Abstract
The liver and pancreas are the prime digestive and metabolic organs in the body. After emerging from the neighboring domains of the foregut endoderm, they turn on distinct differentiation and morphogenesis programs that are regulated by hierarchies of transcription factors. Members of SOX family of transcription factors are expressed in the liver and pancreas throughout development and act upstream of other organ-specific transcription factors. They play key roles in maintaining stem cells and progenitors. They are also master regulators of cell fate determination and tissue morphogenesis. In this review, we summarize the current understanding of SOX transcription factors in mediating liver and pancreas development. We discuss their contribution to adult organ function, homeostasis and injury responses. We also speculate how the knowledge of SOX transcription factors can be applied to improve therapies for liver diseases and diabetes.
Keywords: Sox17, Sox4, Sox9, differentiation, homeostasis, regeneration
1. Introduction
Development of the liver and pancreas has been researched extensively in the past decade, owing to the drastic increase in the incidence of chronic liver diseases and diabetes and the urgent need for treatment other than organ transplant. Characterization of the molecular mechanisms underlying liver and pancreas development leads to the discovery of networks of transcription factors, including members of the sex-determining region on Y box (SOX) family of transcription factors. There are currently 30 SOX transcription factors in mammals, characterized by the possession of an evolutionarily conserved HMG DNA-binding motif [1]. They are further divided into 9 subgroups. Members of the same subgroup have similar structures and overlapping functions, whereas members from different subgroups have little in common outside of the HMG motif. Depending on the binding partners they recruit, SOX transcription factors can act both as transcription activators and repressors by directly binding to the promoter/enhancer region of their downstream targets. Because of this versatility, the same SOX factor may regulate pluripotency as well as cell fate decision during development. In this review, we focus on the recent insights into the function of SOX transcription factors in specification and maintenance of liver and pancreas progenitors, cell-type differentiation, and tissue morphogenesis. We also review the molecular regulation of SOX factor expression and the interactions between SOX factors and their partners in different steps of liver and pancreas development. In addition, we present the emerging investigation into the contribution of SOX factors to liver and pancreas homeostasis and regeneration postnatally.
2. SOX transcription factors in hepatobiliary development, homeostasis, and injury
The liver is the largest internal organ and possesses crucial function in detoxification, digestion, metabolism, and immunity. The basic architectural unit of the liver is the liver lobule [2]. Within the hexagonal-shaped liver lobule, the central vein is located in the center, and a portal triad composed of a portal vein, hepatic artery, and bile duct occupies each of the six corners. In between the central vein and portal triad are cords of hepatocytes that constitute ~80% of the liver mass. Extending along the hepatocyte cords are the sinusoidal capillaries. Mesenchymal cells, including Kupffer cells and hepatic stellate cells, also reside in the sinusoidal space. The liver is derived from the foregut endoderm. At mouse embryonic day 8.5 (E8.5), the ventral domain of the foregut endoderm adopts the hepatic fate upon receiving inductive signals from the neighboring cardiac mesoderm and septum transversum. The liver progenitor cells called hepatoblasts form a diverticulum at E9. They then change into a pseudostratified morphology and invade the surrounding mesenchyme to give rise to the liver bud at E10. Some hepatoblasts near the portal vein differentiate into cholangiocytes to form bile ducts and the rest hepatoblasts form hepatocytes. We will review the roles of SOX17, SOX9, and SOX4 in liver development. Hepatic expression and function of the other SOX factors remain largely unknown.
