Abstract
Rapidly proliferating cells require increased metabolism to allow for the production of nucleic acids, amino acids, and lipids. Pyruvate kinase catalyzes the final step in the glycolysis pathway and different isoforms display vastly different catalytic efficiencies. The M2 isoform of pyruvate kinase (PKM2) is highly expressed in cancer cells and contributes to aerobic glycolysis in what is commonly referred to as the Warburg effect. Here, we show that PKM2 is covalently modified by the lipid electrophiles 4-hydroxy-2-nonenal (HNE) and 4-oxo-2-nonenal (ONE). HNE and ONE modify multiple sites on PKM2 in vitro, including Cys424 and His439, which play a role in protein-protein interactions and fructose 1,6-bis-phosphate binding, respectively. Modification of these sites results in a dose-dependent decrease in enzymatic activity. In addition, high concentrations of electrophile, most notably in the case of ONE, result in substantial protein-protein cross-linking in vitro and in cells. Exposure of RKO cells to electrophiles results in modification of monomeric PKM2 in a dose-dependent manner. There is a concomitant decrease in PKM2 activity in cells upon ONE exposure, but not HNE exposure. Together, our data suggest that modification of PKM2 by certain electrophiles results in kinase inactivation.
Introduction
Early studies by Otto Warburg identified aerobic glycolysis as the main mechanism by which cancer cells generate ATP. Elevated glucose uptake, increased glycolysis, and high lactate production, even in the presence of oxygen, allow for more rapid cell growth and tumorigenesis.1 Malignant tumors can have glycolytic rates up to 200 times that of normal tissues, and the switch from oxidative phosphorylation to aerobic glycolysis is coupled with a switch in isoforms of the glycolytic enzyme, pyruvate kinase.2
Pyruvate kinase catalyzes the final reaction in glycolysis, the transfer of a phosphate group from phosphoenolpyruvate to ADP, to yield one molecule of pyruvate and one molecule of ATP. Expression of pyruvate kinase isoform M2 (PKM2) occurs during fetal development, while the M1 isoform predominates throughout the majority of adult tissues. During tumorigenesis, PKM2 becomes the predominant isoform.2 The lower enzymatic activity of PKM2 relative to that of PKM1 results in increased glycolytic intermediates that can be used for different biosynthetic pathways, thereby contributing to more rapid cell growth.3 PKM2 has also been identified as a regulator of transcription. Translocation of PKM2 to the nucleus allows for coactivation of β-catenin, induction of c-Myc expression, and upregulation of genes which contribute to the Warburg effect.4,5
Regulation of PKM2 activity has been an active area of research for a number of years. PKM2 glycolytic and transcriptional activity is modulated by a number of post-translational modifications, multimerization, and allosteric interactions.6 Fructose 1,6-bis-phosphate allosterically binds PKM2, promoting homotetramer formation and increasing glycolytic activity.2 Acetylation of PKM2 at K433 inhibits this allosteric interaction, resulting in formation of PKM2 dimers that can translocate to the nucleus and transcriptionally regulate genes involved in cell cycle progression and glycolysis.7 These two different functions of PKM2 are thought to further contribute to tumorigenesis.
Lipid electrophiles are generated during periods of oxidative stress and are associated with a multitude of diseases, including cancer. 4-Hydroxy-2-nonenal (HNE) and 4-oxo-2-nonenal (ONE) readily react with Cys, His, and Lys residues to form Michael adducts, whereas ONE displays unique reactivity toward Lys residues to produce the Schiff base and the 4-ketoamide adducts.8,9 Modification of proteins has typically been shown to result in loss of protein function and to promote dysregulation of multiple cell signaling pathways.
Recent work has used alkyne analogues of HNE and ONE, aHNE and aONE, respectively, to selectively isolate adducted proteins via click chemistry for identification by mass spectrometry. Using this approach, Codreanu et al.10 identified >1000 proteins as targets of aHNE or aONE modification. Interestingly, these targets displayed a range of sensitivities to each electrophile. PKM2 was shown to be modified by both aHNE and aONE, and was heavily modified at low concentrations of both electrophiles, revealing it as a preferential target of adduction. Here, we map the sites of PKM2 modification by HNE and ONE and show that these modifications result in PKM2 cross-linking in vitro and in cells. Additionally, cross-linking by ONE produces a fully functioning PKM2 dimer in vitro. In cells, PKM2 is adducted by aHNE and aONE at <10 μM electrophile, though high concentrations of ONE contributes to PKM2 kinase inactivation in cells. Together, our data show that lipid electrophiles can modify PKM2 and inhibit its function.
Methods
Materials and reagents
HNE, ONE, 8,9-alkynyl-HNE (aHNE), and 8,9-alkynyl-ONE (aONE), and the UV-cleavable azido-biotin were synthesized in the laboratory of Dr. Ned Porter at Vanderbilt University as described.11 Cell culture medium was purchased from Invitrogen (Grand Island, NY) and fetal bovine serum (FBS) was purchased from Atlas Biologicals (Ft. Collins, CO). Recombinant PKM2 was purchased from Novus Biologicals (Littleton, CO) or Genway Biosciences (San Diego, CA). Anti-PKM2 antibodies were purchased from Abcam (Boston, MA) or Signaling Antibody (College Park, MD). SDS-PAGE and Western blotting supplies were purchased from Bio-Rad (Hercules, CA) unless otherwise stated. All other reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted.
