Abstract
Oxidation of critical signaling protein cysteines regulated by H2O2 has been considered to involve sulfenic acid (RSOH) formation. RSOH may subsequently form either a sulfenyl amide (RSNHR’) with a neighboring amide, or a mixed disulfide (RSSR’) with another protein cysteine or glutathione. Previous studies have claimed that RSOH can be detected as an adduct (e.g., with 5,5-dimethylcyclohexane-1,3-dione; dimedone). Here, kinetic data are discussed which indicate that few proteins can form RSOH under physiological signaling conditions. We also present experimental evidence that indicates that (1) dimedone reacts rapidly with sulfenyl amides, and more rapidly than with sulfenic acids, and (2) that disulfides can react reversibly with amides to form sulfenyl amides. As some proteins are more stable as the sulfenyl amide than as a glutathionylated species, the former may account for some of the species previously identified as the “sulfenome” - the cellular complement of reversibly-oxidized thiol proteins generated via sulfenic acids.
Graphical abstract
Introduction
This article presents both a brief overview of the chemistry of cysteine oxidation and experimental evidence concerning the detection of sulfenic acids. A large number of recent publications have suggested that that later are formed in a variety of proteins by H2O2 and other oxidants, and that these species are involved in cellular signaling as a result of their reversible formation and reduction. While there is compelling data for the formation of adducts with the chemical trapping agents (e.g. 5,5-dimethylcyclohexane-1,3-dione also known as dimedone) from such oxidant-treated proteins, the kinetic and experimental data presented here suggests that the species that react with the trapping agents are usually sulfenyl amides rather than the sulfenic acid. Kinetic data support the intermediacy of sulfenyl amides as the reactive species, at least in the case of signaling events, as alternative reactions of the sulfenic acid are likely to be too rapid to allow significant reaction with dimedone (or derivatives). The formation of such species can be enhanced by some of the methods used in previous studies to trap reactive sulfur intermediates. These data suggest that there are only a limited number of peroxidases and other select proteins, that can act as sensors and regulators of redox signaling by forming reactive sulfur intermediates (sulfenic acids or sulfenyl amides). On the other hand, proteins can be enzymatically glutathionylated. The disulfide can then reversibly react with an amide forming a cyclic sulfenyl amide and release GSH. These proteins can therefore be easily misidentified as sulfenic acid containing proteins.
Cysteine oxidation in oxidative stress
A vast literature exists on the oxidation of cysteine, either free or in proteins, by one- and two-electron oxidants. In the case of one-electron species (free radicals) the major initial species formed is a thiyl radical (RS.) as a result of hydrogen atom abstraction from the weak RS-H bond. The chemistry of such thiyl radicals has been discussed in detail in a number of excellent reviews and will not be addressed further in this paper [1,2]. It should be noted however, that one electron reactions involving freely diffusible radicals (as opposed to radicals formed in the active site of an enzyme, or on a protein during enzyme catalysis) are very unlikely to participate in signal transduction, which involves regulated enzymatic processes.
In contrast to radical-mediated reactions, enzymatically catalyzed two-electron chemistry of cysteine forms the basis of redox signaling involving hydroperoxides [3,4] – see next section. Major differences in conditions separate the chemistry that can happen under oxidative stress or in a test tube from that which occurs during physiological cell signaling. In this section we describe thiol chemistry that can occur under stress conditions. Evidence has been presented for the formation of sulfenic acids (RS-OH) species on reaction of thiols with H2O2 and other hydroperoxides, sulfenyl chlorides (RS-Cl) on reaction of thiols with HOCl, sulfenyl bromides from reaction with HOBr, sulfenyl thiocyanates (RS-SCN) from HOSCN [5], sulfenamides, sulfinamides and sulfonamides from HOCl-mediated reactions [6–8], sulfenyl amides in the protein PTP1B [9,10], semi-stable S-N derivatives on reaction with nitrosating agents (e.g. NO+, N2O3 [1]) and zwitterions (RS+-OO−) with singlet oxygen (1O2) [11]. There is therefore a large family of RS-X species known, where X can be a range of different heteroatoms. Many of these species show considerable reactivity, particularly in aqueous solution. Reaction with excess or additional oxidant can result in secondary oxidation at the sulfur center thereby giving rise to sulfinic and sulfonic acids, RSO2H and RSO3H. RSO3H can easily eliminate HSO3− producing dehydroalanine from a cysteine residue. Alternatively, these RS-X species can react with available nucleophiles, including water / HO−, thiols, thioethers and amines / amides. Reaction with water / HO− is of particular potential importance in aqueous solution due to the high concentration of this material, with this reaction resulting in the displacement of X− from RSX species. Thus sulfenyl chlorides, bromides and thiocyanates have been reported to undergo rapid hydrolysis in water to give sulfenic acids (RS-OH). Reaction with other nucleophiles can also be very rapid, particularly when the attacking species is a good nucleophile such as the thiolate anion (RS−) or even the neutral species RSH. Reaction of a sulfenic acid with another Cys residues also plays a key role in the catalytic cycles of enzymes – including the peroxiredoxins and cysteine-containing glutathione peroxidases (GPx) – that have suitably placed resolving Cys residues (discussed further below).
Reaction with amines and amides is also known, with these reactions being a source of the sulfenyl amides detected on reaction of thiols with multiple oxidants including H2O2 (for further discussion see below) and HOCl [6–8]. These reactions are particularly facile when the leaving group is very stable (as in the case of sulfenyl chlorides and bromides, where Cl− and Br− are eliminated respectively), or where alternative reactions of the sulfenic acid are prevented for steric or electronic reasons (e.g., isolation at particular sites in complex protein matrices). However, when the leaving group is poor, as is the case of sulfenic acids (where HO− would need to be eliminated), these reactions will not occur unless other groups are present that can enhance the reaction rate by stabilizing the leaving group, or by protonating the putative HO− leaving group to give H2O as the leaving species.
Reversible cysteine oxidation in redox signaling in cells
Redox signaling is believed to be largely due to the oxidation of protein cysteine residues at either the active site of an enzyme, or at sites that modify protein activity via other mechanisms. Here we will focus on reversible oxidation initiated by hydroperoxides, and particularly H2O2, rather than processes induced by other oxidants, or alkylation as observed with redox signaling proteins such as Keap1 [12]. Probably the most studied of the signaling proteins in which the active site Cys is reversibly oxidized is PTP1B [13] in which Cys215 is glutathionylated [14]. Other members of the protein tyrosine phosphatase super family undergo similar reactions, though to different extents and at different rates [15]. An example of hydroperoxide-dependent reversible oxidation of Cys residues that activates an enzyme, but where the critical Cys is not part of the active site, is the activation of the Src family of tyrosine protein kinases [16,17], and conversion of latent matrix metalloproteinases to their active form via the “cysteine switch” mechanism [18,19]. Here we will only consider reversible oxidation of protein Cys to the sulfenic, disulfide (RS-SR), and mixed disulfide (RS-SR’) forms rather than to the sulfinic, sulfonic or other oxidation states that cannot be easily reduced.
The mechanism whereby reversible oxidation of signaling protein Cys occurs is far from certain, but has severe kinetic constraints. The thiol (RSH) group of cysteine is a poor nucleophile and usually reacts very slowly with hydroperoxides. In contrast, the deprotonated thiolate form (RS−) is much more reactive, but in terms of biologically-relevant rate constants reaction with hydroperoxides is still slow, with the apparent second order rate constants, k2, being ~10 M−1s−1 [20], as a result of the leaving group being HO− (or alkoxyl ion, RO−, in the case of alkyl hydroperoxides). As noted above this process only occurs at a significant rate when the leaving group is converted to H2O (or ROH, in the case of alkyl hydroperoxides) by a suitable proton donor [21] (reaction 1). A special case may also exist if the H2O2 is bound to a co-factor in an enzyme active site, with this potentially resulting in enhanced reactivity, or if the oxidant is an alternative species such as a high-oxidation state iron-oxo complex.
| (1) |
For GSH (or Cys), direct reaction with hydroperoxides is dependent on deprotonation of the former and protonation of the latter, which at physiological pH is almost negligible. Nonetheless, the tiny amount of protonation accounts for the slow rate of oxidation. In contrast, rapid oxidation of active site Cys is part of the catalytic mechanism of peroxiredoxins (Prdx) and cysteine-containing glutathione peroxidases (GPx). Related chemistry occurs with selenocysteine-containing glutathione peroxidases, where the selenocysteine takes the place of one of the Cys residues, and is particularly reactive as it is present in its ionized (RSe−) form due to the much lower pKa of Sec (~ 4.8; [22]) compared to 8-9 for most thiols. The catalytic activity of this enzyme has been thoroughly investigated, with the reaction of the active site RSe− (or RS−, in the case of Cys-containing species) with ROOH, occurring simultaneously with the protonation of the leaving group by a neighboring, proton-donating amino acid [23] as in reaction 1. The rate constant for this enzymatically catalyzed first step is ~ 107 M−1s−1 or approximately a million times faster than for the thiolate of free Cys at pH 7. For reviews of this chemistry in the context of redox signaling see [3,4].
