Abstract
4-hydroxy-2-nonenal (HNE), a major non-saturated aldehyde product of lipid peroxidation, has been extensively studied as a signaling messenger. In these studies a wide range of HNE concentrations have been used, ranging from the unstressed plasma concentration to far beyond what would be found in actual pathophysiological condition. In addition, accumulating evidence suggest that signaling protein modification by HNE is specific with only those proteins with cysteine, histidine, and lysine residues located in certain sequence or environments adducted by HNE. HNE-signaling is further regulated through the turnover of HNE-signaling protein adducts through proteolytic process that involve proteasomes, lysosomes and autophagy. This review discusses the HNE concentrations and exposure modes used in signaling studies, the selectivity of the HNE-adduction site, and the turnover of signaling protein adducts.
Keywords: HNE, redox signaling, oxidative stress, concentration, adduct, turnover
1. Introduction
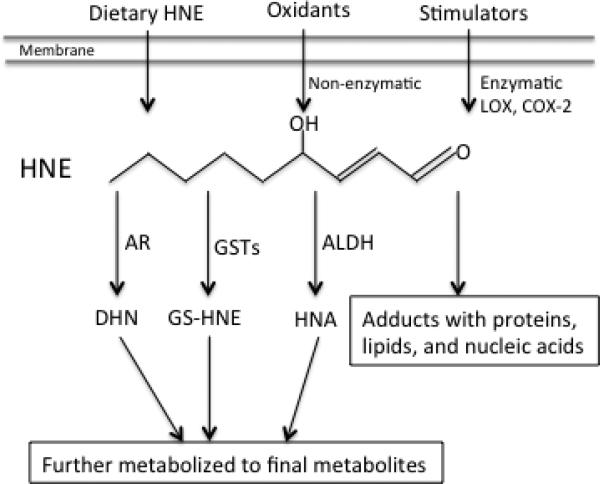
4-hydroxy-2-nonenal (HNE) is a major α, β-unsaturated aldehyde derived from the decomposition of peroxidation products of omega-6 polyunsaturated fatty acids such as arachidonic acid and linoleic acid [1-5]. Besides being produced from the non-enzymatic peroxidation process, which has been well recognized and reviewed [3-6], HNE could also be generated enzymatically by cyclooxygenase-2 and lipoxygenase [7]. In addition, cells/organisms may be exposed to HNE from food. HNE produced from dietary polyunsaturated fatty acids during food processing and storage [8, 9] could result in exposure of cells in the gestrointestinal tract and possibly enter circulation [10] (Fig.1). HNE features two functional groups, its carbonyl (−HC=O) and double bond (C2/C3, −C=C-) groups. This combination in conjugation makes HNE to react readily with bio-molecules including lipids, nucleic acids, and proteins, that can underlie oxidative damage [5]. Therefore, HNE has been widely recognized as both a marker of oxidative stress and the culprit in damage since its discovery in the 1980s [3, 11-13]. Later, numerous studies have clearly shown that at physiological concentration HNE can act as a potent signaling messenger and be involved in the regulation of a variety of signaling pathways [14, 15], cellular processes and functions [16], through forming Michael and/or Schiff base adducts with signaling proteins including receptors, protein kinases, phosphatases, and transcription factors [13, 17].
Figure 1.
HNE formation and pathway of metabolism. LOX, lipoxygenase; COX-2, cyclooxygenase-2; AR, aldo-keto reductase; GSTs, glutathione S-transferases; ALDH, aldehyde dehydrogenase; DHN, 1, 4-dihydroxy-2-nonene; GS-HNE, glutathione-HNE conjugate; HNE HNA, 4-hydroxy-2-nonenoic acid.
Most studies on HNE-induced signaling are conducted in cell models and use a wide range of HNE doses. Some doses are far beyond the pathophysiological concentration of HNE and thus might be irrelevant to what happens in vivo. In addition, although the adduct reaction of HNE with cysteine, histidine, and lysine residues in proteins has been a focus of many studies, the selectivity and turnover of the HNE adducts of signaling proteins have not been summarized. In this review we intend to summarize the concentration of HNE in cells/tissues and in vitro cell models used for cell signaling studies, and discuss the adduction sites of HNE within signaling proteins and the turnover after adduct formation.
2. Tissue concentration of HNE
2.1. Metabolism of HNE is cell/tissue dependent
HNE in the cells is rapidly degraded through several metabolic pathways including conjugation, reduction, and oxidation [18] (Fig.1). Conjugation with GSH, catalyzed by GST alpha isoforms especially GSTA4-4 [19], is the predominant pathway of HNE metabolism [18, 20-22] and responsible for at least 50% of HNE degradation in cells [21, 23, 24]. HNE biotransformation in the other pathways leads to its oxidation to 4-hydroxy-2-nonenoic acid (HNA) by aldehyde dehydrogenase [25, 26], or its reduction to 1, 4-dihydroxy-2-nonene (DHN) by alcohol dehydrogenase [25] and aldo-keto reductase [27]. With reduction, HNE loses its ability to conjugate with proteins [28]. But even though the metabolic removal of HNE is efficient, 2-8% of the HNE in cells appears to form conjugates with proteins [18], and initiates signaling events.
HNE metabolism activities vary among tissues and cell types. Esterbauer et al. reported that rat liver exhibited the highest HNE metabolizing activity among liver, lung, brain, heart, kidney, and intestine. The last had less than 3% of the HNE metabolizing activity of liver, largely due to the lack of alcohol/aldehyde dehydrogenase activity in these tissues [25]. Consistently Zheng et al. also found that lung and brain in rat and mouse showed limited activity of HNE degradation by alcohol/aldehyde dehydrogenase compared to liver [28]. In addition, GST expression level and activity also vary with tissues [28]. Such variation in HNE metabolizing activity in different tissues means that HNE concentration may vary from tissue to tissue in vivo. HNE may have a relatively longer half-life in some tissues and could possibly result in higher and sustained HNE signaling in these tissues. On the other hand, a lower capacity for HNE metabolism may reflect a lower rate of lipid peroxidation and not result in a greater steady state level of free HNE. It is important to note that HNE metabolism may be altered during ontogenesis. For instance, Baradat et al. reported that compared to wild type cells, isogenic colon cells with a mutation on the adenomatous polyposis coli (APC) gene were more efficient in metabolizing HNE, due to a higher expression of HNE metabolizing enzymes [29]. Cancerous cells usually express relatively higher levels of antioxidants including GST [30] and thus potentially metabolize HNE at a faster rate compared to normal cells. The relatively higher HNE metabolism capacity results in a shorter HNE half-life and less HNE toxicity in cancerous cells. On the other hand it suggests that more HNE production may be required to initiate a similar signaling response in cancerous cells. Regardless, the tissue-dependent differences in HNE metabolism may result in variation of HNE levels in different cells/tissues under physiological conditions.
2.2. Tissue concentration
Free HNE remains at very low level in plasma, cells, and tissues under physiological condition. In human plasma it is in the range of 0.28-0.68 μM, similar to that in the plasma of dogs and rats [1, 3, 31]. This persistent existence of free HNE [32] under normal physiological condition may reflect a homeostatic range between its production and metabolism [33]. Plasma HNE may mainly come from tissue cells including vascular endothelial cells and hepatocytes, and blood circulating cells such as lymphocyte and erythrocytes. HNE concentration in rat hepatocytes is in the range of 2.5 μM-3.8 μM, calculated from reports that HNE in rat hepatocytes is 0.86-1.3 nmol/108 cells and that a typical hepatocyte volume is 3.4×10−9 cm3 [34]. HNE in human blood monocytes is 3 times higher than in rat hepatocytes [3]. In other types of cells, HNE concentration is also much higher than its plasma level [5]. Since plasma HNE can reach most tissue cells, it is a fairly good indicator of the exogenous exposure level of cells throughout the body. Due to a tissue-dependent HNE metabolizing capacity as discussed in the above, HNE concentration in different tissues may vary under physiological conditions.
Under conditions of oxidative stress and diseases, HNE level is significantly increased in plasma and tissues [3, 11-13]. In most of these studies, relative level of HNE-protein adduct, instead of free HNE concentration, is usually used as HNE marker, therefore most often only the relative comparison to healthy controls was available. Nonetheless, studies have demonstrated the increase of HNE in diseases and pathologies including Alzheimer disease [35-37], cancer [38], COPD [39], and cardiovascular diseases [40], as summarized in many excellent reviews ([16, 41]).
3. HNE doses used in cell signaling study
A wide range of doses has been used in studying HNE effects on cell signaling pathways. At concentration as low as 0.01 μM HNE was able to reduce endothelial cell junctional communication [42], activate G protein mediated signaling [43], and increase phosphoinositide-specific phospholipase C (PLC) activity and neutrophil migration [44]. On the other end, HNE level as high as 4 mM was used to investigate the effect of HNE on Ca2+-ATPase activity in rat liver plasma membrane [45]. The second highest concentration of HNE in the cell signaling literature was 500 μM, at which it inhibited Ca2+/Mg2+-ATPase activity on erythrocyte membrane [46] and Na+/K+-ATPase activity in rat striatal synaptosomes [47]. Analysis of literatures on HNE-mediated signaling showed that about 17% of studies used 0.1-1 μM of HNE, which is in the physiological range, and that 29% of studies used 1-10 μM of HNE, which is in the range where pathology begins. Another 38% of studies were performed with a pathological level of HNE (10-50 μM). Overall most studies (84%) have used HNE in doses from 0.1-50 μM (Table 1).
Table 1.