Cell Culture and Treatments
The human colorectal cancer RKO cell line was purchased from American Type Culture Collection (ATCC, Manassas, VA) and cultured in Dulbecco's Modified Eagle's Medium (DMEM) with Glutamax (Gibco) with 10% FBS in humidified incubators under 5% CO2 at 37°C. Electrophiles were dissolved in dimethyl sulfoxide (DMSO) and added to cell culture medium with a final concentration of less than 0.1% DMSO.
Treatment of Recombinant PKM2 with Electrophiles
Recombinant PKM2 protein was diluted to 2.5 mg/mL in PBS. HNE, ONE, or DMSO (vehicle control) were added at the indicated concentrations, and the reaction mixture was incubated for 1 h with gentle agitation at 37°C. NaBH4 was added at a final concentration of 20 mM to quench the reaction. Protein was resolved on a 10% NuPAGE Novex Bis-Tris gel and stained with SimplyStain SafeStain (Invitrogen, Waltham, MA). Bands corresponding to PKM2 (monomer, dimer, trimer, or tetramer) were excised from the gel for proteomic analysis.
Analysis of PKM2 Peptides by LC-MS/MS
Gel bands corresponding to monomer, dimer, and tetramer were cut into 1 mm pieces, reduced with 150 μM dithiothreitol for 30 min at 37°C, and alkylated with 750 μM iodoacetamide for 15 min at room temperature in the dark. Gel pieces were destained in 25 mM NH4HCO3 with 50% acetonitrile and subjected to in-gel digestion with 10 ng/μL trypsin overnight. The gel was dehydrated with 60% acetonitrile containing 0.1% trifluoroacetic acid to extract the peptides, which were then dried by vacuum centrifugation and reconstituted in 0.1% formic acid. Using an Eksigent NanoLC Ultra HPLC and autosampler, peptides were loaded onto a capillary reverse-phase analytical column (360 μm O.D. × 100 μm I.D.) packed with 20 cm of C18 reverse-phase material (Jupiter, 3μm beads, 300Å, Phenomenox) with a laser-pulled emitter tip. Peptides were gradient-eluted at a flow rate of 500 nL/min, and the mobile phase solvents consisted of water with 0.1% formic acid (solvent A) and acetonitrile with 0.1% formic acid (solvent B). A 90-minute gradient was performed, consisting of the following: 0-10 min, 2% B; 10-50 min, 2-40% B; 50-60 min, 35-95% B; 60-65 min, 95% B, 65-70 min 95-2% B, 70-90 min, 2% B. Gradient eluted peptides were mass analyzed on a Thermo Scientific LTQ Orbitrap Velos mass spectrometer with a nanoelectrospray ionization source. The mass spectrometer was operated using a data-dependent method with dynamic exclusion enabled. Full scan (m/z 300-2000) spectra were acquired with the Orbitrap as the mass analyzer (resolution 60,000). The ten most abundant ions in each MS scan were selected for fragmentation in the LTQ with an isolation width of 2 m/z, activation time of 10 ms, and 35% normalized collision energy. Tandem mass spectra were searched with Sequest against a human database created from the UniprotKB protein database (www.uniprot.org) to identify modified peptides. Variable mass shifts of +57.0214 m/z on Cys (carbamidomethylation), +15.9949 m/z on Met (oxidation), +158.1306 m/z on Cys, His, and Lys residues (corresponding to the reduced Michael adduct), +141.1279 m/z on Lys and Arg (corresponding to the reduced Schiff base), and +156.1150 m/z on Lys (corresponding to the 4-ketoamide) were included for database searching. Search results were assembled using Scaffold 3.0 (Proteome Software), and sites of electrophile adduction were validated by manual interrogation of tandem mass spectra.
PKM2 Kinase Assays
Recombinant PKM2 (100 ng) was treated with the indicated concentrations of electrophiles in 20 μL Dulbecco's-modified phosphate-buffered saline (DPBS, Gibco; 1X, pH 7.3) for 1 h at 37°C with gentle agitation. Lactate dehydrogenase-coupled PKM2 kinase assays were performed as previously described.12 Briefly, 5 ng of treated PKM2 was incubated with kinase assay buffer (50 mM Tris, pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.6 mM ADP, 0.5 mM phosphoenolpyruvate, 180 μM NADH, 10 μM fructose 1,6-bisphosphate, 8 U lactate dehydrogenase). The decrease in 340 nm absorbance (as a result of NADH oxidation) was recorded over 20 min on a SpectraMax Microplate Reader. Relative PKM2 activity was calculated as change in absorbance at 340 nm per minute per mg protein and normalized to percent of control.