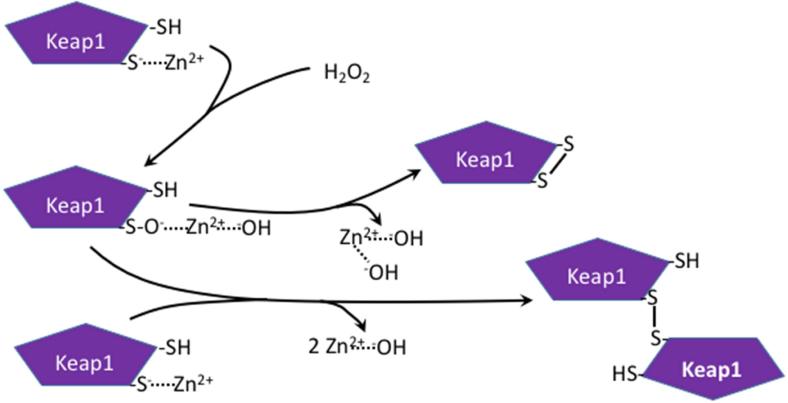
Oxidation to form disulfides has been observed with some proteins in which a zinc ion is bound to the Cys residues resulting in Zn2+-hydroxyl species being the leaving group instead of water or an alcohol. This is shown in Figure 1, which is based on studies showing the involvement of Zn2+ in Keap1 regulation [24] and evidence of both intramolecular disulfide and a dimer formed by a disulfide bond during Keap1 modification by H2O2 [25].
Figure 1.
Proposed mechanism of involvement of Zn2+ in H2O2-mediated oxidation of Cys residues on Keap1 and regulation of protein activity via intra- and inter-molecular disulfide formation. Oxidation of Cys residues in proteins by H2O2 or ROOH, in which a zinc ion is bound to the Cys residues, results in Zn2+-hydroxyl species being the leaving group instead of water or an alcohol, and result in rapid disulfide formation. Adapted from [24,25].
The α and β isoforms of protein kinase C (PKC) contain both active site Cys residues, and Cys bound to zinc in their regulatory domains [26]. Oxidation of the active site Cys requires non-physiological concentrations of H2O2 and results in the inactivation of the enzyme [27]. In contrast, low H2O2 concentrations in the presence of ATP and Mg2+ activate PKC through selective oxidation of the regulatory domain cysteines [28].
The protein tyrosine phosphatase 1B (PTP1B) quandary
As mentioned above, protein tyrosine phosphatase 1B is the prototypic signaling protein that is reversibly inactivated by H2O2 [9]. Early studies demonstrated the formation of a mixed disulfide with the active site Cys [14]. Oxidation at other (non-active site) Cys residues can also affect the activity of the enzyme [29]. The H2O2-dependent formation of a reversibly glutathionylated form of PTP1B was also shown in stimulated macrophages [30] demonstrating its physiological formation. Using purified PTP1B the formation of a sulfenyl amide has been demonstrated in vitro in the absence of GSH [9,10], with the formation of this species clearly consistent with the formation of an intermediate sulfenic acid. It has been proposed that this species is formed in cells stimulated with insulin or epidermal growth factor (i.e., in the presence of GSH) [9]. But, as we will describe below, the sulfenyl amide is far more likely produced by reaction of the mixed disulfide with an amide than direct thiolate oxidation – see below.
This in vitro chemistry is elegant, but the implication that this would explain physiologically reversible PTP1B inactivation does not fit with some of the known chemistry of PTP1B and conditions within the cell. For the in vitro studies, one issue is the absence of GSH that, at millimolar or greater concentration in cells, should react with any sulfenic acid formed thereby inhibiting sulfenyl amide formation. In addition is has been shown that soaking crystals of the purified oxidized PTP1B, containing the sulfenyl amide, with 20 mM GSH (i.e. only slightly higher than physiological levels) reverts the enzyme to the reduced Cys form [10].
A second, and perhaps greater, problem is that the rate constant for oxidation of PTP1B has been reported by several groups, as being 9-43 M−1s−1 [14,31,32], which would be expected to be outcompeted by reaction of the oxidant with GSH (see rate constants above) given these values and the much higher concentration of GSH relative to PTP1B [3,33,34]. Thus, the formation of a long-lived sulfenic acid intermediate from PTP1B under normal physiological signaling conditions is unlikely. One could propose that if the H2O2 required for sulfenic acid formation were generated within a very short distance from the PTP1B, it would be able to reach the active site Cys before being intercepted by GSH. One major problem is that a major source of H2O2 produced by cells for signaling comes from extracellularly generated H2O2 produced by dismutation of O2.− generated by NADPH oxidases. The H2O2 then has to enter through an aquaporin channel before it could encounter PTP1B inside cells. Even if another source of H2O2 such as mitochondria or NOX4 produced the H2O2, probability again suggests that GS− would be a more likely target for the H2O2 by approximately 100,000-fold. The proximity of the oxidant and target is even more of an issue when one considers the catalytic rate and abundance of the peroxiredoxins and glutathione peroxidases that remove H2O2. Thus, the probability of sulfenic acid formation from the active site Cys of PTP1B, and many other proteins that do not have a fast rate constant for reaction with H2O2 is unlikely under physiological conditions.
Clearly there are considerable problems in explaining both the detection of disulfide and mixed glutathione disulfide forms of signaling proteins, and the widespread detection of sulfenic acids on many proteins under physiological conditions. An answer is provided below.
Enzymatic disulfide formation in signaling proteins
As the rate constants for oxidation of protein Cys by H2O2 / ROOH that do not have the assistance of a neighboring proton donor or a bound zinc are clearly low, and unlikely to be competitive, what could account for the observed disulfide (intramolecular and intermolecular) and glutathione mixed disulfides that have been observed in numerous studies of signaling proteins [35–37]? Protein disulfide formation is a very large and important field, particularly as it constitutes an essential part of protein folding. Reversible oxidation of thioredoxin (Trx), described in [35], is the best documented reversible disulfide formation of a signaling protein. But, how are reversible disulfides formed in other signaling proteins?
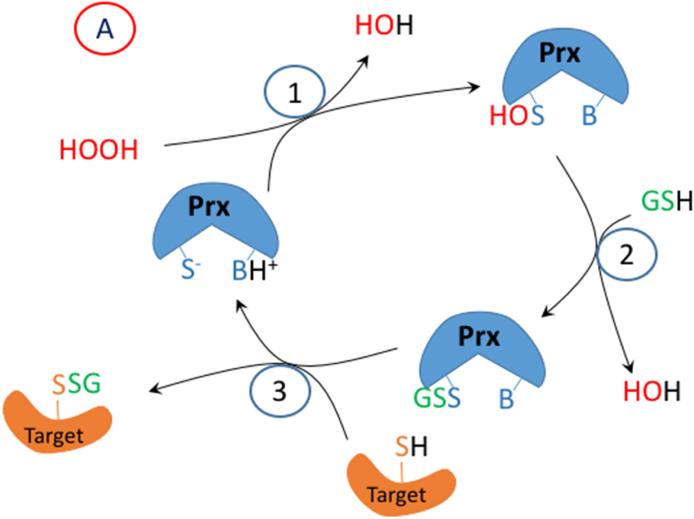
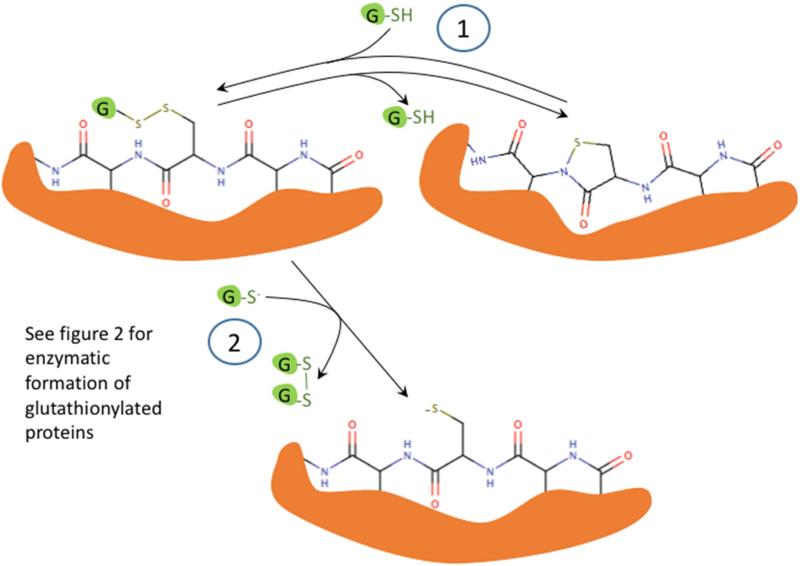
To address this question, we will discuss briefly a few studies that describe enzymatic pathways to disulfide formation in redox signaling. Based on these we have proposed mechanisms that are illustrated in Figure 2. The initial protein thiolate in these reactions forms a sulfenic acid, but these are in proteins, the peroxiredoxins, that have a suitably-placed proton donor to facilitate cleavage of the peroxide (-O-O-) bond. A disulfide in the peroxiredoxin or glutathione peroxidase is formed by reaction between the oxidized peroxidatic Cys and the reduced resolving Cys or GSH. But, then the disulfide in the target signaling protein is formed by disulfide exchange with the peroxidase (Figure 2 A or B). Thus Dick and coworkers have shown that Prdx2, which has a rate constant for sulfenic acid formation of 107 M−1s1 catalyzes the oxidation of STAT3 to form disulfide linked oligomers, thereby resulting in a transient loss of STAT3 function [38]. Similarly, Ledgerwood and coworkers have demonstrated the formation of intermolecular disulfides between Prdx1 and ASK1, a mitogen activated protein kinase kinase kinase that is upstream of both JNK and p38MAPK [39].