Usage of HNE concentration in signaling study
| HNE concentration (μM) | Percentage of studies (%) |
|---|---|
| ≥500 | 1.7 |
| 100<HNE<500 | 1.7 |
| 50<HNE≤100 | 11.7 |
| 10<HNE≤50 | 38.0 |
| 1<HNE≤10 | 29.0 |
| 0.1≤HNE≤1 | 16.8 |
| <0.1 | 1.1 |
Note. HNE concentration was calculated from the amount of HNE added and volume of medium or buffer and was based on 179 studies of HNE effect on signaling molecules/pathways from 1987-2015.
4. Exposure mode of HNE
Exogenous addition of an agent to cultured cells to mimic the in vivo exposure is always a challenge. In a review article Forman discussed the pros and cons of the use in cell model systems of exogenous application of nontoxic concentration of H2O2, a well-recognized second messenger in redox signaling [48]. HNE shares many aspects with H2O2 in terms of production and metabolism in signaling studies. Both are generated in vivo, with a higher loci concentration, and are degraded rapidly, producing a large gradient of concentration. Therefore the challenge of HNE exposure mode in signaling studies is similar as H2O2 -that is to mimic the physiologically relevant intracellular concentration at the loci of target. Indeed, the physiologically relevant target would need to be close to the site of HNE generation in the cell. But, few studies have examined the effect of endogenously generated HNE.
Most HNE signaling studies are performed in cell model systems and a variety of exposure modes have been applied. In this model, cells are usually cultured in medium containing 10% or less fetal bovine serum (FBS), in some cases in FBS free medium or buffers. But in general, HNE is applied to cells through two modes; i.e., bolus one-time addition or repeated addition with intervals for several times. To reach its target signaling proteins, HNE has to escape the scavengers in serum-containing medium, the plasma membrane barrier, and the intracellular degradation system. Bolus-added HNE disappears rapidly in typical 10% FBS cell medium (80% disappears in 30 min) [49, 50], and thus HNE doses much higher than pathophysiologic level were usually used to cause effects in these studies. In other words, HNE reaching substrate-signaling molecules is obviously lower than the initial HNE concentration in the medium.
To mimic a stable HNE level for longer exposure time as is observed in vivo, many studies treat cells with a repeated additions of HNE [50-55]. Such an exposure mode was first used by Barrera et al. to investigate HNE effects on cell differentiation [56]. This group systematically measured and compared HNE concentration in cell medium after bolus or repeated addition of HNE [57]. When 10 μM HNE was added to RPMI medium with 10% FCS, 40% disappeared within 10 min; and after 30 min, HNE concentration in the medium was maintained at 4-6 μM for 1 h, indicating that HNE could be consumed by components in FCS. When 10 μM HNE was added to K562 cell suspension (106 cells/ml) in 10% FCS medium, it disappeared completely in 1h (undetected), with 78% having disappeared in the first 10 min (2.2 μM in medium). On the other hand, when 1 μM of HNE was added every 45 min for 12 times into cell suspension in 10% FCS medium, HNE concentration in the medium remained stable at 1 μM [57]. Similarly, Laurora et al. also measured the HNE concentration in medium with the repeated addition of 1μM HNE to cells in 10% FBS medium every 45 min for 10 times, and found that HNE concentration in the medium could be maintained at around 2 μM [50].
Compared with bolus exposure, the advantages of this exposure mode are obvious. First, a stable HNE level is maintained for a longer period of time, more like the in vivo exposure situation; secondly, potential influence on cellular response resulting from serum-free condition could be avoided; and thirdly, a kinetic response could be detected during the exposure period. In addition, accumulated HNE effects could be studied at lower HNE doses. However, since it takes time to reach a stable HNE level, it is inappropriate to investigate the initial targets and acute effects of HNE. In this case, bolus exposure seems more appropriate.
The biological effects of HNE are closely related to its concentration. At physiological levels, HNE is metabolized efficiently and at low intracellular concentration is maintained. Thus, its biological effects, if any, are barely observed. Under challenging conditions, where its concentration increases, HNE could act as signaling mediator and initiate various signaling cascades and regulate gene expression. At non-lethal but stressful concentrations, HNE induces processes including autophagy, senescence, and cell cycle arrest due to its pathologic modification of proteins and organelles. At lethal concentrations, HNE causes apoptosis or necrosis [58].
5. Selectivity of HNE modification on signaling proteins
HNE mediates cell signaling mainly through forming adducts with signaling protein molecules that results in change of protein activity. Cysteine, histidine, and lysine are the most active amino acids to react with HNE and proteins containing these residues could form Michael and/or Schiff base adducts with HNE [3, 5, 52, 53]. At the beginning Anti-HNE adducts Ab and protein activity assay were used to assess HNE adduct formation and effects, and many HNE targeted proteins were identified with these approaches [59, 60]. The development of mass spectrometry (MS) and proteomic-based approach in the past decade has greatly enhanced the investigation of HNE-protein reaction. With the combination with other technologies such as Click chemistry, MS analysis becomes a power tool to profile HNE targets and spot specific modification sites [54-56].
Protein oxidation, including protein glutathionylation, nitration, and other types of electrophile–protein reactions, exhibits a significant degree of selectivity, which is assumed to be due to protein structure and location [61, 62]. Studies have identified many protein substrates of HNE conjugation, and revealed that HNE-protein reactions do not occur indiscriminately, instead similar to other protein oxidation reactions, exhibit a significant selectivity at several levels.
First, only proteins with residues of cysteine, histidine, or lysine are potential targets, proteins composed of residues other than these are far less likely to form covalent adducts with HNE, as evidenced in studies with model peptides [63] and proteins [64, 65] (Table 2). The covalent modification is mainly through Michael adduction while Schiff base adduction has been less detected.
Table 2.
Modification of proteins by HNEa
| Protein | Adducted Residues | Adducts Type | Reactivity | Detection method | Reference |
|---|---|---|---|---|---|
| Insulin | Two His and one Lys | Michael adduct | His>Lys | HPLC, MS, and amino acid sequencing | [66] |
| GAPDH | Cys, His, and Lys | Michael, Schiff base adducts, and intramolecular and intermolecular crosslink | When [HNE]<0.5 mM, preferentially with Cys and Lys; when [HNE]=2 mM react with all three residues | HPLC | [64] |
| β-lactoglobulin B | Cysteine, histidine, and lysine. | Mainly Michael adducts | Adducts containing from three to nine aldehyde molecules per molecule of protein | Electrospray ionization (ESI) MS | [97] |
| Protein kinas C (PKC) | NA | NA | NA | Anti-HNE adducts Ab | [98] |
| Na (+)-K(+)-ATPase | Cys and Lys | NA | NA | Anti-HNE adducts Ab | [99] |
| Erythrocyte membrane proteins | NA | NA | Mainly with Cys at 0-0.5 mM | Anti-HNE adducts Ab | [100] |
| c-Jun N-terminal kinas (JNK) | NA | NA | NA | Anti-HNE histidine Ab | [49] |
| FR-1 | Cys298 | Michael adduct | NA | ESI-MS | [95] |
| Tau | Lys | NA | NA | Anti-HNE Lysine Ab | [96] |
| Cytochrome c oxidase | NA | NA | NA | Anti HNE-histidine Ab | [101] |
| IκB kinas (Frikke-Schmidt, #312) | NA | NA | NA | Anti-HNE adducts Ab | [102] |
| Bovine cathepsin B | Cys29 and | Michael adducts | NA | Pure protein, tandem MS and Anti-HNE adducts Ab | [103] |
| GAPDH | His-164, Cys-244, Cys-281, His-327, and Lys-331 and revealed | Michael adducts | His-164 and Cys-281 were modified at 5 min, followed by Cys-244 at 15 min and His-327 and Lys-331 at 30 min, Cys-149 modification was not observed | Pure protein, ESI liquid chromatography-mass spectrometry (ESI-LC-MS) | [71] |
| Carnosine | NA | Schiff base | NA | Anti-HNE adducts Ab | [104] |
| Model peptides | Cys, His and Lys are modified by 4HNE; | Michael adducts | Cys>>His>Lys | MALDI-TOF-MS | [63] |
| Alpha 6/C2 subunit of 20s proteasome | NA | NA | NA | MALDI-TOF MS | [105] |
| Actin | Cys374 | NA | Reactivity of Cys374 is due to a significant accessible surface and substantial thiol acidity due to the particular microenvironment | Pure protein LC-ESI-MS/MS | [75] |
| Protein disulfide isomerase (PDI) | Cys | NA | NA | MS | [102] |
| Carnosine | Cys | NA | NA | ESI-MS | [106] |
| Epithelial growth factor receptor (EGFR) | NA | NA | NA | Anti-HNE adducts Ab | [107] |
| Human serum albumin [65] | His-67, His-146, His-242, His-288, His-510, Lys-195, Lys-199, Lys-525 and Cys-34 | 8 Michael Adducts (MA), 3 Schiff Base (SB) | Cys-34 (MA)>Lys-199 (SB)>His-146 (MA) | LC-ESI-MS/MS | [65] |
