Abstract
Imaging genomics is a new field of investigation that seeks to gain insights into the impact of human genetic variation on the structure, chemistry and function of neural systems in health and disease. As publications in this field have increased over the past decade, increasing concerns have been raised about false positive results entering the literature. Here we provide an overview of the field of imaging genomic and genetic approaches, and discuss factors related to research design and analysis that can enhance the informativeness and replicability of these studies. We conclude that imaging genetic studies can provide important insights into the role of human genetic variation on neural systems and circuits, both in the context of normal quantitative variation and in relation to neuropsychiatric disease. We also argue that demonstrating genetic association to imaging-derived traits is subject to the same constraints as any other genetic study, including stringent Type I error control. Adequately powered studies are necessary; however, there are currently limited data available to allow precise estimates of effect sizes for candidate gene studies. Independent replication is necessary before a result can be considered definitive, and for studies with small sample sizes is necessary before publication. Increased transparency of methods and enhanced data sharing will further enhance replicability.
Keywords: Imaging Genomics, Statistical Power, Sample Size, Multiple Comparisons, Replication, Clinical Confounds
Background
Imaging genomics is a relatively new field of investigation that seeks to gain insights into the impact of human genetic variation on the structure, chemistry and function of neural systems in health and disease(1). In general, imaging genomics studies take one of two approaches: 1) candidate gene approaches that attempt to identify an association between a specific genetic variant of a priori interest with quantitative variation in neuroimaging measure(s); 2) discovery based approaches, that seek to discover genes that are associated with variation in specific imaging measures. As such, imaging genomics comprise one element of a range of mechanistic studies which may involve functional, structural, and molecular imaging, as well as cellular and molecular investigations in human tissues or animal model systems, in order to provide converging insights into the mechanisms by which genes may alter brain development and function and may lead to the signs and symptoms of a disorder (2).
During this period there have also been major developments in the genetic tools that are available, from low-cost whole genome sequencing to targeted SNP chips, as well as new methods for imaging brain anatomy, functional and chemistry, and a host of new computational analytic approaches. Additionally, technological innovations, such as those supported by the Brain Research Through Advancing Innovative Neurotechnologies (BRAIN) initiative (http://braininitiative.nih.gov/index.htm), will likely provide a means to more directly connect cellular and molecular mechanisms to specific neural circuitry abnormalities in humans further enhancing the sensitivity of this approach.
In this rapidly advancing field, a range of perspectives on imaging genomics methodologies has evolved, with excitement for the ability to link genetic variations with specific neural systems tempered by a number of concerns. These include the merits of exploratory versus hypothesis-driven approaches, concerns about sample sizes and approaches used to protect against type I error(3) raising concerns about the replicability of many published studies in the field. In a recent review of 40 imaging genomics studies involving 7 GWAS positive genes for schizophrenia it was noted that while most genes showed positive associations with schizophrenia-related imaging phenotypes, only 21% of studies met a previously specified minimum criterion (for an imaging study) of 20 subjects per cell, over 90% had flaws in the clinical/genetic design, correction for multiple comparisons was rare and very few true replications were reported (4). In the present paper we explore these and other issues related to the design, analysis and interpretation of imaging genomics studies and provide suggestions as to how investigators can optimize this approach to understanding the etiology and pathophysiology of brain disorders.
Discovery Based Versus Candidate Gene Approaches
Two general classes of imaging genetics/genomics investigations have emerged: 1) discovery-based studies, that seek to identify genetic associations with brain imaging phenotypes and 2) candidate gene approaches that test specific a priori hypotheses regarding the role of genes in brain biology and disease mechanisms. Discovery-based approaches take advantage of the growing availability of low cost, large-scale genotyping enabling genome-wide analyses. They may identify new genes not previously associated with the phenotype studied or with aspects of brain biology. However, precisely because of their hypothesis-free nature, unbiased correction for multiple testing is required and such studies require very large sample sizes.
Discovery based approaches can be interfaced with imaging in one of two ways. First, a variant discovered in a GWAS or other undirected “forward” genetic study that survives multiple comparison correction and is replicated has strong evidence of being associated with the target phenotype of that study. In that case, prior hypotheses can be formulated with regard to the neural impact of that variant with a higher degree of specificity and confidence, in particular if imaging paradigms are used and neural systems are imaged that have evidence for being intermediate phenotypes (“endophenotypes”) of that disorder (see below). Secondly, imaging data can themselves be the target phenotypes for discovery-based approaches. The ENIGMA consortium has shown that this approach can identify common genetic variants associated with a structural imaging phenotype such as hippocampal volume (5). It is also often suggested that commonly used imaging procedures can provide strict control for false positives in imaging genetics (6).
