Abstract
Serpins are the largest known family of serine proteinase inhibitors and perform a variety of physiological functions in arthropods. Herein, we review the field of serpins in arthropod biology, providing an overview of current knowledge and topics of interest. Serpins regulate insect innate immunity via inhibition of serine proteinase cascades that initiate immune responses such as melanization and antimicrobial peptide production. In addition, several serpins with anti-pathogen activity are expressed as acute-phase serpins in insects upon infection. Parasitoid wasps can downregulate host serpin expression to modulate the host immune system. In addition, examples of serpin activity in development and reproduction in Drosophila have also been discovered. Serpins also function in host-pathogen interactions beyond immunity as constituents of venom in parasitoid wasps and saliva of blood-feeding ticks and mosquitoes. These serpins have distinct effects on immunosuppression and anticoagulation and are of interest for vaccine development. Lastly, the known structures of arthropod serpins are discussed, which represent the serpin inhibitory mechanism and provide a detailed overview of the process.
Keywords: Insect, Tick, Development, Innate immunity, Host-pathogen interactions
1. Introduction
Serpins are a superfamily of proteins, typically around 45 kDa, which generally function as serine proteinase inhibitors and participate in a suicide inhibitory mechanism in which both serpin and proteinase are permanently inactivated. The serpin reactive site that interacts with the target proteinase is part of an exposed loop near the carboxyl-terminal end of the serpin sequence. In an inhibitory reaction between a serpin and proteinase, the reactive center loop (RCL) of the serpin occupies the proteinase active site, and a specific peptide bond in the loop is cleaved (the scissile bond), resulting in a large conformational change in the serpin. However, the hydrolysis reaction is not completed, and the serpin and proteinase are trapped in a covalent complex [1–4]. The scissile bond is defined as the peptide bond between two amino acid residues named P1 and P1’. Residues on the amino-terminal side of the scissile bond are numbered in the C→N direction, and residues on the carboxyl-terminal side of the scissile bond (the “prime” side) are numbered in the N →C direction: (.. ..P5-P4-P3-P2-P1-↓-P1′-P2′-P3′-P4′-P5′.. ..).The sequence of the reactive center loop determines the inhibitory selectivity of a serpin. Detailed information about serpin structure and mechanism from studies of arthropods is provided in Section 5 of this review.
Arthropod serpin sequences are not sufficiently similar to vertebrate serpins to permit assignment of orthology with mammalian serpins. Therefore, physiological proteinase targets of arthropod serpins must be determined experimentally, and not surprisingly, the majority of arthropod serpins are therefore orphans. The known roles of serpins in arthropod biology are summarized in Fig. 1 and Table 1, and discussed in detail in the following sections.
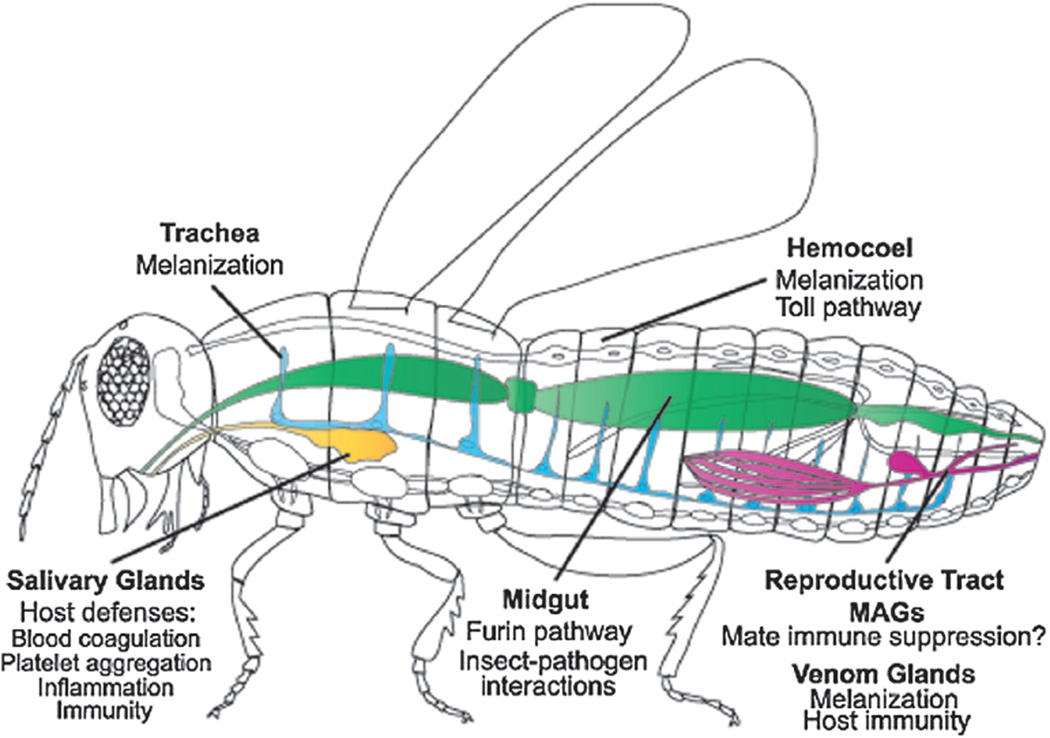
Fig. 1.
The known physiological functions of serpins in arthropod physiology. The majority of insect serpins are produced in fat body and hemocytes and are then secreted into the hemocoel. In addition, serpins are expressed in a number of additional tissues, including tick and mosquito salivary glands (orange), midgut (green), trachea (blue), and in male accessory glands (MAGs), as well as the venom glands of parasitoid wasps (pink). Major known functions are listed and discussed in detail in the different sections of the manuscript.
Table 1.
Arthropod serpins discussed in this review article.
| Gene name | Accession numbera | Species | Site of expression | Functiond | Target proteinase | References |
|---|---|---|---|---|---|---|
| Alboserpin | AAC31158 | Aedes aegypti | Saliva | Anticoagulation | Factor Xa | [101,102] |
| SRPN2 | XP-001651231 | Aedes aegypti | Hemolymph | Immunity, Melanization | TMP, IMP-1 | [52] |
| SRPN1 | XP .001648011 | Aedes aegypti | Hemolymph | Immunity, Melanization | IMP-1, IMP-2, CLIPB10 | [52] |
| SRPN3 | XP .001651232 | Aedes aegypti | Hemolymph | Unknown | Unknown | [52] |
| Alboserpin | AAV90669 | Aedes albopictus | Saliva | Anticoagulation | Factor Xa | [114] |
| Serpin 19 | JAI08902 | Amblyomma americanum | Saliva | Anticoagulation | Factor Xa, Factor XIa, trypsin, plasmin | [113] |
| Serpin 6 (AamS6) | ABS87358 | Amblyomma americanum | Saliva | Anticoagulation, Immunosuppression | elastase, chymase, papain, plasmin | [100] |
| SRPN10A(KRAL) | CAD12784 | Anopheles gambiae | Midgut | Immunity | Unknown | [27,57] |
| SRPN10B (RCM) | CAD12783 | Anopheles gambiae | Midgut | Immunity | Unknown | [27,57] |
| SRPN10C (FCM) | CAD12782 | Anopheles gambiae | Midgut | Immunity | Unknown | [27,57] |
| SRPN10D (CAM) | CAD12781 | Anopheles gambiae | Midgut | Immunity | Unknown | [27,57] |
| SRPN1 | ABL67437.1 | Anopheles gambiae | Hemolymph | Unknown | Unknown | [50] |
| SRPN2 | XP-308845.4 | Anopheles gambiae | Hemolymph | Immunity, Melanization | CLIPB9 | [50,51,126,146] |
| SRPN3 | ABJ52802.1 | Anopheles gambiae | Hemolymph | Unknown | Unknown | [50] |
| SRPN6 | XP_319990.2 | Anopheles gambiae | Midgut, Salivary glands, hemocytes | Immunity | Unknown | [66–69,71] |
| Serpin 1 (SW-AT)-1 | ACT36272 | Bombyx mori | most tissues | Acute phase | Unknown | [26] |
| SW-AT-2 | ACT36273 | Bombyx mori | most tissues | Unknown | Unknown | [26] |
| SW-AT-3 | ACT36274 | Bombyx mori | most tissues | Unknown | Unknown | [26] |
| SW-AT-4 | ACT36275 | Bombyx mori | silk glands, midgut, fat body | Unknown | Unknown | [26] |
| serpin16 | NP-001139708 | Bombyx mori | Silk glands | Unknown | Unknown | [127] |
| serpin18 | ACG61181 | Bombyx mori | Silk glands | Regulation of silk production, Development | fibrinase | [126] |
| (Acp)76A (Spn76A) | NP-524153.1 | Drosophila melanogaster | Seminal fluid | Unknown, possibly hormone transport | Non-inhibitory | [13,82–87] |
| Spn27A | NP-652024.1 | Drosophila melanogaster | Hemolymph | Immunity, Development | MP2 | [46–49,80] |
| Spn28D | NP-609172.1 | Drosophila melanogaster | Hemolymp, trachea | Immunity, Wound healing, Development | Unknown | [13,72–74] |
| Spn28F | NP-524957.2 | Drosophila melanogaster | Seminal fluid | Unknown | Unknown | [13,86] |
| Spn38F | NP-524956.2 | Drosophila melanogaster | Seminal fluid | Antibacterial | Unknown | [13,86] |
| Spn4A (Serpin 42 Da) | NP-724512.1 | Drosophila melanogaster | Midgut epithelia | Immunity, Secretory pathway regulation | Proprotein convertase, furin1, furin2 | [58–61] |
| Spn53F | NP-001036553 | Drosophila melanogaster | Seminal fluid | Unknown | Non-inhibitory | [13,86] |
| Spn75F | NP-001036614 | Drosophila melanogaster | Seminal fluid | Unknown | Non-inhibitory | [13,86] |
| Spn77Bc | NP-649207.1 | Drosophila melanogaster | Seminal fluid | Unknown | Non-inhibitory | [13,86] |
| SRPN10 | GMOY012007b | Glossina morsitans | Midgut, Saliva | Immunosuppression | Complement factor C1 and D | [64] |
| serpin-1 (HLS1) | unknownc | Haemaphysalis longicornis | Midgut | Anticoagulation | Unknown | [116] |
| serpin-2 (HLS2) | BAD11156 | Haemaphysalis longicornis | Hemolymph | Unknown | Unknown | [115] |
| Hdd3 | AAD40672 | Hyphantria cunea | Hemolymph | Immunity, Melanization | PAP | [55,56] |
| Iris | CAB55818 | Ixodes ricinus | Saliva | ImmunosuppressionAnticoagulation | Elastase, tissue plasminogen activator | [105–107] |
| IRS-2 | ABI94056 | Ixodes ricinus | Saliva | Anti-inflammation | Cathepsin G, chymase, thrombin | [108,109] |
| IxscS-1E1 | KF990169 | Ixodes scapularis | Saliva | Anticoagulation | Thrombin, trypsin | [110] |
| LbSPNy | ACQ83466 | Leptopilina boulardi | Venom | Immunosuppression | Unknown | [97] |
| Serpin 1A | AAC47343 | Manduca sexta | Hemolymph | Unknown | Unknown | [20,22,128,129] |
| Serpin 1B | AAC47343 | Manduca sexta | Hemolymph | Unknown | Unknown | [20–22,123,128,129] |
| Serpin 1E | AAC47343 | Manduca sexta | Hemolymph | Unknown | Unknown | [20,22,123,128,129] |
| Serpin 1K | AAC47334 | Manduca sexta | Hemolymph | Unknown | Unknown | [20,22,122,128,129] |
| Serpin-1J | AAC47340 | Manduca sexta | Hemolymph | Immunity, Toll signaling, Melanization | HP8, PAP3 | [20,22,33,40,128,129] |
| Serpin-3 | AAO21505, AAO21506 | Manduca sexta | Hemolymph | Immunity, Melanization | PAP-1, PAP-3, HP8 | [41] |
| Serpin-4 | AAS68503, AAS68504 | Manduca sexta | Hemolymph | Immunity, Melanization | HP-1, HP-6, HP-21 | [53,54] |
| Serpin-5 | AAS68507, AAS68508 | Manduca sexta | Hemolymph | Immunity, Toll signaling | HP-1, HP-6 | [53,54] |
| Serpin-6 | AAV91026 | Manduca sexta | Hemolymph | Immunity, Melanization | PAP3, HP8 | [42] |
| Serpin-7 | ADM86478 | Manduca sexta | Hemolymph | Immunity, Melanization | PAP3 | [44] |
| Serpin-3 | AHA43071 | Ostrinia furnacalis | Hemolymph | Immunity, Melanization | SP13 | [45] |
| Pxserpin-1 | BAD52261 | Plutella xylostella | Hemolymph | Immunity | unknown | [76] |
| Serpin 27 | unknown | Plutella xylostella | Hemolymph | Immunity, AMP production, Melanization | unknown | [78] |
| serpin-1 (RAS-1) | AAK61375 | Rhipicephalus appendiculatus | Unknown | Unknown | Unknown | [118] |
| serpin-2 (RAS-2) | AAK61376 | Rhipicephalus appendiculatus | Unknown | Unknown | Unknown | [118] |
| Rms-15 | AHC98666 | Rhipicephalus microplus | Saliva | Anticoagulation | thrombin | [112] |
| Rms-17 | AHC98658 | Rhipicephalus microplus | Saliva | Anticoagulation | cathepsin G, plasmin | [111] |
| Rms-3 | AHC98654 | Rhipicephalus microplus | Saliva | Anticoagulation | cathepsin G | [111] |
| Rms-6 | AHC98657 | Rhipicephalus microplus | Saliva | Unknown | Unknown | [111] |
| SPN40 | BAI59106 | Tenebrio molitor | Hemolymph | Immunity, Toll signaling, Melanization | MSP | [36] |
| SPN48 | BAI59108 | Tenebrio molitor | Hemolymph | Immunity, Toll signaling, Melanization | SPE | [35] |
| SPN55 | BAI59107 | Tenebrio molitor | Hemolymph | Immunity, Toll signaling, Melanization | SAE | [36] |
Abbreviations: TMP – tissue melanization protease; IMP – immune melanization protease; CLIP – clip-domain containing proteinase; MP – melanization protease; PAP – prophenoloxidase activating proteinase; HP – hemolymph proteinase; SP – serine protease; MSP – modular serine protease; SPE – spätzle-processing enzyme; SAE – spätzle-processing-enzyme-activating enzyme.
Accession numbers according to Genbank (NCBI).
Accession number in VectorBase, no Genbank accession number available.
No accession number identifiable.
Function(s) of serpins that target mammalian host physiologies are italized.
The first arthropod serpins characterized biochemically were from hemolymph of the silkworm, Bombyx mori. Proteins of ~45 kDa purified from larval plasma as inhibitors of trypsin and chymotrypsin were cleaved near their carboxyl-termini and formed SDS-stable complexes with proteinases [5–7] and were speculated to be serpins. Similar inhibitors were isolated from another lepidopteran insect, Manduca sexta [8,9], and amino acid sequences confirmed that the M. sexta and B. mori inhibitors were serpins [8,10]. Serpin sequences have now been identified in many arthropod transcriptomes and genomes, with 30–40 serpin genes in many species, including 34 in B. mori [11], 32 in M. sexta (M. Kanost, unpubished data), 31 in a beetle, Tribolium castaneum [12], 29 in Drosophila melanogaster, and a similar number in other Drosophila species [13]. Other species have significantly fewer serpin genes, including just 7 in the honeybee, Apis mellifera [14] and 10 in the tsetse Glossina morsitans [15]. Mosquito species vary from 18 serpin genes in Anopheles gambiae, 23 in Aedes aegypti, to 31 in Culex quinquefasciatus [16]. Ticks and mites also have considerable variation in the serpin gene content of their genomes, with 45 serpin genes in the blacklegged tick Ixodes scapularis [17], 22 in the cattle tick Rhipicephalus microplus [18], and only 10 in the scabies mite, Sarcoptes scabiei [19].