Click Chemistry
RKO cells were treated with the indicated concentrations of the alkyne analogues of HNE and ONE, aHNE and aONE, respectively, in serum-free medium. Following 1 h treatment, cells were collected by scraping, centrifuged for 5 min at 1000 x g, and washed twice with DPBS. Cells were lysed in RIPA buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 0.5% Igepal) with mammalian protease inhibitor cocktail and phosphatase inhibitor cocktail (Sigma-Aldrich), sonicated for ten 1 s pulses (35% duty cycle, Virsonic Cell Disrupter), and cleared by centrifugation at 16,000 x g for 15 min at 4°C. Adducts were reduced with 20 mM NaBH4 for 30 min on ice. Protein concentration was determined with the bicinchoninic acid (BCA) assay (Thermo Scientific, Waltham, MA). Total cell lysate (800 μg) was subjected to click chemistry as previously described.13
SDS-PAGE and Western Blotting
Protein samples were mixed 1:1 with 2X Laemmeli buffer with 5% β-mercaptoethanol and boiled for 5 min. Proteins were separated by SDS-PAGE on 10% gels, transferred onto 0.45 μm nitrocellulose membranes, and blocked with Odyssey blocking buffer (Li-Cor Bioscience, Superior, NE). Primary antibodies (1:1,000) in Odyssey blocking buffer were added to membranes overnight at 4°C. Blots were washed three times with Tris-buffered saline containing 0.1% Tween-20 (TBST) and secondary antibodies (1:5,000, Li-Cor) in 1:1 Odyssey blocking buffer/TBST were added for 1 h at RT. Blots were developed after three washes with TBST on the Odyssey Infrared Imaging System (Li-Cor).
Results
PKM2 is modified at a number of sites in vitro
Previous proteomic data sets generated by our lab have shown that PKM2 is modified by aHNE and aONE in a number of difference cell types. Among the identified protein targets, PKM2 was shown to be adducted at very low concentrations of electrophile.10 To determine the precise location of HNE and ONE adducts on PKM2, recombinant PKM2 was incubated with increasing concentrations of electrophile in vitro followed by treatment with NaBH4 to reduce any labile adducts, such as the reversible Michael adduct.14 Following digestion with trypsin, peptides were analyzed by LC-MS/MS and data searched for mass shifts corresponding to possible adducts. These analyses revealed the presence of Michael adducts on Cys49, His274, Cys424, and His439 of PKM2 following both HNE and ONE treatments (Table 1, Figure 1, and Supplemental Figure 1A-D). A Michael adduct was observed on Lys256 with HNE treatment only (Table 1 and Supplemental Figure 1E). Adducts were only observed in PKM2 monomers.
Table 1. PKM2 is modified at multiple sites by HNE and ONE.
Recombinant PKM2 was modified in vitro with increasing concentrations of HNE or ONE, adducts were reduced, and proteins resolved by SDS-PAGE prior to in-gel digestion and analysis by LC-MS/MS.
| Peptide | Observed m/z | Theoretical m/z | Charge | Mass Error (ppm) | Residue | Modification | Electrophile |
|---|---|---|---|---|---|---|---|
| NTGIIC*TIGPASR | 730.9112 | 730.9107 | 2 | 0.68 | Cys49 | Michael Adduct | HNE and ONE |
| IENH*EGVR | 556.3110 | 556.3089 | 2 | 3.77 | His274 | Michael Adduct | HNE and ONE |
| CC*SGAIIVLTK | 661.8759 | 661.8747 | 2 | 1.81 | Cys424 | Michael Adduct | HNE and ONE |
| SAH*QVAR | 463.7764 | 463.7745 | 2 | 4.10 | His439 | Michael Adduct | HNE and ONE |
| NIK*IISK | 486.3305 | 486.3286 | 2 | 3.91 | Lys256 | Michael Adduct | HNE |
| VLGEK*GK | 443.7864 | 443.7840 | 2 | 5.41 | Lys261 | Ketoamide | ONE |
Figure 1. Structures of electrophiles and adducts.
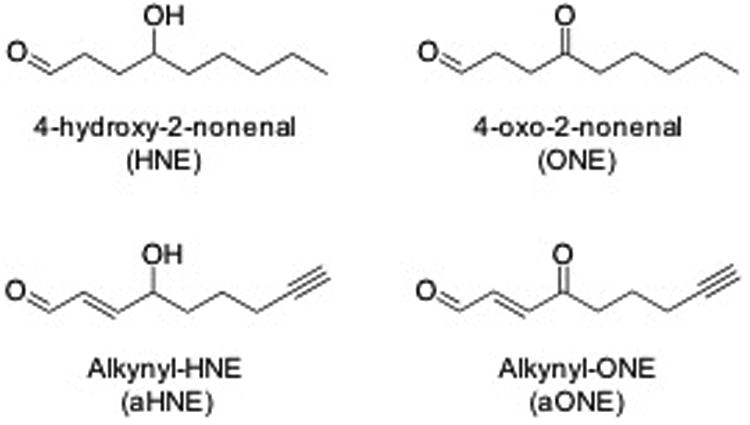
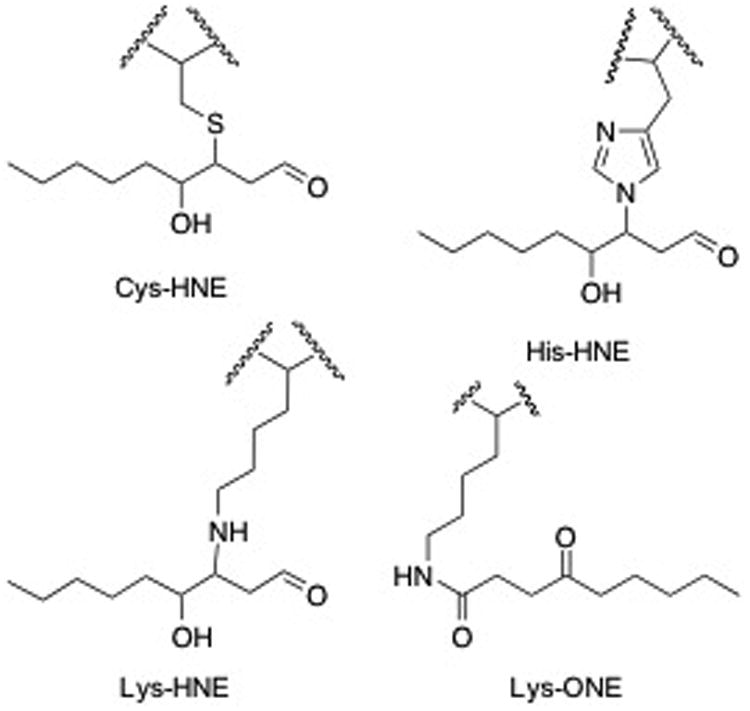
A) Structures of electrophiles used in these studies, HNE and ONE, and the alkyne analogues, aHNE and aONE. B) Structures of adducts found on PKM2. Cys and His adducts were found with in both HNE and ONE. Michael adducts of HNE were found on Lys while ONE formed the 4-ketoamide adduct on Lys.