Since recent developments in GWA studies provide an increasing number of common variants with genome-wide evidence for clinical diagnosis based association, the application of discovery-based strategies in imaging genetics is likely to increase. To date, however, the majority of imaging genomic studies have taken a candidate gene approach, where one or more SNPS for which there is some prior evidence of association with disease are tested against an imaging phenotype/s. Many studies are based on SNPs from candidate gene association studies from early, pre-GWAS era studies. In most instances, the clinical genetics studies that originally identified these candidate gene genetic studies have well detailed problems, including being confounded by sample sizes inadequate for the expected effect sizes, publication bias, and population stratification leading to a lack of consistent reproducibility. Thus, many of these genes are no longer considered among the likely susceptibility genes for the clinical disorders. This does not mean that they will not show association to brain structure or function, but it suggests that the associations in brain, if valid, are not likely related to the psychiatric illness that led to their being of interest in the first place. Imaging studies choosing to focus on candidate genes without strong prior genetic findings would be expected to have higher type I error rates. Additional prior biological evidence may motivate such a particular candidate gene study, such as a known function for the gene in key aspects of brain development or functions that are implicated in the disease. These studies may directly inform hypotheses regarding the cellular and molecular mechanisms underlying the disease in question.
Compelling prior probabilities needed to motivate a candidate gene study could include previous replicated GWAS or CNV findings and/or a biological hypothesis linked to specific mechanisms involved in the structure and function of circuits, specific anatomical regions or cells in the brain.
Studies using polygene risk scores, which can be derived from GWAS or from (for example, meta-analytic) lists of genes associated with a phenotype or from a known molecular pathway offer a simple, omnibus genome-based regressor for analyzing variance in any brain phenotype accounted for by additive effects of common genetic variants. They may have increased sensitivity to establish associations at the genomic level. If derived from GWAS, such scores can inform what - for example - the “(common variant) schizophrenia genome” can do to the brain in a discovery study, but not necessarily establish detailed links across genes, neural circuits and systems affected in brain disorders. To glean insights into neurobiology, the polygenes used to create the score may be further dissected into pathways that have strong neurobiological priors or are informed by expression or other data linking variation to the cellular substrates or circuitry implicated in disease (7).
While most imaging genetics study have examined the association of individual SNPs with a quantitative imaging phenotype, approaches based on multiple variant analyses are increasingly popular, as most complex traits represent genomic variation, not individual gene variation. Yang et al (2011 AJHG)(8) introduced a method called GTCA (genome-wide complex trait analysis) for estimating the variance in a complex trait by multiple SNPs, for example all SNPs on a given chromosome or all SNPs in the genome. This is an algorithm for predicting the narrow range heritability of a complex trait by a large number of common variants genotypes en mass. The accuracy of the statistics is dependent on sample size, and most imaging genetics studies to date have not achieved sufficient power to use this approach with confidence.
Optimizing Imaging Phenotypes to Enhance Replicability
Heritability and Prior Probability of a Neurobiological Mechanism
The underlying principle of any genetic association study is the association of genotype with phenotype. A phenotype is an observable trait that is the manifest expression of genetic and environmental background (and their interaction), and not just anything that can be measured. An ‘intermediate phenotype’ or endophenotype is additionally required to be a trait feature related to a disease process, present to a degree in unaffected relatives, and heritable (9, 10). Establishing the heritability of the measures to be used in imaging genomics is critical for inferring a relationship between genetic association with an imaging phenotype and genetic association with disease pathophysiology. Heritability in studies of normal individual differences can also strengthen the inference that can be made from these studies. However, there is no clear requirement for the level of heritability, and high heritability does not necessarily confer large effect sizes in association studies. For example, IQ is highly heritable, yet finding genes that affect common variation in intellectual abilities across heterogeneous populations is challenging, likely due to its highly polygenic nature. Until recently, successes were largely limited to de novo or inherited mutations with major effects on cognitive function (11). Nevertheless, large-scale investigations suggest that rare CNVs implicated in developmental neuropsychiatric disorders also affect cognition and neuroanatomy even in control carriers, who do not have psychiatric disease or significant intellectual disability. Recently, (12) showed that the presence of schizophrenia-associated CNVs (e.g. chromosome 15q11.2) is associated with reduced cognitive ability and a gene-dosage dependent effect on brain structure in healthy Icelandic individuals.
To establish that a phenotype is related to genetic risk for illness, it is necessary to study individuals who are at increased genetic risk but do not have a clinical diagnosis, such as family members, ideally discordant monozygotic (MZ) twins or siblings. Studying family members across generations requires proper modeling of nonlinear effects of age on brain phenotypes or potentially directly modeling these age-related changes using appropriate genetic models (e.g. (13, 14)). Very few of the phenotypes in the current imaging genetics literature meet all such requirements (15), although evidence for heritability (or at least familiality) has been provided for a few structural MRI phenotypes (14, 16), as well as several functional imaging phenotypes including prefrontal activation(17) hippocampal-prefrontal connectivity (18) during working memory, striatal hypoactivation during reward expectancy(19), cingulate engagement during cognitive control tasks (20) and hippocampal activation during episodic memory (21).