Besides gene duplication, the number of unique serpins encoded by a given arthropod genome can also be increased post-transcriptionally. Some insect serpin genes have a unique structure, in which mutually exclusive alternate splicing of an exon that encodes the RCL results in production of several inhibitors with different inhibitory activities. This phenomenon was first observed in the gene for M. sexta serpin-1, which contains 14 copies of its 9th exon [20] (M. Kanost, unpublished data). Each version of exon 9 encodes a different sequence for the carboxyl-terminal 39–46 residues, including the RCL (Fig. 2), and the resulting serpin variants inhibit a different spectrum of proteinases [21,22]. Orthologous serpin-1 genes from other lepidopteran species, with alternate exons in the same position as in M. sexta serpin-1, have been identified [23–25]. The serpin-1 gene of B. mori, in the same superfamily as M. sexta, has only four alternate versions of exon 9 [11,26], indicating considerable genetic flexibility and relatively recent expansions and divergence of these alternate exons in lepidopteran evolution [24]. Alternative splicing at the same position, to produce serpins with differing RCLs also occurs in An. gambiae SRPN10 [27] and in spn4 orthologs in multiple Drosophila species [28] (discussed more in Section 2.3).
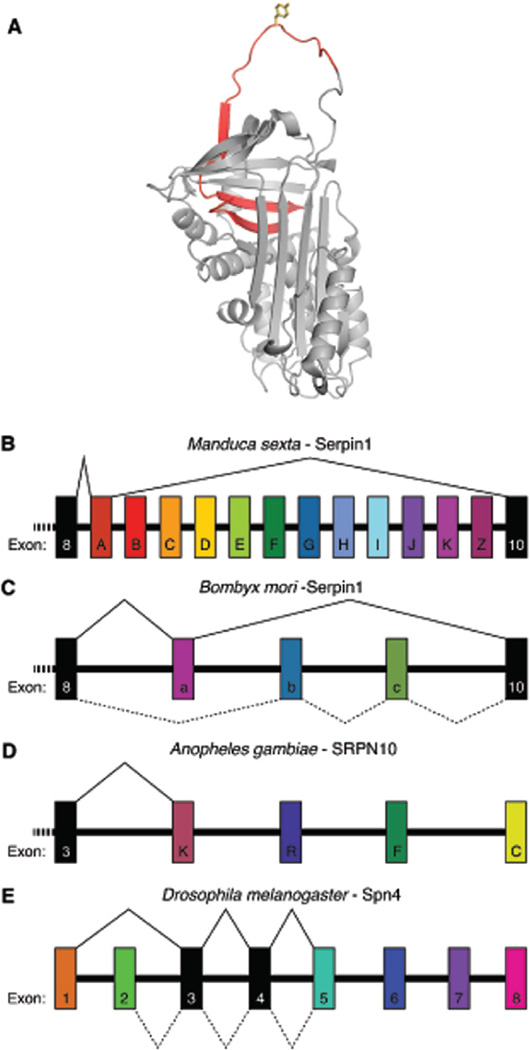
Fig. 2.
Outline of alternative splicing in insect serpins. (A) Structure of M. sexta serpin1K showing the alternatively spliced RCL region (red) with the P1-P1′ residue (yellow). (B) Simplified splice variant diagram in M. sexta serpin-1. Exons that are always expressed are shown in black and alternatively spliced exon 9 variants are colored. Depicted is the splicing diagram of serpin1A, wherein the A isoform of exon 9 is expressed. Expression of B–D, etc. results in expression of serpin1B, −1C, −1D, etc. (C) Simplified splice variant diagram in Bombyx mori serpin-1. The solid line indicates expression of the isoform of exon 9, resulting in isoform 1. Expression of b and c exons results in isoforms 2 and 3, respectively. The dotted line depicts expression of both b and c exons, resulting in isoform 4 expression. (D) Simplified splice variant diagram in An. gambiae SRPN10. The solid line shows expression of the K isoform of exon 9 (KRAL isoform). Expression of R, F, and C exons results in RCM, FCM, and CAM isoforms, respectively. (E) Simplified splice variant diagram in Drosophila melanogaster Spn4. The expression of exon 1 results in Spn4B, D-F, and I isoforms, and expression of exon 2 results in Spn4A, G, H, J, and K isoforms. The solid line depicts the expression of Spn4B and the dotted line depicts expression of Spn4A. Expression of additional Spn4 isoforms arises from alternative splicing of exons 6, 7, and 8.
2. Biological functions of arthropod serpins in insect immunity
Arthropods produce and secrete serpins into their hemolymph to regulate proteinase cascade pathways that amplify signals resulting from detection of pathogens, eliciting innate immune responses. Regulation of such pathways by serpins is an ancient aspect of immune system evolution, occurring in the hemolymph coagulation pathway of horseshoe crabs [29]. The following section will provide specific examples on how insect innate immunity is regulated by serpins.
2.1. Regulation of Toll pathway in hemolymph
The Toll pathway for stimulation of gene expression, particularly of antimicrobial peptides, has been best characterized in D. melanogaster. Binding of pattern recognition proteins in hemolymph to microbial cell wall polysaccharides triggers activation of a serine proteinase pathway in which the final proteinase cleaves proSpätzle, producing the active Spätzle cytokine. Spätzle is the Toll receptor ligand that stimulates an intracellular signal transduction pathway leading to activation of a rel family transcription factor [30]. The Spätzle-processing enzyme in D. melanogaster and other species is a member of the clip domain serine proteinase family, which are serine proteinases with an amino-terminal clip domain, common as immune factors in arthropods [31,32]. Serpins that regulate Spätzle-processing proteinases and their upstream activating proteinases have been identified through biochemical studies in M. sexta and a beetle T. molitor. In M. sexta, hemolymph proteinase-8 (HP8), which can activate proSpätzle, is inhibited by serpin-1J [33]. HP8 is activated by hemolymph proteinase-6 (HP6), which is inhibited by serpin-5 [34]. In T. molitor, the Spätzle-processing enzyme is regulated by SPN48 [35], and upstream proteinases are regulated by SPN40 and SPN55 [36]. An unusual serpin in T. molitor, SPN93, contains dual serpin domains, and can inhibit Spätzle-processing enzyme as well as a modular serine proteinase that initiates the proteinase cascade [37]. In D. melanogaster, a serpin that regulates Spätzle-processing enzyme directly has not yet been identified, but SPN43Ac (necrotic) is a potential inhibitor of an upstream proteinase [32], perhaps the clip proteinase Persephone, an ortholog of M. sexta HP6. Genetic experiments also implicate Spn1 as a regulator of an upstream proteinase in the D. melanogaster Toll pathway [38].
2.2. Regulation of proPO activation
A prominent and broad spectrum arthropod innate immune response that is stimulated by serine proteinase cascade pathways is the activation of prophenoloxidase (proPO) in hemolymph. ProPO activation leads to synthesis of melanin, which localizes to the surface of invading pathogens and parasites or to wound sites [39]. Prophenoloxidase-activating proteinases (PAPs) are clip proteinases that are activated by additional clip proteinases upstream [31]. Four serpins from M. sexta hemolymph (serpin-1J, serpin-3, serpin-6, and serpin-7) can inhibit prophenoloxidase activating proteinases [40–44].
Orthologs of M. sexta serpin-3 regulate proPO activation in other species. In another lepidopteran, Ostrinia furnacalis, serpin-3 inhibits serine proteinase 13, a clip proteinase, and injection of serpin-3 into larvae resulted in decreased melanin synthesis during fungal infection [45]. D. melanogaster Spn27A (M. sexta serpin-3 ortholog) is involved in regulation of melanization [46–48] and was predicted from genetic analyses [46–48] to inhibit PAPs. Recent biochemical analysis demonstrated that D. melanogaster clip proteinase MP2 is a PAP that is inhibited by Spn27 [49]. An. gambiae and Ae. aegypti have three serpins that form an orthologous group with serpin-3: SRPN1, 2, and 3 [50]. An. gambiae SRPN2 is a key regulator of melanization and inhibits a clip proteinase CLIPB9, a prophenoloxidase-activating proteinase [51]. In Ae. aegypti, SRPN1 has an apparent role in regulating the melanization response [52].
Some serpins that regulate clip proteinases upstream from the terminal proteinase have also been identified. In. M. sexta, hemolymph proteinases 21 and 6, which activate PAPs, are inhibited by serpin-4 and serpin-5, respectively [53,54]. In T. molitor, the same proteinase cascade and serpins appear to activate both the Toll pathway and prophenoloxidase activation [36].
2.3. Insect serpins as acute-phase proteins
The expression of immune-responsive serpins can be altered by a number of immune challenges, including viruses, bacteria, fungi, protozoan parasites, and parasitoid wasps. Some immune-responsive serpins are acute-phase proteins, being strongly upregulated in response to infection or injury. The functional role of only a small number of these acute-phase serpin proteins is elucidated, and the molecular underpinnings of their immune responsiveness are known for even fewer. The following section will summarize our knowledge of three such serpins:
The serpin Hdd3 from the fall webworm Hyphantria cunea was identified as an acute-phase protein nearly 20 years ago, showing upregulation upon bacterial challenge and aseptic injury [55]. Initial studies revealed a role in regulation of melanization, as recombinant Hdd3 suppressed PO activity in vitro [56]. Hdd3 is conserved across distantly related insect taxa, and its orthologs include Spn27A from D. melanogaster [46,47], SRPN2 from A gambiae [50] and Ae. aegypti [52], and Serpin-3 from M. sexta [41]. With the exception of Ae. aegypti SRPN2, their molecular function as PAP inhibitors has been confirmed experimentally [41,49,51] (see Section 2.2 above). In Ae. aegypti, the acute phase response of SRPN2 is regulated by the Toll pathway. Knockdown of Cactus, an inhibitor of the Toll pathway, significantly increases SRPN2 levels in Ae. aegypti hemolymph, while the upregulation of SRPN2 after fungal infection is prevented by knockdown of REL1, the transcription factor downstream of the Toll pathway [52]. In contrast, An. gambiae SRPN2 expression is not regulated by the Toll pathway (Michel, unpublished), and basal expression levels of D. melanogaster Spn27A are not affected by depletion of the REL1 ortholog, Dif [47].
Another prominent acute phase serpin is SRPN10. Originally described in An. gambiae, this gene encodes at least four functional serine proteinase inhibitor isoforms, each containing unique RCLs with distinct inhibitory activities [27] (Fig. 2). Interestingly, gene expression of two splice variants is upregulated in midgut cells upon infection with rodent malaria parasites, and the protein isoforms shift in cellular localization from the nucleus to the cytoplasm [57]. All SRPN10 isoforms are currently orphans, with no identified proteinase targets in vivo. Insights into their function may be derived from studies performed with its ortholog, D. melanogaster Spn4 (also referred to Serpin 42 Da, [50]). Like SRPN10, Spn4 is alternatively spliced into eight isoforms, A-H, with four unique RCLs each paired with an intracellular and extracellular isoform [58]. Spn4 is also an acute-phase protein with upregulation documented in D. melanogaster larvae upon Sindbis virus infection [59]. Several lines of evidence suggest that one isoform, Spn4A, inhibits proprotein convertases (PPCs) and thus regulates the secretory pathway. Spn4A contains an ER-retention signal and a P4-P1 sequence consistent with a classical furin cleavage site. Spn4A not only inhibits human furins in vitro [60,61], it also forms inhibitory complexes with D. melanogaster furin1 and 2 [58]. In addition, Spn4A inhibits apolipophorinI/II (apo-LPI/II) processing in a co-expression system utilizing insect cells, further supporting the notion that Spn4A negatively regulates proprotein convertase (PPC) processing. Mosquito lipophorin is a source of lipids for malaria parasite stages within its vector [62], and knockdown of lipophorin gene expression in Ae. aegypti leads to decreased avian malaria development [63]. Together, these data suggest SRPN10 upregulation in parasite-infected midguts transiently reduces maturation of lipophorin, thus limiting lipids required for malaria parasite development. Future experiments are necessary to test whether SRPN10 indeed contributes to parasite reduction by providing the mechanistic link between PPC regulation and lipid resource management.
Co-incidentally, SRPN10 from the tsetse fly Glossina morsitans morsitans, which is not orthologous to An. gambiae SRPN10, is upregulated upon trypanosome parasite infection [64]. Gmm-SRPN10 is expressed in midgut epithelia, but is secreted into the midgut lumen and inhibits the complement cascade in the ingested blood meal. Knockdown of GmmSRPN10 in the midgut causes a decrease of trypanosome infection load, most likely due to increased complement activity in the blood meal that lyses parasites [65].
The final example of acute-phase serpin proteins is SRPN6 from An. gambiae. Like SRPN10, SRPN6 was originally described as a marker for epithelial infection by malaria parasites, both in the midgut [66] and salivary glands [67]. SRPN6 contributes significantly to epithelial immunity against malaria parasites, with SRPN6 knockdown resulting in increased malaria parasite numbers in the midgut [66] and salivary glands [67]. The latter also has impact on transmission, as the extrinsic incubation period is shortened in SRPN6-depleted mosquitoes [67]. Although SRPN6 protein is not detected in normal epithelia, a variety of stimuli, including Gram-positive and negative bacteria, leads to a strong and transient upregulation [66,68]. SRPN6 expression patterns elicited in the midgut by bacterial feeding and parasite infection are temporally and spatially distinct [68]. Therefore, SRPN6 upregulation upon parasite infection is unlikely to be explained by the temporal dysbiosis occurring in the mosquito midgut upon blood feeding. Indeed, SRPN6 upregulation upon rodent malaria parasite infection does not require the presence of gut bacteria, as expression is also induced in midguts from antibiotic-treated mosquitoes [68]. Several lines of evidence have identified the LPS-induced tumor necrosis factor alpha factor (LITAF) as one transcription factor regulating SRPN6 expression. LITAF binds the SRPN6 promoter region and both LITAF and SRPN6 are expressed in parasite-invaded midgut epithelial cells, with LITAF upregulation preceding SRPN6 [69]. Moreover, LITAF knockdown limits SRPN6 parasite-mediated upregulation in mosquito midguts and in cell culture by bacteria [69]. However, the molecular underpinnings of SRPN6 expression are not solved completely, as it is currently unclear how LITAF expression itself is regulated in mosquitoes. In mice, LITAF expression is induced through TLR signaling, suggesting the Toll pathway may be upstream of LITAF in mosquitoes [70]. Future experiments are necessary to determine whether the Toll pathway is indeed involved in acute-phase SRPN6 upregulation.
The basis for SRPN6-mediated malaria parasite killing is currently unclear. The protein encodes a signal peptide, and is secreted from cell lines that naturally express SRPN6, suggesting its target is either present in the secretory pathway or is a hemolymph proteinase [66]. Based on biochemical characterization of recombinant protein, SRPN6 is functional and inhibits kallikrein in vitro [71]. Like most insect serpins however, SRPN6 remains an orphan with no known physiological target. SRPN6 belongs to an orthologous group of serpins [50] with a single ortholog in D. melanogaster (Spn28D) and all other sequenced Drosophila genomes [13]. Spn28D is an acute phase protein, responding mostly to injury rather than infection [72,73]. It regulates melanization in at least two distinct ways: preventing spontaneous melanization in epithelia such as trachea and Malpighian tubules, and limiting hemolymph melanization together with Spn27A, putatively through control of PO release [72]. However, Spn28D and SRPN6 are distinct in most of their bio-logical profile: Spn28D is predominantly expressed in crystal cells [72,74], whereas SRPN6 is mainly expressed in epithelia [66]. Moreover, Spn28D is essential during the larval stage, as null mutants die before pupation [72], whereas SRPN6 is not detectable in immature stages of development [66]. Furthermore, while the RCL sequences of SRPN6 and Spn28D are nearly identical, suggesting similar target proteinase profiles, protein localization is likely distinct, given that the RGD domain is only present in Spn28D. Altogether, it is likely that their functions are not congruent, and contribute to insect immunity in distinct ways.
