals were recruited under protocols approved by the appropriate institutional review boards.
2.2. Procedures
Details of genotyping (including platforms), quality control (including call rates, Hardy-Weinberg equilibrium, discordance with ascertained sex and latent relatedness), and derivation of principal components to adjust for PCs were described previously [1]. Briefly, we used directly measured APOE genotypes. We estimated haplotypes using SHAPEIT [21] and then imputed allele dosages for each dataset separately using IMPUTE2 [22] and 1000 Genomes reference haplotypes (March 2012). We excluded imputed single-nucleotide polymorphisms (SNPs) with an imputation quality estimate of R2 ≤0.40. In the unrelated cohorts, we did not exclude SNPs based on minor allele frequency (MAF) because their inclusion did not lead to P-value inflation. In the family-based cohorts, we excluded SNPs with MAFs <0.05. We derived principal components using EIGENSTRAT [23] separately for each dataset using a set of genotyped SNPs common to all genotyping platforms.
2.3. Calculation of the posterior mean residual liability score
An informed conditioning approach leverages external conditional prevalence data from the epidemiological literature. AA-specific prevalences of LOAD conditioned on LOAD risk factors (age, sex, education, current smoking status, and diabetes) have not been presented in the epidemiological literature. Therefore, we estimated the prevalences of LOAD in AAs >65 years of age, conditioned on a given LOAD risk factor (RF), based on published ancestry-nonspecific relative risks (RRs) of LOAD with respect to the RF [4–6,24,25], published prevalence of each RF (P(RF)) in AAs [26–29] (Supplementary Table 1), and estimated prevalence of LOAD in AAs (P(LOAD)) (Table 1). We assumed that RRs do not differ by ancestry [3,30–34]. The conditional prevalence of LOAD for a given RF, P(LOAD|RF), is related to the RR as follows:
| (1) |
where ~ RF is the absence of the RF.
Table 1.
Estimated conditional prevalence of AD in African Americans .65 years of age
| Trait | Conditional prevalence |
|---|---|
| Age | |
| 65–74 | 0.062 |
| 75–84 | 0.326 |
| ≥85 | 0.598 |
| Sex | |
| Male | 0.172 |
| Female | 0.242 |
| Education | |
| Low | 0.305 |
| High | 0.192 |
| Current smoker | |
| No | 0.190 |
| Yes | 0.302 |
| Diabetes | |
| No | 0.188 |
| Yes | 0.275 |
| Overall | 0.215 |
The law of total probability,
| (2) |
can be used to rewrite the conditional prevalence as follows:
| (3) |
We used equation 3 to calculate the LOAD conditional prevalences in AAs for each of the five LOAD risk factors (Table 1).
An informed conditioning approach, described by Zaitlen et al. [11], models an unobserved underlying quantitative trait, ϕ, called the liability:
| (4) |
where cj is a parameter estimating the effect of a given covariate j on the liability scale, tj is the value of covariate j, and m is a parameter that determines the population prevalence p of LOAD at the covariate mean t̄j such that ϕ is the normal cumulative distribution function and ϕ (−m) = P (x > −m) = p. Finally, ε = γg + N (0,1) where γ is the effect size of the candidate SNP with genotype g normalized to mean 0 and N (0,1) is the standard normal distribution. Multiple covariates are treated independently, but parameters are estimated jointly. A subject is a case if ϕ ≥ 0 and is a control otherwise.
We estimated parameters for two models because the additional risk factor data were missing for a subset of study subjects. The first model included all LOAD risk factors and their corresponding external conditional prevalence estimates, whereas the second model included only age and sex as LOAD risk factors and their corresponding external conditional prevalence estimates. We used these models to calculate the posterior mean residual liability score (hereafter called the LOAD liability score) for each subject given their LOAD and risk factor status. We used the LTSOFT computer program [11,35] for modeling the LOAD liability and for generating the LOAD liability score.
2.4. Association analyses
For mean age, sex, current smoking status, diabetes status, mean education level, APOE ε4 genotype, and rs115550680 MAF, we compared cases with controls in unadjusted models by meta-analyzing the ln odds ratio for categorical variables and standardized mean difference for continuous variables using a fixed-effects model with inverse variance weights. We conducted genome-wide association with the LOAD liability score using linear regression in each of the unrelated cohorts and linear generalized estimating equations (GEEs) in each family-based cohort, as the GEE method is robust to nonindependence of error terms within a family [36]. Association tests were adjusted for three PCs, APOE ε4 dosage, and dosage of the minor allele of ABCA7 SNP rs115550680. Association tests were carried out using R version 3.1.2 software [37]. Results were combined across datasets by meta-analyzing the regression estimates after applying genomic control adjustments using a fixed-effects model with inverse variance weights, as implemented in METAL [38]. The test statistic was the meta-analyzed regression estimate divided by its standard error. Several post hoc analyses were conducted for top-ranked loci. We evaluated several logistic regression models using LOAD case-control status as the outcome with different sets of covariates. In addition, we analyzed a liability model that included the full set of clinical variables but that did not include APOE ε4 genotype and rs115550680 minor allele dosage as covariates.
3. Results
Table 2 shows characteristics for each dataset. In unadjusted models, older age, lower educational attainment, APOE ε4 genotype, and rs115550680 minor allele (G) dosage were associated with LOAD risk. Current smoking and diabetes were associated with reduced risk. Female sex was not associated with LOAD status. The conditional prevalence estimates of LOAD in AAs >65 years of age that were calculated based on published LOAD RF prevalences and LOAD RRs with respect to the RFs (Supplementary Table 1) are listed in Table 1.
Table 2.