| Thioredoxin reductase | Cys-496 | NA | NA | MS | [108] |
| Enolase 3b, aldolase and triosephosphate isomerase 1, creatine kinase, carbonic anyhdrase III, aconitase 2, dihydrolipoamide dehydrogenase, and electron transfer flavoprotein-beta | NA | NA | NA | MS | [106] |
| Type II collagen and MMP-13 | NA | Anti-HNE adducts Ab | [109] | ||
| HSA | 10 His and Lys residues | Michael Adducts | H242 > H510 > H67 > H367 > H247 | LC-MS-MS | [70] |
| α-synuclein | His-50 | NA | NA | LC-MS/MS | [110] |
| ADP/ATP translocase 1 | Cys-256 | Michael adducts | NA | MALDI-MS/MS | [111] |
| Extracellular signal-regulated kinas ½ (ERK1/2) | His-178 | NA | NA | LC-MS/MS | [67] |
| Tubulin | Cys-347, Cys-376, and Cys-303 | Tubulin cross-links are Lys-dependent | NA | LC-MS/MS | [112] |
| HSA | Cys-34 | NA | NA | LC-ESI-MS/MS | [113] |
| HSA | Cys-34 and Lys-199 | Cys-34 (MS) and Lys-199 (SB) | NA | LC-ESI-MS/MS | [72] |
| Akt | NA | NA | NA | Anti-HNE adducts Ab | [114] |
| HSP70 and HSP90 | NA | NA | NA | Click chemistry and proteomics | [115] |
| Liver kinas B1 (LKB1) | NA | NA | NA | Anti-HNE adducts Ab | [92] |
| Toll like receptor 4 (TLR4) | Cys | NA | NA | LC-MS/MS | [116] |
| Trx | Cys-73 and Cys-32 | NA | Cys-73 > Cys-32 | NMR | [73] |
| Type II collagen | NA | NA | NA | Anti-HNE adducts Ab | [117] |
| Glutamate cysteine ligase: catalytic (GCLC) and modifier subunit (GCLM) | Cys-553 on GCLC and Cys-35 on GCLM | NA | NA | Pure protein, MALDI-TOF | [118] |
| Human carboxylesterase1 | Lys-105 and Cys-389 | Only Lys-105 adducted | MS | [74] | |
| SIRT3 deacetylase | Cys-280 | NA | NA | MS/MS | [119] |
| AKT2 | His-196, His-267, and Cys-311 of rat Akt2 | Michael Adducts | NA | Anti-HNE adducts Ab and MALDI-TOF | [94] |
| Phosphatase and tensin homolog (PTEN) | NA | Single Michael adduct | NA | Anti-HNE adducts Ab and MALDI-TOF/TOF | [120] |
| EGFR | NA | NA | NA | Anti-HNE adducts Ab | [121] |
| LKB1 | Lys-97 | NA | NA | Anti-HNE adducts Ab | [68] |
| IκBα | NA | NA | NA | Anti-HNE adducts Ab | [122] |
| PKC | NA | NA | NA | Anti-HNE adducts Ab | [123] |
| Liver fatty acid-binding protein (L-FABP) | Lys-57 and Cys-69 on apo and Lys-6, Lys-31, His-43, Lys-46, Lys-57 and Cys-69) on holo protein | NA | NA | Pure protein, MALDI-TOF/TOF MS | [124] |
| Peptidyl-prolyl cis/trans-isomerase A1 (Pin1) | His-157 and Cys-113 | Michael adducts | Cys-113 is the primary | MALDI-TOF/TOF MS | [125] |
| Mitochondrial aconitase (ACO2) | Cys | Michael adducts | The most reactive sites were Cys-358, Cys-421, Cys-424, Cys-99 and Cys-565 | MS | [126] |
| Angiotensin II (Ang II) | NA | NA | NA | Anti-HNE adducts Ab | [127] |
| GRP78 | Lys and His | NA | Marked propensity for Lys and His adduction within the ATPase domain | MS | [128] |
| Lactate dehydrogenase (LDH) | His-68, Cys-164, Cys-186, and Cys-294 | Michael adducts | NA | Purified protein, MS | [129] |
| 5′ AMP protein kinase (AMPK) | Cys-130, Cys-174, Cys-227, and Cys-304 on AMPKα and Cys-225 on AMPKβ | Michael adducts | NA | Pure protein, MS | [130] |
| Protein kinase A | Cys-199 | NA | NA | MS | [131] |
| Src | Cys-248 | NA | NA | LC-MS/MS | [132] |
| 398 proteins | 386 Cys sites and 12 His sites | Michael adducts | NA | MS | [76] |
| Cyclin-dependent kinas 2 (CDK2) | NA | NA | NA | MS | [133] |
| Apoptosis inducing factor (AIFm2) | His-174 | NA | NA | MS | [134] |
Note. NA, not available; MS, mass spectrometry.
Secondly, reactivity of the three amino acids residues with HNE is different. Among them, Cys is the most preferred, and the reactivity follows the order of Cys>His>Lys [63, 65]. However, this is not absolute and in some signaling proteins, His or Lys is preferred other than Cys, such as in signaling proteins of insulin [66], ERK1/2 [67], and serine/threonine kinase liver kinas B1 (LKB1) [68]. It should be noted that controversy exists on the HNE modification sites and reactivity even in the same protein. For example, Aldini et al. investigated the reactivity of the nine residues of Cys, His, and Lys in human serum albumin [69] with HNE and found it followed an order of Cys-34 (Michael adduction) >Lys-199 (Schiff's base adduction)>His-146 (Michael adduction) [65], while Szapacs et al. reported that the reactivity of these residues followed the order of His-242 > His-510 > His-67 > His-367 > His-247 [70]. Both studies used similar condition (recombinant pure HSA and similar reaction ratio of HSA: HNE) and mass spectrometry analysis. It seems that the selectivity difference is related with experiment conditions such as reaction concentration and time. Uchida et al. reported that HNE preferentially reacted with residues of Cys and Lys in GAPDH protein at concentration of less than 0.5 mM, while it reacted with all three residues at concentration of 2 mM [64]. Ishii et al. on the other hand, reported that the reaction of Cys, His, and Lys in GAPDH were time dependent. Both His-164 and Cys-281 were very rapidly modified at 5 min, followed by Cys-244 at 15 min and His-327 and Lys-331 at 30 min, while the modification of Cys-149 at the catalytic center was not observed [71]. These controversies in reactivity and modification sites suggest that more studies are required to further elucidate the HNE alkylation sites of specific proteins, especially using models with similar condition as in vivo exposure, since most previous data were based on isolated pure protein and used HNE concentration that was not physiologically relevant. Another aspect about selectivity of HNE modification is that specific sequence motifs in proteins or secondary structure may be required for reaction of residues of Cys, His, or Lys with HNE. As observed in HSA [65, 72], Trx[73], human carboxylesterase1 [74] and other proteins that contain several residues of Cys, His, and Lys, only certain residues were able to adduct with HNE, and even these reactive residues exhibited different reactivity. The underlying mechanism of this selectivity remains largely unknown. Using computational modeling analysis, Aldini et al. showed that the reason why Cys374 of actin was the preferred site of HNE adduction was because of its significant accessible surface and substantial thiol acidity due to its particular microenvironment surrounding [75]. Szapacs et al. investigated the reactivity of HNE adduct residues and motif structure of HSA and found that the rate constants of His residues ranged over 4 orders of magnitude with the order of reactivity being His-242 > His-510 > His-67 > His-367 > His-247. The most reactive site H242 was located in a fatty acid- and drug-binding cavity of HSA. Further analysis of adduction kinetics together with HSA structure and pK(a) values of target residues suggested that location in the hydrophobic binding cavity and low predicted pK(a) of His-242 could account for its high reactivity toward HNE [70]. The relation between reactivity (selectivity) and motif sequence is further supported by a study from Doorn et al. [63], in which addition of a methionine to peptides significantly increased the reactivity of HNE reactive residues contained in them. Recently Yang et al. developed a chemoproteomics platform employing a novel, isotope-labeled Az-UV-biotin reagent and analyzed HNE alkylation sites on cysteine and histidine residues in about 400 proteins and revealed a characteristic sequence motif of CxxxK for HNE S-alkylation [76]. This powerful tool has a potential to expand the inventory of HNE modification sites of signaling proteins in complex biological samples. It is important for characterizing the interactions of HNE with redox sensitive cell signaling proteins and understanding how it may modulate their activities under either physiologic or disease conditions. With the combination of computation-based structure analysis, this technology would greatly further the understanding of the site selection of HNE covalent adduction in signaling proteins.
Summarizing this section, it is important to understand that while the rate of adduct formation is much greater for Cys than the other two amino acids, the thiolate form of Cys (S−) is by far a better nucleophile than the thiol (SH) form. Nonetheless Cys, and particularly its thiolate form, are in lower abundance than either His or Lys in proteins. Furthermore, even with its far greater rate constant, Cys in its thiolate form is usually less accessible to adduction than are Lys or His that would tend to be at the protein/solvent interface.
HNE concentration would also affect which amino acids are modified. Aside from the accessibility issue for the Cys thiolate, at low concentrations of HNE, its modification would be greatly favored. When the concentration of HNE is high however, Cys adduction is still the favored reaction kinetically, but the likelihood of adduction to HNE with His and Lys increase. The bottom line is that while kinetics would largely favor Cys modification, geometry and relative abundance limit the modification of Cys, and that higher concentration of HNE makes Lys and His modification increase. Thus, it is important to determine the modifications for each individual target protein at realistic concentrations of HNE.