In summary, transcription activation in response to microbial stimuli is often observed in insect serpins. As acute phase proteins, these serpins contribute to insect immunity against a variety of pathogens, including bacteria and parasites. Besides well-known roles in downregulation of melanization and Toll pathway signaling, other molecular functions of these serpins are emerging, including regulation of protein secretion. In addition, several acute-phase serpins contribute to insect immunity in ways that are little understood. Identification of the physiological targets of orphan serpins will provide a handle on molecular mechanisms underlying these less-characterized immune responses.
2.4. Infection-driven downregulation of serpin expression
Given their central function in suppression of major immunity pathways in arthropods, serpin downregulation in response to infection could function as a means to transiently increase immune responses. Thus far, few examples have been uncovered where serpin downregulation is mediated by infection and consequently leads to increased immune responses. An early description of serpin downregulation upon infection was described in M. sexta infected with the braconid wasp Cotesia congregata [75]. While hemolymph serpin concentration rose in unparasitized aging fifth instar larvae, their concentration remained flat over the same time period in parasitized larvae. Downregulation of serpins in lepidopteran insects infected by parasitoid wasps appears conserved, as Cotesia plutel-lae infection of the diamondback moth Plutella xylostella also leads to Pxserpin-1 depletion from the hemolymph [76]. The physiological consequences of M. sexta or P. xylostella serpin downregulation during parasitization remain unclear. Given that M. sexta PO activity levels were reduced during the same time, downregulated serpins did not result in increased activity of the proPO activation cascade [77]. In addition, precisely how hemolymph serpin concentration is regulated by parasitization in these systems is not understood.
Downregulation of serpin expression is also seen in the diamondback moth, Plutella xylostella parasitized with Diadegma semiclausum [78]. Parasitization affects the miRNA profile of the lepidopteran host, including expression of miR-8, a conserved miRNA that affects antimicrobial peptide production in D. melanogaster [79]. In P. xylostella, miR-8 targets Serpin 27, which regulates the expression of the antimicrobial peptide gloverin [78]. Parasitization by D. semiclausum leads to a miR-8 decrease, which in turn causes an increase in Serpin 27 and thus the downregulation of an antimicrobial peptide. It is thus clear that the infection with D. semiclausum modulates at least part of the immune repertoire of the host through transcriptional modulation of a serpin. Whether downregulation of serpins in parasitized lepidopteran insects has similar consequences on the host’s immune repertoire remains to be tested.
In conclusion, serpins regulate arthropod innate immunity via inhibition of clip proteinase cascades that initiate immune responses such as melanization and the Toll pathway. In addition, several serpins are acute phase proteins that contribute to anti-pathogen immunity in unknown ways. There central role in immunity also make insect serpins targets for pathogens to modulate the host immune system.
3. Insect serpins in development and reproduction
While the majority of characterized insect serpins hold immunity-related functions, other physiological processes regulated by serpins are known. This section summarizes our current knowledge of serpins that regulate insect development and contribute to insect reproduction.
3.1. Drosophila Spn27A regulates embryonic axis formation
Dorsal-ventral axis patterning in D. melanogaster is mediated by Toll pathway activity restricted to the ventral side of the embryo. As in immunity, the Toll pathway is activated by Spätzle. However, proteolytic activation of Spätzle in development is mediated by a distinct cascade consisting of the proteinases (i) Gastrulation defective, (ii) Snake, and (iii) Easter. As in immunity, the Spätzle-activating proteinase cascade that functions in development is regulated by a serpin, although the specific serpin involved is distinct. While necrotic inhibits Toll activation for immune regulation in adult flies (see Section 2.1 above), Spn27A restricts Toll activation in embryos [80]. Female D. melanogaster lacking Spn27A produce embryos with high uniform levels of Toll signaling, and all embryonic cells adopt a ventralized cell fate. Additional genetic evidence suggests that Easter is the inhibitory target of Spn27A during embryonic development [80]. Thus, regulation of proteolytic activity by the same insect serpin can regulate distinct physiological events.
3.2. Serpins in insect seminal fluid
In insects, seminal fluid contains many proteins that are transferred to females during mating and induce a wide range of female post mating responses [81]. D. melanogaster Accessory gland protein (Acp)76A (Spn76A) was the first serpin identified in insect seminal fluid [82]. The protein is transferred during mating to the female uterus where its concentration declines rapidly within several hours [83]. Post mating, Acp76A is detected in females in the mating plug, localizes to sperm storage organs, enters the circulation of the female, and is found on oocytes and oviposited eggs [84]. Based on sequence analysis, Acp76A is a non-inhibitory serpin lacking many conserved residues required for inhibition [13]. Orthologs of Acp76A are only found in three species of the melanogaster subgroup, none of which have been characterized further [13]. While it is likely that Acp76A is not required for sperm storage, its physiological function in D. melanogaster remains elusive. Structural analysis based on homology modeling revealed that Acp76A contains an N-terminally shortened alpha helix 1, resulting in the formation of a hydrophobic pocket, as described for protein C inhibitor [85], suggesting a role of Acp76A in hormone transport [86].
In addition to Acp76A, five more serpins are expressed in male accessory glands in D. melanogaster. These are Spn28F, Spn38F, Spn53F, Spn75F, and Spn77Bc [86]. Of these, only Spn28F and Spn38F, which are the closest paralogs of each other [50], are predicted to be functional serine protease inhibitors [13]. Continuous ectopic expression of individual Acp serpins in females did not affect mating or egg laying rates [87]. However, ectopically expressed Spn28F caused toxicity by an unknown mechanism, potentially contributing to the cost associated with mating [87]. Interestingly, overexpressed Spn38F somewhat limits the growth of the bacterial pathogen Serratia marcescens, suggesting a protective function for sperm against pathogens [87]. However, the mechanism underlying this potential function awaits elucidation.
Serpins have also been found in male reproductive glands of several other insect species, including three mosquito species, An. gambiae [88], Ae. aegypti [89], and Aedes albopictus [90], and the red flour beetle, Tribolium castaneum [91]. In contrast, no serpin transcripts or proteins were detected in the accessory glands and/or seminal fluid of honey bees [92], sandflies [93], or Heliconius butterflies [94]. Instead, proteinase inhibiting activity in the seminal fluid of at least honey bees and sandflies is likely mediated by Pacifastin-like peptides, a distinct class of small proteinase inhibitors found in arthropods [95].
In summary, serpins contribute substantially to the seminal fluid content in some insect species. Limited functional analyses suggest immunoprotective roles through regulation of canonical immune pathways. However, serpins do not seem to play a significant role in proteinase inhibitory activity of insect seminal fluid. Most Acp serpins are predicted to be non-inhibitory and thus carry other functions, potentially in hormone transport.
4. Arthropod serpins at the intersection of host-pathogen interactions
While the vast majority of arthropod serpins are produced and secreted into the hemolymph to regulate internal physiological processes, several serpins are found in extracellular fluids produced specifically to be transferred to other organisms, including saliva and venom. The following section will highlight examples of such arthropod serpins that are used to manipulate the physiology of mammalian or insect hosts to provide a favorable environment for nutrient uptake and survival.
4.1. Serpins in the venom of parasitoid wasps
Parasitoid wasps develop at the cost of their insect host, ultimately killing the host organism. The venom of parasitoid wasps contains several virulence factors that allow the egg to evade the host immune system and develop without being killed by melanotic encapsulation. Several serpins have been identified in the venom of different species of parasitoid wasps through either transcriptomic or proteomic analyses [96–98].The immunosuppressive role for one of these serpins, LbSPNy from the endoparasitoid wasp Leptopilina boulardi, has been evaluated in more detail [97]. This serpin is the most abundant protein in the L. boulardi venom, and venom added to host hemolymph prevents melanization ex vivo as well as in vivo. LbSPNy is at least partly responsible, as recombinant LbSPNy likewise inhibits melanization ex and in vivo, potentially by inhibition of one or more proteinases within the proPO activation cascade. Interestingly, the venom of L boulardi shows strain-specific differences in virulence factor profile, with non-synonymous changes in the RCL sequences of LbSPNy. Furthermore, the ortholog of LbSPNy in a related species, Leptopilina heterotoma, while containing a virtually identical RCL sequence, is very lowly expressed and thus unlikely to contribute to virulence. In summary, these studies reveal a role of serpins as virulence factors in parasitoid wasps and suggest that changes in serpin expression can contribute to the fast evolution of parasitoid venom and a potential arms race between parasitoid and host [97,98].
4.2. Serpins in saliva of arthropod vectors of disease
Blood-feeding arthropods use their saliva to counterbalance the host response to injury. Like their mammalian counterparts [99], arthropod serpins mediate several hemostatic and antiinflammatory effects in mammalian blood (Fig. 3). Thus, successful blood feeding is facilitated by injection of these serpins into the host with saliva. The following section summarizes a few well characterized examples, highlighting the diversity of serpins that contribute to blood feeding.
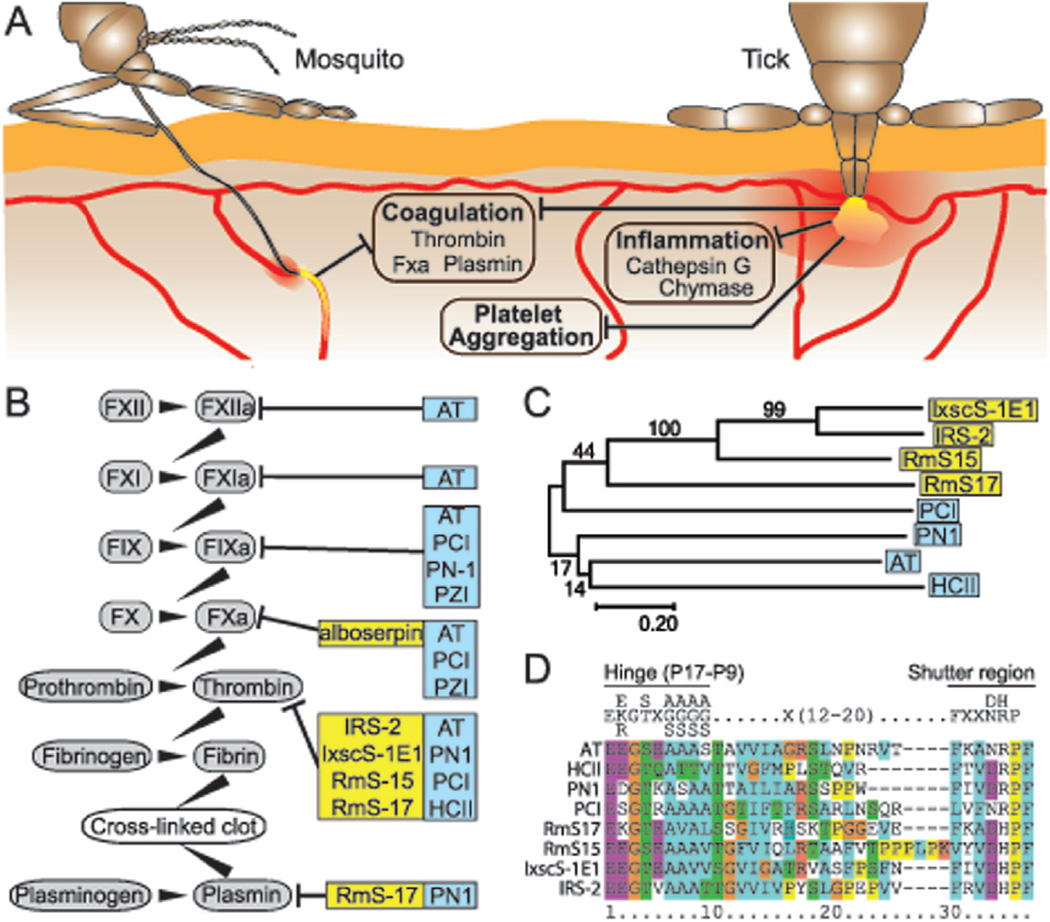
Fig. 3.
Saliva Serpins of Mosquitoes and Ticks Facilitate Blood Feeding on Mammals. (A) Mosquitoes insert their stylets into subcutaneous blood vessels, and obtain a blood meal within minutes. In contrast, ticks use their hypostome to rupture blood vessels causing blood to pool at the bite site, and then take a blood meal over the course of several days. Both ticks and some species of mosquitoes inject serpins as part of their saliva (shown in yellow) to counterbalance the mammalian host response to injury. These saliva serpins are inhibitors of several mammalian proteases required for coagulation, platelet aggregation, and inflammation. (B) Several factors of the intrinsic coagulation pathway (gray ovals) can be inhibited by either mosquito or tick serpins (yellow boxes, see Section 4.2). The mammalian serpins that target the same pathway are shown in comparison (blue boxes) [156–158]. (C) The evolutionary history of tick and mammalian thrombin-inhibiting serpins does not indicate any orthologies between individual tick and human serpin pairs. The ability for serpins to inhibit thrombin has therefore evolved independently in the tick and human lineage. Phylogenetic analysis was performed using the Maximum Likelihood method based on the JTT matrix-based model [159]. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. All evolutionary analyses were conducted in MEGA7 [160]. (D) Alignment of thrombin-inhibiting arthropod and human serpins, reveals significant protein sequence similarity of their reactive center loops (RCL). Based the results of the phyogenetic analysis, this sequence similarity is likely the product of convergent sequence evolution. Alignment was executed using MUSCLE [161], and visualized in Jalview2[162]. Alboserpin, Ae. aegypti (Genbank accession number AAC31158); AT, human antithrombin(NP_000479.1); HCII, human heparin cofactor II (NP_000176.2); IRS-2, Ixodes ricinus (ABI94056); IxscS-1E1, Ixodes scapularis (KF990169); Rms-15, Rhipicephalus microplus (AHC98666); RmS-17, R. microplus (AHC98658); PCI, human Protein C Inhibitor (NP_000615.3); PN1, human Protease nexin 1 (NP_006207.1); ZPI, human Protein Z-dependent protease inhibitor.
The first salivary serpin identified was alboserpin, a major anticoagulant in the saliva of Ae. aegypti and Ae. albopictus [100–102]. This serpin prevents coagulation in vitro and in vivo by inhibition of Factor Xa using an atypical mode of inhibition characterized by reversible interaction of proteinase and inhibitor at a 1:1 stoichiometry [100,101]. Alboserpin also binds phospholipids and heparin, and possibly other glycosaminoglycans. While neither of these binding activities are required nor promote Factor Xa inhibition, they are likely to facilitate alboserpin localization to the site of injury and/or platelet aggregation [100]. However, alboserpin seems to be an evolutionary exception, and the saliva of mosquitoes does not contain a large panel of serpins. Indeed, the sialomes of several anopheline mosquito species suggests that this clade mostly relies on other protease inhibitor classes to prevent blood clotting [103,104].