Characteristics of datasets
| Dataset | N (%) | Mean age (SD) | Women (%) | Current smoker (%) |
Diabetes (%) | Mean education level (SD) |
APOE−/−* (%) | APOE−/4* (%) | APOE 4/4 (%) | rs115550680 MAF† (INFO) |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACT | Case | 32 (32.99) | 83.16 (5) | 24 (75) | 4 (12.5) | 0 (0) | 11.85 (3.67) | 15 (46.88) | 12 (37.5) | 4 (12.5) | 0.16 | (0.94) |
| Control | 65 (67.01) | 79.23 (6.21) | 38 (58.46) | 33 (50.77) | 20 (30.77) | 13.64 (3.56) | 42 (64.62) | 20 (30.77) | 0 (0) | 0.06 | ||
| ADC1&2 | Case | 59 (44.7) | 75.69 (7.3) | 36 (61.02) | 1 (1.69) | 0 (0) | 12.43 (3.68) | 17 (28.81) | 33 (55.93) | 8 (13.56) | 0.08 | (0.91) |
| Control | 73 (55.3) | 72.95 (7.58) | 58 (79.45) | 36 (49.32) | 26 (35.62) | 14.94 (2.97) | 42 (57.53) | 25 (34.25) | 2 (2.74) | 0.05 | ||
| ADC3 | Case | 162 (59.12) | 79.88 (7.22) | 118 (72.84) | 60 (37.04) | 37 (22.84) | 12.13 (3.15) | 39 (24.07) | 78 (48.15) | 17 (10.49) | 0.08 | (0.99) |
| Control | 112 (40.88) | 74.28 (7.55) | 91 (81.25) | 58 (51.79) | 27 (24.11) | 15.06 (3.05) | 62 (55.36) | 39 (34.82) | 4 (3.57) | 0.08 | ||
| CHAP | Case | 114 (20.84) | 81.82 (5.93) | 71 (62.28) | 60 (52.63) | 24 (21.05) | 10.98 (3.57) | 67 (58.77) | 41 (35.96) | 5 (4.39) | 0.09 | (0.88) |
| Control | 433 (79.16) | 78 (6.62) | 290 (66.97) | 230 (53.12) | 50 (11.55) | 12.81 (2.87) | 261 (60.28) | 153 (35.33) | 12 (2.77) | 0.05 | ||
| Indianapolis | Case | 173 (14.72) | 83.6 (6.74) | 108 (62.43) | 90 (52.02) | 43 (24.86) | 9.43 (3.47) | 78 (45.09) | 75 (43.35) | 20 (11.56) | 0.10 | (0.94) |
| Control | 1002 (85.28) | 82.88 (5.31) | 663 (66.17) | 625 (62.38) | 410 (40.92) | 11.25 (2.71) | 670 (66.87) | 298 (29.74) | 34 (3.39) | 0.06 | ||
| ADGC1‡ | Case | 267 (52.56) | 79.29 (7.24) | 185 (69.29) | 123 (46.07) | 50 (18.73) | 10.91 (3.83) | 61 (22.85) | 49 (18.35) | 10 (3.75) | 0.08 | (0.88) |
| Control | 241 (47.44) | 80.68 (6.87) | 169 (70.12) | 121 (50.21) | 50 (20.75) | 13.32 (3.4) | 138 (57.26) | 59 (24.48) | 3 (1.24) | 0.06 | ||
| ADGC2‡ | Case | 554 (28.4) | 77.86 (8.31) | 415 (74.91) | — | — | — | 222 (40.07) | 248 (44.77) | 75 (13.54) | 0.07 | (0.87) |
| Control | 1397 (71.6) | 73.12 (8.22) | 1056 (75.59) | — | — | — | 903 (64.64) | 431 (30.85) | 41 (2.93) | 0.06 | ||
| MIRAGE 300k | Case | 58 (57.43) | 72.69 (7.46) | 40 (68.97) | 27 (46.55) | 12 (20.69) | 11.34 (4.7) | 15 (25.86) | 33 (56.9) | 10 (17.24) | 0.13 | (0.69) |
| Control | 43 (42.57) | 73.12 (8.23) | 30 (69.77) | 23 (53.49) | 9 (20.93) | 11.21 (4.48) | 20 (46.51) | 21 (48.84) | 2 (4.65) | 0.13 | ||
| MIRAGE 660k | Case | 164 (42.82) | 72.95 (6.82) | 119 (72.56) | 56 (34.15) | 45 (27.44) | 9.17 (5.24) | 53 (32.32) | 77 (46.95) | 33 (20.12) | 0.13 | (0.89) |
| Control | 219 (57.18) | 73.39 (8.28) | 159 (72.6) | 72 (32.88) | 48 (21.92) | 10.71 (5.1) | 121 (55.25) | 88 (40.18) | 9 (4.11) | 0.08 | ||
| GenerAAtions | Case | 242 (54.88) | 80.21 (6.6) | 138 (57.02) | — | — | — | 88 (36.36) | 112 (46.28) | 26 (10.74) | 0.09 | (0.88) |
| Control | 199 (45.12) | 78.49 (6.7) | 118 (59.3) | — | — | — | 114 (57.29) | 63 (31.66) | 6 (3.02) | 0.07 | ||
| P value§ | — | 7.23 × 10−28 | 0.11 | 5.01 × 10−4 | 0.11 | 1.03 × 10−45 | 1.43 × 10−53 | 5.90 × 10−30 | 6.12 × 10−5 | |||
Abbreviations: ACT, Adult Changes in Thought; ADC, Alzheimer’s Disease Center; ADGC, Alzheimer’s Disease Genetics Consortium; APOE, apolipoprotein E; CHAP, Chicago Health and Aging Project; GenerAAtions, Genetic and Environmental Risk Factors for Alzheimer’s Disease among African Americans; INFO, information metric for imputation quality from IMPUTE2; MAF, minor allele frequency; MIRAGE, Multi-Institutional Research on Alzheimer Genetic Epidemiology; SD, standard deviation.