A second important requirement for a robust illness related imaging phenotype is a measure that is sensitive to biological processes hypothesized to be affected by a disease and neuroimaging can provide such measures at multiple levels of neural organization from molecular (PET, MRS) to systems (structural and functional MRI).
Unique Issues Related to the Use of Imaging Data as Quantitative Phenotypes
In contrast to clinical phenotypes characterized categorically or by a few values, neuroimaging provides a magnitude of data on a scale comparable to genomics itself. Current methods of data analysis are often aimed at reducing this dimensionality by focusing on linear models of variation in regions of interest (ROI). However, there is no assurance that reducing dimensionality of the neuroimaging data “carves the joints” in a manner that is most productive for integration with genomic data, or representative of underlying biology. As even larger samples are essential for discovery-based integration of neuroimaging and genomics, across heterogeneous samples, multi-site collaborations have increasingly emerged. Such efforts had first to demonstrate that measures of brain structure and function are reproducible. Advanced high-throughput image processing and automated segmentation methods were developed and demonstrated high intra- and inter scanner reliability of measuring brain volumes in multiple structures (22, 23). Several collaborations have integrated structural data from multiple sites and accepted methods for “harmonizing” scanners have been developed. The Alzheimer's Disease Neuroimaging Initiative (24), has developed a standardized approach to structural MRI and the functional Brain Imaging Research Network (25) has developed methods for functional MRI studies (26). The ENIGMA Consortium has paved the way for participation of multiple institutions and a systematic meta-analytic approach (27).
There are additional issues that are unique to imaging phenotypes. Even within structural imaging, while progress has been made in standardizing volumetric measurements, there is still considerable uncertainty about the nature of variation in structural measures with MRI. MRI is a biochemical assay, not an assay of physical structure, per se. Indeed, many environmental factors have been found to influence structural MRI measures, including subject motion, exercise history, weight, hydration, medications, alcohol consumption, smoking, and other substance use, making interpretation of structural differences between cases and controls and even with genetic association problematic. DTI based connectivity measures are promising but understanding the origins of variation in white matter microstructure, as assessed with DTI, in clinical populations is challenging (28). Functional neuroimaging enables the measurement of brain responses during cognitive demands that can be quantified in terms of both activation and functional or effective connectivity. In contrast to structural and DTI measures which are derived single values, functional imaging involves multiple measures on the same individual over time. Comparison across sites and studies of functional imaging data may be limited by variation in the tasks used and in the approaches to analysis, as well as reliability of the measure. Drawing upon efforts such as CNTRICS (29, 30) in which a set of specific cognitive imaging paradigms were recommended based upon their construct validity can help bring consistency to the field. Following similar recommendations developed by the NIMH RDoC initiative (31, 32) these approaches have the potential to bring more coherence to the field and facilitate data pooling and meta-analyses. In both Europe and the US, standardized functional imaging paradigms that fulfill at least some of these requirements have been developed and their reliability quantified (33, 34). It must be noted, however, that new paradigms will be introduced as cognitive and affective neuroscience continues to progress, and it will be important to ensure that the somewhat arduous work involved in optimizing these paradigms and establishing their measurement reliability is undertaken.
Specific Issues Related to the Design and Analysis of fMRI-based Phenotypes
Typically, fMRI/genomic studies use behavioral paradigms that have construct validity as sensitive measures of cognitive or emotional functions that are impaired by the illness. It is important that subjects’ behavior be monitored during scanning. A question that was raised early in the design and interpretation of clinical functional neuroimaging studies is how to address the likelihood that a clinical group will perform worse than controls? On the one hand, this is to be expected if paradigms are valid and sensitive measures targeting illness-related deficits. On the other hand, impaired performance may confound the interpretation of results as patients may not be engaged in the task to the same degree as controls (35). This is especially a problem in blocked design studies, where data are acquired over multiple trials and lapses off task may result in a contrast of brain activity associated with different brain states. One solution is to match patients and controls on performance. This ensures that both groups are comparably engaged in the task. A disadvantage of this approach is that it may require administering different versions of a task in order to match performance, or the selection of an atypical patient cohort or a poorly functioning control group.
Another concern about this approach is that the absence of behavioral differences does not allow as robust a link to be established at the individual subject level between genes, brain activity and behavior. An alternative approach, widely adopted since the advent of event-related fMRI, is to constrain the fMRI analysis to correct trials, ensuring that all subjects are on task during the acquisition. This approach also permits individual task performance to be used as an additional behavioral correlate of illness status and genotype, further strengthening the link between dysfunctional neural circuitry and cognitive impairment at the individual subject level.