6. Stability and turnover of HNE-adducted signaling proteins
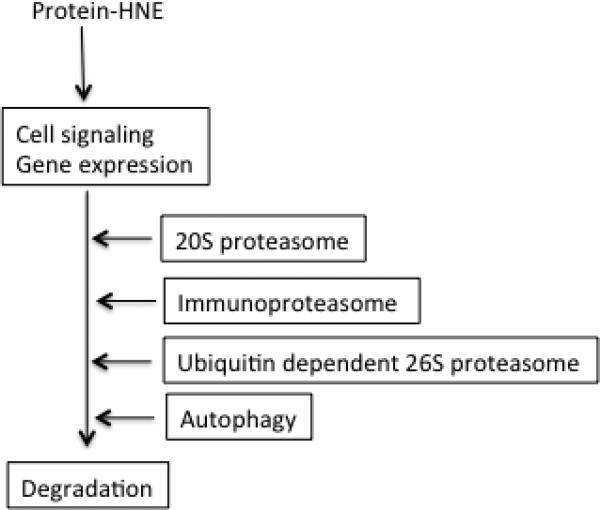
Protein modification by HNE is often associated with conformation change and loss of normal function. The accumulation of these non-native proteins could lead to harmful effects and is associated with pathologic changes. Therefore, HNE-adducted proteins are labeled as abnormal by cell protein quality control system, and are either repaired, removed, or accumulate as aggregated proteins in cells. Although the underlying mechanism of how cells discriminate between native and oxidized form of proteins including HNE-adducts remains to be further elucidated, a growing body of evidence suggests that oxidation of amino acid residues could cause protein unfolding and exposure of hydrophobic regions, which are normally buried in native form. The exposed hydrophobic patches could serve as signals for molecular chaperones and proteolytic system, and result in the refolding or degradation of target proteins [77]. A majority of studies indicate that oxidized proteins are mainly degraded by 20S proteasome [78-80], while there are also evidence suggest that ubiquitin-dependent 26S proteasome, immunoproteasome [81, 82] and lysosome [77] are also involved in the degradation of oxidized proteins. Accumulating evidence suggests that autophagy may play a significant role in the degradation of heavily oxidized and aggregated proteins, which are poor substrate of proteasome system [83, 84]. It is generally considered that like other types of protein oxidation, HNE-modified proteins are degraded through proteasomes especially 20S proteasome. In addition, lysosome [85] and autophagy may also play a key role in the degradation and recycling of HNE-protein adducts [86, 87] (Fig.2). HNE-mediated signaling is usually transient and turned on and off in a time-dependent manner, and this is obviously related to the removal of bolus-added HNE and turnover of the signaling proteins initially adducted by HNE. Studies suggest that the appearance of HNE-signaling protein adducts is substrate dependent, some occur within minutes while others occur at as late as several hours after HNE exposure. For instance, IκB-HNE adduct level in response to uniaxial cyclic stretch reached the highest level at 2 min [88] and HNE inhibited protein tyrosine phosphatase 1B (PTP1B ) activity in 5 min [89], while platelet derived growth factor (PDGF)-HNE adduct was detected only after 2 h of HNE exposure [81]. This time difference in the occurrence of HNE-adducts may be related to the local HNE concentration. Rinna et al. investigated the response of SHP1 activity to different HNE concentrations and found that at 15μM, HNE inhibited SHP1 as early as in 5 min but at 5 μM it needed 15 min to reach the same degree of SHP1 inhibition. In addition, reactivity and the location of the protein substrates may also play important roles in this time-dependent difference in adduct occurrence. To clarify the exact mechanism however, further studies are needed.
Figure 2.
Turnover of HNE-modified signaling molecules.
The stability or the turnover rate of HNE-signaling protein adducts also exhibit variation among target proteins. After formation, IκB-HNE adduct returned to basal level in 2 minutes [88], while HNE adducts with epithelial growth factor (EGFR) and insulin receptor substrate (IRS) remained detectable for at least 5 h [90, 91]. The presence of HNE-protein adduct however, seems to be due to accumulation other than resistance to degradation, as evidenced by both Dolinsky et al. [92] and Ma et al. [93] who observed that LKB1-HNE adduct level was higher in 1h of HNE exposure, but the total LKB1 protein was decreased. Consistently, Demozay et al. also found that even though the increase of HNE-IRS adducts level lasted for at least 6 h, the total IRS protein was significantly decreased [91]. However, it remains unclear whether other mechanisms such as increased secretion or inhibited translation are involved in the decrease in total proteins in the above cases. In contrast, Shearn et al. reported that AKT2-HNE level increased by 40 times in 2 h, and the total AKT2 protein did not change [94]. In summary, these limited data suggest that the turnover of HNE-signaling protein adducts may be substrate dependent. This point is supported by a recent study by Yang et al., who investigated the turnover of 398 HNE-protein adducts using a mass spectrometry-based quantitative chemoproteomics platform and found that the adduct turnover rates varied in a site-specific manner [76]. In contrast, studies found that HNE-modified proteins usually decreased rapidly after removing HNE from medium and then remained at a stable level for longer time [85, 95]. For instance, Liu et al. reported that overall HNE protein adducts level decreased by 60% in 2 min of HNE exposure and then remained stable for at least 1h [87]. It remains unclear if proteasome was responsible for the rapid degradation of HNE-adducts, as it is hard to explain why the left 40% could be stable for 1 h. It is worthy to note that most HNE signaling studies are performed in cell models, which usually means short observation period and transient HNE exposure, compared to in vivo situation where HNE exposure persists and a possible balance between adduct formation and turnover could be reached. Further investigation of HNE-adduct turnover under these conditions would further our understanding of the mechanism of HNE-mediated signaling.
7. Conclusion and future studies
As a potent signaling mediator, HNE plays important roles in maintaining cellular homeostasis and in oxidative stress-implicated pathological changes [12, 96]. Most studies on HNE signaling in cultured cells were performed with HNE concentrations in the pathophysiologic range and whether HNE contributes to signaling effects at physiological concentration in vivo remains largely unknown. In addition, future studies on HNE signaling should consider the exposure mode; i.e., bolus-addition and repeat exposure, which is closely related to the effective HNE doses and reaction time with targets.
Selectivity of HNE adduction reaction is an important topic in the research of HNE signaling, and is relatively less studied. Most previous studies on HNE target sites and selectivity assay were conducted on recombinant protein instead of living cells, in which different HNE concentration, protein structure, and microenvironment may exist. More studies with in vivo models, and with mass spectrometry based technologies [76], are needed before a clear and general rule in the selectivity of HNE target can be drawn
Highlight.
A wide range of HNE concentrations have been used in studies of HNE signaling
Bolus and repeated addition of HNE are useful, but do not exactly mimic physiologic exposure
HNE conjugates with and modifies signaling proteins selectively
HNE-signaling protein adducts are subsequently degraded
Acknowledgement
This work was supported by NIH grant ES023864.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Niki E. Lipid peroxidation: physiological levels and dual biological effects. Free Radic Biol Med. 2009;47(5):469–84. doi: 10.1016/j.freeradbiomed.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 2.Semchyshyn HM. Reactive carbonyl species in vivo: generation and dual biological effects. ScientificWorldJournal. 2014;2014:417842. doi: 10.1155/2014/417842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11(1):81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 4.Spickett CM. The lipid peroxidation product 4-hydroxy-2-nonenal: Advances in chemistry and analysis. Redox Biol. 2013;1:145–52. doi: 10.1016/j.redox.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaur RJ, Siems W, Bresgen N, Eckl PM. 4-Hydroxy-nonenal-A Bioactive Lipid Peroxidation Product. Biomolecules. 2015;5(4):2247–337. doi: 10.3390/biom5042247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider C, Tallman KA, Porter NA, Brash AR. Two distinct pathways of formation of 4-hydroxynonenal. Mechanisms of nonenzymatic transformation of the 9 and 13-hydroperoxides of linoleic acid to 4-hydroxyalkenals. J Biol Chem. 2001;276(24):20831–8. doi: 10.1074/jbc.M101821200. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Allen TD, Yang Y, Moore DR, Huycke MM. Cyclooxygenase-2 generates the endogenous mutagen trans-4-hydroxy-2-nonenal in Enterococcus faecalis-infected macrophages. Cancer Prev Res (Phila) 2013;6(3):206–16. doi: 10.1158/1940-6207.CAPR-12-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goicoechea E, Van Twillert K, Duits M, Brandon ED, Kootstra PR, Blokland MH, Guillen MD. Use of an in vitro digestion model to study the bioaccessibility of 4-hydroxy-2-nonenal and related aldehydes present in oxidized oils rich in omega-6 acyl groups. J Agric Food Chem. 2008;56(18):8475–83. doi: 10.1021/jf801212k. [DOI] [PubMed] [Google Scholar]