(–) refers to all non–APOE ε4-containing genotypes (APOE 3/3, APOE 2/3, APOE 2/2).
Imputed single-nucleotide polymorphism, minor allele = G, major allele = A.
Samples genotyped by the ADGC for this project were received from the African American Alzheimer’s Disease Genetics Study, the ADCs, CHAP, Mayo Clinic, Mount Sinai School of Medicine, Religious Orders Study/Rush Memory and Aging Project/Minority Aging Research Study/Rush Clinical Core, University of Miami/Vanderbilt University, University of Pittsburgh, Washington Heights Columbia Aging Project, and Washington University.
Comparison of cases with controls; results were combined across datasets by meta-analyzing the ln odds ratio for categorical variables and standardized mean difference for continuous variables using a fixed-effects model with inverse variance weights.
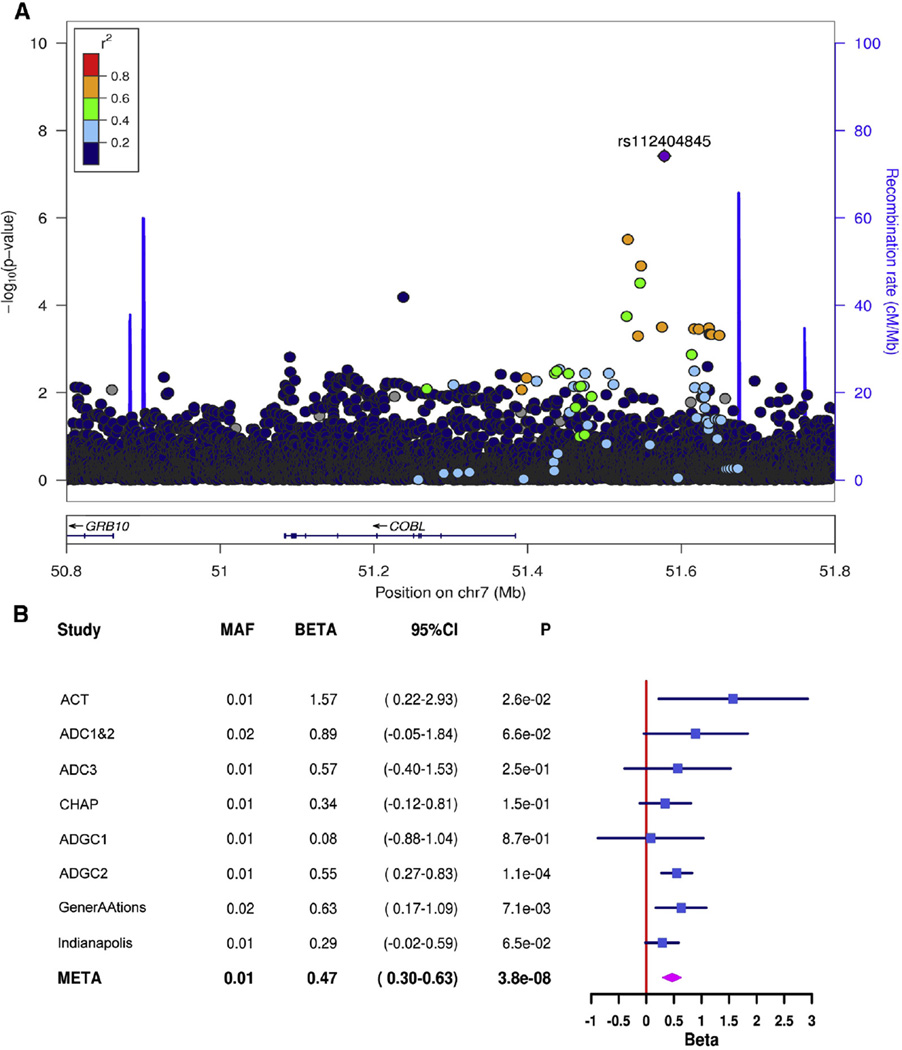
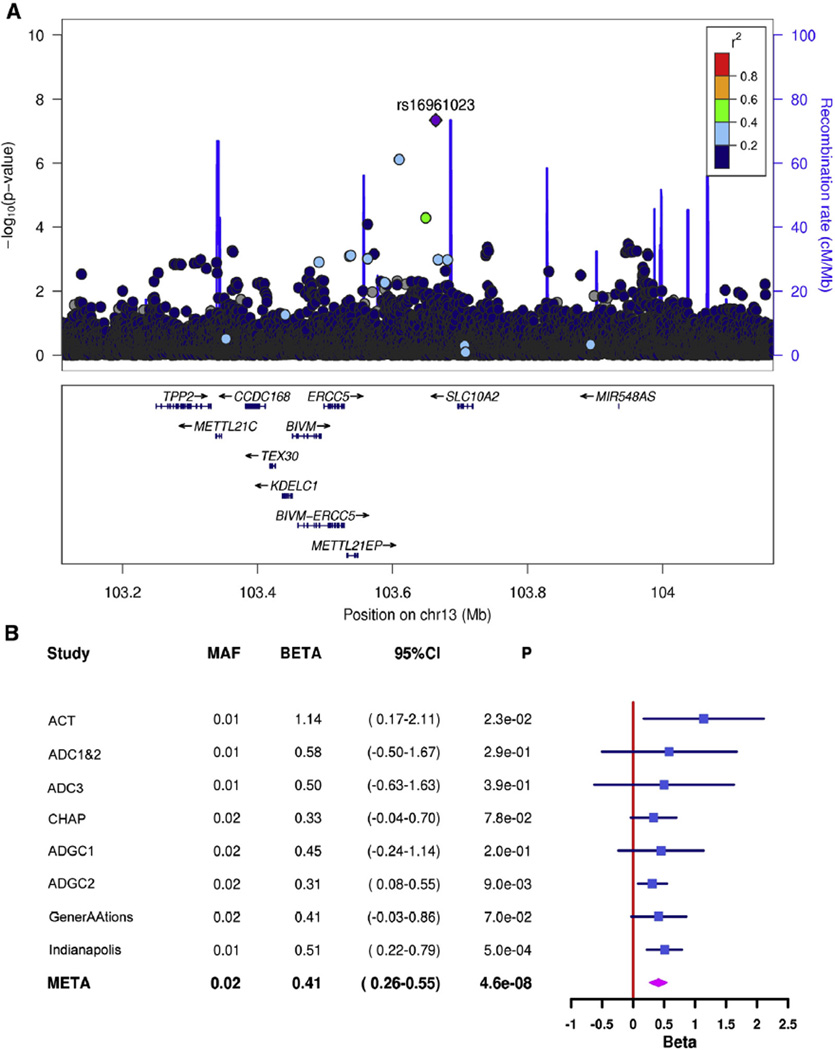
A total of 19,725,427 SNPs passed quality control and were included in the GWAS. For individual datasets, the genomic inflation factor λ ranged from 0.984 to 1.127. There was no evidence of inflation of test statistics in the meta-analyzed LOAD liability model (λ = 0.993, Supplementary Fig. 1). We found genome-wide significant associations (P < = × 10−8) using informed conditioning in two distinct regions, 200-kb upstream of cordon-bleu WH2 repeat protein (COBL) and 30-kb downstream of solute carrier family 10, member 2 (SLC10A2) (Table 3). For the top SNP in each region (COBL: rs112404845, SLC10A2: rs16961023), the effect was in the same direction for all datasets (Figs. 1 and 2) and the final liability model showed a smaller P value than the logistic models by one to two orders of magnitude (Table 3).
Table 3.
Logistic and liability models for the top independent SNPs to achieve genome-wide significance in the liability model
| Chrom | Gene | SNP | BP | Minor allele* |
Major allele |
MAF | Model | Covariates | Effect size† |
95% CI | P value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | COBL | rsl12404845 | 51578022 | T | A | 0.01 | Logistic | Age, sex, three principal components | 3.28 | 1.71–4.85 | 1.22 × 10−6 |
| Logistic | Age, sex, smoking, diabetes, education, three principal components |
3.59 | 1.76–5.41 | 8.70 × 10−7 | |||||||
| Liability: age, sex, smoking, diabetes, education |
Three principal components | 0.46 | 0.28–0.64 | 1.28 × 10−7 | |||||||
| Liability: age, sex, smoking, diabetes, education |
APOE ε4, rsl 15550680, three principal components |
0.47 | 0.29–0.65 | 3.82 × 10−8 | |||||||
| 13 | SLC10A2 | rsl6961023 | 103663945 | G | C | 0.02 | Logistic | Age, sex, three principal components | 2.77 | 1.65–3.89 | 8.01 × 10−7 |
| Logistic | Age, sex, smoking, diabetes, education, three principal components |