Figure 2.
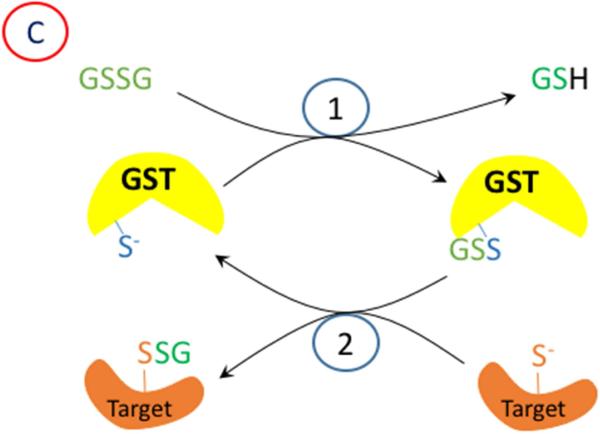
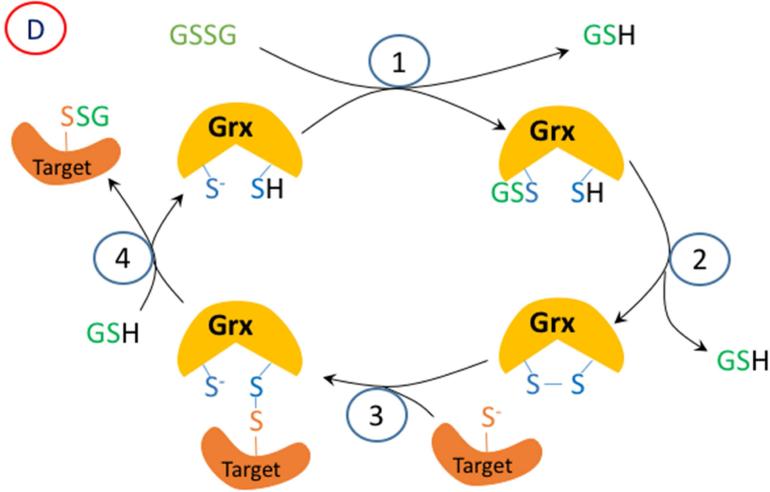
A proposed mechanism for enzymatic glutathionylation of target signaling proteins by peroxiredoxin (Prx), glutathione S-transferase π (GST), or glutaredoxin (Grx). Similar reactions may also occur with GPx. In (A), 1- H2O2 oxidized Prx; 2- GSH forms a disulfide with Prx; 3- disulfide exchange occurs with the target protein thiolate. In (B), steps 2 and 3 are in reverse sequence from (A). In (C), 1- GSSG, which must be formed by a closely associated peroxidase undergoes disulfide exchange with the thiolate in the active site of GST π; 2- disulfide exchange occurs with the target protein thiolate. In (D), GSSG, which must be formed by a closely associated peroxidase undergoes disulfide exchange with the thiolate in the active site of Grx; 2- An intramolecular disulfide forms in the active site of Grx; 3- disulfide exchange occurs with the target protein thiolate; 4- disulfide exchange occurs with GSH.
Recently, Tew and Janssen-Heininger and their colleagues have demonstrated that glutathione S-transferase π (GSTP) can glutathionylate IKKβ with GSSG, and thereby activate NF-κB [40]. This process requires a source of GSSG, which is likely to be a GPx or Prdx6 closely associated with the target protein, so that a relatively high concentration of GSSG is present in the immediate location. Interestingly, while Prdxs (other than Prdx6) supposedly use Trx specifically, rather than GSH as a substrate, Winterbourn and coworkers recently described the glutathionylation of Prdx2 and its recycling by glutaredoxin [41]. It is conceivable that the GSH could be transferred to another protein as proposed in these other studies. While GSTP may function by catalyzing disulfide exchange between a protein thiolate and GSSG, a much better known enzyme with this function is glutaredoxin-1. Mailloux and Treberg have demonstrated that glutathionylation of several signaling proteins can be catalyzed by glutaredoxins [42]. For this to occur in a cell where the total GSSG is only about 1% of the total glutathione (i.e. 20 - 100 μM), the source of the GSSG would again need to be very close spatially to the GSTP or glutaredoxin, and the target species.
Proposed mechanisms for the glutathionylation of target signaling proteins is given Figure 2. At this point in time, which of the mechanisms described above may be responsible is uncertain, but it seems clear that widespread non-enzymatic reaction of H2O2 with protein Cys residues to form sulfenic acids is unlikely under physiological conditions. However, disulfide formation is known, at least in some cases, to be enzymatic, which would fit the expectation of signal transduction being a tightly regulated process.
Other possible routes to sulfenic acid and sulfenyl amide
Before discussing how sulfenic acids or a sulfenyl amides can be detected and quantified under physiological conditions, another essential question needs to be answered: can sulfenic acids be formed by other processes (in addition to Cys oxidation proposed in many of the above studies) given the clear kinetic and biochemical constraints on their formation? Some evidence is available in the literature that this can be the case.
It was proposed over a century ago that disulfides hydrolyze to yield a thiol and sulfenic acid where the R groups may be the same or different [43,44] (reaction 2).
| (2) |
The thiol product was easy to identify, but the RSOH was elusive. Usually, the more extensively oxidized sulfinic or sulfonic acid was found. Indeed, the labile nature of RSOH has always resulted in problems in detecting this species. Nonetheless, the only conceivable way for the disulfide and water to yield a thiol and a more oxidized form of the other sulfur atom requires a sulfenic acid intermediate. While experimental evidence fails to demonstrate the formation of sulfenic acid, hydrolysis can be shown to occur at very low yield with production of sulfinic and sulfonic acids and the beta cleavage product (data not shown). Thus, hydrolysis of disulfides occurs, but cannot account for the detection of dimedone adducts.
Several decades after the initial studies with disulfides, researchers began to investigate the oxidation of cysteine in proteins. As described earlier, addition of exogenous (and often high levels) of H2O2 to proteins has allowed what was thought to be sulfenic acids to be detected using reagents such as 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole (NBD-Cl) that forms different products with sulfenic acids and thiols [45]. Sulfenic acids have also been detected with other hydroperoxides using this method [46]. However, at a similar time, 5,5-dimethyl-1,3-cyclohexanedione (dimedone) was introduced, which was purported to react specifically with sulfenic acids [45]. Dimedone and its derivatives react slowly with sulfenic acids, and it was not widely employed until studies on the cell permeability of these compounds highlighted the greater penetration of dimedone in to cells [47]. The authors who devised many of the protocols for measuring sulfenic acid in proteins using dimedone [48,49], appear to have ruled out direct reaction of dimedone with disulfides. On the other hand, the potential reaction of dimedone with sulfenyl amides has been largely ignored with regard to protein chemistry.
Experimental evidence for multiple routes and sources of dimedone-reactive species
Methods for the experiments described here follow the discussion.
Incubation of either lipoic acid or GSSG (100 μM) with dimedone (200 μM, from a 10 mM stock solution in 95% ethanol) for short periods of time (e.g. 30 min) at pH 7.4, before analysis using a QTOF mass spectrometer, did not provide any evidence for the formation of dimedone adducts, though low levels of reduced GSH from GSSG were detected. Similar data were obtained when either GSSG or dithiodiglycolic acid (both 1 mM) were incubated for 2-18 h with a much higher concentration of dimedone (12 mM), before analysis by MS. No evidence for dimedone adducts were seen, and only very low concentrations of GSH and reduced thioglycolic acid were detected.
Similarly, when the disulfide (1 mM) formed on cross-linking of the Cys residues in the peptides TAVGPCPASGK and TAVGGCGASGK, was incubated for 2 h with 12 mM dimedone, the majority of the dipeptide remained in its disulfide form. Traces of the reduced (thiol-containing) compounds were detected (presumably originating from hydrolysis) together with limited amounts of oxidation products (data not shown). No dimedone adducts were detected. In particular, the sulfonic acid forms (RSO3H) of both peptides were observed, together with dehydroalanine (DHA), which is known to arise from β-elimination of sulfite from the sulfonic acid. These data indicate that disulfide hydrolysis can occur on extended incubation, with consequent formation of both the reduced species and oxidation products, but not dimedone adducts. Furthermore, it is clear that any sulfenic acids formed undergo other reactions (e.g., further oxidation) in preference to reaction with dimedone, even when the latter is present at high levels.