2.1 SOX17 in segregation of ventral foregut endodermal organs
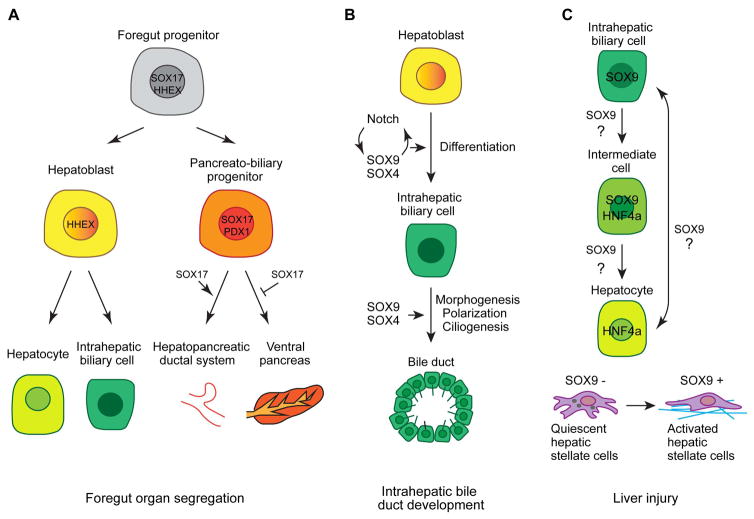
Sox17 of SOXF family is expressed in the endoderm from the onset of gastrulation and serves as an important intrinsic regulator of endoderm formation across vertebrate species [3–6]. Sox17−/− mutant mice show severe deficit in gut endoderm [6]. During later endoderm development, Sox17 mediates segregation of the liver, biliary system, and ventral pancreas (Fig. 1A) [7, 8]. At the beginning of hepatopancreatic specification, Sox17 is co-expressed with hematopoietically expressed homeobox Hhex and pancreas/duodenum homeobox protein 1/Pdx1 in the ventral foregut endoderm [8]. The first segregation occurs at E8.5 when the presumptive liver primordium downregulates Sox17 but continues expressing Hhex. The co-expression domain of Sox17 and Pdx1 in the posterior ventral foregut segregates at E9.5 so that the Sox17+ domain forms the extrahepatobiliary system, while the Pdx1+ domain develops into the ventral pancreas. Both global and ventral foregut-specific knockout of Sox17 causes a complete loss of gallbladder and cystic duct, confirming its role in specification of the extrahepatobiliary system [7, 8]. Depletion of Sox17 in the ventral foregut from E8.5 onwards results in an expansion of Pdx1 expression throughout the ventral foregut. Pdx1+ cells are aberrantly localized in the liver bud and ectopic pancreatic tissue is present in the common bile duct. Conversely, prolonged Sox17 expression in the Pdx1+ domain suppresses pancreas development and results in formation of ectopic ductal tissue in the stomach and duodenum [7]. Ectopic expression of Sox17 in the Pdx1+ domain does not alter Pdx1 expression but reduces expression of pancreatic transcription factor Nkx2.2. This result suggests that SOX17 acts upstream of NKX2.2 to define pancreatic fate. Meanwhile, SOX17 does not directly suppress PDX1 expression during segregation of the biliary and ventral pancreas lineage. A negative feedback loop between SOX17 and hairy enhancer of split-1 (HES1), a transcriptional effector of Notch signaling, has been proposed to regulate segregation of the SOX17/PDX1 lineages [7]. SOX17 promotes high Hes1 expression in the Sox17+Pdx1+ progenitors. HES1 in turn restricts Sox17+ cells to the presumptive biliary domain to facilitate segregation of the Sox17+ biliary lineage and Pdx1+ ventral pancreas lineage. The proposed interactions between SOX17 and HES1/HHEX/PDX1 as well as the repressive effect of SOX17 on NKX2.2 are largely based on how these factors changes expression in Sox17 gain- and loss-of-function embryos. The direct targets and binding partners of SOX17 in these processes have not been identified.
Fig. 1.
Overview of SOX4, SOX9, and SOX17 in hepatobilliary development and injury.
(A) A proposed model of how SOX17 regulates the segregation of foregut endodermal organs. Initially foregut progenitors co-express SOX17 and HHEX. The first segregation occurs when SOX17 becomes downregulated in the hepatic progenitors. Next SOX17 and PDX1 expression segregates so that SOX17+ cells form the extrahepatobiliary primordium and PDX1+ cells generate the ventral pancreas primordium [modified from 7]. (B) A schematic showing that SOX9 and SOX4 cooperate to regulate differentiation of intrahepatic biliary cells and morphogenesis of bile ducts. (C) During liver injury, SOX9+ intrahepatic biliary cells can convert into HNF4a+ hepatocytes and vice versa. SOX9 is also upregulated in activated hepatic stellate cells that secrete extracellular matrix proteins (blue) to cause liver fibrosis.
Although SOX17 is not required for liver specification, it is expressed in part of the liver bud [9, 10]. In mouse, SOX17 cooperates with another SOXF family member SOX18 to mediate neovascularization of the liver [9]. In zebrafish, Sox17 is thought to label a progenitor population that is responsible for the resumption of liver formation in wnt2bb mutant in which the initial liver formation is blocked due to impaired Wnt signaling [10].
In line with its role in extrahepatobiliary specification, dysregulation of SOX17 has been linked to congenital biliary atresia (BA), a severe progressive cholangiopathy of infancy due to defective biliary morphology and function. Sox17 heterozygous mice in C57BL/6 background develop BA-like phenotype as the gallbladder epithelium becomes detached from the luminal wall [11]. Treating cholangiocyte spheroids with a plant toxin biliatresone induces BA-like syndrome in newborn lambs [12]. The expression of Sox17 is significantly decreased in the biliatresone-treated spheroids and knocking down Sox17 in the spheroids mimics the effect of biliatresone treatment [13]. It will be interesting to examine whether SOX17 is associated with BA pathogenesis in patients.