2.68 | 1.52–3.84 | 7.92 × 10−6 | |||||||
| Liability: age, sex, smoking, diabetes, education |
Three principal components | 0.41 | 0.25–0.57 | 1.03 × 10−7 | |||||||
| Liability: age, sex, smoking, diabetes, education |
APOE ε4, rsl 15550680, three principal components |
0.41 | 0.27–0.55 | 4.59 × 10−8 |
Abbreviations: APOE, apolipoprotein E; BP, base pair position; Chrom, chromosome; CI, confidence interval; COBL, cordon-bleu WH2 repeat protein; MAF, minor allele frequency; SLC10A2, solute carrier family 10, member 2; SNP, single-nucleotide polymorphism.
Effect allele.
Odds ratios (ORs) for logistic models and beta coefficients for liability models. Note ORs and beta coefficients are not on the same scale and cannot be compared. Effect is for the minor allele.
Fig. 1.
(A) Regional association plot of the COBL region on chromosome 7 and (B) forest plots for rs112404845, the top single-nucleotide polymorphism in the region. Abbreviations: ACT, Adult Changes in Thought; ADC, Alzheimer’s Disease Center; ADGC, Alzheimer’s Disease Genetics Consortium; CHAP, Chicago Health and Aging Project; CI, confidence interval; COBL, Cordon-Bleu WH2 Repeat Protein; GenerAAtions, Genetic and Environmental Risk Factors for Alzheimer Disease among African Americans; and MAF, minor allele frequency.
Fig. 2.
(A) Regional association plot of the SLC10A2 region on chromosome 13 and (B) forest plots for rs16961023, the top single-nucleotide polymorphism in the region. Abbreviations: ACT, Adult Changes in Thought; ADC, Alzheimer’s Disease Center; ADGC, Alzheimer’s Disease Genetics Consortium; CHAP, Chicago Health and Aging Project; CI, confidence interval; GenerAAtions, Genetic and Environmental Risk Factors for Alzheimer Disease among African Americans; MAF, minor allele frequency; and SLC10A2, solute carrier family 10, member 2.
Because both variants were imputed and relatively rare, we compared allele dosages from imputation with direct genotyping. For rs112404845 (imputation quality range [using the IMPUTE2 information metric]: 0.905–1.039), we Sanger sequenced 20 predicted risk-allele carriers and an equal number of noncarriers. We found perfect correlation between the imputed dosage and direct genotype. For rs16961023 (imputation quality range: 0.598–0.917), we used a Taqman assay to directly genotype 35 predicted risk allele carriers and 1720 noncarriers from the ADGC1 and ADGC2 datasets. As expected based on the imputation quality (ADGC1 = 0.704, ADGC2 = 0.679), the correlation between direct genotype and imputed dosage (ADGC1 = 0.736, ADGC2 = 0.565) was adequate. When we repeated our association analysis across all datasets, among subjects with imputed posterior probabilities >0.8, using best-guess genotype, there was a reduction in effect size from 0.41 to 0.33.