Additionally, we searched for the +156.1150 m/z mass shift, corresponding to the 4-ketoamide adduct on Lys residues, and the +141.1279 m/z mass shift, corresponding to the Schiff base adduct on Lys and Arg residues. While no Schiff base adducts were found, the 4-ketoamide adduct was observed on Lys261 (Table 1, Figure 1, and Supplementary Figure 2). As expected, the 4-ketoamide adduct was only observed with ONE treatment. These data suggest that PKM2 is susceptible to multiple adducts at distinct sites.
PKM2 cross-links occur upon exposure to HNE and ONE
PKM2 is found in two conformations, a dimer and a tetramer, which exhibit different rates of glycolytic activity and are typically regulated by multiple post-translational modifications.15 During in vitro modification and preparation of the samples for LC-MS/MS, we observed multimerization of recombinant PKM2 with increasing concentrations of HNE (Figure 2A) and ONE (Figure 2B), likely the result of electrophile-induced cross-linking. Treatment with ONE induced cross-linking at much lower concentrations compared to treatment with HNE. This observation is consistent with previous reports on the relative reactivities of each electrophile and the chemistry of cross-link formation.16 Notably, cross-linking of PKM2 to a dimer occurred at very low concentrations of ONE (0.3 μM) and very little monomeric PKM2 was observed at ONE concentrations >10 μM (Figure 2B). These data suggest that ONE-mediated cross-linking of PKM2 may contribute to altered activity in the cell.
Figure 2. PKM2 cross-links occur upon exposure to HNE and ONE.
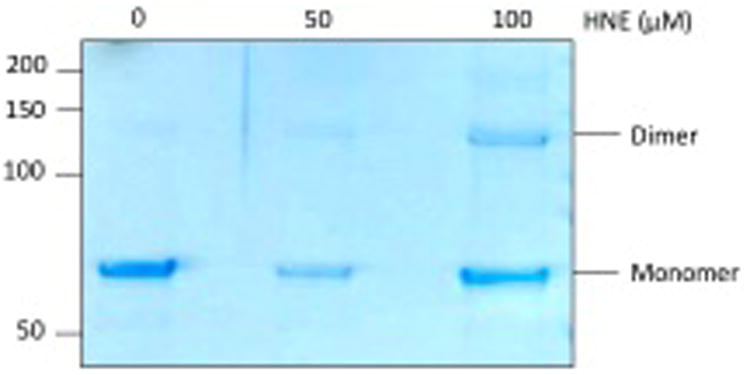
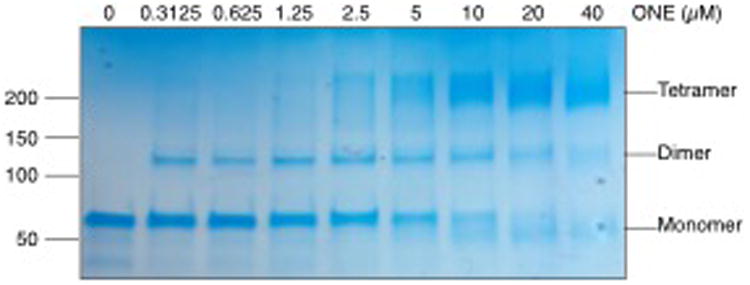
Recombinant PKM2 was modified in vitro with increasing concentrations of A) HNE or B) ONE for 1 h at 37°C. Staining of SDS-PAGE with Coomassie blue shows cross-linking of PKM2 monomers with increasing concentrations of electrophile.
HNE and ONE inhibit kinase activity in recombinant PKM2 protein
Due to the ability of HNE and ONE to cross-link PKM2 monomers, we assessed the impact of electrophile treatment and subsequent protein adduction on PKM2 kinase activity. Recombinant PKM2 protein was incubated with increasing concentrations of HNE or ONE that were known to contribute to dimer, trimer, and tetramer formation, and PKM2 kinase activity was assessed by a lactate dehydrogenase-coupled assay to measure the oxidation of NADH to NAD+.12 In vitro adduction of PKM2 with HNE resulted in a significant decrease in PKM2 kinase activity at HNE concentrations greater than 40 μM (Figure 3A). These concentrations only contributed to a low percentage of cross-linked dimer at similar concentrations. These data suggest that PKM2 activity decreases with increasing HNE adduction.
Figure 3. Cross-linking inhibits kinase activity in recombinant PKM2 protein.