- 9.Guillen MD, Goicoechea E. Toxic oxygenated alpha,beta-unsaturated aldehydes and their study in foods: a review. Crit Rev Food Sci Nutr. 2008;48(2):119–36. doi: 10.1080/10408390601177613. [DOI] [PubMed] [Google Scholar]
- 10.Beretta G, Furlanetto S, Regazzoni L, Zarrella M, Facino RM. Quenching of alpha,beta-unsaturated aldehydes by green tea polyphenols: HPLC-ESI-MS/MS studies. J Pharm Biomed Anal. 2008;48(3):606–11. doi: 10.1016/j.jpba.2008.05.036. [DOI] [PubMed] [Google Scholar]
- 11.Schaur RJ. Basic aspects of the biochemical reactivity of 4-hydroxynonenal. Molecular aspects of medicine. 2003;24(4-5):149–59. doi: 10.1016/s0098-2997(03)00009-8. [DOI] [PubMed] [Google Scholar]
- 12.Zarkovic N, Cipak A, Jaganjac M, Borovic S, Zarkovic K. Pathophysiological relevance of aldehydic protein modifications. J Proteomics. 2013;92:239–47. doi: 10.1016/j.jprot.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Poli G, Schaur RJ, Siems WG, Leonarduzzi G. 4-hydroxynonenal: a membrane lipid oxidation product of medicinal interest. Medicinal research reviews. 2008;28(4):569–631. doi: 10.1002/med.20117. [DOI] [PubMed] [Google Scholar]
- 14.Forman HJ, Fukuto JM, Miller T, Zhang H, Rinna A, Levy S. The chemistry of cell signaling by reactive oxygen and nitrogen species and 4-hydroxynonenal. Archives of biochemistry and biophysics. 2008;477(2):183–95. doi: 10.1016/j.abb.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaganjac M, Cacev T, Cipak A, Kapitanovic S, Gall Troselj K, Zarkovic N. Even stressed cells are individuals: second messengers of free radicals in pathophysiology of cancer. Croat Med J. 2012;53(4):304–9. doi: 10.3325/cmj.2012.53.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoeb M, Ansari NH, Srivastava SK, Ramana KV. 4-Hydroxynonenal in the pathogenesis and progression of human diseases. Curr Med Chem. 2014;21(2):230–7. doi: 10.2174/09298673113209990181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ullery JC, Marnett LJ. Protein modification by oxidized phospholipids and hydrolytically released lipid electrophiles: Investigating cellular responses. Biochim Biophys Acta. 2012;1818(10):2424–35. doi: 10.1016/j.bbamem.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siems W, Grune T. Intracellular metabolism of 4-hydroxynonenal. Molecular aspects of medicine. 2003;24(4-5):167–75. doi: 10.1016/s0098-2997(03)00011-6. [DOI] [PubMed] [Google Scholar]
- 19.Balogh LM, Le Trong I, Kripps KA, Shireman LM, Stenkamp RE, Zhang W, Mannervik B, Atkins WM. Substrate specificity combined with stereopromiscuity in glutathione transferase A4-4-dependent metabolism of 4-hydroxynonenal. Biochemistry. 2010;49(7):1541–8. doi: 10.1021/bi902038u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spitz DR, Sullivan SJ, Malcolm RR, Roberts RJ. Glutathione dependent metabolism and detoxification of 4-hydroxy-2-nonenal. Free Radic Biol Med. 1991;11(4):415–23. doi: 10.1016/0891-5849(91)90159-z. [DOI] [PubMed] [Google Scholar]
- 21.Tjalkens RB, Cook LW, Petersen DR. Formation and export of the glutathione conjugate of 4-hydroxy-2, 3-E-nonenal (4-HNE) in hepatoma cells. Archives of biochemistry and biophysics. 1999;361(1):113–9. doi: 10.1006/abbi.1998.0946. [DOI] [PubMed] [Google Scholar]
- 22.Alary J, Gueraud F, Cravedi JP. Fate of 4-hydroxynonenal in vivo: disposition and metabolic pathways. Molecular aspects of medicine. 2003;24(4-5):177–87. doi: 10.1016/s0098-2997(03)00012-8. [DOI] [PubMed] [Google Scholar]
- 23.Srivastava S, Dixit BL, Cai J, Sharma S, Hurst HE, Bhatnagar A, Srivastava SK. Metabolism of lipid peroxidation product, 4-hydroxynonenal (HNE) in rat erythrocytes: role of aldose reductase. Free Radic Biol Med. 2000;29(7):642–51. doi: 10.1016/s0891-5849(00)00351-8. [DOI] [PubMed] [Google Scholar]
- 24.Hartley DP, Ruth JA, Petersen DR. The hepatocellular metabolism of 4-hydroxynonenal by alcohol dehydrogenase, aldehyde dehydrogenase, and glutathione S-transferase. Archives of biochemistry and biophysics. 1995;316(1):197–205. doi: 10.1006/abbi.1995.1028. [DOI] [PubMed] [Google Scholar]
- 25.Esterbauer H, Zollner H, Lang J. Metabolism of the lipid peroxidation product 4-hydroxynonenal by isolated hepatocytes and by liver cytosolic fractions. Biochem J. 1985;228(2):363–73. doi: 10.1042/bj2280363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell DY, Petersen DR. The oxidation of alpha-beta unsaturated aldehydic products of lipid peroxidation by rat liver aldehyde dehydrogenases. Toxicol Appl Pharmacol. 1987;87(3):403–10. doi: 10.1016/0041-008x(87)90245-6. [DOI] [PubMed] [Google Scholar]
- 27.Martin HJ, Maser E. Role of human aldo-keto-reductase AKR1B10 in the protection against toxic aldehydes. Chem Biol Interact. 2009;178(1-3):145–50. doi: 10.1016/j.cbi.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 28.Zheng R, Dragomir AC, Mishin V, Richardson JR, Heck DE, Laskin DL, Laskin JD. Differential metabolism of 4-hydroxynonenal in liver, lung and brain of mice and rats. Toxicol Appl Pharmacol. 2014;279(1):43–52. doi: 10.1016/j.taap.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baradat M, Jouanin I, Dalleau S, Tache S, Gieules M, Debrauwer L, Canlet C, Huc L, Dupuy J, Pierre FH, Gueraud F. 4-Hydroxy-2(E)-nonenal metabolism differs in Apc(+/+) cells and in Apc(Min/+) cells: it may explain colon cancer promotion by heme iron. Chem Res Toxicol. 2011;24(11):1984–93. doi: 10.1021/tx2003036. [DOI] [PubMed] [Google Scholar]
- 30.Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30(6):445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 31.Spies-Martin D, Sommerburg O, Langhans CD, Leichsenring M. Measurement of 4-hydroxynonenal in small volume blood plasma samples: modification of a gas chromatographic-mass spectrometric method for clinical settings. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;774(2):231–9. doi: 10.1016/s1570-0232(02)00242-8. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida Y, Saito Y, Hayakawa M, Habuchi Y, Imai Y, Sawai Y, Niki E. Levels of lipid peroxidation in human plasma and erythrocytes: comparison between fatty acids and cholesterol. Lipids. 2007;42(5):439–49. doi: 10.1007/s11745-007-3037-5. [DOI] [PubMed] [Google Scholar]
- 33.Ursini F, Maiorino M, Forman HJ. Redox homeostasis: The Golden Mean of healthy living. Redox Biol. 2016;8:205–15. doi: 10.1016/j.redox.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell JE. Molecular Cell Biology. Fifth Edition W. H. Freeman and Company; New York: 2000. [Google Scholar]
- 35.Lovell MA, Ehmann WD, Mattson MP, Markesbery WR. Elevated 4-hydroxynonenal in ventricular fluid in Alzheimer's disease. Neurobiology of aging. 1997;18(5):457–61. doi: 10.1016/s0197-4580(97)00108-5. [DOI] [PubMed] [Google Scholar]
- 36.Fukuda M, Kanou F, Shimada N, Sawabe M, Saito Y, Murayama S, Hashimoto M, Maruyama N, Ishigami A. Elevated levels of 4-hydroxynonenal-histidine Michael adduct in the hippocampi of patients with Alzheimer's disease. Biomed Res. 2009;30(4):227–33. doi: 10.2220/biomedres.30.227. [DOI] [PubMed] [Google Scholar]
- 37.Bradley MA, Markesbery WR, Lovell MA. Increased levels of 4-hydroxynonenal and acrolein in the brain in preclinical Alzheimer disease. Free Radic Biol Med. 2010;48(12):1570–6. doi: 10.1016/j.freeradbiomed.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong H, Yin H. Role of lipid peroxidation derived 4-hydroxynonenal (4-HNE) in cancer: focusing on mitochondria. Redox Biol. 2015;4:193–9. doi: 10.1016/j.redox.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H, Xu JY. [The role of 4-hydroxynonenal in assessment of chronic obstructive pulmonary disease severity] Zhonghua Jie He He Hu Xi Za Zhi. 2012;35(10):758–61. [PubMed] [Google Scholar]
- 40.Leonarduzzi G, Chiarpotto E, Biasi F, Poli G. 4-Hydroxynonenal and cholesterol oxidation products in atherosclerosis. Mol Nutr Food Res. 2005;49(11):1044–9. doi: 10.1002/mnfr.200500090. [DOI] [PubMed] [Google Scholar]
- 41.Csala M, Kardon T, Legeza B, Lizak B, Mandl J, Margittai E, Puskas F, Szaraz P, Szelenyi P, Banhegyi G. On the role of 4-hydroxynonenal in health and disease. Biochim Biophys Acta. 2015;1852(5):826–38. doi: 10.1016/j.bbadis.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 42.Radu A, Moldovan N. 4-Hydroxynonenal reduces junctional communication between endothelial cells in culture. Exp Cell Res. 1991;196(1):121–6. doi: 10.1016/0014-4827(91)90463-5. [DOI] [PubMed] [Google Scholar]