Supplementary Table 2 shows SNPs with suggestive associations (P < = × 10−6) using informed conditioning. No loci identified as risk factors for LOAD in GWAS in white non-Hispanics showed suggestive associations. As a positive control, we compared the effect of the ABCA7 rs115550680 variant in a logistic model that controlled for age, sex, current smoking status, diabetes status, educational attainment, and PCs with the liability model. Using the informed conditioning approach, the P value decreased by a half order of magnitude in the liability model compared with a logistic model (Supplementary Table 3).
4. Discussion
In this study, we conducted a GWAS in AAs, employing informed conditioning on LOAD status and well-established LOAD risk factors including age, sex, diabetes status, current smoking status, and educational attainment. Our model is informed by external prevalence data from the epidemiological literature. Using this approach, which has been shown to outperform standard case-control association tests [11,12], we identified two genome-wide significant novel LOAD loci, upstream of COBL and downstream of SLC10A2.
COBL is predominantly expressed in brain, and its protein product regulates neuron morphogenesis. It mediates actin nucleation, ensuring that neurites form, elongate, and branch correctly to produce functional neuronal networks. In Cobl-deficient dissociated hippocampal neurons, neurite and dendritic branch point numbers were significantly reduced [39]. An SNP approximately 500-kb upstream of COBL was implicated in posttraumatic stress disorder in cohorts with European [40] and African ancestry [41]. Rs112404845, the top SNP in the COBL region in our study, is located 200-kb upstream of COBL and is present only in persons with African ancestry (MAF = 0.012 in the 1000 Genomes reference panel). This may explain why COBL has not been recognized previously as an AD risk gene. Variation at rs112404845 leads to a Pax-4 regulatory motif change. Rs113739092, an SNP in linkage disequilibrium with rs112404845 (r2 = 0.64) and which achieved a P value of 1.3 × 10−5, is an enhancer histone mark in brain [42].
SLC10A2 encodes a sodium/bile acid cotransporter that is essential for cholesterol homeostasis. Mutations in SLC10A2 have been found in cases of familial hypercholesterolemia [43]. Several other genes implicated by GWAS in LOAD pathogenesis function in cholesterol metabolism include APOE, CLU, ABCA7, and SORL1 [44]. Although its function is best understood in the small intestine [45], SLC10A2 also is expressed in brain [46]. Resveratrol, a chief constituent of red wine, inhibits SLC10A2 expression and function through a Sirt1 (sirtuin 1)–independent manner [47]. Potentially an exciting therapy for LOAD, resveratrol reduces amyloid plaque pathology in AD animal models [48] and has been shown to be safe and well-tolerated in a large phase 2 LOAD clinical trial [49]. Although resveratrol’s antiamyloidogenic effects have been suggested to be mediated by Sirt1 [50], our findings indirectly suggest that resveratrol may affect AD through multiple mechanisms. Rs16961023, the most significantly associated SNP in the SLC10A2 region in our study, is located 30-kb downstream of SLC10A2. Variation at rs16961023 leads to an Egr-1 regulatory motif change [42]. In the 1000 Genomes reference panel, the rs16961023 minor allele is infrequent among persons with African ancestry (MAF = 0.02) and rare among persons with European ancestry (MAF = 0.004), but is common in East Asians (MAF = 0.15).We previously conducted a GWAS for LOAD case/control status in a Japanese cohort [51] but did not find any nominally associated SNPs at this locus. Genetic association findings may be specific to a particular ethnic background, especially when variants are rare [52].
Current smoking has been found to increase LOAD risk in meta-analyses [24]; however, in this study, it was more frequent in LOAD controls than in LOAD cases when we combined the datasets. Ascertainment bias may explain this finding because our datasets are a mixture of clinic-and community-based studies. Cases disproportionately come from the clinic-based studies, whereas controls disproportionately come from the community-based studies. Typically, clinic-based cohorts have fewer vascular risk factors, including smoking, than their community-based counterparts [53]. Alternatively, survival bias may explain this finding, as smokers with LOAD may have died before entering the study [54]. Although early cross-sectional case-control studies observed that smoking was associated with a reduced risk of LOAD, a meta-analysis that included 23 longitudinal studies found that smoking increased risk [55]. That smoking occurs in our combined cohort at such a different relative rate than what is observed in the epidemiological literature emphasizes the importance of the informed conditioning approach, which makes use of external prevalence data.
Our study has several potential caveats. Because LOAD conditional prevalence data for AAs have not been reported, we estimated these values using available relative risk data that are not ancestry specific and therefore assumed that LOAD relative risks do not differ by ancestral population. Support for this assumption comes from review of the literature that shows the relative risks of dementia, LOAD, and/or cognitive decline for age, sex, diabetes status, smoking status, and educational attainment do not appear to differ significantly by ancestry [3,30–34]; however, this warrants further investigation. This concern is further lessened by a simulation study showing that moderate misspecification of model parameters did not reduce power and that even when parameters were misspecified by a large amount (up to 100%), the model still performed at least and logistic regression [11].