In contrast, ticks rely mostly on serpins to control host hemostasis and immunity to enable their hematophagic life style (Fig. 3). The best characterized tick saliva serpin is the Ixodes ricinus immunosuppressor, Iris, from the major vector of lyme disease in Europe. Iris is an active serine proteinase inhibitor with a predicted P1 methionine. Iris is upregulated during blood meal [105], and displays pleitropic effects on host immunity and hemostasis. Iris acts as an anticoagulant and delays fibrinolysis by inhibiting serine proteinases, especially elastase and tissue plasminogen activator [106]. Iris also interferes with hemostasis by hindering platelet adhesion via an unknown mechanism [106]. Besides its actions on hemostasis, Iris also inhibits the secretion of pro-inflammatory cytokines, especially Interferon-γ and Interleukin(IL)-6, from both T cells and macrophages, as well as Tumor necrosis factor (TNF) α from macrophages [105]. This anti-inflammatory action is at least in part mediated by physical binding of Iris to monocyte/macrophage cells and does not require inhibitory activity. Based on preliminary analysis using peptide antibodies, this function of Iris rather seems to rely on several exosites in helices D and E [107].
In contrast to Iris, the anti-inflammatory action of a second serpin from I. ricinus, called IRS-2 [108], is based solely on its function as a serine proteinase inhibitor. Like Iris, IRS-2 is secreted into the saliva and injected into the host during blood feeding. IRS-2 also prevents edema formation and neutrophil influx in inflamed tissues of a mouse acute inflammation model. The P1 site of IRS-2 was confirmed to be tyrosine 341, indicative of inhibitory activity against chymotrypsin-like serine proteinases [108]. Indeed, IRS-2 inhibits mainly the proinflammatory proteinases cathepsin G and mast cell chymase, and at higher concentrations, thrombin. Inhibition of cathepsin G and thrombin also contribute to the blocking of platelet aggregation by IRS-2 [108]. Furthermore, recent data demonstrate that IRS-2 can interfere with adaptive immune responses [109]. IRS-2 inhibits IL-6 production by dendritic cells stimulated with Borellia spirochetes, and primary neutrophils. One consequence of this lower production of IL-6 can be reduced IL-6/STAT3 signaling, which is required for the differentiation of IL-17 producing CD4-positive T lymphocytes. Together these data suggest that IRS-2 is capable to suppress the differentiation of Th17 cells, and thus may modulate in part the infection-induced immunopathology observed in Lyme disease [109].
In contrast to I. ricinus, little is known about the biochemical and functional properties of serpins in the black-legged tick, Ixodes scapularis, the major vector of lyme disease in North America. The only characterized serpin from this species is IxscS-1E1, which is a functional serine proteinase inhibitor with a predicted arginine at its P1 site. Recombinant IxscS-1E1 inhibits thrombin and trypsin in vitro, and inhibits platelet aggregation as well as plasma clotting time, potentially mediated by thrombin inhibition in vivo [110]. Given that the I. scapularis genome encodes putative orthologs of Iris and IRS-2 [17], it is likely that tick saliva inhibits several non-redundant steps in hemostasis to ensure blood feeding for extended time periods (Fig. 3B).
Most recently, additional salivary serpins from other tick species have been functionally analyzed, including RmS–3, −6, −7, and −15 from the cattle tick Rhipicephalus microplus [111,112]. These inhibitory serpins can inhibit platelet aggregation (RmS-3, −17) and plasma clotting (RmS-15, −17), consistent with their ability to inhibit cathepsin G and/or thrombin in vitro. In addition, the saliva serpin 6 and −19 from the lone star tick Amblyomma americanum [113,114], contribute to host hemostasis dysregulation, facilitating blood feeding of this important vector species of ehrlichiosis in the US. At high molecular excess, recombinant Serpin19 inhibits several serine proteinases of the blood clotting cascade, and forms inhibitory complexes with Factor Xa, XIa, and trypsin [114]. However, high stoichiometries of inhibition and unknown rates of inhibition of these Serpin19-proteinase interactions make conclusions on physiological targets of these serpins difficult.
The important role of arthropod saliva serpins for successful blood feeding is further highlighted in studies that explore the consequences of anti-tick serpin antibodies in the host on feeding success. Injection of recombinant serpin-1 (HLS1) from the tick Haemaphysalis longicornis, a common ectoparasite of cattle in East Asian countries, into rabbits resulted in significant mortalities in nymphs and adult ticks when fed on the immunized rabbits [115]. Similar observations were obtained using recombinant H. longicornis serpin-2 (HLS2) [116], and recombinant Iris protein from I. ricinus [117]. Importantly, a protective effect was also observed when a natural host was vaccinated. Injection of cattle with two serine proteinase inhibitors (RAS-1 and −2) from the tick Rhipicephalus appendiculutus, an ectoparasite of cattle and other domestic animals in Africa, conferred partial protection against tick feeding on cattle [118].
While these examples highlight the diverse, unique, and crucial roles of saliva serpins in host-pathogen interactions, our understanding of their physiological and biochemical functions is just emerging. For example, the salivary glands of I. scapularis express 19 extracellular serpins, mostly with predicted trypsin-like specificity. Eleven of these serpins are specifically induced upon blood feeding, and presumably all 19 are present in the saliva and injected into the host [17]. Likewise, transcriptomic and/or proteomic analyses of sialomes from other tick species, including A americanum [119], the cattle tick Rhipicephalus microplus [111], and Rhipicephaluss anguineus [120] revealed a significant number of serpins as part of tick saliva.
In summary, arthropod saliva serpins are functionally diverse and important factors for successful blood feeding, acting as inhibitors of blood coagulation, platelet aggregation, inflammation, and host immunity. In addition, given the large number of still uncharacterized saliva serpins, the roles they occupy at the parasite-host interface are likely even more diverse than currently understood. Their functional analysis should therefore be a research priority and will contribute substantially our understanding of the pharmacological space occupied by serpins.
5. Structure/function aspects of arthropod serpins
Serpins have a conserved tertiary structure that is key to understanding their mechanism of proteinase inhibition [1–3,121]. Currently, the structures of seven different arthropod serpins have been solved from M. sexta [122,123], D. melanogaster [124], An. gambiae [125], B. mori [126], I. ricinus [108], and T. molitor [35]. These arthropod serpins participate in diverse physiological processes, including immunity [35,36,50,51], blood feeding [108,109], and silk production [126,127]. However, the solved insect serpin structures are highly representative of the overall structural basis of the serpin inhibitory mechanism.
The most important functional regions participating in the classical serpin inhibitory mechanism of proteinase inhibition are β-sheet A and the RCL [1–4]. The RCL extends into the solvent and acts as bait for target proteinases, which forms a non-covalent Michaelis-Menten interaction with the RCL. The proteinase will then cleave the P1-P1′ scissile bond within the RCL, forming an acylenzyme intermediate. At this point the proteinase may simply disassociate from the complex, with the proteinase remaining unaffected but the serpin inactivated. Alternatively, inhibition is achieved when the RCL is inserted into β-sheet A as an additional β-strand and the proteinase is translocated to the opposite end of the serpin. The proteinase active site is disrupted, and both serpin and proteinase remain inactive within an SDS-stable complex. Each of these steps is represented within the determined arthropod serpin structures, which highlight the molecular nuances of the complex inhibitory event.
5.1. Structural analyses of insect serpins in the native fold
The native fold of active inhibitory serpins is represented by the structure of M. sexta serpin 1 K (pdb code: 1SEK) [122]. Serpin 1K is a chymotrypsin inhibitor and one of 12 serpins found in M. sexta hemolymph expressed via alternative splicing of the RCL region from a single gene (see Section 1) [128,129]. Serpin 1K was only the fourth determined serpin structure containing the native, active fold and provided pioneering insights into the native serpin conformation. Like most other serpins, serpin 1K folds into three β-sheets (designated A-C) surrounded by nine α-helices (designated A-I) (Fig. 4A) [122]. Although the RCL is often disordered and unresolved in serpin structures, electron density was observed for all residues of the RCL in serpin 1K (P17-P5’). Its RCL extends farther from the surface of the β-sheet B/C barrel relative to α1-antitrypsin [130,131], due to two additional residues near the P2′ position. The N-terminal portion of the RCL is referred to as the hinge region (P17-P13), due to its central importance in the conformational transition from the native to the cleaved, inserted state [132,133]. The serpin 1K hinge region makes several contacts that stabilize the closed form of β-sheet A. Due to the metastable native serpin conformation, the RCL of some serpins will spontaneously insert into β-sheet A, forming a latent, inactive form [134–136]. Therefore, the extension of the RCL away from the body of the serpin and the stabilization of the closed form of β-sheet A in serpin 1K illustrates the structural maintenance of the active state and prevents inactivation. The active serpin state, exemplified by serpin 1K, therefore increases the likelihood of proteinase inhibition while limiting spontaneous inactivation.
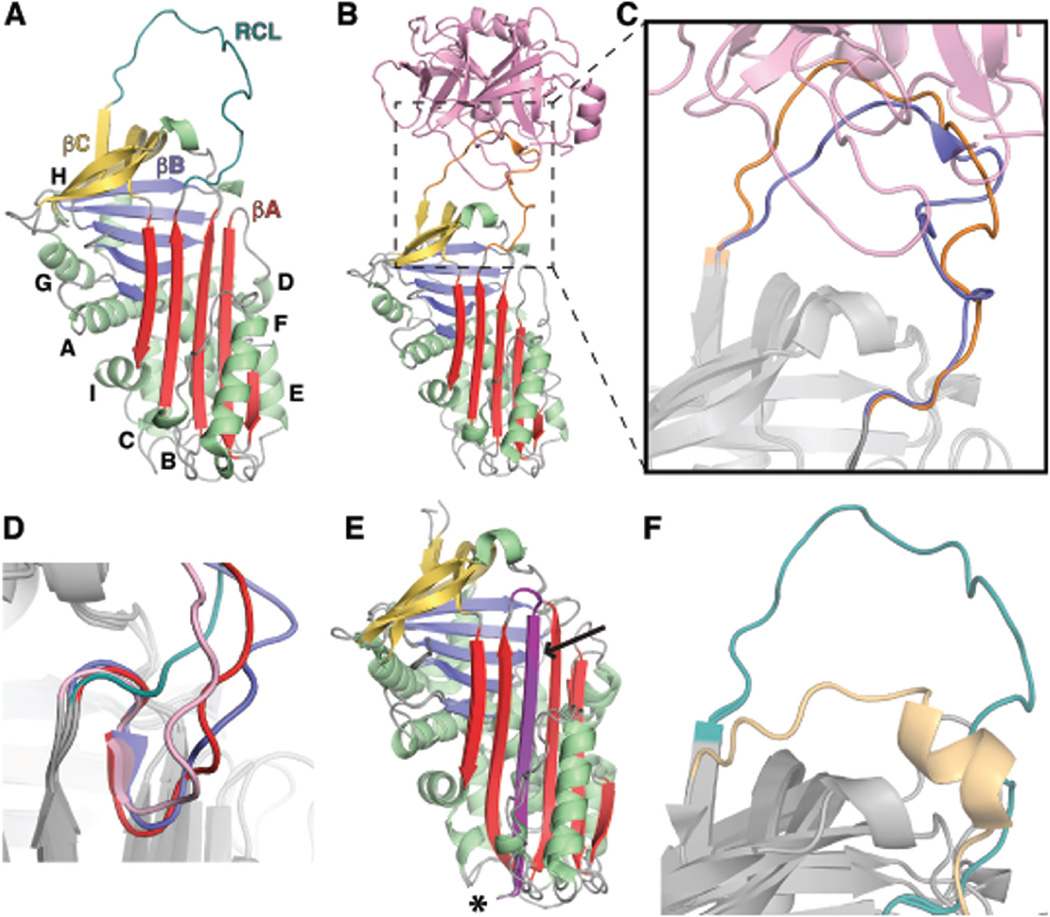
Fig. 4.
Structures of Insect Serpins. (A) Structure of M. sexta serpin1K(pdb code: 1SEK) [122], showing the conserved serpin fold. The serpin contains three β-sheets (βA, red; βB, blue; βC, yellow) surrounded by nine conserved helices (A-I, green). The Reactive Center Loop (RCL, teal) is located above βC. (B) The structure of M. sexta serpin1B A353K co-crystallized in a Michaelis-Menten complex with rat trypsin (pdb code: 1I99) [123]. The serpin1B RCL (orange) interacts with trypsin (pink) at the active site. (C) Close-up the structural alignment of M. sexta serpin1K (teal) and serpin 1B A353K (orange) RCLs interacting with trypsin (pink) indicating the movement of the serpin in response to interaction with trypsin. (D) Alignment of the hinge region of antithrombin (pink, pdb code: 1ATH) [138], T. molitor SPN48 (red, pdb code: 3OZQ) [35], An. gambiae SRPN2 (blue, pdb code: 3PZF) [125], and M. sexta serpin 1K (teal) demonstrating the partial hinge insertion found in antithrombin, SPN48 and SRPN2. (E) Alignment of the cleaved D. melanogaster Serpin 42 Da (pdb codes: 4P0O) [124] and I. ricinus IRS-2 (pdb code 3NDA) [108] structures indicating complete insertion of the RCL (magenta and purple, respectively), indicated by an arrow. Although neither protein was crystallized in an inhibitory complex with a proteinase, the presumed position of the inactivated proteinase in such a complex is marked with an asterisk. (F) Close-up of the RCL of B. moriserpin18(pdbcode: 4R9I) [126] and M. sexta serpin1K indicating the close proximity of the Serpin18 RCL to the serpin body compared to normal serine proteinase inhibitory serpins.
5.2. The Michaelis-Menten complex of M. sexta serpin 1B and trypsin
The shift in serpin conformation upon proteinase interaction was partly established via the structure of another M. sexta serpin, serpin 1B A353K, which was co-crystallized in a Michaelis-Menten complex with rat trypsin (pdb code: 1I99) [123]. The co-crystallized trypsin contains an active site mutation permitting investigation of the interaction before formation of the acyl-enzyme intermediate. The trypsin molecule interacts with the serpin 1B RCL above the β-sheet B/C barrel (Fig. 4B). Serpin 1B A353K is closely related in sequence and structure to serpin 1K [21,128]. Therefore, comparison of the complexed serpin 1B structure with serpin 1K provides important insights into conformational changes that occur due to proteinase interaction prior to RCL insertion. Although there are few conformational changes in the overall serpin 1B fold, significant movements were observed in the RCL, the hinge region, and β-sheet A. The P16-P5 RCL region moves up to 10 Å to interact with the trypsin, with the P13-P3 region forming extensive interactions with the trypsin active site (Fig. 4C). This movement loosens previous interactions that stabilize the closed conformation. Moreover, the βA1 strand moves away from βA2, resulting in an overall β-sheet A conformation closer to the cleaved structure. Therefore, the Michaelis-Menten serpin 1B structure indicates that non-covalent serpin-proteinase interaction initiates a structural transition towards RCL insertion and formation of the inhibitory complex.