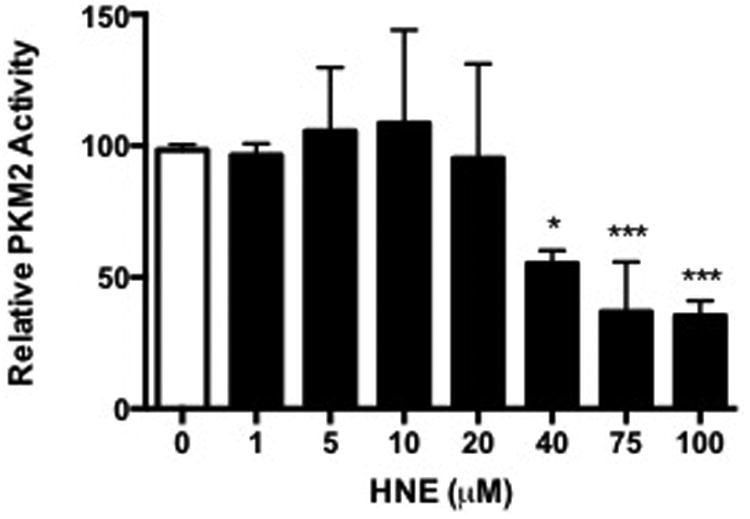
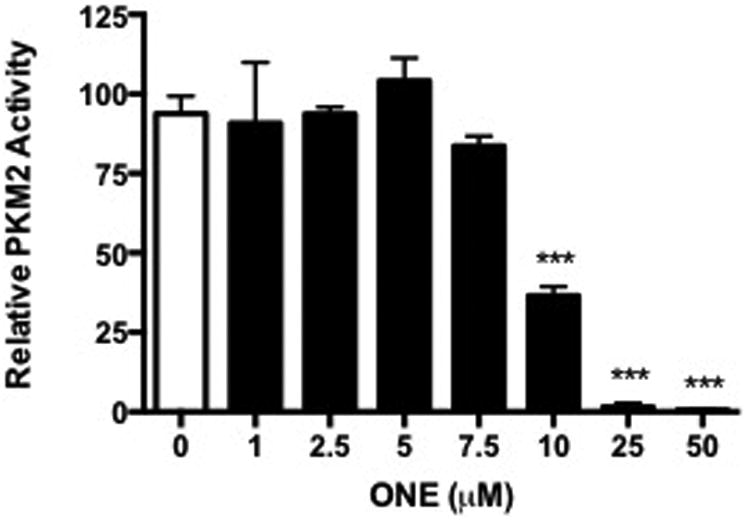
Recombinant PKM2 was modified in vitro with increasing concentrations of A) HNE or B) ONE for 1 h at 37°C. PKM2 activity was assessed indirectly by the lactate dehydrogenase-coupled enzyme assay. Data represent an n=3 ± SD, *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA.
Conversely, treatment with ONE resulted in a significant decrease in PKM2 kinase activity at low electrophile concentration. When recombinant PKM2 protein was treated with increasing concentrations of ONE in vitro, activity assays showed >50% reduction in PKM2 kinase activity at 10 μM ONE, with complete inhibition at higher concentrations (Figure 3B). Interestingly, there was little difference in activity at low concentrations of ONE, which were shown to cross-link PKM2 monomers to dimers. It is possible that ONE-induced cross-linking of PKM2 alone does not result in inhibition of activity.
PKM2 is adducted by electrophiles in RKO cells
To verify modification of PKM2 in RKO cells, cells were treated with aHNE and aONE to enable selective tagging and isolation of adducted proteins by click chemistry.11 RKO cells were treated with aHNE or aONE for 1 h, lysed, and the protein extract reduced and subjected to click chemistry with a UV-cleavable N3-biotin. Photo-elution from streptavidin beads allowed for selective elution of adducted proteins. Western blot analysis of eluates revealed that PKM2 was modified by both aHNE (Figure 4A) and aONE (Figure 4B) in a dose-dependent manner. Only PKM2 monomers were recovered at the concentrations of aHNE and aONE that we investigated, which suggests that multimerization of PKM2 only occurs at higher concentrations.
Figure 4. PKM2 is adducted by electrophiles in RKO cells.

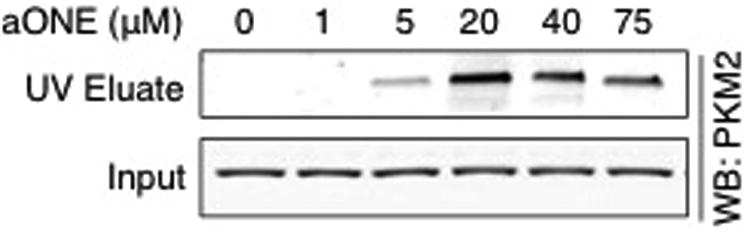
RKO cells were treated with increasing concentrations of aHNE or aONE for 1 h. Modified proteins were conjugated with UV-cleavable N3-biotin via click chemistry, bound to streptavidin beads, and eluted under 365 nm UV light. Western blot analysis of UV eluates and whole cell lysate (Input, 0.1%) showed a dose-dependent increase in modification of PKM2 by both electrophiles.