It should also be noted that the genome-wide significant variants near COBL and SLC10A2 have low frequency (0.01–0.02). By comparison, SNPs with MAF <0.05 were not analyzed in the GWAS of Reitz et al. [1] because genotype imputation quality was poor for low-frequency SNPs using the older 1000 Genomes reference panel. Using a newer reference panel, we were able to include low-frequency variants with improved imputation quality. For rs112404845, the top SNP in the COBL region, imputation quality, and correlation between imputed and direct genotype dosages were excellent, suggesting that our association findings were unlikely to be influenced by imputation quality. However, for rs16961023, the top SNP in the SLC10A2 region, imputation quality, and correlation between imputed and direct genotype dosages were similar, but only adequate, and there was a reduction in effect size when we repeated our association analysis among subjects with imputed posterior probabilities >0.8. It appears that subjects with less certain genotype probabilities may contribute disproportionately to the association signal, and, therefore, the SLC10A2 finding warrants cautious optimism. Finally, inclusion of low-frequency variants in GWAS can increase genomic inflation [56]. However, despite inclusion of low-frequency variants in our study, we did not see evidence of inflation (λ = 0.993, Supplementary Fig. 1).
Although COBL and SLC10A2 are attractive candidate genes, variants identified through GWAS may not be causal and do not necessarily act at the gene closest to them. Therefore, it is only speculative that these genes are the causal risk factors for LOAD in AAs and that the identified variants have an effect on AD via the proposed regulatory mechanisms. Finally, our findings should be regarded with measured enthusiasm until they are confirmed in independent samples of AAs. Unfortunately, to our knowledge, additional large AA LOAD cohorts with genotype data are not currently available. Therefore, validation of the role of these loci in AD will likely require experimental evidence.
Taken together, these findings suggest that an informed conditioning approach can be used to identify new genetic associations for complex genetic traits where risk is a mix of genetic and environmental factors. Our success in using informed conditioning to identify new risk loci for AD mirrors the success of informed conditioning in GWASs of other phenotypes [11,12]. This work furthers our understanding of the biological underpinnings of AD in AAs. Functional studies are needed to determine whether COBL and SLC10A2 are suitable targets for development of novel therapies.
Supplementary Material
RESEARCH IN CONTEXT.
Systematic review: The authors are members of the Alzheimer’s Disease Genetics Consortium and therefore are familiar with emerging pertinent literature. PubMed searches were conducted to identify other relevant publications. References that informed the novel approach and that support the significance of the identified risk loci are cited.
Interpretation: Although >20 late-onset Alzheimer’s disease (LOAD) risk genes have been identified from genome-wide association studies (GWASs) for non-Hispanic whites, this report identifies only the third and fourth loci from GWAS associated with LOAD for African Americans (AAs). The COBL and SLC10A2 loci provide further evidence that axonal integrity and cholesterol homeostasis underlie LOAD pathophysiology.
Future directions: The study findings should be confirmed in independent AA samples. Unfortunately, additional large AA LOAD cohorts with genotype data are not currently available. Functional studies are needed to determine whether COBL and SLC10A2 are suitable targets for development of novel therapies.
Acknowledgments
Additional members of the Alzheimer’s Disease Genetics Consortium who Contributed to this study: Guiqing Cai, PhD, Laura B. Cantwell, MPH, Philip L. De Jager, MD, PhD, Rodney C. P. Go, PhD, Patrick Griffith, MD, Rosalyn Lang, PhD, Oscar L. Lopez, MD, Thomas O. Obisesan, MD, Towfique Raj, PhD, and Beth Dombroski, PhD. Funding support: this work has been supported by National Institute on Aging/National Institutes of Health grants K23-AG046377 (Dr Mez); R01-AG09029, R01-AG025259, P30-AG13846 (Dr Farrer); U01-AG032984, RC2-AG036528, U01-AG016976 (Dr Kukull); U24-AG026395, U24-AG026390, R01-AG037212, R37-AG015473 (Dr Mayeux); U24-AG021886 (Dr Foroud); R01-AG20688 (Dr Fallin); P50-AG005133, AG041718, AG030653 (Dr Kamboh); R01-AG019085 (Dr Haines); R01-AG1101, R01-AG030146, RC2-AG036650 (Dr Evans); P30-AG10161, R01-AG15819, R01-AG17917 (Dr Bennett); R01AG028786 (Dr Manly); R01-AG22018, P30-AG10161 (Dr Barnes); P50-AG016574, R01-AG032990, R01-NS080820 (Dr Ertekin-Taner); R01-AG027944, R01-AG028786 (Dr Pericak-Vance); P20-MD000546, R01-AG28786 (Dr Byrd); AG005138 (Dr Buxbaum); P50-AG05681, P01-AG03991, P01-AG026276 (Dr Goate); U01-AG06781 (Drs Larson and Crane); R01-AG009956, RC2-AG036650 (Dr Hall); U01-AG032984 (Dr Schellenberg); and U24-AG041689-01 (National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site at the University of Pennsylvania).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jalz.2016.09.002.