Multiple comparison statistical correction is standard for structural and functional imaging studies with a variety of valid methods in general use including family-wise, cluster level and false discovery rate based approaches (36, 37). An additional challenge for imaging genetics is that the measures obtained are often not independent of each other. From a statistical standpoint, this brings up the issue of how to determine the appropriate statistical threshold. When applied to neuroimaging data, Bonferroni correction may inappropriately eliminate both false and true positive results. Thus, other approaches such as false discovery rate (FDR) or permutation testing have been advocated in order to better control the expected proportion of falsely rejected hypotheses. There is variation, however, in the manner in which these methods are applied, and studies sometimes perform multiple analyses that can inflate type I error. One example that can be readily addressed is the use of multiple small-volume corrected (SVC) regions of interest, where correction is at the region level rather than across all voxels analyzed. If an a priori hypothesis is about a network (i.e. multiple ROI's) Type I error can be appropriately controlled by correcting for all voxels in the network (combined ROI's). The situation becomes more complex when a study involves multiple contrasts, and correlations between clinical or performance measures and fMRI data, and the field struggles with when and how to correct for these multiple analyses. It is critical that authors clearly articulate the number of comparisons that were conducted, and clearly describe the methods and rationale for their approach to controlling type I error. A more daunting issue arises when one considers the likelihood that data may be analyzed using different models and thresholds in a manner that may increase type I error. The development of online data repositories where data are available for re-analysis is one solution to this.
Clinical Factors That May Influence Results of Imaging Genomic Studies
Many clinical factors can complicate the interpretation of imaging data, with direct implications for imaging genomic and genetic studies. These include, including the well-documented effects of antipsychotics and lithium on measures of brain structure as well as the cardiovascular and substance related factors listed in the preceding section on MRI based phenotypes. For large population-based studies these factors are difficult to control. Focusing on young, recent onset patients and the use of rigorous exclusions can address some of these concerns but this is a practical limitation for sample sizes. While including chlorpromazine equivalents as a covariate is often done to address medication effects, it is difficult to capture the duration and extent of treatment accurately, and it is unlikely that this will correct for the biological effects of long-term treatment. One solution is to study healthy siblings of patients who share increased risk but not clinical confounders. Another is the study of individuals in the prodromal phase of illness, before psychoactive medications are applied.
A number of studies have reported an effect of a particular variant on an imaging phenotype in a patient group, but an absence of an effect in controls. A possible explanation for association in patients not present in controls is because of illness-relevant contexts that interact with genotype, or the effect in patients may be linked to clinical epiphenoma, such as medication effects that interact with genotype. For example, if a gene influences drug metabolism or the impact of a drug on brain structural or functional measurements, this effect will appear in patients, but not in the controls, yet be unrelated to the neural substrates of risk. Imaging associations observed only in patients should be viewed with caution.
Optimizing Candidate Gene Based Analyses to Enhance Replicability
Addressing key analytic issues will enhance the replicability of imaging genomic studies. Population stratification can be partially addressed through appropriate statistical modeling, such as multivariate indices of lineage informative markers, in large samples. In small samples this can be addressed by focusing on homogeneous subgroups, though the generalizability of these results will be limited.
Correction for multiple comparisons is essential to protect against type I error. Because imaging phenotypes are often complex and multimodal, e.g. behavior and brain activation across various contrasts, and because the same cohort is often used across multiple genetic studies, it is critical to specify how many comparisons were actually conducted and how they were controlled for. An additional challenge for imaging genetics is that the measures obtained are often not independent of each other. From a statistical standpoint, this brings up the issue of how to determine the appropriate statistical threshold. When applied to neuroimaging data, Bonferroni correction may inappropriately eliminate both false and true positive results Thus, other approaches such as false discovery rate (FDR) or permutation testing have been advocated in order to better control the expected proportion of falsely rejected hypotheses.
Data reduction can increase power to detect associations; a candidate gene approach could potentially serve this purpose when it is well motivated. In the absence of robust priors this approach is prone to increased Type I error due to lower statistical thresholds typically used in candidate gene studies compared to GWAS. The prior probability of a particular hypothesis or research question is reflected by the answer to the question “What is the probability of a meaningful association between a genetic variant and a disease” and can include previous GWAS association or a compelling neurobiological mechanism? (38).