In contrast, when BSA (final concentration, 0.22 mg/mL) which was preincubated without or with GSH to form the glutathionylated species (with the excess GSH subsequently removed), was reacted with dimedone (5 mM) for 30 min, a peptide (GLVLIAFSQYLQQCPFDEHVK) was detected with dimedone attached to the Cys, as shown by a characteristic mass addition of 138 Da. However, this oxidation product was also detected with BSA incubated for 72 h without GSH (data not shown). These data indicate that long term, but not short term, incubation of proteins can result in the formation of species that react with dimedone even in the absence of any added oxidant such as H2O2. Slow reaction with low levels of peroxide formed during the extended incubation cannot however be excluded as a source of the dimedone reactive species in this case. These data indicate that considerable care needs to be taken to avoid artefactual dimedone adduct formation.
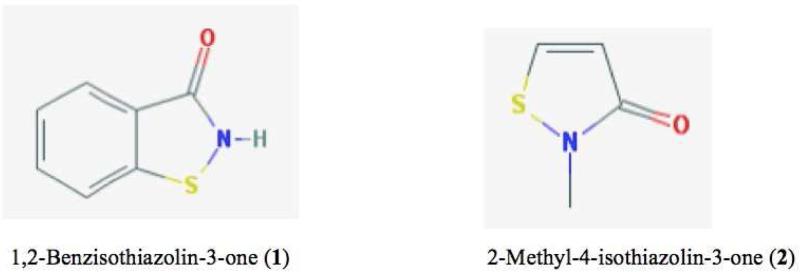
The second question as to whether other species, apart from sulfenic acids, may react with dimedone has been addressed experimentally by carrying out studies with two purified, commercially-available sulfenyl amides: 1,2-benzisothiazolin-3-one (1) and 2-methyl-4-isothiazolin-3-one (2) (for structures see Figure 3). The former is generated from a primary amide, whereas the latter arises from a secondary amide and more closely models sulfenyl amides that may be formed in proteins. For compound (1), incubation of this compound at 1 mM with dimedone (1.2 mM) in aqueous buffer-CH3CN mixtures (75:25 v/v), or 0.5 M (1) with 0.5 M dimedone in CH3CN, for 0.1 – 18 h with subsequent analysis by MS, did not provide any evidence for dimedone adducts (data not shown). These data indicate that this sulfenyl amide does not undergo ready hydrolysis even in alkaline conditions to give dimedone reactive species.
Figure 3.
Structures of sulfenyl amides examined.
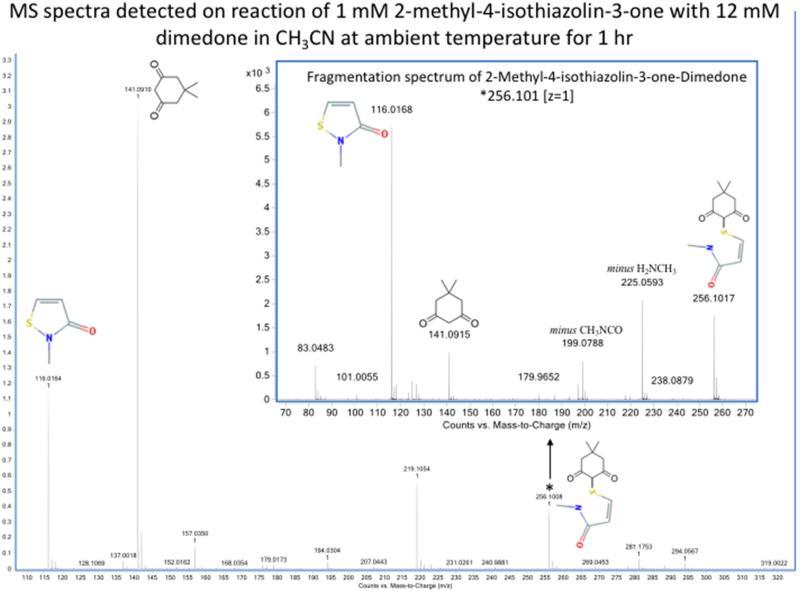
In contrast, incubation of compound (2) (1 mM or 0.5 M) with 1.2 mM or 0.5 M dimedone respectively in aqueous buffer for 0.1 – 18 h gave high yields of dimedone adducts (Table 1). Similar reactions in CH3CN (1 mM (2) with 1.2 mM dimedone) also gave high yields of dimedone adducts (Figure 4), with higher levels detected in this aprotic solvent compared to the aqueous buffer system (Table 1). In neither case were any hydrolysis products detected. These date are consistent with direct reaction of the sulfenyl amide with dimedone without the intermediacy of a sulfenic acid. The increased yield detected under aprotic conditions compared to aqueous buffer (Table 1) is consistent with the occurrence of a direct sulfenyl amide – dimedone reaction.
Table 1.
2-Methyl-4-isothiazolin-3-one reaction ratios with dimedone (dimedone adduct m/z 256.1; unreacted sulfenyl amide m/z 116.016)
| Condition | Sulfenyl amide* | Dimedone | Reaction time** | Intensity ratio % (m/z 256.1:116.016) |
|---|---|---|---|---|
| Buffer | 1 mM | 1.2 mM | 1 h | 4.4 |
| CH3CN | 1 mM | 1.2 mM | 1 h | 8.1 |
| Buffer | 0.5 M | 0.5 M | 1 h | 13.3 |
| CH3CN | 0.5 M | 0.5 M | 1 h | 26.0 |
| CH3CN | 0.5 M | 0.5 M | 18 h*** | 34.0 |
Buffer: 75:25 v/v ammonium bicarbonate 40 mM, pH 8 : CH3CN
All data refer to 2-methyl-4-isothiazolin-3-one, since no reaction of 1,2-benzisothiazolin-3-one was detected in any of these conditions
Results obtained from 10 to 120 min were not significantly different (data not shown).
All reactions were carried out at ambient temperature. Long term reaction was performed at 37 °C.
Figure 4.
Detection by mass spectrometry of dimedone adducts of sulfenyl amides arising from incubation of compound (2) with dimedone. MS spectra showing the formation of the dimedone adduct of 2-methyl-4-isothiazolin-3-one corresponding to m/z 256.101 (z = 1). The identity of this adduct has been confirmed by the MS/MS spectra obtained by CID fragmentation (insert). Samples were analyzed by direct infusion as described in the Material and methods. Resolution 10000, accuracy 5 ppm.
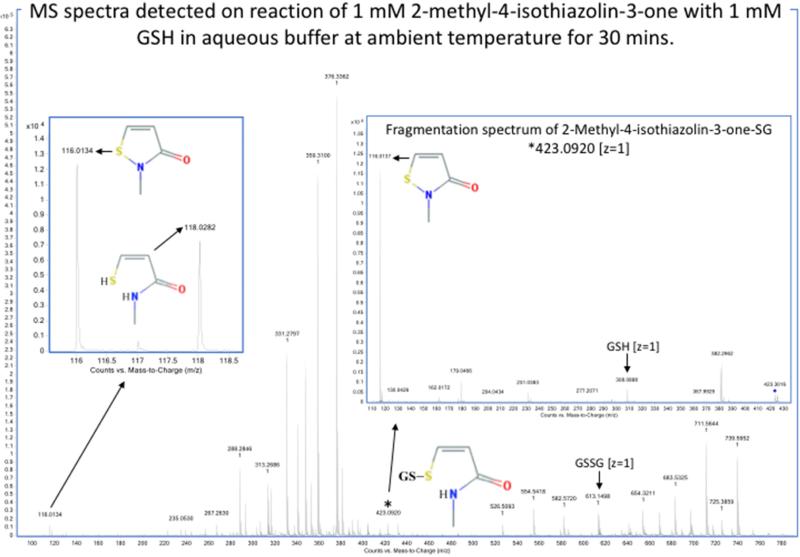
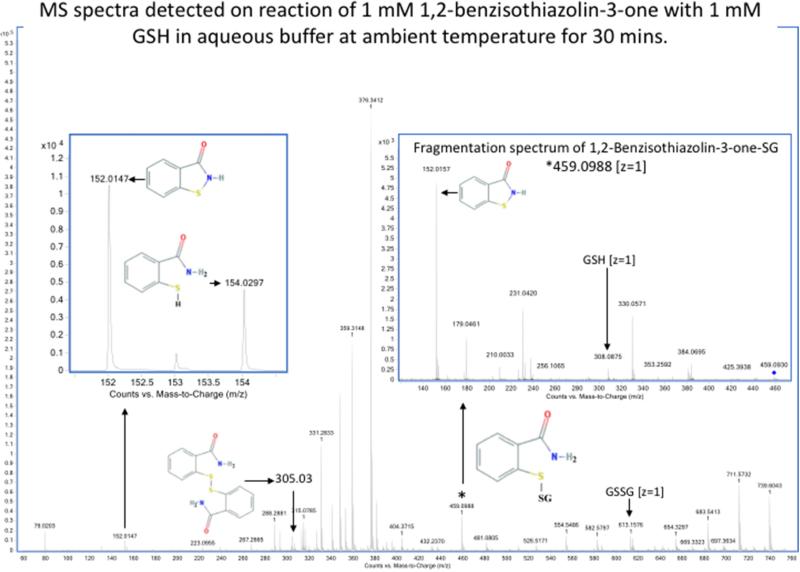
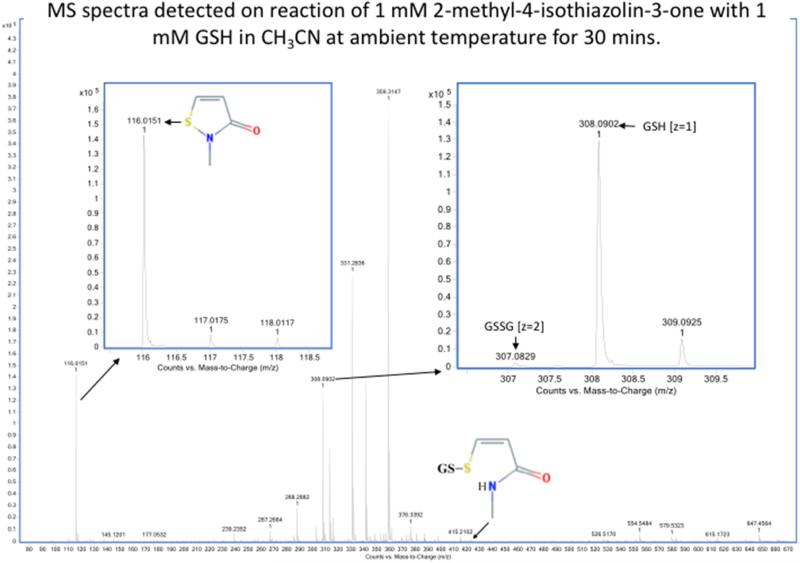
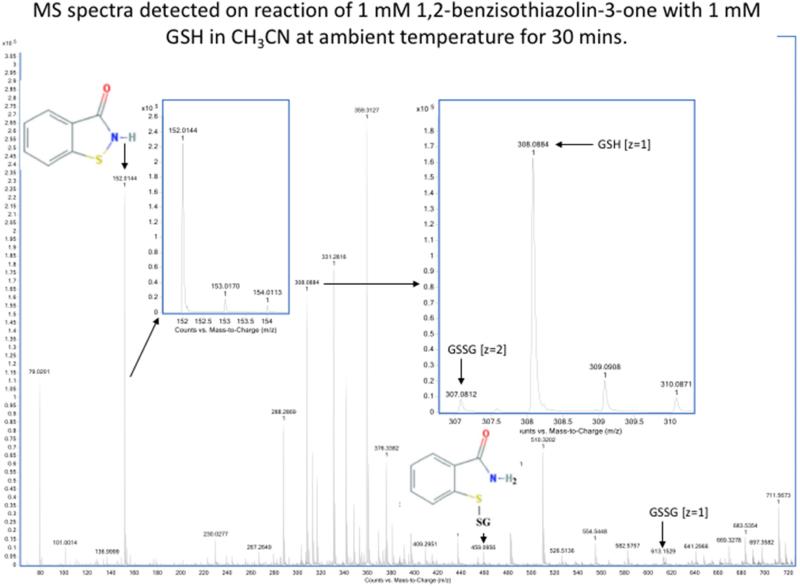