References
- 1.Reitz C, Jun G, Naj A, Rajbhandary R, Vardarajan BN, Wang L-S, et al. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E ε4,and the risk of late-onset Alzheimer disease in African Americans. JAMA. 2013;309:1483–1492. doi: 10.1001/jama.2013.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachman DL, Green RC, Benke KS, Cupples LA, Farrer LA. MIRAGE Study Group. Comparison of Alzheimer’s disease risk factors in white and African American families. Neurology. 2003;60:1372–1374. doi: 10.1212/01.wnl.0000058751.43033.4d. [DOI] [PubMed] [Google Scholar]
- 3.Barnes LL, Bennett DA. Alzheimer’s disease in African Americans: risk factors and challenges for the future. Health Aff (Millwood) 2014;33:580–586. doi: 10.1377/hlthaff.2013.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demirovic J, Prineas R, Loewenstein D, Bean J, Duara R, Sevush S, et al. Prevalence of dementia in three ethnic groups: the South Florida program on aging and health. Ann Epidemiol. 2003;13:472–478. doi: 10.1016/s1047-2797(02)00437-4. [DOI] [PubMed] [Google Scholar]
- 5.Plassman BL, Langa KM, Fisher GG, Heeringa SG, Weir DR, Ofstedal MB, et al. Prevalence of dementia in the United States: the aging, demographics, and memory study. Neuroepidemiology. 2007;29:125–132. doi: 10.1159/000109998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potter GG, Plassman BL, Burke JR, Kabeto MU, Langa KM, Llewellyn DJ, et al. Cognitive performance and informant reports in the diagnosis of cognitive impairment and dementia in African Americans and whites. Alzheimers Dement. 2009;5:445–453. doi: 10.1016/j.jalz.2009.04.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 8.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson LD, Jewell NP. Some surprising results about covariate adjustment in logistic regression models. Int Stat Rev. 1991;59:227. [Google Scholar]
- 10.Pirinen M, Donnelly P, Spencer CC. Including known covariates can reduce power to detect genetic effects in case-control studies. Nat Genet. 2012;44:848–851. doi: 10.1038/ng.2346. [DOI] [PubMed] [Google Scholar]
- 11.Zaitlen N, Lindström S, Pasaniuc B, Cornelis M, Genovese G, Pollack S, et al. Informed conditioning on clinical covariates increases power in case-control association studies. PLoS Genet. 2012;8:e1003032. doi: 10.1371/journal.pgen.1003032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Traylor M, Mäkelä KM, Kilarski LL, Holliday EG, Devan WJ, Nalls MA, et al. A novel MMP12 locus is associated with large artery atherosclerotic stroke using a genome-wide age-at-onset informed approach. PLoS Genet. 2014;10:e1004469. doi: 10.1371/journal.pgen.1004469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kukull WA, Higdon R, Bowen JD, McCormick WC, Teri L, Schellenberg GD, et al. Dementia and Alzheimer disease incidence: a prospective cohort study. Arch Neurol. 2002;59:1737–1746. doi: 10.1001/archneur.59.11.1737. [DOI] [PubMed] [Google Scholar]
- 14.Morris JC, Weintraub S, Chui HC, Cummings J, Decarli C, Ferris S, et al. The Uniform Data Set (UDS): clinical and cognitive variables and descriptive data from Alzheimer Disease Centers. Alzheimer Dis Assoc Disord. 2006;20:210–216. doi: 10.1097/01.wad.0000213865.09806.92. [DOI] [PubMed] [Google Scholar]
- 15.Bienias JL, Beckett LA, Bennett DA, Wilson RS, Evans DA. Design of the Chicago Health and Aging Project (CHAP) J Alzheimers Dis. 2003;5:349–355. doi: 10.3233/jad-2003-5501. [DOI] [PubMed] [Google Scholar]
- 16.Hendrie HC, Ogunniyi A, Hall KS, Baiyewu O, Unverzagt FW, Gureje O, et al. Incidence of dementia and Alzheimer disease in 2 communities: Yoruba residing in Ibadan, Nigeria, and African Americans residing in Indianapolis, Indiana. JAMA. 2001;285:739–747. doi: 10.1001/jama.285.6.739. [DOI] [PubMed] [Google Scholar]
- 17.Farrer LA, Cupples LA, Blackburn S, Kiely DK, Auerbaeh S, Growdon JH, et al. Interrater agreement for diagnosis of Alzheimer’s disease: the MIRAGE study. Neurology. 1994;44:652–656. doi: 10.1212/wnl.44.4.652. [DOI] [PubMed] [Google Scholar]
- 18.Bennett DA, Schneider JA, Buchman AS, Mendes de Leon C, Bienias JL, Wilson RS. The Rush Memory and Aging Project: study design and baseline characteristics of the study cohort. Neuroepidemiology. 2005;25:163–175. doi: 10.1159/000087446. [DOI] [PubMed] [Google Scholar]
- 19.Bennett DA, Wilson RS, Schneider JA, Evans DA, Beckett LA, Aggarwal NT, et al. Natural history of mild cognitive impairment in older persons. Neurology. 2002;59:198–205. doi: 10.1212/wnl.59.2.198. [DOI] [PubMed] [Google Scholar]
- 20.Barnes LL, Shah RC, Aggarwal NT, Bennett DA, Schneider JA. The Minority Aging Research Study: ongoing efforts to obtain brain donation in African Americans without dementia. Curr Alzheimer Res. 2012;9:734–745. doi: 10.2174/156720512801322627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 22.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 24.Barnes DE, Yaffe K. The projected effect of risk factor reduction on Alzheimer’s disease prevalence. Lancet Neurol. 2011;10:819–828. doi: 10.1016/S1474-4422(11)70072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang MX, Cross P, Andrews H, Jacobs DM, Small S, Bell K, et al. Incidence of AD in African-Americans, Caribbean Hispanics, and Caucasians in northern Manhattan. Neurology. 2001;56:49–56. doi: 10.1212/wnl.56.1.49. [DOI] [PubMed] [Google Scholar]
- 26.Centers for Disease Control and Prevention. National Diabetes Statistics Report: Estimates of Diabetes and Its Burden in the United States, 2014. Atlanta, GA: US Department of Health and Human Services; 2014. [Accessed October 20, 2016]. Available at: http://www.cdc.gov/diabetes/data/statistics/2014StatisticsReport.html. [Google Scholar]
- 27.Agaku IT, King BA, Dube SR. Centers for Disease Control and Prevention: current cigarette smoking among adults—United States, 2005–2012. MMWR Morb Mortal Wkly Rep. 2014;63:29–34. [PMC free article] [PubMed] [Google Scholar]
- 28.United States Census Bureau: American Community Survey Data on Educational Attainment n.d. [Accessed August 26, 2015]; Available at: http://www.census.gov/hhes/socdemo/education/data/acs/index.html.
- 29.United States Census Bureau: The Black Alone Population in the United States: 2012 n.d. [Accessed August 26, 2015]; Available at: http://www.census.gov/population/race/data/ppl-ba12.html.