- 43.Rossi MA, Di Mauro C, Dianzani MU. Experimental studies on the mechanism of phospholipase C activation by the lipid peroxidation products 4-hydroxynonenal and 2-nonenal. Int J Tissue React. 2001;23(2):45–50. [PubMed] [Google Scholar]
- 44.Rossi MA, Fidale F, Di Mauro C, Esterbauer H, Dianzani MU. Effect of 4-hydroxy-2,3-trans-nonenal and related aldehydes on phospholipase C activity of rat neutrophils. Int J Tissue React. 1993;15(5):201–5. [PubMed] [Google Scholar]
- 45.Parola M, Albano E, Autelli R, Barrera G, Biocca ME, Paradisi L, Dianzani MU. Inhibition of the high affinity Ca2(+)-ATPase activity in rat liver plasma membranes following carbon tetrachloride intoxication. Chem Biol Interact. 1990;73(1):103–19. doi: 10.1016/0009-2797(90)90111-y. [DOI] [PubMed] [Google Scholar]
- 46.Raess BU, Keenan CE, McConnell EJ. Effects of 4-OH-2,3-trans-nonenal on human erythrocyte plasma membrane Ca2+ pump and passive Ca2+ permeability. Biochemical and biophysical research communications. 1997;235(3):451–4. doi: 10.1006/bbrc.1997.6819. [DOI] [PubMed] [Google Scholar]
- 47.Morel P, Tallineau C, Pontcharraud R, Piriou A, Huguet F. Effects of 4-hydroxynonenal, a lipid peroxidation product, on dopamine transport and Na+/K+ ATPase in rat striatal synaptosomes. Neurochem Int. 1998;33(6):531–40. doi: 10.1016/s0197-0186(98)00062-x. [DOI] [PubMed] [Google Scholar]
- 48.Forman HJ. Use and abuse of exogenous H2O2 in studies of signal transduction. Free Radic Biol Med. 2007;42(7):926–32. doi: 10.1016/j.freeradbiomed.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parola M, Robino G, Marra F, Pinzani M, Bellomo G, Leonarduzzi G, Chiarugi P, Camandola S, Poli G, Waeg G, Gentilini P, Dianzani MU. HNE interacts directly with JNK isoforms in human hepatic stellate cells. The Journal of clinical investigation. 1998;102(11):1942–50. doi: 10.1172/JCI1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laurora S, Tamagno E, Briatore F, Bardini P, Pizzimenti S, Toaldo C, Reffo P, Costelli P, Dianzani MU, Danni O, Barrera G. 4-Hydroxynonenal modulation of p53 family gene expression in the SK-N-BE neuroblastoma cell line. Free Radic Biol Med. 2005;38(2):215–25. doi: 10.1016/j.freeradbiomed.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 51.Rinaldi M, Barrera G, Aquino A, Spinsanti P, Pizzimenti S, Farace MG, Dianzani MU, Fazio VM. 4-Hydroxynonenal-induced MEL cell differentiation involves PKC activity translocation. Biochemical and biophysical research communications. 2000;272(1):75–80. doi: 10.1006/bbrc.2000.2691. [DOI] [PubMed] [Google Scholar]
- 52.Pizzimenti S, Briatore F, Laurora S, Toaldo C, Maggio M, De Grandi M, Meaglia L, Menegatti E, Giglioni B, Dianzani MU, Barrera G. 4-Hydroxynonenal inhibits telomerase activity and hTERT expression in human leukemic cell lines. Free Radic Biol Med. 2006;40(9):1578–91. doi: 10.1016/j.freeradbiomed.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 53.Cerbone A, Toaldo C, Laurora S, Briatore F, Pizzimenti S, Dianzani MU, Ferretti C, Barrera G. 4-Hydroxynonenal and PPARgamma ligands affect proliferation, differentiation, and apoptosis in colon cancer cells. Free Radic Biol Med. 2007;42(11):1661–70. doi: 10.1016/j.freeradbiomed.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 54.Zhang H, Court N, Forman HJ. Submicromolar concentrations of 4-hydroxynonenal induce glutamate cysteine ligase expression in HBE1 cells. Redox report : communications in free radical research. 2007;12(1):101–6. doi: 10.1179/135100007X162266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pizzimenti S, Menegatti E, Berardi D, Toaldo C, Pettazzoni P, Minelli R, Giglioni B, Cerbone A, Dianzani MU, Ferretti C, Barrera G. 4-hydroxynonenal, a lipid peroxidation product of dietary polyunsaturated fatty acids, has anticarcinogenic properties in colon carcinoma cell lines through the inhibition of telomerase activity. J Nutr Biochem. 2010;21(9):818–26. doi: 10.1016/j.jnutbio.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 56.Barrera G, Di Mauro C, Muraca R, Ferrero D, Cavalli G, Fazio VM, Paradisi L, Dianzani MU. Induction of differentiation in human HL-60 cells by 4-hydroxynonenal, a product of lipid peroxidation. Exp Cell Res. 1991;197(2):148–52. doi: 10.1016/0014-4827(91)90416-r. [DOI] [PubMed] [Google Scholar]
- 57.Barrera FBG, Fazio VM, Paradisi L, Dianzani MU. Repeated Treatments with a Low HNE Concentration Affect K562 Cell Proliferation. Springer; US, New York: 1991. [Google Scholar]
- 58.Ayala A, Munoz MF, Arguelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev. 2014;2014:360438. doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Progress in lipid research. 2003;42(4):318–43. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 60.Petersen DR, Doorn JA. Reactions of 4-hydroxynonenal with proteins and cellular targets. Free Radic Biol Med. 2004;37(7):937–45. doi: 10.1016/j.freeradbiomed.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 61.Dalle-Donne I, Aldini G, Carini M, Colombo R, Rossi R, Milzani A. Protein carbonylation, cellular dysfunction, and disease progression. J Cell Mol Med. 2006;10(2):389–406. doi: 10.1111/j.1582-4934.2006.tb00407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.LoPachin RM, Gavin T. Molecular mechanisms of aldehyde toxicity: a chemical perspective. Chem Res Toxicol. 2014;27(7):1081–91. doi: 10.1021/tx5001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doorn JA, Petersen DR. Covalent adduction of nucleophilic amino acids by 4-hydroxynonenal and 4-oxononenal. Chem Biol Interact. 2003;143-144:93–100. doi: 10.1016/s0009-2797(02)00178-3. [DOI] [PubMed] [Google Scholar]
- 64.Uchida K, Stadtman ER. Covalent attachment of 4-hydroxynonenal to glyceraldehyde-3-phosphate dehydrogenase. A possible involvement of intra-and intermolecular cross-linking reaction. J Biol Chem. 1993;268(9):6388–93. [PubMed] [Google Scholar]
- 65.Aldini G, Gamberoni L, Orioli M, Beretta G, Regazzoni L, Maffei Facino R, Carini M. Mass spectrometric characterization of covalent modification of human serum albumin by 4-hydroxy-trans-2-nonenal. J Mass Spectrom. 2006;41(9):1149–61. doi: 10.1002/jms.1067. [DOI] [PubMed] [Google Scholar]
- 66.Uchida K, Stadtman ER. Modification of histidine residues in proteins by reaction with 4-hydroxynonenal. Proc Natl Acad Sci U S A. 1992;89(10):4544–8. doi: 10.1073/pnas.89.10.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sampey BP, Carbone DL, Doorn JA, Drechsel DA, Petersen DR. 4-Hydroxy-2-nonenal adduction of extracellular signal-regulated kinase (Erk) and the inhibition of hepatocyte Erk-Est-like protein-1-activating protein-1 signal transduction. Mol Pharmacol. 2007;71(3):871–83. doi: 10.1124/mol.106.029686. [DOI] [PubMed] [Google Scholar]
- 68.Calamaras TD, Lee C, Lan F, Ido Y, Siwik DA, Colucci WS. Post-translational modification of serine/threonine kinase LKB1 via Adduction of the Reactive Lipid Species 4-Hydroxy-trans-2-nonenal (HNE) at lysine residue 97 directly inhibits kinase activity. J Biol Chem. 2012;287(50):42400–6. doi: 10.1074/jbc.M112.385831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ohsawa I, Nishimaki K, Murakami Y, Suzuki Y, Ishikawa M, Ohta S. Age-dependent neurodegeneration accompanying memory loss in transgenic mice defective in mitochondrial aldehyde dehydrogenase 2 activity. J Neurosci. 2008;28(24):6239–49. doi: 10.1523/JNEUROSCI.4956-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Szapacs ME, Riggins JN, Zimmerman LJ, Liebler DC. Covalent adduction of human serum albumin by 4-hydroxy-2-nonenal: kinetic analysis of competing alkylation reactions. Biochemistry. 2006;45(35):10521–8. doi: 10.1021/bi060535q. [DOI] [PubMed] [Google Scholar]
- 71.Ishii T, Tatsuda E, Kumazawa S, Nakayama T, Uchida K. Molecular basis of enzyme inactivation by an endogenous electrophile 4-hydroxy-2-nonenal: identification of modification sites in glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 2003;42(12):3474–80. doi: 10.1021/bi027172o. [DOI] [PubMed] [Google Scholar]
- 72.Aldini G, Vistoli G, Regazzoni L, Gamberoni L, Facino RM, Yamaguchi S, Uchida K, Carini M. Albumin is the main nucleophilic target of human plasma: a protective role against pro-atherogenic electrophilic reactive carbonyl species? Chem Res Toxicol. 2008;21(4):824–35. doi: 10.1021/tx700349r. [DOI] [PubMed] [Google Scholar]