When these two sulfenyl amides (1 and 2) are treated with stoichiometric concentrations of GSH, rapid conversion to the mixed disulfides was detected, together with the disulfide linked dimer and reduced form of the compound (i.e., with a free –SH and amide) (Figure 5a and Table 2). This is consistent with GSH-catalyzed conversion of the sulfenyl amide back to an open chain state. This conversion occurs to only a very minor extent in CH3CN (Figure 5b and Table 2), but occurs readily in the presence of traces of water or buffer, and is rapid in aqueous buffer. These data indicate that the reaction of the cyclic sulfenyl amide with GSH occurs predominantly via the thiolate anion (GS−). With equimolar concentration of the sulfenyl amides and GSH, the extent of conversion to products was significant, and the reaction occurred rapidly with both sulfenyl amides. Moreover it was more effective with (1) than (2) (Table 2), and contrasts with the results obtained with dimedone, which failed to react with (1). The results cast further doubt on the role of dimedone as a specific trap for sulfenic acids.
Figure 5a.
Detection by mass spectrometry of products of reaction of sulfenyl amides arising from incubation with GSH in aqueous buffer. MS spectra showing the compounds arising from (1)1,2-benzisothiazolin-3-one (m/z 152.0165, z = 1) (upper panel) and (2) 2-methyl-4-isothiazolin-3-one (m/z 116.0165, z = 1) (lower panel) after 30 minutes of incubation with equimolar reduced glutathione in aqueous buffer (75:25 v/v, ammonium bicarbonate 40 mM, pH 8.0, and CH3CN). The main products observed are: the glutathionylated form (1) m/z 459.0988 (z =1) and (2) m/z 423.092 (z =1); the reduced form (1) m/z 154.0321 (z =1) and (2) m/z 118.0321 (z =1), the disulfide linked dimer of sulfenyl amides (1) m/z 305.039 (z =1) and the oxidized glutathione (GSSG) m/z 613.159 (z =1). The identity of the glutathionylated form was confirmed on the basis of the MS/MS spectra obtained by CID fragmentation (inserts). Samples were analyzed by direct infusion as described in the Material and methods. Resolution 10000, accuracy 5 ppm.
Table 2.
Sulfenyl amide reactions ratio with glutathione. MS monoisotopic intensities of all detected forms of each compound and glutathione are presented.
| Compound | Condition | Compound forms detected by MS | GSH forms | ||||
|---|---|---|---|---|---|---|---|
| Sulfenyl amide | Reduced | Dimer | GSH adduct | GSH | GSSG | ||
| 2-Methyl-4-isothiazolin-3-one | Buffer, 30 min | 40 % | 20 % | 2 % | 38 % | 1.5 % | 98.5 % |
| 1,2-Benzisothiazolin-3-one | Buffer, 30 min | 13 % | 5 % | 39 % | 43 % | 1.3 % | 98.7 % |
| 1,2-Benzisothiazolin-3-one | Buffer, 48 h | 34 % | 10 % | 25 % | 31 % | 2.1 % | 97.9 % |
| 2-Methyl-4-isothiazolin-3-one | CH3CN, 30 min | 98.6 % | -- | -- | 1.4 % | 99 % | < 1 % |
| 1,2-Benzisothiazolin-3-one | CH3CN, 30 min | 96.5% | -- | -- | 3.5 % | 99 % | < 1 % |
Buffer: 75:25 v/v ammonium bicarbonate 40 mM, pH 8 and CH3CN
Figure 5b.
Detection by mass spectrometry of products of reaction of sulfenyl amides arising from incubation with GSH in aprotic buffer. MS spectra showing the compounds arising from (1) 1,2-benzisothiazolin-3-one (m/z 152.0165, z = 1) (upper panel) and (2) 2-methyl-4-isothiazolin-3-one (m/z 116.0165, z = 1) (lower panel) after 30 minutes of incubation with equimolar reduced glutathione in 100% CH3CN. The main products observed are: the glutathionylated form (1) m/z 459.0988 (z =1) and (2) m/z 423.092 (z =1) and very small amount of the oxidized glutathione (GSSG) m/z 613.159 (z =1) and m/z 307.09 (z =2) as evidenced from comparison with reduced glutathione (GSH) m/z 308.091 in right inserts. No reduced forms of (1) and (2) were detected as shown in the left-hand inserts. Samples were analyzed by direct infusion as described in the Material and methods. Resolution 10000, accuracy 5 ppm.
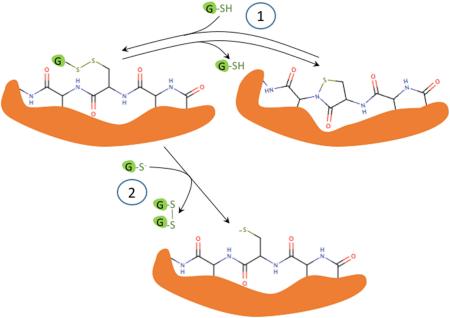
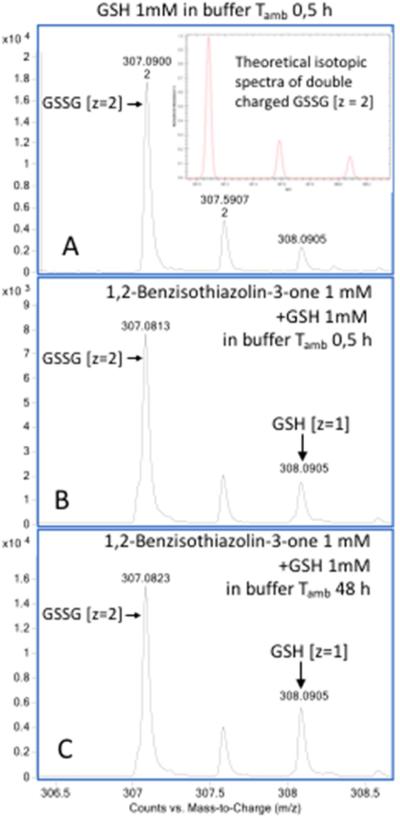
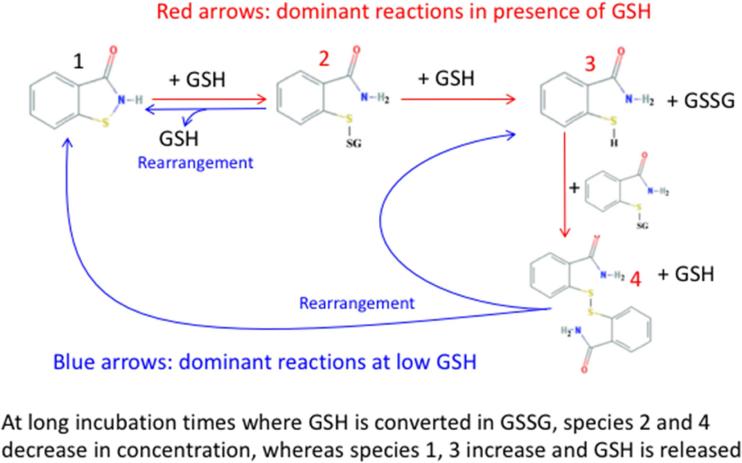
Interestingly, on prolonged incubation (48 h) after complete oxidation of added GSH (Figure 6 panel A), a decrease in the levels of the glutathionylated sulfenyl amides and disulfide linked dimer of sulfenyl amide was detected, with a concurrent increase of both reduced and oxidized (cyclic) sulfenyl amides (Table 2). This is consistent with a rearrangement of the glutathionylated sulfenyl amides and disulfide-linked dimer of sulfenyl amides as proposed in Figure 7. This interpretation is also supported by a parallel increase in GSH levels as expected from the proposed rearrangement (Figure 6, panels B and C; Table 2).