2.2 Sox9 in biliary development
SOX9 of SOXE family is one of the most studied SOX factors as haploinsufficiency of SOX9 in human is associated with Campomelic dysplasia (CD), a disorder characterized by severe skeletal malformation and sex reversals [14, 15]. During mouse liver development, Sox9 is first expressed in the endodermal cells lining the lumen of the liver diverticulum at E10.5 [16]. Its expression is lost as the hepatoblasts invade the septum transversum, but re-emerges in the hepatoblasts near the portal vein at E11.5. Responding to signals from the portal mesenchyme, these Sox9+ hepatoblasts arrange into a single layer of cells surrounding the branches of the portal vein to form a ductal plate at E15.5. The primary ductal structures have an asymmetric composition of Sox9+ biliary cells on the portal side and Sox9− undifferentiated hepatoblasts on the parenchymal side. Such asymmetry resolves as some of the hepatoblasts on the parenchymal side differentiate into biliary cells and join the periportal biliary cells to form a mature bile duct. The ductal plate cells that do not make bile duct differentiate into periportal hepatocytes [16]. Although SOX9 is the earliest marker for the intrahepatic biliary cells, it is dispensable for biliary differentiation. Mice with liver-specific inactivation of SOX9 show a delay in the resolution of the asymmetric primary ductal structures [16], indicating that SOX9 controls the timing of intrahepatic bile duct morphogenesis. In zebrafish, the intrahepatic bile ducts are formed by direct joining of cellular processes among individual biliary cells rather than through ductal plate [17]. Zebrafish has two orthologs of mammalian Sox9 [18], but only sox9b is expressed in the intrahepatic biliary cells [19, 20]. Consistent with the mouse Sox9 mutant data, the intrahepatic biliary cells are specified in zebrafish sox9b homozygous mutant. However, they fail to undergo proper morphogenesis and remain clustered, resulting in a primitive ductal network [19, 20].
In human, mutations in Notch ligand JAGGED1 and receptor NOTCH2 cause Alagille syndrome, characterized by intrahepatic bile duct paucity and cholestasis [21, 22]. Notch signaling interacts with SOX9 in a positive feedback loop to regulate bile duct morphogenesis [16, 19, 23]. Sox9 and Hes1 mutant mice have similar defects in ductal plate remodeling [16, 24]. JAGGED1 produced by the portal vein mesenchyme induces Sox9 expression in periportal biliary cells [23, 24]. SOX9 is likely a direct target of Notch signaling as its promoter contains ten consensus Rbpj binding sites [25]. Conversely, SOX9 positively regulates the expression of Hes1 in biliary cells [24].
Sox9 expression is also detected in the extrahepatic biliary tract in mouse embryos at E13.5 and persists through adulthood [26]. Yet it is difficult to investigate the function of SOX9 in this tissue as Sox9 homozygous mutant mice die at E11.5 [16] and there is no molecular marker to specifically target the extrahepatic biliary tract. Characterization of zebrafish sox9b mutant reveals that sox9b is required for the patterning and differentiation of the hepatopancreatic ductal (HPD) system. The mutant forms a dysmorphic HPD system with a smaller gallbladder and no clear morphological distinction between the cystic duct, extrahepatic duct, and common bile duct [19, 20]. It also contains misdifferentiated cells that express hepatic marker hnf4α or pancreatic marker pax6 [20], indicating that Sox9b is involved in the segregation of foregut-derived organs in zebrafish.