- 73.Wakita C, Maeshima T, Yamazaki A, Shibata T, Ito S, Akagawa M, Ojika M, Yodoi J, Uchida K. Stereochemical configuration of 4-hydroxy-2-nonenal-cysteine adducts and their stereoselective formation in a redox-regulated protein. J Biol Chem. 2009;284(42):28810–22. doi: 10.1074/jbc.M109.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Borazjani A, Edelmann MJ, Hardin KL, Herring KL, Allen Crow J, Ross MK. Catabolism of 4-hydroxy-2-trans-nonenal by THP1 monocytes/macrophages and inactivation of carboxylesterases by this lipid electrophile. Chem Biol Interact. 2011;194(1):1–12. doi: 10.1016/j.cbi.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aldini G, Dalle-Donne I, Vistoli G, Maffei Facino R, Carini M. Covalent modification of actin by 4-hydroxy-trans-2-nonenal (HNE): LC-ESI-MS/MS evidence for Cys374 Michael adduction. J Mass Spectrom. 2005;40(7):946–54. doi: 10.1002/jms.872. [DOI] [PubMed] [Google Scholar]
- 76.Yang J, Tallman KA, Porter NA, Liebler DC. Quantitative chemoproteomics for site-specific analysis of protein alkylation by 4-hydroxy-2-nonenal in cells. Anal Chem. 2015;87(5):2535–41. doi: 10.1021/ac504685y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shang F, Taylor A. Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free Radic Biol Med. 2011;51(1):5–16. doi: 10.1016/j.freeradbiomed.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grune T, Reinheckel T, Davies KJ. Degradation of oxidized proteins in mammalian cells. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1997;11(7):526–34. [PubMed] [Google Scholar]
- 79.Ferrington DA, Sun H, Murray KK, Costa J, Williams TD, Bigelow DJ, Squier TC. Selective degradation of oxidized calmodulin by the 20 S proteasome. J Biol Chem. 2001;276(2):937–43. doi: 10.1074/jbc.M005356200. [DOI] [PubMed] [Google Scholar]
- 80.Grune T, Davies KJ. The proteasomal system and HNE-modified proteins. Molecular aspects of medicine. 2003;24(4-5):195–204. doi: 10.1016/s0098-2997(03)00014-1. [DOI] [PubMed] [Google Scholar]
- 81.Pickering AM, Koop AL, Teoh CY, Ermak G, Grune T, Davies KJ. The immunoproteasome, the 20S proteasome and the PA28alphabeta proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem J. 2010;432(3):585–94. doi: 10.1042/BJ20100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raynes R, Pomatto LC, Davies KJ. Degradation of oxidized proteins by the proteasome: Distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Molecular aspects of medicine. 2016 doi: 10.1016/j.mam.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dunlop RA, Brunk UT, Rodgers KJ. Oxidized proteins: mechanisms of removal and consequences of accumulation. IUBMB Life. 2009;61(5):522–7. doi: 10.1002/iub.189. [DOI] [PubMed] [Google Scholar]
- 84.Pajares M, Jimenez-Moreno N, Dias IH, Debelec B, Vucetic M, Fladmark KE, Basaga H, Ribaric S, Milisav I, Cuadrado A. Redox control of protein degradation. Redox Biol. 2015;6:409–20. doi: 10.1016/j.redox.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marques C, Pereira P, Taylor A, Liang JN, Reddy VN, Szweda LI, Shang F. Ubiquitin-dependent lysosomal degradation of the HNE-modified proteins in lens epithelial cells. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2004;18(12):1424–6. doi: 10.1096/fj.04-1743fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hill BG, Haberzettl P, Ahmed Y, Srivastava S, Bhatnagar A. Unsaturated lipid peroxidation-derived aldehydes activate autophagy in vascular smooth-muscle cells. Biochem J. 2008;410(3):525–34. doi: 10.1042/BJ20071063. [DOI] [PubMed] [Google Scholar]
- 87.Bonet-Ponce L, Saez-Atienzar S, da Casa C, Sancho-Pelluz J, Barcia JM, Martinez-Gil N, Nava E, Jordan J, Romero FJ, Galindo MF. Rotenone Induces the Formation of 4-Hydroxynonenal Aggresomes. Role of ROS-Mediated Tubulin Hyperacetylation and Autophagic Flux Disruption. Mol Neurobiol. 2015 doi: 10.1007/s12035-015-9509-3. [DOI] [PubMed] [Google Scholar]
- 88.Amma H, Naruse K, Ishiguro N, Sokabe M. Involvement of reactive oxygen species in cyclic stretch-induced NF-kappaB activation in human fibroblast cells. Br J Pharmacol. 2005;145(3):364–73. doi: 10.1038/sj.bjp.0706182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rinna A, Forman HJ. SHP-1 inhibition by 4-hydroxynonenal activates Jun N-terminal kinase and glutamate cysteine ligase. Am J Respir Cell Mol Biol. 2008;39(1):97–104. doi: 10.1165/rcmb.2007-0371OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suc I, Meilhac O, Lajoie-Mazenc I, Vandaele J, Jurgens G, Salvayre R, Negre-Salvayre A. Activation of EGF receptor by oxidized LDL. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1998;12(9):665–71. doi: 10.1096/fasebj.12.9.665. [DOI] [PubMed] [Google Scholar]
- 91.Demozay D, Mas JC, Rocchi S, Van Obberghen E. FALDH reverses the deleterious action of oxidative stress induced by lipid peroxidation product 4-hydroxynonenal on insulin signaling in 3T3-L1 adipocytes. Diabetes. 2008;57(5):1216–26. doi: 10.2337/db07-0389. [DOI] [PubMed] [Google Scholar]
- 92.Dolinsky VW, Chan AY, Robillard Frayne I, Light PE, Des Rosiers C, Dyck JR. Resveratrol prevents the prohypertrophic effects of oxidative stress on LKB1. Circulation. 2009;119(12):1643–52. doi: 10.1161/CIRCULATIONAHA.108.787440. [DOI] [PubMed] [Google Scholar]
- 93.Ma H, Guo R, Yu L, Zhang Y, Ren J. Aldehyde dehydrogenase 2 (ALDH2) rescues myocardial ischaemia/reperfusion injury: role of autophagy paradox and toxic aldehyde. Eur Heart J. 2011;32(8):1025–38. doi: 10.1093/eurheartj/ehq253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shearn CT, Fritz KS, Reigan P, Petersen DR. Modification of Akt2 by 4-hydroxynonenal inhibits insulin-dependent Akt signaling in HepG2 cells. Biochemistry. 2011;50(19):3984–96. doi: 10.1021/bi200029w. [DOI] [PubMed] [Google Scholar]
- 95.Liu W, Akhand AA, Kato M, Yokoyama I, Miyata T, Kurokawa K, Uchida K, Nakashima I. 4-hydroxynonenal triggers an epidermal growth factor receptor-linked signal pathway for growth inhibition. J Cell Sci. 1999;112(Pt 14):2409–17. doi: 10.1242/jcs.112.14.2409. [DOI] [PubMed] [Google Scholar]
- 96.Pizzimenti S, Ciamporcero E, Daga M, Pettazzoni P, Arcaro A, Cetrangolo G, Minelli R, Dianzani C, Lepore A, Gentile F, Barrera G. Interaction of aldehydes derived from lipid peroxidation and membrane proteins. Frontiers in physiology. 2013;4:242. doi: 10.3389/fphys.2013.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bruenner BA, Jones AD, German JB. Maximum entropy deconvolution of heterogeneity in protein modification: protein adducts of 4-hydroxy-2-nonenal. Rapid Commun Mass Spectrom. 1994;8(7):509–12. doi: 10.1002/rcm.1290080703. [DOI] [PubMed] [Google Scholar]
- 98.Schwarzer E, Muller O, Arese P, Siems WG, Grune T. Increased levels of 4-hydroxynonenal in human monocytes fed with malarial pigment hemozoin. A possible clue for hemozoin toxicity. FEBS Lett. 1996;388(2-3):119–22. doi: 10.1016/0014-5793(96)00523-6. [DOI] [PubMed] [Google Scholar]
- 99.Siems WG, Hapner SJ, van Kuijk FJ. 4-hydroxynonenal inhibits Na(+)-K(+)-ATPase. Free Radic Biol Med. 1996;20(2):215–23. doi: 10.1016/0891-5849(95)02041-1. [DOI] [PubMed] [Google Scholar]
- 100.Uchida K, Hasui Y, Osawa T. Covalent attachment of 4-hydroxy-2-nonenal to erythrocyte proteins. J Biochem. 1997;122(6):1246–51. doi: 10.1093/oxfordjournals.jbchem.a021888. [DOI] [PubMed] [Google Scholar]
- 101.Chen J, Henderson GI, Freeman GL. Role of 4-hydroxynonenal in modification of cytochrome c oxidase in ischemia/reperfused rat heart. J Mol Cell Cardiol. 2001;33(11):1919–27. doi: 10.1006/jmcc.2001.1454. [DOI] [PubMed] [Google Scholar]
- 102.Ji C, Kozak KR, Marnett LJ. IkappaB kinase, a molecular target for inhibition by 4-hydroxy-2-nonenal. J Biol Chem. 2001;276(21):18223–8. doi: 10.1074/jbc.M101266200. [DOI] [PubMed] [Google Scholar]
- 103.Crabb JW, O'Neil J, Miyagi M, West K, Hoff HF. Hydroxynonenal inactivates cathepsin B by forming Michael adducts with active site residues. Protein Sci. 2002;11(4):831–40. doi: 10.1110/ps.4400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu Y, Xu G, Sayre LM. Carnosine inhibits (E)-4-hydroxy-2-nonenal-induced protein cross-linking: structural characterization of carnosine-HNE adducts. Chem Res Toxicol. 2003;16(12):1589–97. doi: 10.1021/tx034160a. [DOI] [PubMed] [Google Scholar]
- 105.Ferrington DA, Kapphahn RJ. Catalytic site-specific inhibition of the 20S proteasome by 4-hydroxynonenal. FEBS Lett. 2004;578(3):217–23. doi: 10.1016/j.febslet.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 106.Orioli M, Aldini G, Beretta G, Facino RM, Carini M. LC-ESI-MS/MS determination of 4-hydroxy-trans-2-nonenal Michael adducts with cysteine and histidine-containing peptides as early markers of oxidative stress in excitable tissues. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;827(1):109–18. doi: 10.1016/j.jchromb.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 107.Robbesyn F, Auge N, Vindis C, Cantero AV, Barbaras R, Negre-Salvayre A, Salvayre R. High-density lipoproteins prevent the oxidized low-density lipoprotein-induced epidermal [corrected] growth factor receptor activation and subsequent matrix metalloproteinase-2 upregulation. Arterioscler Thromb Vasc Biol. 2005;25(6):1206–12. doi: 10.1161/01.ATV.0000164805.73558.80. [DOI] [PubMed] [Google Scholar]