PKM2 is cross-linked by aONE in RKO cells
While we did not observe multimers of PKM2 in the click elution (Figure 4), we wanted to definitively assess if PKM2 can be cross-linked by electrophiles in the cell. Following treatment of RKO cells with increasing concentrations of HNE or ONE, we performed western blot analysis to assess if dimers of PKM2 were visible. Interestingly, high concentrations of ONE (>100 μM) resulted in the appearance of PKM2 dimers in whole cell lysates (Figure 5). Dimers were not observed in lysates that had been treated with HNE (data not shown). These data show that cross-linking of PKM2 can occur in living cells; however high levels of exogenous ONE produce only low levels of cross-linking.
Figure 5. PKM2 is cross-linked by aONE in RKO cells.
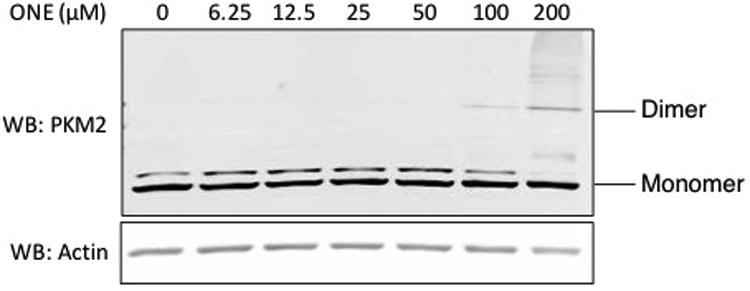
RKO cells were treated with increasing concentrations of ONE for 1 h. After reduction of lysates with NaBH4, proteins were separated by SDS-PAGE and PKM2 levels were assessed with western blot. High concentrations of ONE resulted in formation of PKM2 cross-linked dimers.
ONE inhibits kinase activity in cellular PKM2
To begin to elucidate the biological impact of electrophile modification of PKM2, kinase assays were performed on RKO cell lysates. RKO cells were treated with increasing concentrations of HNE and ONE, and lysates were reduced with NaBH4. In cells treated with HNE, no differences were observed in PKM2 activity at any of the concentrations we tested (Figure 6A). Interestingly, cells treated with ONE displayed a significant decrease in PKM2 activity at high concentrations (Figure 6B). It is notable that this altered activity of PKM2 in response to ONE correlates with the cross-linking that is observed at high cellular ONE concentrations (Figure 5).
Figure 6. Oligomer formation inhibits kinase activity in cellular PKM2.
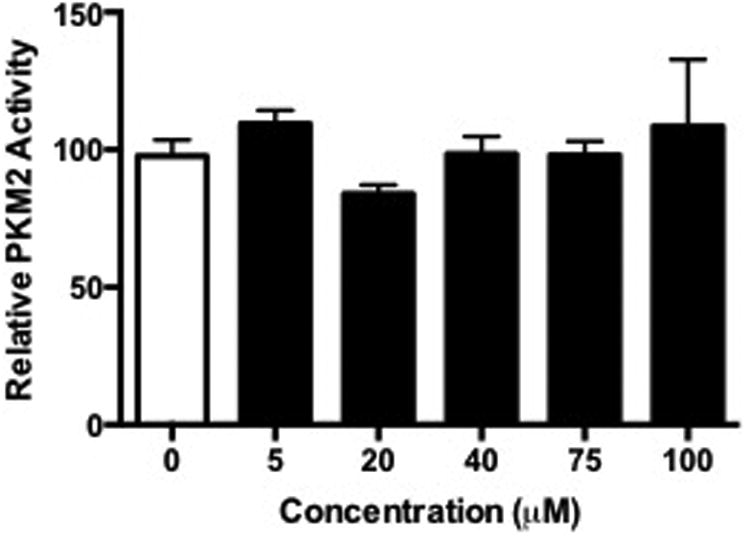
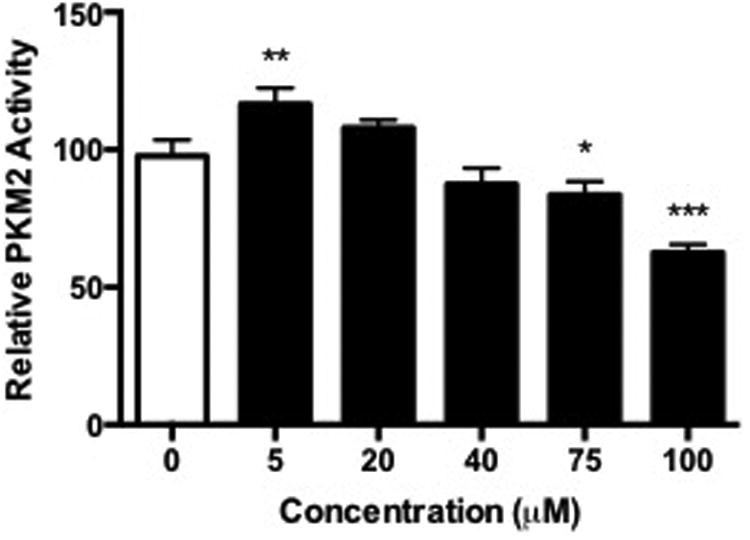
RKO cells were treated with increasing concentrations of HNE or ONE for 1 h, lysed, reduced with NaBH4, and subjected to in vitro PKM2 kinase assays. Data represent the mean ± SD (n = 3), *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA.