5.3. Partial RCL insertion in insect serpins is not indicative of allosteric activation
The M. sexta structures clearly demonstrate the link between the RCL, the hinge region, and β-sheet A in the serpin inhibitory mechanism. This structural connection is highly significant in instances where the native state of the serpin contains a partial RCL hinge insertion into β-sheet A. The partial hinge insertion is involved in cofactor-mediated allosteric regulation of inhibitory activity of several mammalian serpins, including antithrombin III (ATIII) and heparin cofactor II (HCII) [137–139]. Briefly, heparin binding to these serpins causes inhibitory activity to increase up to 1000-fold due to, a) allosteric conformational changes resulting in expulsion of the hinge region causing increased flexibility and accessibility of the RCL, and/or b) a bridging mechanism that increases the interaction between the serpin and proteinase [139–146]. Two insect serpin structures were determined containing a partial insertion of the hinge region: T. molitor SPN48 (pdb code: 3OZQ) [35] and A. gambiae SRPN2 (pdb code: 3PZF) [125]. Both SPN48 and SRPN2 are involved in innate immune responses (see Sections 2.1 and 2.2). The partial insertion of the hinge region in both serpins, reminiscent of ATIII, suggested a similar mechanism of allosteric regulation (Fig. 4D) [35,125]. SPN48 is capable of binding heparin and the structure revealed a distinct lysine-rich region appropriate for heparin binding, although in a different region than in ATIII [35]. Full-length heparin did significantly accelerate SPN48 inhibition of SPE, though lower weight heparin molecules did not. These data indicate the SPN48 hinge region likely does not participate in allosteric activation, and its heparin-mediated activation is likely due to a bridging effect. The partial hinge insertion in SPN48 may be explained by subsequent investigations into SRPN2. The SRPN2 structure did not reveal a heparin-binding site, making allosteric regulation unlikely [125]. To understand its function, three SRPN2 mutants containing either constitutively expelled or inserted hinge regions were developed, their structures were solved, and their activity against CLIPB9 was measured. SRPN2 hinge expulsion did not significantly increase CLIPB9 inhibition, but stable hinge insertion did not decrease the rate of RCL cleavage [147]. These data suggest the SRPN2 hinge insertion represents a molecular trade-off between RCL accessibility and rapid formation of the inhibitory complex, since the partial insertion provides an advantage towards complete RCL insertion following cleavage. Although the connection between this structural feature in both SPN48 and SRPN2 is unknown, together the two structures provide insights into the biophysical balance between the RCL, hinge region, and β-sheet A in serpins important for insect immunity.
5.4. Arthropod serpin structures with complete RCL insertion
The final conformation of the serpin inhibitory mechanism is the complete insertion of the RCL into β-sheet A, resulting in deformation of the proteinase active site and permanent inactivation. Two arthropod serpin structures have been determined in their cleaved, inactivated form: D. melanogaster Spn4 (Serpin 42 Da) (pdb codes: 4P0O and 4P0F, [124]) and I. ricinus IRS-2 (pdb code 3NDA, [148]) (Fig. 4E), previously discussed in Sections 2.3 and 4.2, respectively. The Spn4A/B isoform structures were determined, which contain the same furin-directed RCL [60,61,108,149]. Both structures were nearly identical, with the RCL fully inserted into β-sheet A as a fourth strand in the six stranded β-sheet and evidence of cleavage after the P1 arginine. The structure of IRS-2 also shows the serpin containing a typical serpin fold but with the RCL inserted into β-sheet A due to cleavage at the P1 tyrosine residue. The shutter region, a collection of conserved residues within the center of β-sheet A [3], is displaced in both structures allowing complete insertion of the RCL. Structural insights into the cleaved IRS-2 structure are particularly interesting, as drugs could be developed to block insertion and mitigate the immunosuppressive activity of the serpin [108]. Both the Serpin 42 Da and IRS-2 structures provide insights into the ultimate step of the serpin inhibitory mechanism and clearly demonstrate the final, relaxed state of the serpin upon formation of the inhibitory complex.
5.5. Structure of a cysteine-proteinase inhibitory serpin
In addition to the serpin suicide inhibitory mechanism, directed at serine proteinases, some serpins also have highly important non-inhibitory functions, and a small number have the ability to inhibit cysteine proteinases using an alternative mechanism [150]. Cysteine proteinase inhibitory serpins include human squamous cell carcinoma antigen 1 (SCCA1) which inhibits cathepsins K, L, and S [151,152], as well as the viral serpin crmA and human P19, which inhibit certain caspases [153,154]. B. mori Serpin18 specifically inhibits cysteine proteinases, including a B. mori fibrinase involved in silk production during development [126]. The Ser-pin18 structure (pdb code: 4R9I) indicated that it contains a typical serpin fold, although the three β-sheets (particularly β-sheet C) are bent more towards the serpin core than typical serpins [126]. The most surprising structural feature of Serpin18 is its RCL, which is located closer than usual to the body of the serpin and makes multiple, specific contacts with β-sheet C (Fig. 4F). This close interaction between the RCL and the serpin body is due to the extremely short RCL length. The typical RCL is 20 residues from P17 to P3′, however the Serpin18 RCL contains only 13 residues [126]. This short length, coupled with several non-canonical RCL residues, would preclude the complete insertion of the RCL and the ability to form the standard inhibitory complex [155]. Previous mutagenesis studies have shown that shortening of the RCL in α1-antitrypsin by two residues decreases inhibitory efficiency while increasing the stability of the intermediate complex [155]. Therefore, shortening of the Serpin18 RCL by six residues would allow it to form a stable thioacyl-intermediate complex with target cysteine proteinases, allowing a prolonged susceptibility to proteolytic degradation and effective inhibition.
Together, the currently solved arthropod serpin structures form a strong representation of the mechanism involved in serine proteinase inhibition as well as structures elucidating atypical mechanisms associated with cysteine proteinase inhibition.
6. Conclusions and future prospects
Serpins are a critical superfamily of proteinase inhibitors that are essential to a diverse range of physiological functions. Arthropod genomes typically contain 10–40 serpin genes with increased functional diversity due to alternative splicing of a sequence encoding the RCL. Investigations into arthropod serpins have provided fundamental understanding of the molecular basis of their activity, and have provided insights into multiple aspects of arthropod biology, including immunity, development, reproduction, parasitism, and blood-feeding. However, we are just at the beginning of understanding the diverse roles of serpins in arthropod biology, as the vast majority of serpins identified in arthropod genomes have not been characterized. Comprehensive phylogenetic studies across arthropod serpins should help with initial assignment of putative function based on orthology to known regulators of extracellular proteinase cascades, and would be especially useful within the group of Ixodidae.
Furthermore, most arthropod serpins with known physiological effects remain orphans whose proteinase targets are unknown. Genetic or biochemical approaches that have been used traditionally to investigate individual serpins are often difficult to apply in a given arthropod species and are time consuming. Proteomic approaches to identify target proteinases are becoming available that will allow over the coming years to gain a more global understanding of specific proteinase-serpin pairings. With the exception of D. melanogaster Spn4, the function of any non-secreted serpins from arthropods remains unknown. In addition the molecular function of non-inhibitory serpins, such Acp76A are unknown.
Serpins are often essential components of the arthropod immune response, with their main role being regulatory control over Toll pathway and proPO activation by inhibiting serine proteinase cascades. However, these pathways are only partially understood, and additional unidentified serpin-proteinase pairs that regulate immune responses remain to be discovered. Acute-phase serpins, expressed upon infection, can function in insect immunity. However, the molecular mechanisms of acute-phase serpins, such as An. gambiae SRPN6 and SRPN10, are unknown and will require additional studies to identify their physiological targets and understand their mechanisms for parasite antagonism. In addition, the basis for regulating the expression of acute-phase serpins must be more completely understood. Lastly, serpins can be downregulated in the context of parasitism, leading to alterations in the immune response. Further details are needed to fully understand the pathways through which these events are mediated and how serpin downregulation is linked to wider effects through the immune repertoire.
Limited studies have shown that serpins also play a role in development and reproduction. Drosophila Spn27A provides a link between immunity and development, regulating dorsal-ventral axis patterning in larvae via inhibition of Toll pathway signaling cascades. However, additional serpins in arthropod development have not yet been elucidated. In addition, multiple serpins have been identified in either Drosophila seminal fluid or male accessory glands, but their role in reproduction is currently unclear. Putative function of reproduction-related serpins in hormone transport (D. melanogaster Acp76A) or microbial protection (D. melanogaster Spn38F) will require additional studies.
Some arthropod serpins are secreted in extracellular venom and saliva to modulate host immunity. The immunosuppressive role of serpins via inhibition of immune signaling cascades is a key function in the venom of parasitoid wasps, which require down-regulation of host immunity to allow survival and development of eggs. Additional studies, beyond L. boulardi, may reveal more diverse effects of serpins excreted with venom. In addition, serpins secreted in the saliva in several tick and mosquito species have generated significant interest due to their critical role anticoagulation and immunosuppression during blood feeding, and their potential as vaccine targets. Specifically, emerging anti-tick vaccine development would be significantly aided by the structures of additional saliva serpins involved in blood feeding, which would facilitate antigen mapping.
The structures of insect serpins have been fundamental in determining the basis of serpin activity in general and continued studies will provide important insights into their specific functions within arthropod physiology. Absent, however, are published structures of non-inhibitory insect serpins, e.g. alboserpin, which would shed light on their mechanisms of activity. Moreover, the overall number of arthropod serpin structures is low. Determination of additional serpins structures, including acute phase serpins involved in immunity and serpins present in seminal fluid, venom, and saliva, will provide additional insights into their function, targets, and possibly mechanisms for their regulation and targeting of tissue. Such data would be bolstered further by additional serpin structures complexed with proteinase targets and/or cofactors involved in their activity.
Acknowledgments
The authors acknowledge gratefully funding through NIH grants R01AI095842 to K. Michel, and R37GM041247 to M. Kanost. DA Meekins was supported by a KINBRE Postdoctoral fellowship through the NIGMS, NIH under grant number P20GM103418. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. This is Contribution 16-268-J from the Kansas Agricultural Experiment Station.
References
- 1.Huntington JA. Serpin structure, function and dysfunction. J. Thromb. Haemost. 2011;9(Suppl. 1):26–34. doi: 10.1111/j.1538-7836.2011.04360.x. http://dx.doi.org/10.1111/j.1538-7836.2011.04360.x. [DOI] [PubMed] [Google Scholar]
- 2.Gettins PG. Serpin structure, mechanism, and function. Chem. Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12475206. [DOI] [PubMed] [Google Scholar]
- 3.Whisstock JC, Bottomley SP. Molecular gymnastics: serpin structure, folding and misfolding. Curr. Opin. Struct. Biol. 2006;16:761–768. doi: 10.1016/j.sbi.2006.10.005. http://dx.doi.org/10.1016/j.sbi.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–9236. doi: 10.1038/35038119. http://dx.doi.org/10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki T, Kobayashi K. Isolation of two novel proteinase inhibitors from hemolymph of silkworm larva Bombyx mori. Comparison with human serum proteinase inhibitors. J. Biochem. 1984;95:1009. doi: 10.1093/oxfordjournals.jbchem.a134688. [DOI] [PubMed] [Google Scholar]
- 6.Sasaki T, Kohara A, Shimidzu T, Kobayashi K. Single site proteolysis in silkworm antitrypsin causes structural changes in behavior against denaturing reagents. Agric. Biol. Chem. 1990;54:139–145. [PubMed] [Google Scholar]
- 7.Sasaki T. The reactive site of silkworm hemolymph antichymotrypsin is located at the COOH-terminal region of the molecule. Biochem. Biophys. Res. Commun. 1985;132:320–326. doi: 10.1016/0006-291x(85)91025-3. [DOI] [PubMed] [Google Scholar]
- 8.Kanost MR, Prasad SV, Wells MA. Primary structure of a member of the serpin superfamily of proteinase inhibitors from an insect, Manduca sexta. J. Biol. Chem. 1989;264:965–972. http://www.ncbi.nlm.nih.gov/pubmed/2463253. [PubMed] [Google Scholar]
- 9.Kanost MR. Isolation and characterization of four serine proteinase inhibitors (serpins) from hemolymph of Manduca sexta. Insect Biochem. 1990;20:141–147. http://dx.doi.org/10.1016/0020-1790(90)90006-G. [Google Scholar]
- 10.Takagi H, Narumi H, Nakamura K, Sasaki T. Amino acid sequence of silkworm (Bombyx mori) hemolymph antitrypsin deduced from its cDNA nucleotide sequence: confirmation of its homology with serpins. J. Biochem. 1990;108:372–378. doi: 10.1093/oxfordjournals.jbchem.a123208. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=2277028. [DOI] [PubMed] [Google Scholar]