- 108.Fang J, Holmgren A. Inhibition of thioredoxin and thioredoxin reductase by 4-hydroxy-2-nonenal in vitro and in vivo. J Am Chem Soc. 2006;128(6):1879–85. doi: 10.1021/ja057358l. [DOI] [PubMed] [Google Scholar]
- 109.Morquette B, Shi Q, Lavigne P, Ranger P, Fernandes JC, Benderdour M. Production of lipid peroxidation products in osteoarthritic tissues: new evidence linking 4-hydroxynonenal to cartilage degradation. Arthritis Rheum. 2006;54(1):271–81. doi: 10.1002/art.21559. [DOI] [PubMed] [Google Scholar]
- 110.Trostchansky A, Lind S, Hodara R, Oe T, Blair IA, Ischiropoulos H, Rubbo H, Souza JM. Interaction with phospholipids modulates alpha-synuclein nitration and lipid-protein adduct formation. Biochem J. 2006;393(Pt 1):343–9. doi: 10.1042/BJ20051277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Han B, Stevens JF, Maier CS. Design, synthesis, and application of a hydrazide functionalized isotope-coded affinity tag for the quantification of oxylipid-protein conjugates. Anal Chem. 2007;79(9):3342–54. doi: 10.1021/ac062262a. [DOI] [PubMed] [Google Scholar]
- 112.Stewart BJ, Doorn JA, Petersen DR. Residue-specific adduction of tubulin by 4-hydroxynonenal and 4-oxononenal causes cross-linking and inhibits polymerization. Chem Res Toxicol. 2007;20(8):1111–9. doi: 10.1021/tx700106v. [DOI] [PubMed] [Google Scholar]
- 113.Aldini G, Regazzoni L, Orioli M, Rimoldi I, Facino RM, Carini M. A tandem MS precursor-ion scan approach to identify variable covalent modification of albumin Cys34: a new tool for studying vascular carbonylation. J Mass Spectrom. 2008;43(11):1470–81. doi: 10.1002/jms.1419. [DOI] [PubMed] [Google Scholar]
- 114.Guedes RP, Araujo AS, Janner D, Bello-Klein A, Ribeiro MF, Partata WA. Increase in reactive oxygen species and activation of Akt signaling pathway in neuropathic pain. Cell Mol Neurobiol. 2008;28(8):1049–56. doi: 10.1007/s10571-008-9279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vila A, Tallman KA, Jacobs AT, Liebler DC, Porter NA, Marnett LJ. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem Res Toxicol. 2008;21(2):432–44. doi: 10.1021/tx700347w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim YS, Park ZY, Kim SY, Jeong E, Lee JY. Alteration of Toll-like receptor 4 activation by 4-hydroxy-2-nonenal mediated by the suppression of receptor homodimerization. Chem Biol Interact. 2009;182(1):59–66. doi: 10.1016/j.cbi.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 117.El-Bikai R, Welman M, Margaron Y, Cote JF, Macqueen L, Buschmann MD, Fahmi H, Shi Q, Maghni K, Fernandes JC, Benderdour M. Perturbation of adhesion molecule-mediated chondrocyte-matrix interactions by 4-hydroxynonenal binding: implication in osteoarthritis pathogenesis. Arthritis Res Ther. 2010;12(5):R201. doi: 10.1186/ar3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Backos DS, Fritz KS, Roede JR, Petersen DR, Franklin CC. Posttranslational modification and regulation of glutamate-cysteine ligase by the alpha,beta-unsaturated aldehyde 4-hydroxy-2-nonenal. Free Radic Biol Med. 2011;50(1):14–26. doi: 10.1016/j.freeradbiomed.2010.10.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fritz KS, Galligan JJ, Smathers RL, Roede JR, Shearn CT, Reigan P, Petersen DR. 4-Hydroxynonenal inhibits SIRT3 via thiol-specific modification. Chem Res Toxicol. 2011;24(5):651–62. doi: 10.1021/tx100355a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shearn CT, Smathers RL, Stewart BJ, Fritz KS, Galligan JJ, Hail N, Jr., Petersen DR. Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) inhibition by 4-hydroxynonenal leads to increased Akt activation in hepatocytes. Mol Pharmacol. 2011;79(6):941–52. doi: 10.1124/mol.110.069534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Uno K, Kato K, Kusaka G, Asano N, Iijima K, Shimosegawa T. The balance between 4-hydroxynonenal and intrinsic glutathione/glutathione S-transferase A4 system may be critical for the epidermal growth factor receptor phosphorylation of human esophageal squamous cell carcinomas. Mol Carcinog. 2011;50(10):781–90. doi: 10.1002/mc.20699. [DOI] [PubMed] [Google Scholar]
- 122.Dou X, Li S, Wang Z, Gu D, Shen C, Yao T, Song Z. Inhibition of NF-kappaB activation by 4-hydroxynonenal contributes to liver injury in a mouse model of alcoholic liver disease. Am J Pathol. 2012;181(5):1702–10. doi: 10.1016/j.ajpath.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Harry RS, Hiatt LA, Kimmel DW, Carney CK, Halfpenny KC, Cliffel DE, Wright DW. Metabolic impact of 4-hydroxynonenal on macrophage-like RAW 264.7 function and activation. Chem Res Toxicol. 2012;25(8):1643–51. doi: 10.1021/tx3001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Smathers RL, Fritz KS, Galligan JJ, Shearn CT, Reigan P, Marks MJ, Petersen DR. Characterization of 4-HNE modified L-FABP reveals alterations in structural and functional dynamics. PLoS One. 2012;7(6):e38459. doi: 10.1371/journal.pone.0038459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Aluise CD, Rose K, Boiani M, Reyzer ML, Manna JD, Tallman K, Porter NA, Marnett LJ. Peptidyl-prolyl cis/trans-isomerase A1 (Pin1) is a target for modification by lipid electrophiles. Chem Res Toxicol. 2013;26(2):270–9. doi: 10.1021/tx300449g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu Q, Simpson DC, Gronert S. Carbonylation of mitochondrial aconitase with 4-hydroxy-2-(E)-nonenal: localization and relative reactivity of addition sites. Biochim Biophys Acta. 2013;1834(6):1144–54. doi: 10.1016/j.bbapap.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rudic M, Milkovic L, Zarkovic K, Borovic-Sunjic S, Sterkers O, Waeg G, Ferrary E, Bozorg Grayeli A, Zarkovic N. The effects of angiotensin II and the oxidative stress mediator 4-hydroxynonenal on human osteoblast-like cell growth: possible relevance to otosclerosis. Free Radic Biol Med. 2013;57:22–8. doi: 10.1016/j.freeradbiomed.2012.11.023. [DOI] [PubMed] [Google Scholar]
- 128.Galligan JJ, Fritz KS, Backos DS, Shearn CT, Smathers RL, Jiang H, MacLean KN, Reigan PR, Petersen DR. Oxidative stress-mediated aldehyde adduction of GRP78 in a mouse model of alcoholic liver disease: functional independence of ATPase activity and chaperone function. Free Radic Biol Med. 2014;73:411–20. doi: 10.1016/j.freeradbiomed.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ramanathan R, Mancini RA, Suman SP, Beach CM. Covalent binding of 4-hydroxy-2-nonenal to lactate dehydrogenase decreases NADH formation and metmyoglobin reducing activity. J Agric Food Chem. 2014;62(9):2112–7. doi: 10.1021/jf404900y. [DOI] [PubMed] [Google Scholar]
- 130.Shearn CT, Backos DS, Orlicky DJ, Smathers-McCullough RL, Petersen DR. Identification of 5' AMP-activated kinase as a target of reactive aldehydes during chronic ingestion of high concentrations of ethanol. J Biol Chem. 2014;289(22):15449–62. doi: 10.1074/jbc.M113.543942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Baker MA, Weinberg A, Hetherington L, Villaverde AI, Velkov T, Baell J, Gordon CP. Defining the mechanisms by which the reactive oxygen species by-product, 4-hydroxynonenal, affects human sperm cell function. Biol Reprod. 2015;92(4):108. doi: 10.1095/biolreprod.114.126680. [DOI] [PubMed] [Google Scholar]
- 132.Jang EJ, Jeong HO, Park D, Kim DH, Choi YJ, Chung KW, Park MH, Yu BP, Chung HY. Src Tyrosine Kinase Activation by 4-Hydroxynonenal Upregulates p38, ERK/AP-1 Signaling and COX-2 Expression in YPEN-1 Cells. PLoS One. 2015;10(10):e0129244. doi: 10.1371/journal.pone.0129244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Camarillo JM, Rose KL, Galligan JJ, Xu S, Marnett LJ. Covalent modification of CDK2 by 4-hydroxynonenal as a mechanism of inhibition of cell cycle progression. Chem Res Toxicol. 2016 doi: 10.1021/acs.chemrestox.5b00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Miriyala S, Thippakorn C, Chaiswing L, Xu Y, Noel T, Tovmasyan A, Batinic-Haberle I, Vander Kooi CW, Chi W, Latif AA, Panchatcharam M, Prachayasittikul V, Allan Butterfield D, Vore M, Moscow J, St Clair DK. Novel role of 4-hydroxy-2-nonenal in AIFm2-mediated mitochondrial stress signaling. Free Radic Biol Med. 2016;91:68–80. doi: 10.1016/j.freeradbiomed.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]