Employing sample sizes that are adequate to detect the expected effect size is essential for any genetic study. For imaging genetic association studies a frequent assumption is that brain-based phenotypes will be more sensitive to detecting association than is the case for clinical disorders. The fact that some small sample studies have been published with replications (39) provides some limited evidence in support of this premise. However recent critiques of neuroscience research in general and imaging genetics specifically have argued that the small sample size studies are inherently prone to both type I and type II error, and lead to inflated estimates in effect sizes [18, 19]. These studies have suggested that up to 50% of published associations may be false positives (40, 41). Whether the mean effect size of loci that contribute to variation in intermediate phenotypes is larger than the mean effect size of loci contributing to psychiatric disease, an assumption that has often been made in imaging genomics, has been an issue of recent controversy. However based upon the existing literature it is unlikely that effect sizes for imaging phenotypes are dramatically larger than those for psychiatric disease (42) (43), and are also highly likely to be polygenic. Based on hundreds of studies in the National Human Genome Research Institute (NHGRI) Catalog of published GWAS )(44), for quantitative traits the amount of variance explained by any one locus is well under 0.5% (45). An appropriately conservative approach to this problem, that addresses the trade-offs between an unknown effect size (limiting the ability to estimate the power of a given study) and the practicalities of scanning very large samples is to require independent replication for small sample studies. Replication is defined as studying the same phenotype and the same genetic variant, which has an effect in the same direction (46).
The widespread requirement for the US genetics community to share all data through repositories such as dbGAP has been critical to the success currently being enjoyed in psychiatric genetics using the GWA approach to syndromal disorders. Given the resource intensity of imaging genomic studies, creating structures that increase the feasibility of reanalysis and independent replication is essential. Enhanced methods for data sharing and public access to imaging genomic data sets would be particularly helpful in this regard. Recent examples for this include the ENIGMA consortium of brain structure (27).
Specific issues related to imaging genetics studies of common versus rare variants
The literature to date has predominantly focused on the investigation of common allelic variation in relation to variation in brain imaging parameters. This is because high-throughput methods were developed first for common variants and current imaging sample sizes are generally too small to focus on rare variants. However, it has long been appreciated that rare variants also play a role in the etiology of neurodevelopmental disorders, particularly intellectual disability, autism and schizophrenia (47). In addition, data are rapidly accumulating that rare variants have a large cumulative effect on normal variation in brain function (.e.g cognition) (48-50). Given this context, there is a growing recognition of the value of neuroimaging investigations of large numbers of individuals with the same CNV that confers elevated risk for neuropsychiatric disorders and/or cognitive impairment, in order to better understand underlying mechanisms (51).
Summary, Recommendations and Conclusions
Imaging Genomics leverages recent advances in both genetics and the non-invasive measurement of brain structure, function and chemistry. The approach has the potential to provide valuable insights into the molecular and cellular etiology and pathophysiology of brain disorders by linking genetic variation with changes at cellular, systems and behavioral levels of measurement. Many issues related to the use of imaging phenotypes are identical to those for other quantitative traits, including heritability, heterogeneity, population stratification effects, the statistical challenges of controlling type I error in the face of multiple comparisons and publication biases. Operational solutions to those challenges have been adopted in the broader field of genetics and many of these should be consistently applied to imaging genetics. For example:
A well-designed study will use a well-validated phenotype, measured using tools that have construct and neural validity, sensitivity(52) and reliability (33).
Examination of genetic association should be based on prior evidence of involvement in the illness or the quantitative phenotype and, if the genetic evidence pertains to a disorder, linked to a pathophysiological mechanism related to the disease.
The study should apply established methods for addressing population stratification and other forms of heterogeneity.
Imaging analysis methods should be transparent, with an explicit statement regarding the number of comparisons conducted, and appropriate correction for multiple comparisons.
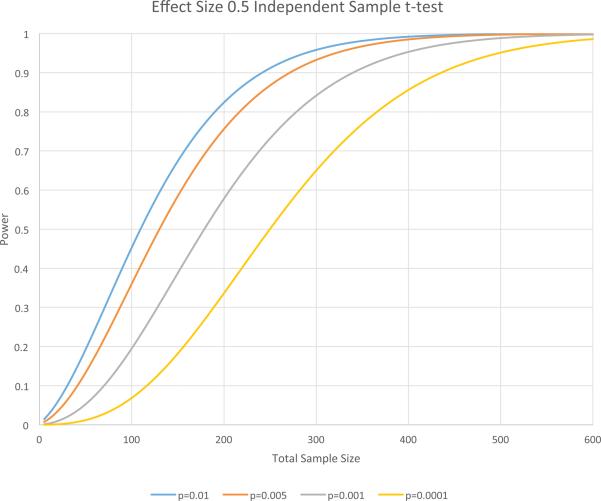
The study should be well-powered, with an empirical justification for the selected sample size. All small sample studies (n's in the 50-100 range which will have reasonable power to detect effect sizes of 0.5 or greater, see Figure 1) should be accompanied by independent replication where replication is strictly defined as the same phenotype, same variant, and same direction of effect.
Mechanisms for broader data sharing need to be developed and implemented.
Figure 1.
Power calculations for an independent sample t-test for an effect size of 0.5 and different height thresholds typically used in imaging genetic studies. The simulations assume equal sample sizes for each of the groups. For typically used height thresholds and this effect size having 50-100 subjects in each group will generally have only moderate power to reject the null hypothesis.