Figure 6.
Evaluation by mass spectrometry of reduced glutathione (GSH) levels following incubation of (1) in aqueous buffer containing an equimolar concentration of GSH. The amount of GSH has been estimated by the intensity of its monoisotopic mass (m/z 308.091, z =1) corrected for the intensity of the isobaric component present in the isotopic pattern of concurrent double charged GSSG (m/z 307.09, z =2), equivalent to 14.7 % of the monoisotopic mass intensity (insert in panel A). GSH (1 mM) was incubated 0.5 hours in aqueous buffer (75:25 v/v ammonium bicarbonate 40 mM, pH 8.0, CH3CN) with the resulting analysis showing > 99% oxidation to GSSG, as deduced from isotopic ratio of the double charged GSSG (panel A). Meanwhile the presence of (1) 1,2-benzisothiazolin-3-one gave rise to significant concentrations of GSH that increased over 48 hours (panel B, C and table 2). Samples were analyzed by direct infusion as described in the Material and methods. Resolution 10000, accuracy 5 ppm.
Figure 7.
Proposed interconversion between sulfenyl amide and glutathionylated forms.
Conclusions
The kinetic, mechanistic and experimental data outlined above, suggest that much of the data that has been published in the literature as the cellular “sulfenome” (the cellular complement of reversibly oxidized thiol proteins generated via sulfenic acids) actually arises from reaction of proteins containing a sulfenyl amide that can be trapped with dimedone. These data indicate that dimedone is less selective than initially reported, and reacts relatively slowly with these species. However whilst new probes are being developed that show higher reactivity with sulfenic acids than dimedone, an underlying kinetic problems remains. Thus the rate constants available for the reactions of H2O2, together with information on the physiological concentrations of GSH, and Cys residues on target proteins, indicate that direct oxidation of most protein thiols by H2O2 in cells to give sulfenic acids is unlikely. Reaction with protein thiolates (i.e., reaction with ionized Cys residues) is somewhat more rapid, but is still – in many cases – not competitive when compared to reaction with GSH, and the enzymatic systems that remove H2O2 and hydroperoxides within cells. There are however, significant and important exceptions, with these predominantly being protein thiolates present in the active site of peroxidases, and zinc-bound cysteines, where special structural and electronic conditions exist that enhance the rate of reaction by facilitating cleavage of the peroxide bond.
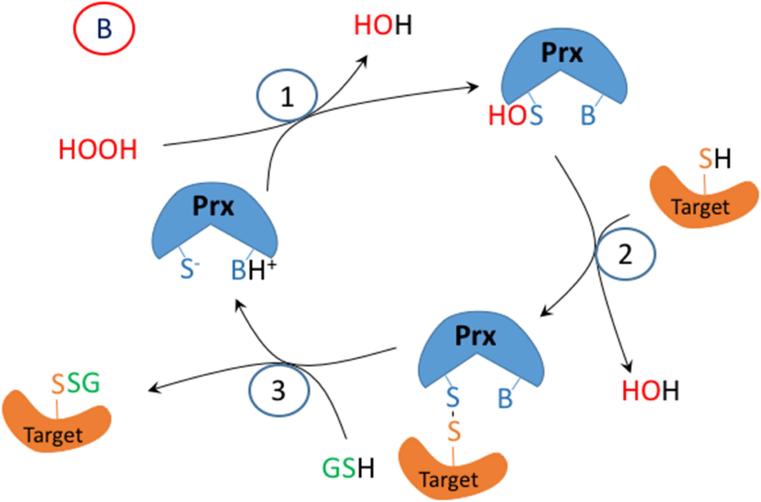
The experimental evidence suggests firstly, that hydrolysis of disulfides can occur to form a thiol and sulfenic acid, but that the sulfenic acid once formed does not have a sufficiently long lifetime to react efficiently with dimedone, with the sulfenic acid preferentially undergoing further oxidation to give sulfinic and sulfonic acids, and dehydro species from sulfite elimination (i.e., β-cleavage). Secondly, some sulfenyl amides react with dimedone to give adducts that are identical to those formed by reaction with a sulfenic acid. Thus, they may be mistakenly assumed to arise from a sulfenic acid. Thirdly, when sulfenyl amides undergo hydrolysis the yield of the dimedone adduct is significantly decreased, suggesting that dimedone reacts more rapidly with the sulfenyl amide rather than the sulfenic acid. Finally, sulfenyl amides react rapidly and reversibly with GSH to produce the amide and thiolate, which would account for the restoration of oxidized signaling proteins to their original state. Thus, we propose that the major pathway for sulfenyl amide formation in proteins that cannot protonate H2O2 (or otherwise facilitate its decomposition) or contain zinc-thiolate groups, is enzyme-catalyzed glutathionylation followed by reaction of the disulfide with an amide (Figure 8). Such a pathway would offer significant advantages in terms of “control” of signaling processes due to the ready enzymatic reversibility of glutathionylation reactions.
Figure 8.
Proposed mechanism for sulfenyl amide formation and reduction in proteins. Step 1 shows the reversible reaction between the sulfenyl amide- and disulfide-containing protein. Step 2 shows the reversible reaction between the glutathionylated and completely reduced-protein.
So, the question then arises as to the reasons for the widespread literature reports of sulfenic acid formation on proteins in intact cells. The data presented here indicates a number of possibilities. Firstly, data has been obtained for direction reaction of sulfenyl amides – which appear to have considerably longer lifetimes than the sulfenic acids – with dimedone. These sulfenyl amides may be generated in preference to sulfenic acids, due to their greater stability, during the degradation of disulfides (e.g., glutathionylated proteins). Formation of sulfenyl amides over sulfenic acids may have a considerable evolutionary pressure, as a result of the tendency of sulfenic acids to be readily oxidized to sulfinic and sulfonic acids that cannot be easily repaired, unlike the sulfenyl amides. The rapidity of these over-oxidation reactions, account for the detection of oxy acids and dehydroalanine from GSSG and peptides, without significant formation of sulfenic acids and dimedone adducts.
It has been claimed that the “sulfenome” differs from the disulfideome based on comparison of reported proteomes and a published Venn diagram [50]. However, examination of the specific proteins discussed in the “Signal Transduction” and “Metabolism” sections of this previous study, indicates that all of these (so-called) sulfenic acid-containing proteins can also exist in a glutathionylated form: aldolase and enolase [51], pyruvate kinase [52], glyceraldehyde phosphate dehydrogenase [53], phosphoinositide-3-kinase [54], IQGAP1 [54] and PP1 [54]. Thus the “sulfenome” may actually consist of proteins containing sulfenyl amides. Furthermore, we have provided evidence for the reversible formation of sulfenyl amides from a mixed disulfide and an amide. Thus, differences between the “sulfenyl amidome” and the “disulfidome” are likely to be due to some proteins in cells being more stable as the sulfenyl amide than the glutathionylated form. Perhaps, a more consistent and accurate term, if one insists on using “ome,” would be the “reversibly oxidizable thiolateome.”