- 30.Barnes LL, Wilson RS, Li Y, Aggarwal NT, Gilley DW, McCann JJ, et al. Racial differences in the progression of cognitive decline in Alzheimer disease. Am J Geriatr Psychiatry. 2005;13:959–967. doi: 10.1176/appi.ajgp.13.11.959. [DOI] [PubMed] [Google Scholar]
- 31.Wessels AM, Lane KA, Gao S, Hall KS, Unverzagt FW, Hendrie HC. Diabetes and cognitive decline in elderly African Americans: a 15-year follow-up study. Alzheimers Dement. 2011;7:418–424. doi: 10.1016/j.jalz.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yaffe K, Falvey C, Hamilton N, Schwartz AV, Simonsick EM, Satterfield S, et al. Diabetes, glucose control, and 9-year cognitive decline among older adults without dementia. Arch Neurol. 2012;69:1170–1175. doi: 10.1001/archneurol.2012.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reitz C, Luchsinger J, Tang MX, Mayeux R. Effect of smoking and time on cognitive function in the elderly without dementia. Neurology. 2005;65:870–875. doi: 10.1212/01.wnl.0000176057.22827.b7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shadlen MF, Siscovick D, Fitzpatrick AL, Dulberg C, Kuller LH, Jackson S. Education, cognitive test scores, and black-white differences in dementia risk. J Am Geriatr Soc. 2006;54:898–905. doi: 10.1111/j.1532-5415.2006.00747.x. [DOI] [PubMed] [Google Scholar]
- 35.Zaitlen N, Pasaniuc B, Patterson N, Pollack S, Voight B, Groop L, et al. Analysis of case-control association studies with known risk variants. Bioinformatics. 2012;28:1729–1737. doi: 10.1093/bioinformatics/bts259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manichaikul A, Chen WM, Williams K, Wong Q, Sale MM, Pankow JS, et al. Analysis of family- and population-based samples in cohort genome-wide association studies. Hum Genet. 2012;131:275–287. doi: 10.1007/s00439-011-1071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 38.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, et al. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie P, Kranzler HR, Yang C, Zhao H, Farrer LA, Gelernter J. Genome-wide association study identifies new susceptibility loci for posttraumatic stress disorder. Biol Psychiatry. 2013;74:656–663. doi: 10.1016/j.biopsych.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Almli LM, Srivastava A, Fani N, Kerley K, Mercer KB, Feng H, et al. Follow-up and extension of a prior genome-wide association study of posttraumatic stress disorder: gene × environment associations and structural magnetic resonance imaging in a highly traumatized African-American civilian population. Biol Psychiatry. 2014;76:e3–e4. doi: 10.1016/j.biopsych.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MW, Craddock AL, Angelin B, Brunzell JD, Duane WC, Dawson PA. Analysis of the ileal bile acid transporter gene, SLC10A2, in subjects with familial hypertriglyceridemia. Arterioscler Thromb Vasc Biol. 2001;21:2039–2045. doi: 10.1161/hq1201.100262. [DOI] [PubMed] [Google Scholar]
- 44.Karch CM, Goate AM. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77:43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Craddock AL, Love MW, Daniel RW, Kirby LC, Walters HC, Wong MH, et al. Expression and transport properties of the human ileal and renal sodium-dependent bile acid transporter. Am J Physiol. 1998;274:G157–G169. doi: 10.1152/ajpgi.1998.274.1.G157. [DOI] [PubMed] [Google Scholar]
- 46.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chothe PP, Swaan PW. Resveratrol promotes degradation of the human bile acid transporter ASBT (SLC10A2) Biochem J. 2014;459:301–312. doi: 10.1042/BJ20131428. [DOI] [PubMed] [Google Scholar]
- 48.Karuppagounder SS, Pinto JT, Xu H, Chen HL, Beal MF, Gibson GE. Dietary supplementation with resveratrol reduces plaque pathology in a transgenic model of Alzheimer’s disease. Neurochem Int. 2009;54:111–118. doi: 10.1016/j.neuint.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turner RS, Thomas RG, Craft S, van Dyck CH, Mintzer J, Reynolds BA, et al. A randomized, double-blind, placebo-controlled trial of resveratrol for Alzheimer disease. Neurology. 2015;85:1383–1391. doi: 10.1212/WNL.0000000000002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pasinetti GM, Wang J, Ho L, Zhao W, Dubner L. Roles of resveratrol and other grape-derived polyphenols in Alzheimer’s disease prevention and treatment. Biochim Biophys Acta. 2015;1852:1202–1208. doi: 10.1016/j.bbadis.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyashita A, Koike A, Jun G, Wang L-S, Takahashi S, Matsubara E, et al. SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS One. 2013;8:e58618. doi: 10.1371/journal.pone.0058618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11:356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Massoud F, Devi G, Stern Y, Lawton A, Goldman JE, Liu Y, et al. A clinicopathological comparison of community-based and clinic-based cohorts of patients with dementia. Arch Neurol. 1999;56:1368–1373. doi: 10.1001/archneur.56.11.1368. [DOI] [PubMed] [Google Scholar]
- 54.Sabbagh MN, Tyas SL, Emery SC, Hansen LA, Alford MF, Reid RT, et al. Smoking affects the phenotype of Alzheimer disease. Neurology. 2005;64:1301–1303. doi: 10.1212/01.WNL.0000156912.54593.65. [DOI] [PubMed] [Google Scholar]
- 55.Peters R, Poulter R, Warner J, Beckett N, Burch L, Bulpitt C. Smoking, dementia and cognitive decline in the elderly, a systematic review. BMC Geriatr. 2008;8:36. doi: 10.1186/1471-2318-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathieson I, McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat Genet. 2012;44:243–246. doi: 10.1038/ng.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.