There are a number of unique issues related to the use of imaging measures as quantitative traits in genetic studies.
Well-justified selection of imaging measures in specific brain disorders, as well as motivated selection of candidate genes will enhance the sensitivity and replicability of these studies.
Careful implementation of best practices in the design and analysis of imaging studies, including appropriate correction for the number of comparisons and the number of analyses, will have a similar positive impact.
There is a consensus that independent replication and confirmation of findings in independent samples are important going forward; there is a need for the development of methodology and data sharing tools to enhance the feasibility of this. Avenues for the publication of negative studies should be supported. Increased attention to the publication of replication studies will also have a positive impact.
This review was intended to identify and offer potential solutions to a number of challenging issues related to the design, analysis and interpretation of imaging genomic studies. We undertook this effort in the context of a larger concern about replication in the biological sciences along with the recognition that imaging genomics is an approach that will continue to develop as a major focus of psychiatric research. We believe that attention to the issues raised and solutions offered in this paper will enhance the replicability and interpretability of future imaging genomic studies and stimulate needed method development to optimize the contribution of this approach to understanding the neurobiology of psychiatric disease.
Glossary
- GWAS
Genome Wide Association Study
- BOLD
Blood Oxygenation Level Dependent
- ENIGMA
Enhancing NeuroImaging Genetics through Meta-Analysis
- SNP
single nucleotide polymorphism
- PET
Positron Emission Tomography
- MRI
Magnetic Resonance Imaging
- DTI
Diffusion Tensor Imaging
- CNTRICS
Cognitive Neuroscience Approaches to Treatment Development of Cognition in Schizophrenia
- RDoC
Research Domain Criteria
- CNV
Copy Number Variant
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial disclosures: Drs. Carter, Bearden, and Glahn report no biomedical financial interests or potential conflicts of interest. Dr Bullmore is a part time employee of Glaxo-Smith-Kline and owns stock in that company. Dr Gur is on the advisory board of Otsuka Phamaceuticals. Dr Geschwind is on the advisory board of Synapdx. Dr Meyer-Lindenberg Consultancy: Astra Zeneca, Bristol Myers Squibb, Defined Health, Desitin Arzneimittel, Elsevier, F. Hoffmann-La Roche, Gerson Lehrman Group, Lundbeck, Outcome Europe Sárl, Outcome Sciences, Pricespective, Roche Pharma, Servier International, Thieme Verlag, Lectures including travel fees: Abbott, Alexza Pharmaceuticals, Astra Zeneca, Aula Médica Congresos, BASF, Groupo Ferrer International, Janssen-Cilag, Lilly Deutschland, LVR Klinikum Düsseldorf, Pfizer Pharma, Servier Deutschland, Dr. Weinberger serves on the Scientific Advisory Council of Sunovion.
Contributor Information
Cameron S. Carter, University of California at Davis, 4701 X Street Sacramento, CA 95816, phone 916 7348883 fax 916 7347884, cameron.carter@ucdmc.ucdavis.edu
Carrie E. Bearden, University of California at Los Angeles
Edward T. Bullmore, Cambridge University
Daniel H. Geschwind, University of California at Los Angeles
David C. Glahn, Yale University
Raquel E. Gur, University of Pennsylvania
Andreas Meyer-Lindenberg, University of Heidelberg Central Institute of Mental Health.