Experimental methods
BSA (Sigma A9647) at a final concentration of 2.8 mg mL−1 was incubated at 4 °C for 72 h with or without GSH (Sigma G4251) at a final concentration of 0.22 mg mL−1. The reaction pH (7.4) was maintained using sodium phosphate buffer (0.1 M). Subsequently excess GSH and salt were removed using a PD10 column, and the samples incubated for 30 min with dimedone (5 mM, from a stock solution in 95% ethanol), before analysis by LC-MS/MS as described previously [55]. Briefly, 10 μg of the protein was dried down in a SpeedVac RVC 2-33 (Christ, Germany), denaturated using 8 M urea, reduced with 40 mM DTT for 45 min, and alkylated with iodoacetamide (77 mM) for 60 min in the dark. The samples were then diluted 4-fold with ammonium bicarbonate buffer (10 mM) and digested overnight at 37 °C with trypsin (Promega) at a proteinase : protein ratio of 1:50 ratio. Reaction was stopped by adding 0.2% trifluoroacetic acid (TFA), then desalted using c18 Ziptips (Merck Millipore, Germany). Samples were separated using a Dionex 3000 UPLC system (Thermo Scientific, Sunnyvale, CA) interfaced with a Bruker Impact 2 QTOF mass spectrometer equipped with a Bruker Apollo II Electro source (Bruker, Bremen, Germany). Peptides (20 μL injections) were separated at a flow rate of 0.2 mL min−1 using an Aeris XB-C18 LC column (2.6 μm, 250 × 2.1 mm; Phenomenex, Torrance, CA), using gradient elution. Solvent A was H2O containing 0.1% TFA, and solvent B 90% acetonitrile, and 0.1% aqueous TFA. Over the first 30 min solvent B was gradually increased from 2% to 45%, then increased to 90% over 1 min, held at this level for 5 min, then. decreased to 2% and the column re-equilibrated, with a total run time of 50 min. Maxquant proteomics software (Max Planck institute, Germany) was used for data analysis. Dimedone addition to cysteine (+ 138 Da) and carbamidomethylation of cysteine (+ 57 Da) were specified as variable modifications. Lipoic acid (Sigma T5625) and GSSG (Sigma G4376) were dissolved in 50% CH3CN (Sigma 34998) at a concentration of 100 μM and incubated for 30 min with or without 200 μM dimedone (from a 10 mM stock solution in 95% ethanol), before direct injection into the QTOF mass spectrometer at a flow rate of 160 μL h−1. The samples containing lipoic acid were run in a negative ion mode, whereas the GSSG samples were acidified with 1% formic acid and run in positive ion mode.
For studies of reactivity of disulfide with dimedone, disulfide linked synthetic di-peptides TAVGPCPASGK and TAVGGCGASGK with the final concentration of 1 mM were incubated for 2 h at ambient temperature with dimedone (final concentration 12 mM, from a stock solution 1 M in DMSO). The pH of 8.0 was controlled by ammonium bicarbonate buffer (40 mM). Similarly, for studies of reactivity of sulfenyl amides with dimedone, 2-Methyl-4-isothiazolin-3-one (stock solution 1 M in CH3CN) or 1,2-Benzisothiazolin-3-one (stock solution 1 M in DMSO) with the final concentration of 1 mM or 0.5 M, were incubated for up to 18 hours at ambient temperature with dimedone (final concentration respectively of 1 mM or 0.5 M from a stock solution 1 M in DMSO). All reactions were carried out both in aqueous buffer (75:25 v/v ammonium bicarbonate 40 mM, pH 8.0 : CH3CN) or 100% CH3CN in order to evaluate the effect of aprotic solvent on reactivity. The reactivity of sulfenyl amides was also tested against glutathione, by incubating 2-methyl-4-isothiazolin-3-one or 1,2-benzisothiazolin-3-one with the final concentration of 1 mM with equimolar glutathione (final concentration 1 mM from a stock solution 250 mM in H2O mQ). Reactivity of sulfenyl amides with glutathione was assayed both in aqueous buffer (75:25 v/v ammonium bicarbonate 40 mM, pH 8.0 : CH3CN) or 100% CH3CN for up to 48 h at ambient temperature.
MS analysis of the peptides TAVGPCPASGK and TAVGGCGASGK after reaction with dimedone was carried out by separating the materials by means of reversed-phase chromatography on a nano-fluidic HPLC-Chip apparatus, with subsequent analysis using a coupled quadrupole ion trap and time of flight mass spectrometer, using the 6520 Accurate-Mass Q-TOF LC/MS system (Agilent Technologies, Santa Clara, CA, USA) equipped with MassHunter Workstation Software Qualitative Analysis B.02.00 as graphical interface for data handling. A 1200 Rapid Resolution system (Agilent Technologies, Santa Clara, CA, USA) was used containing a binary pump and degasser and a well-plate autosampler associated to the HPLC-Chip interface connected to ionization source. Loaded samples were enriched on a 160 nL enrichment column and separated with an acetonitrile gradient (50% CH3CN in 60 min, 80% CH3CN in 80 min, and re-equilibration to 10% CH3CN in 120 min) on a 75 μm × 150 mm separation column packed with 3 μm Polaris C18 at a flow rate of 0.4 μL min−1. Source parameters were: gas temperature 325 °C and drying gas 4.8 L min−1 while voltage values for capillary, fragmentor, skimmer and octapole were respectively 1650 V, 170 V, 65 V and 750 V. Scan rate was 4 spectra/second for MS and 3 spectra/second (2378 transient spectrum) for MS/MS (3213 transient spectrum). All samples from sulfenyl amides reacted with both dimedone or glutathione were diluted 1:100 in 80% CH3CN, 20% diluted formic acid (0.1% in water) and directly infused into the Chip Cube ion source for analysis. Selected ion fragmentation was performed at 20 eV and accumulated for 1 second with an isolation width of 1.3 m/z.
Highlights.
Dimedone has been proposed as a selective sulfenic acid (RSOH) trapping agent; this is incorrect
Dimedone reacts rapidly with sulfenyl amides as well as sulfenic acids
Sulfenyl amides are formed either from disulfides or sulfenic acids, and amides
Sulfenyl amides may account for some of the species identified as the “sulfenome.”
These data contribute to rationalizing problems in understanding H2O2-mediated cell signaling
Acknowledgements
This work was supported by grants from the National Institutes of Health, USA (ES023864 to HJF), the Novo Nordisk Foundation (Laureate grant: NNF13OC0004294 to MJD) and Human Frontier Science Program, Italy Grant (RGP0013/2014 to FU).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Trujillo M, Alvarez B, Radi R. One- and two-electron oxidation of thiols: mechanisms, kinetics and biological fates. Free Radic. Res. 2016;50:150–171. doi: 10.3109/10715762.2015.1089988. [DOI] [PubMed] [Google Scholar]
- 2.Schöneich C. Thiyl radicals and induction of protein degradation. Free Radic. Res. 2016 doi: 10.3109/10715762.2015.1077385. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forman HJ, Maiorino M, Ursini F. Signaling Functions of Reactive Oxygen Species. Biochemistry. 2010;49:835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forman HJ, Ursini F, Maiorino M. An overview of mechanisms of redox signaling. J. Mol. Cell. Cardiol. 2014;73:2–9. doi: 10.1016/j.yjmcc.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett TJ, Pattison DI, Leonard SE, Carroll KS, Davies MJ, Hawkins CL. Inactivation of thiol-dependent enzymes by hypothiocyanous acid: role of sulfenyl thiocyanate and sulfenic acid intermediates. Free Radic. Biol. Med. 2012;52:1075–1085. doi: 10.1016/j.freeradbiomed.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu X, Mueller DM, Heinecke JW. Generation of intramolecular and intermolecular sulfenamides, sulfinamides, and sulfonamides by hypochlorous acid: a potential pathway for oxidative cross-linking of low-density lipoprotein by myeloperoxidase. Biochemistry. 2002;41:1293–1301. doi: 10.1021/bi015777z. [DOI] [PubMed] [Google Scholar]
- 7.Harwood DT, Kettle AJ, Winterbourn CC. Production of glutathione sulfonamide and dehydroglutathione from GSH by myeloperoxidase-derived oxidants and detection using a novel LC-MS/MS method. Biochem J. 2006;399:161–168. doi: 10.1042/BJ20060978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pullar JM, Vissers MC, Winterbourn CC. Glutathione oxidation by hypochlorous acid in endothelial cells produces glutathione sulfonamide as a major product but not glutathione disulfide. J. Biol. Chem. 2001;276:22120–22125. doi: 10.1074/jbc.M102088200. [DOI] [PubMed] [Google Scholar]
- 9.Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 10.van Montfort RL, Congreve M, Tisi D, Carr R, Jhoti H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature. 2003;423:773–777. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- 11.Foote CS, Clennan EL. Properties and reactions of singlet oxygen. In: Foote CS, Valentine JS, Greenberg A, Liebman JF, editors. Act. Oxyg. Chem. Blackie Academic and Professional; London: 1995. pp. 105–140. [Google Scholar]
- 12.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SR, Kwon KS, Kim SR, Rhee SG. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J Biol Chem. 1998;273:15366–15372. doi: 10.1074/jbc.273.25.15366. [DOI] [PubMed] [Google Scholar]
- 14.Barrett WC, DeGnore JP, Konig S, Fales HM, Keng YF, Zhang ZY, Yim MB, Chock PB. Regulation of PTP1B via glutathionylation of the active site cysteine 215. Biochem. 1999;38:6699–6705. doi: 10.1021/bi990240v. [DOI] [PubMed] [Google Scholar]