- 11.Zou Z, Picheng Z, Weng H, Mita K, Jiang H. A comparative analysis of serpin genes in the silkworm genome. Genomics. 2009;93:367–375. doi: 10.1016/j.ygeno.2008.12.010. http://dx.doi.org/10.1016/j.ygeno.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou Z, Evans JD, Lu Z, Zhao P, Williams M, Sumathipala N, et al. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 2007;8:R177. doi: 10.1186/gb-2007-8-8-r177. http://dx.doi.org/10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrett M, Fullaondo A, Troxler L, Micklem G, Gubb D. Identification and analysis of serpin-family genes by homology and synteny across the 12 sequenced Drosophilid genomes. BMC Genom. 2009;10:489. doi: 10.1186/1471-2164-10-489. http://dx.doi.org/10.1186/1471-2164-10-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans JD, Aronstein K, Chen YP, Hetru C, Imler J-L, Jiang H, et al. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006;15:645–656. doi: 10.1111/j.1365-2583.2006.00682.x. http://dx.doi.org/10.1111/j.1365-2583.2006.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mwangi S, Murungi E, Jonas M, Christoffels A. Evolutionary genomics of Glossina morsitans immune-related CLIP domain serine proteases and serine protease inhibitors. Infect. Genet. Evol. 2011;11:740–745. doi: 10.1016/j.meegid.2010.10.006. http://dx.doi.org/10.1016/j.meegid.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Gulley MM, Zhang X, Michel K. The roles of serpins in mosquito immunology and physiology. J. Insect Physiol. 2013;59:138–147. doi: 10.1016/j.jinsphys.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulenga A, Khumthong R, Chalaire KC. Ixodes scapularis tick serine proteinase inhibitor (serpin) gene family; annotation and transcriptional analysis. BMC Genom. 2009;10:217. doi: 10.1186/1471-2164-10-217. http://dx.doi.org/10.1186/1471-2164-10-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez-Valle M, Xu T, Kurscheid S, Lew-Tabor AE. Rhipicephalus microplus serine protease inhibitor family: annotation, expression and functional characterisation assessment. Parasites Vectors. 2015;8:7. doi: 10.1186/s13071-014-0605-4. http://dx.doi.org/10.1186/s13071-014-0605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rider SD, Morgan MS, Arlian LG. Draft genome of the scabies mite. Parasites Vectors. 2015;8:585. doi: 10.1186/s13071-015-1198-2. http://dx.doi.org/10.1186/s13071-015-1198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang H, Wang Y, Huang Y, Mulnix AB, Kadel J, Cole K, et al. Organization of serpin gene-1 from Manduca sexta evolution of a family of alternate exons encoding the reactive site loop. J. Biol. Chem. 1996;271:28017–28023. doi: 10.1074/jbc.271.45.28017. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8910411. [DOI] [PubMed] [Google Scholar]
- 21.Jiang H, Mulnix AB, Kanost MR. Expression and characterization of recombinant Manduca sexta serpin-1B and site-directed mutants that change its inhibitory selectivity. Insect Biochem. Mol. Biol. 1995;25:1093–1100. doi: 10.1016/0965-1748(95)00042-9. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8580909. [DOI] [PubMed] [Google Scholar]
- 22.Ragan EJ, An C, Yang CT, Kanost MR. Analysis of mutually exclusive alternatively spliced serpin-1 isoforms and identification of serpin-1 proteinase complexes in Manduca sexta hemolymph. J. Biol. Chem. 2010;285:29642–29650. doi: 10.1074/jbc.M110.125419. http://dx.doi.org/10.1074/jbc.M110.125419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasaki T. Patchwork-structure serpins from silkworm (Bombyx mori) larval hemolymph. Eur. J. Biochem. 1991;202:255. doi: 10.1111/j.1432-1033.1991.tb16370.x. [DOI] [PubMed] [Google Scholar]
- 24.Hegedus DD, Erlandson M, Baldwin D, Hou X, Chamankhah M. Differential expansion and evolution of the exon family encoding the serpin-1 reactive centre loop has resulted in divergent serpin repertoires among the Lepidoptera. Gene. 2008;418:15–21. doi: 10.1016/j.gene.2008.03.015. http://dx.doi.org/10.1016/j.gene.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 25.Zheng YP, He W-Y, Béliveau C, Nisole A, Stewart D, Zheng S-C, et al. Cloning, expression and characterization of four serpin-1 cDNA variants from the spruce budworm, Choristoneura fumiferana. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 2009;154:165–173. doi: 10.1016/j.cbpb.2009.05.016. http://dx.doi.org/10.1016/j.cbpb.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Li Y, Jia R, Cui W, Mu Z, Zhang Z. Alternative splicing of the antitrypsin gene in the silkworm, Bombyx mori. Mol. Biol. Rep. 2011;38:2793–27939. doi: 10.1007/s11033-010-0424-4. http://dx.doi.org/10.1007/s11033-010-0424-4. [DOI] [PubMed] [Google Scholar]
- 27.Danielli A, Kafatos FC, Loukeris TG. Cloning and characterization of four Anopheles gambiae serpin isoforms, differentially induced in the midgut by Plasmodium berghei invasion. J. Biol. Chem. 2003;278:4184–4193. doi: 10.1074/jbc.M208187200. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12456678. [DOI] [PubMed] [Google Scholar]
- 28.Börner S, Ragg H. Functional diversification of a protease inhibitor gene in the genus Drosophila and its molecular basis. Gene. 2008;415:23–31. doi: 10.1016/j.gene.2008.02.004. http://dx.doi.org/10.1016/j.gene.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Iwanaga S, Kawabata S. Evolution and phylogeny of defense molecules associated with innate immunity in horseshoe crab. Front. Biosci. 1998;3:D973–D984. doi: 10.2741/a337. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9727083. [DOI] [PubMed] [Google Scholar]
- 30.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. http://dx.doi.org/10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 31.Kanost MR, Jiang H. Clip-domain serine proteases as immune factors in insect hemolymph. Curr. Opin. Insect Sci. 2015;11:47–55. doi: 10.1016/j.cois.2015.09.003. http://dx.doi.org/10.1016/j.cois.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veillard F, Troxler L, Reichhart J-M. Drosophila melanogaster clip-domain serine proteases: structure, function and regulation. Biochimie. 2015 doi: 10.1016/j.biochi.2015.10.007. http://dx.doi.org/10.1016/j.biochi.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 33.An C, Ragan EJ, Kanost MR. Serpin-1 splicing isoform J inhibits the proSpätzle-activating proteinase HP8 to regulate expression of antimicrobial hemolymph proteins in Manduca sexta. Dev. Comp. Immunol. 2011;35:135–141. doi: 10.1016/j.dci.2010.09.004. http://dx.doi.org/10.1016/j.dci.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An C, Kanost MR. Manduca sexta serpin-5 regulates prophenoloxidase activation and the Toll signaling pathway by inhibiting hemolymph proteinase HP6. Insect Biochem. Mol. Biol. 2010;40:683–689. doi: 10.1016/j.ibmb.2010.07.001. http://dx.doi.org/10.1016/j.ibmb.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SH, Jiang R, Piao SF, Zhang B, Kim EH, Kwon HM, et al. Structural and functional characterization of a highly specific serpin in the insect innate immunity. J. Biol. Chem. 2011;286:1567–1575. doi: 10.1074/jbc.M110.144006. http://dx.doi.org/10.1074/jbc.M110.144006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang R, Kim EH, Gong JH, Kwon HM, Kim CH, Ryu KH, et al. Three pairs of protease-serpin complexes cooperatively regulate the insect innate immune responses. J. Biol. Chem. 2009;284:35652. doi: 10.1074/jbc.M109.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang R, Zhang B, Kurokawa K, So Y-I, Kim E-H, Hwang HO, et al. 93-kDa twin-domain serine protease inhibitor (Serpin) has a regulatory function on the beetle Toll proteolytic signaling cascade. J. Biol. Chem. 2011;286:35087–35095. doi: 10.1074/jbc.M111.277343. http://dx.doi.org/10.1074/jbc.M111.277343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fullaondo A, García-Sánchez S, Sanz-Parra A, Recio E, Lee SY, Gubb D. Spn1 regulates the GNBP3-dependent Toll signaling pathway in Drosophila melanogaster. Mol. Cell. Biol. 2011;31:2960–2972. doi: 10.1128/MCB.01397-10. http://dx.doi.org/10.1128/MCB.01397-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanost M, Gorman MJ. Phenoloxidase in insect immunity. In: Beckage NE, editor. Insect Immunol. San Diego: Academic Press/Elsevier; 2008. pp. 69–96. [Google Scholar]
- 40.Jiang H, Wang Y, Yu XQ, Zhu Y, Kanost M. Prophenoloxidase-activating proteinase-3 (PAP-3) from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem. Mol. Biol. 2003;33:1049–1060. doi: 10.1016/s0965-1748(03)00123-1. [DOI] [PubMed] [Google Scholar]
- 41.Zhu Y, Wang Y, Gorman MJ, Jiang H, Kanost MR. Manduca sexta serpin-3 regulates prophenoloxidase activation in response to infection by inhibiting prophenoloxidase-activating proteinases. J. Biol. Chem. 2003;278:46556–46564. doi: 10.1074/jbc.M309682200. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12966082. [DOI] [PubMed] [Google Scholar]
- 42.Zou Z, Jiang H. Manduca sexta serpin-6 regulates immune serine proteinases PAP-3 and HP8 cDNA cloning, protein expression, inhibition kinetics, and function elucidation. J. Biol. Chem. 2005;280:14341–14348. doi: 10.1074/jbc.M500570200. http://dx.doi.org/10.1074/jbc.M500570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christen JM, Hiromasa Y, An C, Kanost MR. Identification of plasma proteinase complexes with serpin-3 in Manduca sexta. Insect Biochem. Mol. Biol. 2012;42:946–955. doi: 10.1016/j.ibmb.2012.09.008. http://dx.doi.org/10.1016/j.ibmb.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suwanchaichinda C, Ochieng R, Zhuang S, Kanost MR. Manduca sexta serpin-7, a putative regulator of hemolymph prophenoloxidase activation. Insect Biochem. Mol. Biol. 2013;43:555–561. doi: 10.1016/j.ibmb.2013.03.015. http://dx.doi.org/10.1016/j.ibmb.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu Y, Zhou F, Liu Y, Hong F, Wang G, An C. Ostrinia furnacalis serpin-3 regulates melanization cascade by inhibiting a prophenoloxidase-activating protease. Insect Biochem. Mol. Biol. 2015;61:53–61. doi: 10.1016/j.ibmb.2015.03.007. http://dx.doi.org/10.1016/j.ibmb.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 46.De Gregorio E, Han SJ, Lee WJ, Baek MJ, Osaki T, Kawabata S, et al. An immune-responsive serpin regulates the melanization cascade in Drosophila. Dev. Cell. 2002;3:581–592. doi: 10.1016/s1534-5807(02)00267-8. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12408809. [DOI] [PubMed] [Google Scholar]
- 47.Ligoxygakis P, Pelte N, Ji C, Leclerc V, Duvic B, Belvin M, et al. A serpin mutant links Toll activation to melanization in the host defence of Drosophila. EMBO J. 2002;21:6330–6337. doi: 10.1093/emboj/cdf661. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12456640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nappi AJ, Frey F, Carton Y. Drosophila serpin 27A is a likely target for immune suppression of the blood cell-mediated melanotic encapsulation response. J. Insect Physiol. 2005;51:197–205. doi: 10.1016/j.jinsphys.2004.10.013. http://dx.doi.org/10.1016/j.jinsphys.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 49.An C, Zhang M, Chu Y, Zhao Z. Serine protease MP2 activates prophenoloxidase in the melanization immune response of Drosophila melanogaster. PLoS One. 2013;8:e79533. doi: 10.1371/journal.pone.0079533. http://dx.doi.org/10.1371/journalpone.0079533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michel K, Budd A, Pinto S, Gibson TJ, Kafatos FC. Anopheles gambiae SRPN2 facilitates midgut invasion by the malaria parasite Plasmodium berghei. EMBO Rep. 2005;6:891–897. doi: 10.1038/sj.embor.7400478. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16113656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.An C, Budd A, Kanost MR, Michel K. Characterization of a regulatory unit that controls melanization and affects longevity of mosquitoes. Cell. Mol. Life Sci. 2011;68:1929–1939. doi: 10.1007/s00018-010-0543-z. http://dx.doi.org/10.1007/s00018-010-0543-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zou Z, Shin SW, Alvarez KS, Kokoza V, Raikhel AS. Distinct melanization pathways in the Mosquito Aedes aegypti. Immunity. 2010;32:41–53. doi: 10.1016/j.immuni.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 53.Tong Y, Jiang H, Kanost MR. Identification of plasma proteases inhibited by Manduca sexta serpin-4 and serpin-5 and their association with components of the prophenol oxidase activation pathway. J. Biol. Chem. 2005;280:14932–14942. doi: 10.1074/jbc.M500532200. http://dx.doi.org/10.1074/jbc.M500532200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tong Y, Kanost MR. Manduca sexta serpin-4 and serpin-5 inhibit the prophenol oxidase activation pathway: cDNA cloning, protein expression, and characterization. J. Biol. Chem. 2005;280:14923–14931. doi: 10.1074/jbc.M500531200. http://dx.doi.org/10.1074/jbc.M500531200. [DOI] [PubMed] [Google Scholar]
- 55.Woon Shin S, Park S-S, Park D-S, Gwang Kim M, Brey PT, Park H-Y. Isolation and characterization of immune-related genes from the fall webworm, Hyphantria cunea, using PCR-based differential display and subtractive cloning. Insect Biochem. Mol. Biol. 1998;28:827–837. doi: 10.1016/s0965-1748(98)00077-0. http://dx.doi.org/10.1016/S0965-1748(98)00077-0. [DOI] [PubMed] [Google Scholar]
- 56.Park DS, Shin SW, Hong SD, Park HY. Immunological detection of serpin in the fall webworm, Hyphantria cunea and its inhibitory activity on the prophenoloxidase system. Mol. Cells. 2000;10:186–192. doi: 10.1007/s10059-000-0186-2. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10850660. [DOI] [PubMed] [Google Scholar]
- 57.Danielli A, Barillas-Mury C, Kumar S, Kafatos FC, Loukeris TG. Overexpression and altered nucleocytoplasmic distribution of Anopheles ovalbumin-like SRPN10 serpins in Plasmodium-infected midgut cells. Cell. Microbiol. 2005;7:181–190. doi: 10.1111/j.1462-5822.2004.00445.x. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15659062. [DOI] [PubMed] [Google Scholar]
- 58.Brüning M, Lummer M, Bentele C, Smolenaars MMW, Rodenburg KW, Ragg H. The Spn4 gene from Drosophila melanogaster is a multipurpose defence tool directed against proteases from three different peptidase families. Biochem. J. 2007;401:325–331. doi: 10.1042/BJ20060648. http://dx.doi.org/10.1042/BJ20060648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Graveley BR, May G, Brooks AN, Carlson JW, Cherbas L, Davis CA, et al. The D. melanogaster transcriptome: modENCODE RNA-Seq data for differing treatment conditions. 2011 http://www.modencode.org/celniker/ [Google Scholar]
- 60.Richer MJ, Keays CA, Waterhouse J, Minhas J, Hashimoto C, Jean F. The Spn4 gene of Drosophila encodes a potent furin-directed secretory pathway serpin. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10560–10565. doi: 10.1073/pnas.0401406101. http://dx.doi.org/10.1073/pnas.0401406101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oley M, Letzel MC, Ragg H. Inhibition of furin by serpin Spn4A from Drosophila melanogaster. FEBS Lett. 2004;577:165–169. doi: 10.1016/j.febslet.2004.10.003. http://dx.doi.org/10.1016/j.febslet.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 62.Atella GC, Bittencourt-Cunha PR, Nunes RD, Shahabuddin M, Silva-Neto MAC. The major insect lipoprotein is a lipid source to mosquito stages of malaria parasite. Acta Trop. 2009;109:159–162. doi: 10.1016/j.actatropica.2008.10.004. http://dx.doi.org/10.1016/j.actatropica.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 63.Cheon HM, Shin SW, Bian G, Park JH, Raikhel AS. Regulation of lipid metabolism genes, lipid carrier protein lipophorin, and its receptor during immune challenge in the mosquito Aedes aegypti. J. Biol. Chem. 2006;281:8426–8435. doi: 10.1074/jbc.M510957200. http://dx.doi.org/10.1074/jbc.M510957200, M510957200[pii] [DOI] [PubMed] [Google Scholar]