Danile R. Weinberger, Lieber Institute for Brain Development
References
- 1.Hariri AR, Weinberger DR. Imaging genomics. Br Med Bull. 2003;65:259–270. doi: 10.1093/bmb/65.1.259. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Z, Zhu G, Hariri AR, Enoch MA, Scott D, Sinha R, et al. Genetic variation in human NPY expression affects stress response and emotion. Nature. 2008;452:997–1001. doi: 10.1038/nature06858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Button KS, Ioannidis JP, Mokrysz C, Nosek BA, Flint J, Robinson ES, et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat Rev Neurosci. 2013;14:365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- 4.Gurung R, Prata DP. What is the impact of genome-wide supported risk variants for schizophrenia and bipolar disorder on brain structure and function? A systematic review. Psychol Med. 2015;45:2461–2480. doi: 10.1017/S0033291715000537. [DOI] [PubMed] [Google Scholar]
- 5.Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer-Lindenberg A, Nicodemus KK, Egan MF, Callicott JH, Mattay V, Weinberger DR. False positives in imaging genetics. Neuroimage. 2008;40:655–661. doi: 10.1016/j.neuroimage.2007.11.058. [DOI] [PubMed] [Google Scholar]
- 7.Lencz T, Knowles E, Davies G, Guha S, Liewald DC, Starr JM, et al. Molecular genetic evidence for overlap between general cognitive ability and risk for schizophrenia: a report from the Cognitive Genomics consorTium (COGENT). Molecular psychiatry. 2014;19:168–174. doi: 10.1038/mp.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesman, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 10.Bearden CE, Freimer NB. Endophenotypes for psychiatric disorders: ready for primetime? Trends Genet. 2006;22:306–313. doi: 10.1016/j.tig.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 11.de Geus EJ, Wright MJ, Martin NG, Boomsma DI. Genetics of brain function and cognition. Behav Genet. 2001;31:489–495. doi: 10.1023/a:1013360909048. [DOI] [PubMed] [Google Scholar]
- 12.Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505:361–366. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- 13.Glahn DC, Kent JW, Jr., Sprooten E, Diego VP, Winkler AM, Curran JE, et al. Genetic basis of neurocognitive decline and reduced white-matter integrity in normal human brain aging. Proc Natl Acad Sci U S A. 2013;110:19006–19011. doi: 10.1073/pnas.1313735110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fears SC, Service SK, Kremeyer B, Araya C, Araya X, Bejarano J, et al. Multisystem Component Phenotypes of Bipolar Disorder for Genetic Investigations of Extended Pedigrees. JAMA Psychiatry. 2014 doi: 10.1001/jamapsychiatry.2013.4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasetti R, Weinberger DR. Intermediate phenotypes in psychiatric disorders. Curr Opin Genet Dev. 2011;21:340–348. doi: 10.1016/j.gde.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roalf DR, Vandekar SN, Almasy L, Ruparel K, Satterthwaite TD, Elliott MA, et al. Heritability of Subcortical and Limbic Brain Volume and Shape in Multiplex-Multigenerational Families with Schizophrenia. Biol Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callicott JH, Egan MF, Mattay VS, Bertolino A, Bone AD, Verchinksi B, et al. Abnormal fMRI response of the dorsolateral prefrontal cortex in cognitively intact siblings of patients with schizophrenia. Am J Psychiatry. 2003;160:709–719. doi: 10.1176/appi.ajp.160.4.709. [DOI] [PubMed] [Google Scholar]
- 18.Rasetti R, Sambataro F, Chen Q, Callicott JH, Mattay VS, Weinberger DR. Altered cortical network dynamics: a potential intermediate phenotype for schizophrenia and association with ZNF804A. Arch Gen Psychiatry. 2011;68:1207–1217. doi: 10.1001/archgenpsychiatry.2011.103. [DOI] [PubMed] [Google Scholar]
- 19.Grimm O, Heinz A, Walter H, Kirsch P, Erk S, Haddad L, et al. Striatal Response to Reward Anticipation: Evidence for a Systems-Level Intermediate Phenotype for Schizophrenia. JAMA Psychiatry. 2014 doi: 10.1001/jamapsychiatry.2014.9. [DOI] [PubMed] [Google Scholar]
- 20.Sambataro F, Mattay VS, Thurin K, Safrin M, Rasetti R, Blasi G, et al. Altered cerebral response during cognitive control: a potential indicator of genetic liability for schizophrenia. Neuropsychopharmacology. 2013;38:846–853. doi: 10.1038/npp.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erk S, Meyer-Lindenberg A, Schmierer P, Mohnke S, Grimm O, Garbusow M, et al. Hippocampal and Frontolimbic Function as Intermediate Phenotype for Psychosis: Evidence from Healthy Relatives and a Common Risk Variant in CACNA1C. Biol Psychiatry. 2013 doi: 10.1016/j.biopsych.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 22.Holmes AJ, Lee PH, Hollinshead MO, Bakst L, Roffman JL, Smoller JW, et al. Individual differences in amygdala-medial prefrontal anatomy link negative affect, impaired social functioning, and polygenic depression risk. J Neurosci. 2012;32:18087–18100. doi: 10.1523/JNEUROSCI.2531-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nugent AC, Luckenbaugh DA, Wood SE, Bogers W, Zarate CA, Jr., Drevets WC. Automated subcortical segmentation using FIRST: test-retest reliability, interscanner reliability, and comparison to manual segmentation. Hum Brain Mapp. 2013;34:2313–2329. doi: 10.1002/hbm.22068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer's Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2012;8:S1–68. doi: 10.1016/j.jalz.2011.