- 15.Groen A, Lemeer S, van der Wijk T, Overvoorde J, Heck AJ, Ostman A, Barford D, Slijper M, den Hertog J. Differential oxidation of protein-tyrosine phosphatases. J Biol Chem. 2005;280:10298–10304. doi: 10.1074/jbc.M412424200. [DOI] [PubMed] [Google Scholar]
- 16.Giannoni E, Taddei ML, Chiarugi P. Src redox regulation: again in the front line. Free Radic Biol Med. 2010;49:516–527. doi: 10.1016/j.freeradbiomed.2010.04.025. [DOI] [PubMed] [Google Scholar]
- 17.Parasassi T, Brunelli R, Costa G, De Spirito M, Krasnowska E, Lundeberg T, Pittaluga E, Ursini F. Thiol redox transitions in cell signaling: a lesson from N-acetylcysteine. ScientificWorldJournal. 2010;10:1192–1202. doi: 10.1100/tsw.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu X, Kassim SY, Parks WC, Heinecke JW. Hypochlorous acid oxygenates the cysteine switch domain of pro- matrilysin (MMP-7). A mechanism for matrix metalloproteinase activation and atherosclerotic plaque rupture by myeloperoxidase. J. Biol. Chem. 2001;276:41279–141287. doi: 10.1074/jbc.M106958200. [DOI] [PubMed] [Google Scholar]
- 19.Van Wart HE, Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc Natl Acad Sci U S A. 1990;87:5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winterbourn CC, Metodiewa D. Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radic. Biol. Med. 1999;27:322–328. doi: 10.1016/s0891-5849(99)00051-9. [DOI] [PubMed] [Google Scholar]
- 21.Strukul Giorgio., editor. Catalytic Oxidations with Hydrogen Peroxide as Oxidant. Springer Science. 1992 [Google Scholar]
- 22.Stadtman TC. Selenocysteine. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 23.Orian L, Mauri P, Roveri A, Toppo S, Benazzi L, Bosello-Travain V, De Palma A, Maiorino M, Miotto G, Zaccarin M, Polimeno A, Flohé L, Ursini F. Selenocysteine oxidation in glutathione peroxidase catalysis: An MS-supported quantum mechanics study. Free Radic. Biol. Med. 2015;87:1–14. doi: 10.1016/j.freeradbiomed.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Dinkova-Kostova AT, Holtzclaw WD, Wakabayashi N. Keap1, the sensor for electrophiles and oxidants that regulates the phase 2 response, is a zinc metalloprotein. Biochemistry. 2005;44:6889–6899. doi: 10.1021/bi047434h. [DOI] [PubMed] [Google Scholar]
- 25.Fourquet S, Guerois R, Biard D, Toledano MB. Activation of NRF2 by nitrosative agents and H2O2 involves KEAP1 disulfide formation. J Biol Chem. 2010;285:8463–8471. doi: 10.1074/jbc.M109.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gopalakrishna R, McNeill TH, Elhiani AA, Gundimeda U. Methods for studying oxidative regulation of protein kinase C. Methods Enzym. 2013;528:79–98. doi: 10.1016/B978-0-12-405881-1.00005-7. [DOI] [PubMed] [Google Scholar]
- 27.Gopalakrishna R, Anderson WB. Susceptibility of protein kinase C to oxidative inactivation: loss of both phosphotransferase activity and phorbol diester binding. FEBS Lett. 1987;225:233–237. doi: 10.1016/0014-5793(87)81164-x. [DOI] [PubMed] [Google Scholar]
- 28.Gopalakrishna R, Anderson WB. Ca2+- and phospholipid-independent activation of protein kinase C by selective oxidative modification of the regulatory domain. Proc. Natl. Acad. Sci. USA . 1989;86:6758–6762. doi: 10.1073/pnas.86.17.6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen SK, Cancilla MT, Shiau TP, Kung J, Chen T, Erlanson DA. Allosteric inhibition of PTP1B activity by selective modification of a non-active site cysteine residue. Biochemistry. 2005;44:7704–7712. doi: 10.1021/bi047417s. [DOI] [PubMed] [Google Scholar]
- 30.Rinna A, Torres M, Forman HJ. Stimulation of the alveolar macrophage respiratory burst by ADP causes selective glutathionylation of protein tyrosine phosphatase 1B. Free Radic. Biol. Med. 2006;41:86–91. doi: 10.1016/j.freeradbiomed.2006.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denu JM, Tanner KG. Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochem. 1998;37:5633–5642. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 32.Sohn J, Rudolph J. Catalytic and chemical competence of regulation of cdc25 phosphatase by oxidation/reduction. Biochem. 2003;42:10060–10070. doi: 10.1021/bi0345081. [DOI] [PubMed] [Google Scholar]
- 33.Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 34.Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem . 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 36.Ghezzi P. Regulation of protein function by glutathionylation. 2005;39:573–580. doi: 10.1080/10715760500072172. [DOI] [PubMed] [Google Scholar]
- 37.Shelton MD, Chock PB, Mieyal JJ. Glutaredoxin: role in reversible protein S-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid Redox Signal. 2005;7:348–366. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- 38.Sobotta MC, Liou W, Stöcker S, Talwar D, Oehler M, Ruppert T, Scharf AND, Dick TP. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 2014:1–16. doi: 10.1038/nchembio.1695. [DOI] [PubMed] [Google Scholar]
- 39.Jarvis RM, Hughes SM, Ledgerwood EC. Peroxiredoxin 1 functions as a signal peroxidase to receive, transduce, and transmit peroxide signals in mammalian cells. Free Radic. Biol. Med. 2012;53:1522–1530. doi: 10.1016/j.freeradbiomed.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Jones JT, Qian X, van der Velden JLJ, Chia SB, McMillan DH, Flemer S, Hoffman SM, Lahue KG, Schneider RW, Nolin JD, Anathy V, van der Vliet A, Townsend DM, Tew KD, Janssen-Heininger YMW. Glutathione S-transferase pi modulates NF-κB activation and pro-inflammatory responses in lung epithelial cells. Redox Biol. 2016;8:375–382. doi: 10.1016/j.redox.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peskin AV, Pace PE, Behring JB, Paton LN, Soethoudt M, Bachschmid MM, Winterbourn CC. Glutathionylation of the active site cysteines of peroxiredoxin 2 and recycling by glutaredoxin. J. Biol. Chem. 2016;291:3053–3062. doi: 10.1074/jbc.M115.692798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mailloux RJ, Treberg JR. Protein S-glutathionlyation links energy metabolism to redox signaling in mitochondria. Redox Biol. 2016;8:110–118. doi: 10.1016/j.redox.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prescott WG, Smiles S. LXXII.-The Interaction of Aromatic Disulphides with Sulphuric Acid. J. Chem. Soc. Trans. 1911;99:640–649. [Google Scholar]
- 44.Schoberl Alfons. Modellversuche zum oxydativen Abbau biologisch wichtiger organischer Schwefelverbindungen. Justus Liebigs Ann. Chem. 1933;507:111–127. [Google Scholar]
- 45.Ellis HR, Poole LB. Novel application of 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole to identify cysteine sulfenic acid in the AhpC component of alkyl hydroperoxide reductase. Biochem. 1997;36:15013–15018. doi: 10.1021/bi972191x. [DOI] [PubMed] [Google Scholar]
- 46.Headlam HA, Gracanin M, Rodgers KJ, Davies MJ. Inhibition of cathepsins and related proteases by amino acid, peptide, and protein hydroperoxides. Free Radic. Biol. Med. 2006;40:1539–1548. doi: 10.1016/j.freeradbiomed.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 47.Paulsen CE, Carroll KS. Chemical Dissection of an Essential Redox Switch in Yeast. Chem. Biol. 2009;16:217–225. doi: 10.1016/j.chembiol.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 48.Yang J, Gupta V, Tallman K, Porter N, Carroll KS, Liebler DC. Global, in situ, site-specific analysis of protein S-sulfenylation. Nat. Protoc. 2015;10:1022–37. doi: 10.1038/nprot.2015.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klomsiri C, Nelson KJ, Bechtold E, Soito L, Johnson LC, Lowther WT, Ryu S-E, King SB, Furdui CM, Poole LB. Use of dimedone-based chemical probes for sulfenic acid detection evaluation of conditions affecting probe incorporation into redox-sensitive proteins. Methods Enzymol. 2010;473:77–94. doi: 10.1016/S0076-6879(10)73003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leonard SE, Reddie KG, Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem. Biol. 2009;4:783–799. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- 51.Fratelli M, Demol H, Puype M, Casagrande S, Eberini I, Salmona M, Bonetto V, Mengozzi M, Duffieux F, Miclet E, Bachi A, Vandekerckhove J, Gianazza E, Ghezzi P. Identification by redox proteomics of glutathionylated proteins in oxidatively stressed human T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3505–10. doi: 10.1073/pnas.052592699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seo M, Lee Y-H. PFKFB3 regulates oxidative stress homeostasis via its S-glutathionylation in cancer. J. Mol. Biol. 2014;426:830–42. doi: 10.1016/j.jmb.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hildebrandt T, Knuesting J, Berndt C, Morgan B, Scheibe R. Cytosolic thiol switches regulating basic cellular functions: GAPDH as an information hub? Biol. Chem. 2015;396:523–37. doi: 10.1515/hsz-2014-0295. [DOI] [PubMed] [Google Scholar]
- 54.dbGSH database of cysteine S-glutathionylation. 2014 http://csb.cse.yzu.edu.tw/dbGSH/index.php.
- 55.Krämer AC, Thulstrup PW, Lund MN, Davies MJ. Key role of cysteine residues and sulfenic acids in thermal- and H2O2-mediated modification of β-lactoglobulin. Free Radic. Biol. Med. 2016;97:544–565. doi: 10.1016/j.freeradbiomed.2016.07.010. [DOI] [PubMed] [Google Scholar]