Discussion
While the chemistry of lipid peroxidation and subsequent protein adduction has been extensively investigated, the number of protein targets and the potential biological implications are only recently being elucidated. HNE was identified prior to ONE, thus it has been the most highly studied product of autoxidation of polyunsaturated fatty acids. Upon the discovery of ONE, studies began to focus on the increased reactivity of ONE relative to HNE.17 These studies led to the identification of a number of new ONE-derived modifications, including cross-links formed between Cys or His Michael adducts and Schiff base-derived adducts on Lys residues.
Previous work has investigated the protein targets of different lipid electrophiles in multiple cell types. While these large-scale proteomic data sets have provided insight into the signaling pathways that could be disrupted as a result of electrophile modification, the precise impact on protein function remains unknown for the majority of modified proteins. Our results have not only investigated the effect on kinase activity of a key glycolytic protein, but also provided insight into how its modification could influence cancer progression.
Using HNE and ONE treatment in vitro, we have shown by mass spectrometry that PKM2 is adducted at Cys, His, and Lys residues in the form of Michael adducts, as well as a Lys residue in the form of the 4-ketoamide. Electrophile treatment, most notably with ONE, results in cross-linking of PKM2 monomers. ONE cross-links can occur between Cys/Lys and His/Lys,18,19 and the susceptibility of these residues to cross-linking has been exemplified with other α,β- unsaturated carbonyls, such as cis-2-butene-1,4-dial.20 In all these examples, the cross-link is an N-substituted pyrrole, but observation of this structure was only on individual amino acids or single tryptic peptides. Other cross-link structures, such as a Lys-Lys cross-link, have also been proposed to occur with HNE and ONE.21,22 Technology limitations make it difficult to determine inter-peptide cross-links by mass spectrometry. While SDS-PAGE and Coomassie blue staining show that treatment with either HNE or ONE results in cross-linking between PKM2 monomers in vitro, manual investigation of the proteomic data did not yield any potential peptide cross-links. Additionally, these proteomic data show adducts occurring on digested monomers, but not on multimers, suggesting that the majority of adducts on multimers may be within cross-links.
PKM2 activity in vitro is significantly decreased in response to both HNE and ONE. The loss of activity correlates with the increase in cross-linking, and it is possible that this loss of activity is the direct result of cross-linking. Additional hypotheses can also explain the loss of activity. Adduction of specific sites could contribute to the disruption of substrate binding, thereby reducing catalytic activity independent of cross-linking. While none of the sites of adduction we found are known to directly interact with substrate binding, it is possible that adduction could disrupt the local structure to alter binding.
Consistent with observations in vitro, our results in RKO cells show that PKM2 is highly susceptible to electrophile adduction. Click chemistry and selective isolation of adducted proteins shows measurable adduction of PKM2 monomers at concentrations as low as 5-10 μM. Increased PKM2 adduction in cells by aONE is likely due to the higher reactivity of aONE versus aHNE. High ONE levels resulted in formation of cross-linked PKM2 dimers and contributed to decreased kinase activity. While the levels of ONE required to induce cross-link formation and loss of activity in cells are much higher than in vitro, these results are likely due the additional protein targets in cells and the possibility that not all the exogenous electrophile penetrates the cell.
ONE is a more potent cross-linker than HNE, and the cross-links generated are thought to be chemically stable.18,23 The lack of cross-linking and altered activity of PKM2 in response to HNE suggests different residues of modification in cells between the two electrophiles. In other words, HNE may target sites that do not play a role in substrate, allosteric, or protein binding, thereby making these adducts benign to PKM2 activity. ONE appears to disrupt PKM2 activity in cells, so the modified residues may play a more significant role in catalysis or protein binding. Ultimately, ONE has a more potent inhibition of PKM2 activity, and we propose that inhibition of PKM2 glycolytic activity is due to the cross-linking of active dimers to tetramers (Figure 7). Methods to determine sites of cross-links is an area of further development and investigation.
Figure 7. Proposed mechanism of PKM2 inhibition.
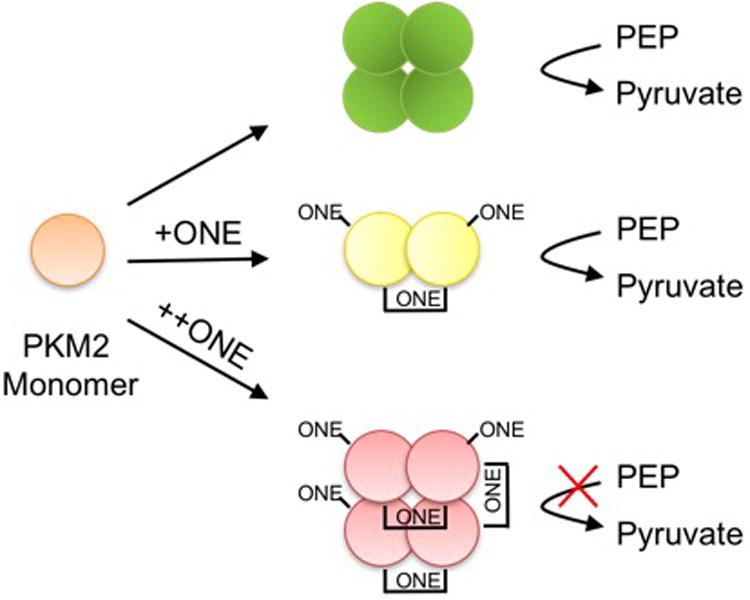
Under normal conditions, PKM2 monomoer form tetramers to convert PEP to pyruvate. In vitro and in cells, ONE forms cross-links between PKM2 monomers. Dimer formation allows for catalytic activity while formation of tetramers contributes to enzyme inactivation.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Carol Rouzer for critical review of the manuscript.