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potkin SG, Ford JM. Widespread cortical dysfunction in schizophrenia: the FBIRN imaging consortium. Schizophr Bull. 2009;35:15–18. doi: 10.1093/schbul/sbn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glover GH, Mueller BA, Turner JA, van Erp TG, Liu TT, Greve DN, et al. Function biomedical informatics research network recommendations for prospective multicenter functional MRI studies. J Magn Reson Imaging. 2012;36:39–54. doi: 10.1002/jmri.23572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, et al. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014 doi: 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erus G, Battapady H, Satterthwaite TD, Hakonarson H, Gur RE, Davatzikos C, et al. Imaging Patterns of Brain Development and their Relationship to Cognition. Cereb Cortex. 2014 doi: 10.1093/cercor/bht425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter CS, Barch DM, Bullmore E, Breiling J, Buchanan RW, Butler P, et al. Cognitive Neuroscience Treatment Research to Improve Cognition in Schizophrenia II: developing imaging biomarkers to enhance treatment development for schizophrenia and related disorders. Biol Psychiatry. 2011;70:7–12. doi: 10.1016/j.biopsych.2011.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter CS, Barch DM. Imaging biomarkers for treatment development for impaired cognition: report of the sixth CNTRICS meeting: Biomarkers recommended for further development. Schizophr Bull. 2012;38:26–33. doi: 10.1093/schbul/sbr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuthbert BN, Insel TR. Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med. 2013;11:126. doi: 10.1186/1741-7015-11-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. American Journal of Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 33.Plichta MM, Schwarz AJ, Grimm O, Morgen K, Mier D, Haddad L, et al. Test-retest reliability of evoked BOLD signals from a cognitive-emotive fMRI test battery. Neuroimage. 2012;60:1746–1758. doi: 10.1016/j.neuroimage.2012.01.129. [DOI] [PubMed] [Google Scholar]
- 34.Forsyth JK, McEwen SC, Gee DG, Bearden CE, Addington J, Goodyear B, et al. Reliability of functional magnetic resonance imaging activation during working memory in a multi-site study: Analysis from the North American Prodrome Longitudinal Study. Neuroimage. 2014;97:41–52. doi: 10.1016/j.neuroimage.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter CS, Heckers S, Nichols T, Pine DS, Strother S. Optimizing the design and analysis of clinical functional magnetic resonance imaging research studies. Biol Psychiatry. 2008;64:842–849. doi: 10.1016/j.biopsych.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Poldrack RA, Fletcher PC, Henson RN, Worsley KJ, Brett M, Nichols TE. Guidelines for reporting an fMRI study. Neuroimage. 2008;40:409–414. doi: 10.1016/j.neuroimage.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carter CS, Barch DM, Buchanan RW, Bullmore E, Krystal JH, Cohen J, et al. Identifying cognitive mechanisms targeted for treatment development in schizophrenia: an overview of the first meeting of the Cognitive Neuroscience Treatment Research to Improve Cognition in Schizophrenia Initiative. Biol Psychiatry. 2008;64:4–10. doi: 10.1016/j.biopsych.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wacholder S, Hartge P, Prentice R, Garcia-Closas M, Feigelson HS, Diver WR, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362:986–993. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callicott JH, Feighery EL, Mattay VS, White MG, Chen Q, Baranger DA, et al. DISC1 and SLC12A2 interaction affects human hippocampal function and connectivity. J Clin Invest. 2013;123:2961–2964. doi: 10.1172/JCI67510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munafo MR, Freimer NB, Ng W, Ophoff R, Veijola J, Miettunen J, et al. 5-HTTLPR genotype and anxiety-related personality traits: a meta-analysis and new data. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:271–281. doi: 10.1002/ajmg.b.30808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munafo MR, Bowes L, Clark TG, Flint J. Lack of association of the COMT (Val158/108 Met) gene and schizophrenia: a meta-analysis of case-control studies. Molecular psychiatry. 2005;10:765–770. doi: 10.1038/sj.mp.4001664. [DOI] [PubMed] [Google Scholar]
- 42.Munafo MR, Flint J. The genetic architecture of psychophysiological phenotypes. Psychophysiology. 2014;51:1331–1332. doi: 10.1111/psyp.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivieres S, Jahanshad N, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flint J, Timpson N, Munafo M. Assessing the utility of intermediate phenotypes for genetic mapping of psychiatric disease. Trends Neurosci. 2014;37:733–741. doi: 10.1016/j.tins.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan PF. Spurious genetic associations. Biol Psychiatry. 2007;61:1121–1126. doi: 10.1016/j.biopsych.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 47.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 49.De La Vega FM, Bustamante CD, Leal SM. Genome-Wide Association Mapping and Rare Alleles: From Population Genomics to Personalized Medicine. Pac Symp Biocomput. 2011:74–75. doi: 10.1142/9789814335058_0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 51.Simons Vip C. Simons Variation in Individuals Project (Simons VIP): a genetics-first approach to studying autism spectrum and related neurodevelopmental disorders. Neuron. 2012;73:1063–1067. doi: 10.1016/j.neuron.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 52.Carter CS, Barch DM. Cognitive neuroscience-based approaches to measuring and improving treatment effects on cognition in schizophrenia: the CNTRICS initiative. Schizophr Bull. 2007;33:1131–1137. doi: 10.1093/schbul/sbm081. [DOI] [PMC free article] [PubMed] [Google Scholar]