- 64.Telleria EL, Benoit JB, Zhao X, Savage AF, Regmi S, Alves e Silva TL, et al. Insights into the trypanosome-host interactions revealed through transcriptomic analysis of parasitized tsetse fly salivary glands. PLoS Negl. Trop. Dis. 2014;8:e2649. doi: 10.1371/journal.pntd.0002649. http://dx.doi.org/10.1371/journal.pntd.0002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ooi C-P, Haines LR, Southern DM, Lehane MJ, Acosta-Serrano A. Tsetse GmmSRPN10 has anti-complement activity and is important for successful establishment of trypanosome infections in the fly midgut. PLoS Negl Trop. Dis. 2015;9:e3448. doi: 10.1371/journal.pntd.0003448. http://dx.doi.org/10.1371/journal.pntd.0003448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abraham EG, Pinto SB, Ghosh A, Vanlandingham DL, Budd A, Higgs S, et al. An immune-responsive serpin, SRPN6, mediates mosquito defense against malaria parasites. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16327–16332. doi: 10.1073/pnas.0508335102. http://dx.doi.org/10.1073/pnas.0508335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pinto SB, Kafatos FC, Michel K. The parasite invasion marker SRPN6 reduces sporozoite numbers in salivary glands of Anopheles gambiae. Cell. Microbiol. 2008;10:891–898. doi: 10.1111/j.1462-5822.2007.01091.x. http://dx.doi.org/10.1111/j.1462-5822.2007.01091.x. [DOI] [PubMed] [Google Scholar]
- 68.Eappen AG, Smith RC, Jacobs-Lorena M. Enterobacter-activated mosquito immune responses to Plasmodium involve activation of SRPN6 in Anopheles stephensi. PLoS One. 2013;8:e62937. doi: 10.1371/journal.pone.0062937. http://dx.doi.org/10.1371/journal.pone.0062937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith RC, Eappen AG, Radtke AJ, Jacobs-Lorena M. Regulation of anti-Plasmodium immunity by a LITAF-like transcription factor in the malaria vector Anopheles gambiae. PLoS Pathog. 2012;8:e1002965. doi: 10.1371/journal.ppat.1002965. http://dx.doi.org/10.1371/journal.ppat.1002965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang X, Metzger D, Leeman S, Amar S. LPS-induced TNF-alpha factor (LITAF)-deficient mice express reduced LPS-induced cytokine: evidence for LITAF-dependent LPS signaling pathways. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13777–13782. doi: 10.1073/pnas.0605988103. http://dx.doi.org/10.1073/pnas.0605988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.An C, Hiromasa Y, Zhang X, Lovell S, Zolkiewski M, Tomich JM, et al. Biochemical characterization of Anopheles gambiae SRPN6, a malaria parasite invasion marker in mosquitoes. PLoS One. 2012;7:e48689. doi: 10.1371/journal.pone.0048689. http://dx.doi.org/10.1371/journal.pone.0048689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scherfer C, Tang H, Kambris Z, Lhocine N, Hashimoto C, Lemaitre B. Drosophila serpin-28D regulates hemolymph phenoloxidase activity and adult pigmentation. Dev. Biol. 2008;323:189–196. doi: 10.1016/j.ydbio.2008.08.030. http://dx.doi.org/10.1016/j.ydbio.2008.08.030, S0012-1606(08)01151-2 [pii] [DOI] [PubMed] [Google Scholar]
- 73.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11606746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Irving P, Ubeda JM, Doucet D, Troxler L, Lagueux M, Zachary D, et al. New insights into Drosophila larval haemocyte functions through genome-wide analysis. Cell. Microbiol. 2005;7:335–350. doi: 10.1111/j.1462-5822.2004.00462.x. http://dx.doi.org/10.1111/j.1462-5822.2004.00462.x, CMI462 [pii] [DOI] [PubMed] [Google Scholar]
- 75.Beckage NE, Kanost MR. Effects of parasitism by the braconid wasp Cotesia congregata on host hemolymph proteins of the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 1993;23:643–653. doi: 10.1016/0965-1748(93)90038-t. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8353522. [DOI] [PubMed] [Google Scholar]
- 76.Song K-H, Jung M-K, Eum J-H, Hwang I-C, Han SS. Proteomic analysis of parasitized Plutellaxylostella larvae plasma. J. Insect Physiol. 2008;54:1270–1280. doi: 10.1016/j.jinsphys.2008.06.010. http://dx.doi.org/10.1016/jjinsphys.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 77.Beckage NE, Metcalf JS, Nesbit DJ, Schleifer KW, Zetlan SR, de Buron I. Host hemolymph monophenoloxidase activity in parasitized Manduca sexta larvae and evidence for inhibition by wasp polydnavirus. Insect Biochem. 1990;20:285–294. http://dx.doi.org/10.1016/0020-1790(90)90046-W. [Google Scholar]
- 78.Etebari K, Asgari S. Conserved microRNA miR-8 blocks activation of the Toll pathway by upregulating serpin 27 transcripts. RNA Biol. 2014;10:1356–1364. doi: 10.4161/rna.25481. http://dx.doi.org/10.4161/rna.25481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee GJ, Hyun S. Multiple targets of the microRNA miR-8 contribute to immune homeostasis in Drosophila. Dev. Comp. Immunol. 2014;45:245–251. doi: 10.1016/j.dci.2014.03.015. http://dx.doi.org/10.1016/j.dci.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 80.Ligoxygakis P, Roth S, Reichhart JM. A serpin regulates dorsal-ventral axis formation in the Drosophila embryo. Curr. Biol. 2003;13:2097–2102. doi: 10.1016/j.cub.2003.10.062. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14654000. [DOI] [PubMed] [Google Scholar]
- 81.Avila FW, Sirot LK, LaFlamme BA, Rubinstein CD, Wolfner MF. Insect seminal fluid proteins: identification and function. Annu. Rev. Entomol. 2011;56:21–40. doi: 10.1146/annurev-ento-120709-144823. http://dx.doi.org/10.1146/annurev-ento-120709-144823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wolfner MF, Harada HA, Bertram MJ, Stelick TJ, Kraus KW, Kalb JM, et al. New genes for male accessory gland proteins in Drosophila melanogaster. Insect Biochem. Mol. Biol. 1997;27:825–834. doi: 10.1016/s0965-1748(97)00056-8. http://dx.doi.org/10.1016/S0965-1748(97)00056-8. [DOI] [PubMed] [Google Scholar]
- 83.Coleman S, Drahn B, Petersen G, Stolorov J, Kraus K. A Drosophila male accessory gland protein that is a member of the serpin superfamily of proteinase inhibitors is transferred to females during mating. Insect Biochem. Mol. Biol. 1995;25:203–207. doi: 10.1016/0965-1748(94)00055-m. http://www.ncbi.nlm.nih.gov/pubmed/7711750. [DOI] [PubMed] [Google Scholar]
- 84.Ravi Ram K, Ji S, Wolfner MF. Fates and targets of male accessory gland proteins in mated female Drosophila melanogaster. Insect Biochem. Mol. Biol. 2005;35:1059–1071. doi: 10.1016/j.ibmb.2005.05.001. http://dx.doi.org/10.1016/j.ibmb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 85.Huntington JA, Kjellberg M, Stenflo J. Crystal structure of protein C inhibitor provides insights into hormone binding and heparin activation. Structure. 2003;11:205–215. doi: 10.1016/s0969-2126(02)00944-9. http://dx.doi.org/10.1016/S0969-2126(02)00944-9. [DOI] [PubMed] [Google Scholar]
- 86.Mueller JL, Ripoll DR, Aquadro CF, Wolfner MF. Comparative structural modeling and inference of conserved protein classes in Drosophila seminal fluid. Proc. Natl. Acad. Sci. U. S. A. 2004;101:13542–13547. doi: 10.1073/pnas.0405579101. http://dx.doi.org/10.1073/pnas.0405579101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mueller JL, Page JL, Wolfner MF. An ectopic expression screen reveals the protective and toxic effects of Drosophila seminal fluid proteins. Genetics. 2007;175:777–783. doi: 10.1534/genetics.106.065318. http://dx.doi.org/10.1534/genetics.106.065318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dottorini T, Nicolaides L, Ranson H, Rogers DW, Crisanti A, Catteruccia F. A genome-wide analysis in Anopheles gambiae mosquitoes reveals 46 male accessory gland genes, possible modulators of female behavior. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16215–16220. doi: 10.1073/pnas.0703904104. http://dx.doi.org/10.1073/pnas.0703904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sirot LK, Poulson RL, McKenna MC, Girnary H, Wolfner MF, Harrington LC. Identity and transfer of male reproductive gland proteins of the dengue vector mosquito, Aedes aegypti: potential tools for control of female feeding and reproduction. Insect Biochem. Mol. Biol. 2008;38:176–189. doi: 10.1016/j.ibmb.2007.10.007. http://dx.doi.org/10.1016/j.ibmb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boes KE, Ribeiro JMC, Wong A, Harrington LC, Wolfner MF, Sirot LK. Identification and characterization of seminal fluid proteins in the Asian tiger mosquito, Aedes albopictus. PLoS Negl. Trop. Dis. 2014;8:e2946. doi: 10.1371/journal.pntd.0002946. http://dx.doi.org/10.1371/journal.pntd.0002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu J, Baulding J, Palli SR. Proteomics of Tribolium castaneum seminal fluid proteins: identification of an angiotensin-converting enzyme as a key player in regulation of reproduction. J. Proteom. 2013;78:83–93. doi: 10.1016/j.jprot.2012.11.011. http://dx.doi.org/10.1016/j.jprot.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 92.Baer B, Heazlewood JL, Taylor NL, Eubel H, Millar AH. The seminal fluid proteome of the honeybee Apis mellifera. Proteomics. 2009;9:2085–2097. doi: 10.1002/pmic.200800708. http://dx.doi.org/10.1002/pmic.200800708. [DOI] [PubMed] [Google Scholar]
- 93.Azevedo RVDM, Dias DBS, Bretãs JAC, Mazzoni CJ, Souza NA, Albano RM, et al. The transcriptome of Lutzomyia longipalpis (Diptera: Psychodidae) male reproductive organs. PLoS One. 2012;7:e34495. doi: 10.1371/journal.pone.0034495. http://dx.doi.org/10.1371/journal.pone.0034495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Walters JR, Harrison RG. Combined EST and proteomic analysis identifies rapidly evolving seminal fluid proteins in Heliconius butterflies. Mol. Biol. Evol. 2010;27:2000–2013. doi: 10.1093/molbev/msq092. http://dx.doi.org/10.1093/molbev/msq092. [DOI] [PubMed] [Google Scholar]
- 95.Breugelmans B, Simonet G, van Hoef V, Van Soest S, Vanden Broeck J. Pacifastin-related peptides: structural and functional characteristics of a family of serine peptidase inhibitors. Peptides. 2009;30:622–632. doi: 10.1016/j.peptides.2008.07.026. http://dx.doi.org/10.1016/j.peptides.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 96.Dorémus T, Urbach S, Jouan V, Cousserans F, Ravallec M, Demettre E, et al. Venom gland extract is not required for successful parasitism in the polydnavirus-associated endoparasitoid Hyposoter didymator (Hym. Ichneumonidae) despite the presence of numerous novel and conserved venom proteins. Insect Biochem. Mol. Biol. 2013;43:292–307. doi: 10.1016/j.ibmb.2012.12.010. http://dx.doi.org/10.1016/j.ibmb.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 97.Colinet D, Dubuffet A, Cazes D, Moreau S, Drezen J-M, Poirié M. Aserpin from the parasitoid wasp Leptopilina boulardi targets the Drosophila phenoloxidase cascade. Dev. Comp. Immunol. 2009;33:681–689. doi: 10.1016/j.dci.2008.11.013. http://dx.doi.org/10.1016/j.dci.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 98.Colinet D, Deleury E, Anselme C, Cazes D, Poulain J, Azema-Dossat C, et al. Extensive inter- and intraspecific venom variation in closely related parasites targeting the same host: the case of Leptopilina parasitoids of Drosophila. Insect Biochem. Mol. Biol. 2013;43:601–611. doi: 10.1016/j.ibmb.2013.03.010. http://dx.doi.org/10.1016/j.ibmb.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 99.Rau JC, Beaulieu LM, Huntington JA, Church FC. Serpins in thrombosis, hemostasis and fibrinolysis. J. Thromb. Haemost. 2007;5(Suppl. 1):102–115. doi: 10.1111/j.1538-7836.2007.02516.x. http://dx.doi.org/10.1111/j.1538-7836.2007.02516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Calvo E, Mizurini DM, Sa-Nunes A, Ribeiro JMC, Andersen JF, Mans BJ, et al. Alboserpin, a factor Xa inhibitor from the mosquito vector of yellow fever, binds heparin and membrane phospholipids and exhibits antithrombotic activity. J. Biol. Chem. 2011;286:27998–28010. doi: 10.1074/jbc.M111.247924. http://.doi.org/10.1074/jbc.M111.247924dx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stark KR, James AA. Isolation and characterization of the gene encoding a novel factor Xa-directed anticoagulant from the yellow fever mosquito, Aedes aegypti. J. Biol. Chem. 1998;273:20802–20809. doi: 10.1074/jbc.273.33.20802. http://www.ncbi.nlm.nih.gov/pubmed/9694825. [DOI] [PubMed] [Google Scholar]
- 102.Stark KR, James AA. A factor Xa-directed anticoagulant from the salivary glands of the yellow fever mosquito Aedes aegypti. Exp. Parasitol. 1995;81:321–331. doi: 10.1006/expr.1995.1123. http://dx.doi.org/10.1006/expr.1995.1123. [DOI] [PubMed] [Google Scholar]
- 103.Calvo E, Dao A, Pham VM, Ribeiro JM. An insight into the sialome of Anopheles funestus reveals an emerging pattern in anopheline salivary protein families. Insect Biochem. Mol. Biol. 2007;37:164–175. doi: 10.1016/j.ibmb.2006.11.005. http://dx.doi.org/10.1016/j.ibmb.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Francischetti IM, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JM. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J. Exp. Biol. 2002;205:2429–2451. doi: 10.1242/jeb.205.16.2429. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12124367. [DOI] [PubMed] [Google Scholar]
- 105.Leboulle G, Crippa M, Decrem Y, Mejri N, Brossard M, Bollen A, et al. Characterization of a novel salivary immunosuppressive protein from Ixodes ricinus ticks. J. Biol. Chem. 2002;277:10083–10089. doi: 10.1074/jbc.M111391200. http://dx.doi.org/10.1074/jbc.M111391200. [DOI] [PubMed] [Google Scholar]
- 106.Prevot PP, Adam B, Boudjeltia KZ, Brossard M, Lins L, Cauchie P, et al. Anti-hemostatic effects of a serpin from the saliva of the tick Ixodes ricinus. J. Biol. Chem. 2006;281:26361–26369. doi: 10.1074/jbc.M604197200. http://dx.doi.org/10.1074/jbc.M604197200. [DOI] [PubMed] [Google Scholar]
- 107.Prevot PP, Beschin A, Lins L, Beaufays J, Grosjean A, Bruys L, et al. Exosites mediate the antiinflammatory effects of a multifunctional serpin from the saliva of the tick Ixodes ricinus. FEBS J. 2009;276:3235–3246. doi: 10.1111/j.1742-4658.2009.07038.x. http://dx.doi.org/10.1111/j.1742-4658.2009.07038.x. [DOI] [PubMed] [Google Scholar]
- 108.Chmelar J, Oliveira CJ, Rezacova P, Francischetti IMB, Kovarova Z, Pejler G, et al. A tick salivary protein targets cathepsin G and chymase and inhibits host inflammation and platelet aggregation. Blood. 2011;117:736–744. doi: 10.1182/blood-2010-06-293241. http://dx.doi.org/10.1182/blood-2010-06-293241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Páleníková J, Lieskovská J, Langhansová H, Kotsyfakis M, Chmelar J, Kopecky J. Ixodes ricinus salivary serpin IRS-2 affects Th17 differentiation via inhibition of the interleukin-6/STAT-3 signaling pathway. Infect. Immun. 2015;83:1949–1956. doi: 10.1128/IAI.03065-14. http://dx.doi.org/10.1128/IAI.03065-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ibelli AMG, Kim TK, Hill CC, Lewis LA, Bakshi M, Miller S, et al. A blood meal-induced Ixodes scapularis tick saliva serpin inhibits trypsin and thrombin, and interferes with platelet aggregation and blood clotting. Int. J. Parasitol. 2014;44:369–379. doi: 10.1016/j.ijpara.2014.01.010. http://dx.doi.org/10.1016/j.ijpara.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tirloni L, Kim TK, Coutinho ML, Ali A, Seixas A, Termignoni C, et al. The putative role of Rhipicephalus microplus salivary serpins in the tick-host relationship. Insect Biochem. Mol. Biol. 2016;71:12–28. doi: 10.1016/j.ibmb.2016.01.004. http://dx.doi.org/10.1016/j.ibmb.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu T, Lew-Tabor A, Rodriguez-Valle M. Effective inhibition of thrombin by Rhipicephalus microplus serpin-15 (RmS-15) obtained in the yeast Pichia pastoris. Ticks Tick Borne Dis. 2016;7:180–187. doi: 10.1016/j.ttbdis.2015.09.007. http://dx.doi.org/10.1016/j.ttbdis.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 113.Mulenga A, Kim T, Ibelli AMG. Amblyomma americanum tick saliva serine protease inhibitor 6 is across-class inhibitor of serine proteases and papain-like cysteine proteases that delays plasma clotting and inhibits platelet aggregation. Insect Mol. Biol. 2013;22:306–319. doi: 10.1111/imb.12024. http://dx.doi.org/10.1111/imb.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim TK, Tirloni L, Radulovic Z, Lewis L, Bakshi M, Hill C, et al. Conserved Amblyomma americanum tick serpin19, an inhibitor of blood clotting factors Xa and XIa, trypsin and plasmin, has anti-haemostatic functions. Int. J. Parasitol. 2015;45:613–627. doi: 10.1016/j.ijpara.2015.03.009. http://dx.doi.org/10.1016/j.ijpara.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sugino M, Imamura S, Mulenga A, Nakajima M, Tsuda A, Ohashi K, et al. A serine proteinase inhibitor (serpin) from ixodidtick Haemaphysalis longicornis; cloning and preliminary assessment of its suitability as a candidate for a tick vaccine. Vaccine. 2003;21:2844–2851. doi: 10.1016/s0264-410x(03)00167-1. http://dx.doi.org/10.1016/S0264-410X(03)00167-1. [DOI] [PubMed] [Google Scholar]