Funding Information: This work was supported by the NCI F31 CA192861 (J.M.C.) and R37 CA087819 (L.J.M.).
Abbreviations
- HNE
4-Hydroxy-2-nonenal
- ONE
4-Oxo-2-nonenal
- DMSO
Dimethyl Sulfoxide
- RT
Room Temperature
- aHNE
Alkynyl 4-Hydroxy-2-nonenal
- aONE
Alkynyl 4-Oxo-2-nonenal
- PKM2
Pyruvate Kinase Isoform M2
- TBST
Tris-Buffered Saline with Tween-20
- DPBS
Dulbecco's Phosphate Buffered Saline
- FBS
Fetal Bovine Serum
Footnotes
Supporting Information: MS2 spectra of adducted peptides. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Seminars in cancer biology. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Chaneton B, Hillmann P, Zheng L, Martin ACL, Maddocks ODK, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, Frezza C, O/'Reilly M, Gottlieb E. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong N, Ojo D, Yan J, Tang D. PKM2 contributes to cancer metabolism. Cancer Letters. 2015;356:184–191. doi: 10.1016/j.canlet.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 7.Lv L, Xu YP, Zhao D, Li FL, Wang W, Sasaki N, Jiang Y, Zhou X, Li TT, Guan KL, Lei QY, Xiong Y. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol Cell. 2013;52:340–352. doi: 10.1016/j.molcel.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free radical biology & medicine. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 9.Ullery JC, Marnett LJ. Protein modification by oxidized phospholipids and hydrolytically released lipid electrophiles: Investigating cellular responses. Biochim Biophys Acta. 2012;1818:2424–2435. doi: 10.1016/j.bbamem.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Codreanu SG, Ullery JC, Zhu J, Tallman KA, Beavers WN, Porter NA, Marnett LJ, Zhang B, Liebler DC. Alkylation damage by lipid electrophiles targets functional protein systems. Mol Cell Proteomics. 2014;13:849–859. doi: 10.1074/mcp.M113.032953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vila A, Tallman KA, Jacobs AT, Liebler DC, Porter NA, Marnett LJ. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem Res Toxicol. 2008;21:432–444. doi: 10.1021/tx700347w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 13.Camarillo JM, Rose KL, Galligan JJ, Xu S, Marnett LJ. Covalent Modification of CDK2 by 4-Hydroxynonenal as a Mechanism of Inhibition of Cell Cycle Progression. Chem Res Toxicol. 2016;29:323–332. doi: 10.1021/acs.chemrestox.5b00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uchida K, Stadtman ER. Modification of histidine residues in proteins by reaction with 4-hydroxynonenal. Proc Natl Acad Sci U S A. 1992;89:4544–4548. doi: 10.1073/pnas.89.10.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gui DY, Lewis CA, Vander Heiden MG. Allosteric Regulation of PKM2 Allows Cellular Adaptation to Different Physiological States. Science Signaling. 2013;6:pe7–pe7. doi: 10.1126/scisignal.2003925. [DOI] [PubMed] [Google Scholar]
- 16.Stewart BJ, Doorn JA, Petersen DR. Residue-Specific Adduction of Tubulin by 4-Hydroxynonenal and 4-Oxononenal Causes Cross-Linking and Inhibits Polymerization. Chemical Research in Toxicology. 2007;20:1111–1119. doi: 10.1021/tx700106v. [DOI] [PubMed] [Google Scholar]
- 17.Doorn JA, Petersen DR. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem Res Toxicol. 2002;15:1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 18.Aluise CD, Camarillo JM, Shimozu Y, Galligan JJ, Rose KL, Tallman KA, Marnett LJ. Site-specific, intramolecular cross-linking of Pin1 active site residues by the lipid electrophile 4-oxo-2-nonenal. Chem Res Toxicol. 2015;28:817–827. doi: 10.1021/acs.chemrestox.5b00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oe T, Arora JS, Lee SH, Blair IA. A novel lipid hydroperoxide-derived cyclic covalent modification to histone H4. The Journal of biological chemistry. 2003;278:42098–42105. doi: 10.1074/jbc.M308167200. [DOI] [PubMed] [Google Scholar]
- 20.Chen LJ, Hecht SS, Peterson LA. Characterization of Amino Acid and Glutathione Adducts of cis-2-Butene-1,4-dial, a Reactive Metabolite of Furan. Chemical Research in Toxicology. 1997;10:866–874. doi: 10.1021/tx9700174. [DOI] [PubMed] [Google Scholar]
- 21.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein adducts generated from products of lipid oxidation: focus on HNE and one. Drug Metab Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 22.Nadkarni DV, Sayre LM. Structural definition of early lysine and histidine adduction chemistry of 4-hydroxynonenal. Chem Res Toxicol. 1995;8:284–291. doi: 10.1021/tx00044a014. [DOI] [PubMed] [Google Scholar]
- 23.Oe T, Lee SH, Silva Elipe MV, Arison BH, Blair IA. A novel lipid hydroperoxide-derived modification to arginine. Chem Res Toxicol. 2003;16:1598–1605. doi: 10.1021/tx034178l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