- 116.Imamura S, da SilvaVaz Junior I, Sugino M, Ohashi K, Onuma M. A serine protease inhibitor (serpin) from Haemaphysalis longicornis as an anti-tick vaccine. Vaccine. 2005;23:1301–1311. doi: 10.1016/j.vaccine.2004.08.041. http://dx.doi.org/10.1016/j.vaccine.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 117.Prevot PP, Couvreur B, Denis V, Brossard M, Vanhamme L, Godfroid E. Protective immunity against Ixodes ricinus induced by a salivary serpin. Vaccine. 2007;25:3284–3292. doi: 10.1016/j.vaccine.2007.01.008. http://dx.doi.org/10.1016/j.vaccine.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 118.Imamura S, Namangala B, Tajima T, Tembo ME, Yasuda J, Ohashi K, et al. Two serine protease inhibitors (serpins) that induce a bovine protective immune response against Rhipicephalus appendiculatus ticks. Vaccine. 2006;24:2230–2237. doi: 10.1016/j.vaccine.2005.10.055. http://dx.doi.org/10.1016/j.vaccine.2005.10.055. [DOI] [PubMed] [Google Scholar]
- 119.Porter L, Radulovic Z, Kim T, Braz GRC, DaSilva Vaz I, Mulenga A. Bioinformatic analyses of male and female Amblyomma americanum tick expressed serine protease inhibitors (serpins) Ticks Tick Borne Dis. 2015;6:16–30. doi: 10.1016/j.ttbdis.2014.08.002. http://dx.doi.org/10.1016/j.ttbdis.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oliveira CJ, Anatriello E, de Miranda-Santos IK, Francischetti IM, Sá-Nunes A, Ferreira BR, et al. Proteome of Rhipicephalus sanguineus tick saliva induced by the secretagogues pilocarpine and dopamine. Ticks Tick Borne Dis. 2013;4:469–477. doi: 10.1016/j.ttbdis.2013.05.001. http://dx.doi.org/10.1016/j.ttbdis.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Law RHP, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, et al. An overview of the serpin superfamily. Genome Biol. 2006;7:216. doi: 10.1186/gb-2006-7-5-216. http://dx.doi.org/10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li J, Wang Z, Canagarajah B, Jiang H, Kanost M, Goldsmith EJ. The structure of active serpin 1 K from Manduca sexta. Struct. Fold Des. 1999;7:103–109. doi: 10.1016/s0969-2126(99)80013-6. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10368276. [DOI] [PubMed] [Google Scholar]
- 123.Ye S, Cech AL, Belmares R, Bergstrom RC, Tong Y, Corey DR, et al. The structure of a Michaelis serpin-protease complex. Nat. Struct. Biol. 2001;8:979–983. doi: 10.1038/nsb1101-979. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11685246. [DOI] [PubMed] [Google Scholar]
- 124.Ellisdon AM, Zhang Q, Henstridge MA, Johnson TK, Warr CG, Law RH, et al. High resolution structure of cleaved serpin 42 Da from Drosophila melanogaster. BMC Struct. Biol. 2014;14:14. doi: 10.1186/1472-6807-14-14. http://dx.doi.org/10.1186/1472-6807-14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.An C, Lovell S, Kanost MR, Battaile KP, Michel K. Crystal structure of native Anopheles gambiae serpin-2, a negative regulator of melanization in mosquitoes. Proteins. 2011;79:1999–2003. doi: 10.1002/prot.23002. http://dx.doi.org/10.1002/prot.23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guo P-C, Dong Z, Zhao P, Zhang Y, He H, Tan X, et al. Structural insights into the unique inhibitory mechanism of the silkworm protease inhibitor serpin18. Sci. Rep. 2015;5:11863. doi: 10.1038/srep11863. http://dx.doi.org/10.1038/srep11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guo P-C, Dong Z, Xiao L, Li T, Zhang Y, He H, et al. Silk gland-specific proteinase inhibitor serpin16 from the Bombyx mori shows cysteine proteinase inhibitory activity. Biochem. Biophys. Res. Commun. 2015;457:31–36. doi: 10.1016/j.bbrc.2014.12.056. http://dx.doi.org/10.1016/j.bbrc.2014.12.056. [DOI] [PubMed] [Google Scholar]
- 128.Jiang H, Kanost MR. Characterization and functional analysis of 12 naturally occurring reactive site variants of serpin-1 from Manduca sexta. J. Biol. Chem. 1997;272:1082–1087. doi: 10.1074/jbc.272.2.1082. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8995406. [DOI] [PubMed] [Google Scholar]
- 129.Jiang H, Wang Y, Kanost MR. Mutually exclusive exon use and reactive center diversity in insect serpins. J. Biol. Chem. 1994;269:55–58. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8276850. [PubMed] [Google Scholar]
- 130.Elliott PR, Lomas DA, Carrell RW, Abrahams JP. Inhibitory conformation of the reactive loop of alpha 1-antitrypsin. Nat. Struct. Biol. 1996;3:676–681. doi: 10.1038/nsb0896-676. http://www.ncbi.nlm.nih.gov/pubmed/8756325. [DOI] [PubMed] [Google Scholar]
- 131.Ryu SE, Choi HJ, Kwon KS, Lee KN, Yu MH. The native strains in the hydrophobic core and flexible reactive loop of a serine protease inhibitor: crystal structure of an uncleaved alpha1-antitrypsin at 2.7 A. Structure. 1996;4:1181–1192. doi: 10.1016/s0969-2126(96)00126-8. http://www.ncbi.nlm.nih.gov/pubmed/8939743. [DOI] [PubMed] [Google Scholar]
- 132.Davis AE, Aulak K, Parad RB, Stecklein HP, Eldering E, Hack CE, et al. C1 inhibitor hinge region mutations produce dysfunction by different mechanisms. Nat. Genet. 1992;1:354–358. doi: 10.1038/ng0892-354. http://dx.doi.org/10.1038/ng0892-354. [DOI] [PubMed] [Google Scholar]
- 133.Hopkins PC, Carrell RW, Stone SR. Effects of mutations in the hinge region of serpins. Biochemistry. 1993;32:7650–7657. doi: 10.1021/bi00081a008. http://www.ncbi.nlm.nih.gov/pubmed/8347575. [DOI] [PubMed] [Google Scholar]
- 134.Mottonen J, Strand A, Symersky J, Sweet RM, Danley DE, Geoghegan KF, et al. Structural basis of latency in plasminogen activator inhibitor-1. Nature. 1992;355:270–273. doi: 10.1038/355270a0. http://dx.doi.org/10.1038/355270a0. [DOI] [PubMed] [Google Scholar]
- 135.Wang Z, Mottonen J, Goldsmith EJ. Kinetically controlled folding of the serpin plasminogen activator inhibitor 1. Biochemistry. 1996;35:16443–16448. doi: 10.1021/bi961214p. http://dx.doi.org/10.1021/bi961214p. [DOI] [PubMed] [Google Scholar]
- 136.Bruch M, Weiss V, Engel J. Plasma serine proteinase inhibitors (serpins) exhibit major conformational changes and a large increase in conformational stability upon cleavage at their reactive sites. J. Biol. Chem. 1988;263:16626–16630. http://www.ncbi.nlm.nih.gov/pubmed/3263371. [PubMed] [Google Scholar]
- 137.McCoy AJ, Pei XY, Skinner R, Abrahams JP, Carrell RW. Structure of beta-antithrombin and the effect of glycosylation on antithrombin’s heparin affinity and activity. J. Mol. Biol. 2003;326:823–833. doi: 10.1016/s0022-2836(02)01382-7. S0022283602013827 [pii] [DOI] [PubMed] [Google Scholar]
- 138.Schreuder HA, de Boer B, Dijkema R, Mulders J, Theunissen HJ, Grootenhuis PD, et al. The intact and cleaved human antithrombin III complex as a model for serpin-proteinase interactions. Nat. Struct. Biol. 1994;1:48–54. doi: 10.1038/nsb0194-48. http://www.ncbi.nlm.nih.gov/pubmed/7656006. [DOI] [PubMed] [Google Scholar]
- 139.Baglin TP, Carrell RW, Church FC, Esmon CT, Huntington JA. Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multistep allosteric mechanism. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11079–11084. doi: 10.1073/pnas.162232399. http://dx.doi.org/10.1073/pnas.162232399162232399 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jin L, Abrahams JP, Skinner R, Petitou M, Pike RN, Carrell RW. The anticoagulant activation of antithrombin by heparin. Proc. Natl. Acad. Sci. U. S. A. 1997;94:14683–14688. doi: 10.1073/pnas.94.26.14683. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9405673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huntington JA, Olson ST, Fan B, Gettins PG. Mechanism of heparin activation of antithrombin Evidence for reactive center loop preinsertion with expulsion upon heparin binding. Biochemistry. 1996;35:8495–8503. doi: 10.1021/bi9604643. http://dx.doi.org/10.1021/bi9604643bi9604643 [pii] [DOI] [PubMed] [Google Scholar]
- 142.Dementiev A, Swanson R, Roth R, Isetti G, Izaguirre G, Olson ST, et al. The allosteric mechanism of activation of antithrombin as an inhibitor of factor IXa and factor Xa: heparin-independent full activation through mutations adjacent to helix D. J. Biol. Chem. 2013;288:33611–33619. doi: 10.1074/jbc.M113.510727. http://dx.doi.org/10.1074/jbc.M113.510727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Olson ST, Richard B, Izaguirre G, Schedin-Weiss S, Gettins PGW. Molecular mechanisms of antithrombin-heparin regulation of blood clotting proteinases. A paradigm for understanding proteinase regulation by serpin family protein proteinase inhibitors. Biochimie. 2010;92:1587–1596. doi: 10.1016/j.biochi.2010.05.011. http://dx.doi.org/10.1016/j.biochi.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Izaguirre G, Olson ST. Residues Tyr253 and Glu255 in strand 3 of beta-sheet C of antithrombin are key determinants of an exosite made accessible by heparin activation to promote rapid inhibition of factors Xa and IXa. J. Biol. Chem. 2006;281:13424–13432. doi: 10.1074/jbc.M600415200. http://dx.doi.org/10.1074/jbc.M600415200. [DOI] [PubMed] [Google Scholar]
- 145.Futamura A, Gettins PG. Serine 380 (P14) ->glutamate mutation activates antithrombin as an inhibitor of factor Xa. J. Biol. Chem. 2000;275:4092–4098. doi: 10.1074/jbc.275.6.4092. http://www.ncbi.nlm.nih.gov/pubmed/10660568. [DOI] [PubMed] [Google Scholar]
- 146.Johnson DJD, Li W, Adams TE, Huntington JA. Antithrombin-S195A factor Xa-heparin structure reveals the allosteric mechanism of antithrombin activation. EMBO J. 2006;25:2029–2037. doi: 10.1038/sj.emboj.7601089. http://dx.doi.org/10.1038/sj.emboj.7601089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang X, Meekins DA, An C, Zolkiewski M, Battaile KP, Kanost MR, et al. Structural and inhibitory effects of hinge loop mutagenesis in serpin-2 from the malaria vector Anopheles gambiae. J. Biol. Chem. 2015;290:2946–2956. doi: 10.1074/jbc.M114.625665. http://dx.doi.org/10.1074/jbc.M114.625665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chmelar J, Oliveira CJ, Rezacova P, Francischetti IM, Kovarova Z, Pejler G, et al. A tick salivary protein targets cathepsin G and chymase and inhibits host inflammation and platelet aggregation. Blood. 2011;117:736–744. doi: 10.1182/blood-2010-06-293241. http://dx.doi.org/10.1182/blood-2010-06-293241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Osterwalder T, Kuhnen A, Leiserson WM, Kim YS, Keshishian H. Drosophila serpin 4 functions as a neuroserpin-like inhibitor of subtilisin-like proprotein convertases. J. Neurosci. 2004;24:5482–5491. doi: 10.1523/JNEUROSCI.5577-03.2004. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15201320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Whisstock JC, Silverman GA, Bird PI, Bottomley SP, Kaiserman D, Luke CJ, et al. Serpins flex their muscle: II. Structural insights into target peptidase recognition, polymerization, and transport functions. J. Biol. Chem. 2010;285:24307–24312. doi: 10.1074/jbc.R110.141408. http://dx.doi.org/10.1074/jbc.r110.141408, R110.141408 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Schick C, Pemberton PA, Shi GP, Kamachi Y, Cataltepe S, Bartuski AJ, et al. Cross-class inhibition of the cysteine proteinases cathepsins K, L, and S by the serpin squamous cell carcinoma antigen 1: a kinetic analysis. Biochemistry. 1998;37:5258–5266. doi: 10.1021/bi972521d. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9548757. [DOI] [PubMed] [Google Scholar]
- 152.Schick C, Brömme D, Bartuski AJ, Uemura Y, Schechter NM, Silverman GA. The reactive site loop of the serpin SCCA1 is essential for cysteine proteinase inhibition. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13465–13470. doi: 10.1073/pnas.95.23.13465. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=24842&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhou Q, Snipas S, Orth K, Muzio M, Dixit VM, Salvesen GS. Target protease specificity of the viral serpin CrmA: analysis of five caspases. J. Biol. Chem. 1997;272:7797–7800. doi: 10.1074/jbc.272.12.7797. http://dx.doi.org/10.1074/jbc.272.12.7797. [DOI] [PubMed] [Google Scholar]
- 154.Annand RR, Dahlen JR, Sprecher CA, De Dreu P, Foster DC, Mankovich JA, et al. Caspase-1 (interleukin-1beta-converting enzyme) is inhibited by the human serpin analogue proteinase inhibitor 9. Biochem. J. 1999;342(Pt. 3):655–665. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1220507&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- 155.Zhou A, Carrell RW, Huntington JA. The serpin inhibitory mechanism is critically dependent on the length of the reactive center loop. J. Biol. Chem. 2001;276:27541–27547. doi: 10.1074/jbc.M102594200. http://dx.doi.org/10.1074/jbc.M102594200. [DOI] [PubMed] [Google Scholar]
- 156.Rau JC, Beaulieu LM, Huntington JA, Church FC. Serpins in thrombosis, hemostasis and fibrinolysis. J. Thromb. Haemost. 2007;5:102–115. doi: 10.1111/j.1538-7836.2007.02516.x. http://dx.doi.org/10.1111/j.1538-7836.2006.02283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Huntington JA. Natural inhibitors of thrombin. Thromb. Haemost. 2014;111:583–589. doi: 10.1160/TH13-10-0811. http://dx.doi.org/10.1160/TH13-10-0811. [DOI] [PubMed] [Google Scholar]
- 158.Bhakuni T, Ali MF, Ahmad I, Bano S, Ansari S, Jairajpuri MA. Role of heparin and non heparin binding serpins in coagulation and angiogenesis: a complex interplay. Arch. Biochem. Biophys. 2016;604:128–142. doi: 10.1016/j.abb.2016.06.018. http://dx.doi.org/10.1016/j.abb.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 159.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. http://www.ncbi.nlm.nih.gov/pubmed/1633570. [DOI] [PubMed] [Google Scholar]
- 160.Kumar S, Stecher G, Tamura K. MEGA7 molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. http://dx.doi.org/10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. http://dx.doi.org/10.1093/nar/gkh34032/5/1792 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. http://dx.doi.org/10.1093/bioinformatics/btp033, bt033 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]