2.3 SOX4 and SOX9 cooperate to control bile duct development
The modest intrahepatic bile duct phenotype seen in Sox9 mutant mice implicate that other SOX family members may compensate for the loss of SOX9 function. Members of the same SOX subfamily often bind to the same DNA sequence and compensate for each other’s function [1]. However, the other two SOXE family members SOX8 and SOX10 are expressed at low levels in the developing liver [27]. Meanwhile, the SOXC family member SOX4 exhibits similar expression pattern in the ductal plate and developing bile duct as SOX9 [27]. Liver-specific inactivation of SOX4 results in delayed bile duct differentiation and morphogenesis, suggesting that both SOX4 and SOX9 control the timing of bile duct development. Strikingly, biliary differentiation and morphogenesis are completely blocked in Sox4/Sox9 double mutants (Fig. 1B). All the parenchymal cells surrounding the portal vein persistently express hepatoblast/hepatocyte marker HNF4. They fail to establish apico-basal polarity and lack primary cilia. Whereas Sox4 and Sox9 individual mutants resume bile duct development after birth, the double mutants exhibit dilated and truncated hilar ducts and develop cholestasis, fibrosis, and ductular reaction at 6 weeks postnatally. During normal biliary development, TGFβ, Notch, and Yap signaling pathways that are originated from the portal vein promote biliary differentiation and control bile duct formation [28]. Transforming growth factor β receptor II (TβRII) is initially expressed in developing hepatoblasts and becomes repressed as hepatoblasts differentiate into cholangiocytes. In Sox4 individual mutant and Sox4/Sox9 double mutant, TβRII is not repressed in biliary cells, indicating that SOX4 and SOX9 control TGFβ signaling during biliary development. SOX4 and SOX9 may repress TβRII expression through transcription factor HNF6, which is a known repressor of TBRII expression [27, 29, 30]. SOX4 and SOX9 also regulate Notch signaling as loss of SOX4 and/or SOX9 reduces HES1 expression in the developing biliary cells. SOX4 and SOX9 may act directly on Notch signaling. Alternatively, since abnormal HES1 expression occurs at a later stage than abnormal TβRII expression, perturbation of Notch signaling in Sox4/Sox9 mutants could also be secondary to impaired TGFβ signaling [27]. Lastly, Sox4/Sox9 double mutants show decreased expression of some Hippo/Yap target genes, yet the nuclear localization of YAP is not altered [27]. This suggests that SOX4 and SOX9 are mediators of Hippo/YAP signaling, but Hippo/YAP signaling is not completely dependent on SOX4 and SOX9 [27].
2.4 SOX9 in liver homeostasis
Sox9 is continuously expressed in the adult liver [16, 26, 31]. Furuyama et al. crossed the Sox9IRES-CreERT2 knockin mice into the R26R reporter mice to lineage trace Sox9-expressing cells. They reported that Sox9+ cells supply hepatocytes and intrahepatic biliary cells during normal liver homeostasis. Sox9+ cells that are located in the canals of Hering and glands of intrahepatic bile ducts express stem cell markers and are proposed to be hepatic progenitor cells (HPCs) [32, 33]. Sox9+ cells in the peribiliary glands of the extrahepatic biliary system have also been shown to differentiate into hepatocytes and cholangiocytes in vitro and after being transplanted into the liver [32]. Other groups conducted lineage tracing using the Sox9CreERT2 transgenic mice in which the Cre recombinase expression is driven by the Sox9 regulatory sequence from a bacterial artificial chromosome (BAC) [31, 34–37]. They did not detect colonization of Sox9+ cell in the liver over time. The contradictory results can be attributed to the different Sox9CreERT2 strains used: the Sox9IRES-CreERT2 knockin mice were made by insertion of IRES-CreERT2 into the 3′UTR of the Sox9 gene, thus may affect the endogenous Sox9 expression [26]. The BAC Sox9CreERT2 transgenic mice, on the other hand, maintain the intact genomic Sox9 locus [36]. High dosage of tamoxifen may cause hepatic toxicity and induce ectopic expression of Sox9 in the hepatocytes [34, 37], raising the question whether the newly formed hepatocytes seen in the knockin mice are from the endogenous Sox9-expressing cells or from the hepatocytes that normally did not express Sox9. Two recent studies used adeno-associated virus (AAV) to drive hepatocyte-specific expression of Cre without the use of tamoxifen [38, 39]. They lineage traced hepatocytes and showed that adult hepatocytes are generated by self-duplication, rather than differentiation from HPCs.
2.5 SOX9 in liver injury and regeneration
The liver has remarkable regenerative capacity. Under most circumstances, liver regeneration is driven by proliferation of preexisting hepatocytes [40, 41]. However, when the proliferative capability of hepatocytes is severely compromised, facultative HPCs/stem cells may be activated to replenish the lost hepatocytes [42]. The presumptive HPCs/stem cells have duct-like appearance and express Sox9. Lineage tracing of Sox9+ cells in mouse liver injury models has yielded mixed results regarding whether they contain HPCs that contribute to regenerating hepatocytes [26, 31, 34, 35, 37]. It is likely due to the differences in lineage-tracing methodology and the type of liver injury. Lineage tracing of hepatocytes using AAV-Cre indicated that preexisting hepatocytes are the main source of new hepatocytes in most classic mouse liver injury models, arguing against the contribution of HPCs, biliary cells or mesenchymal cells to hepatocyte regeneration [38, 39, 43]. One propable explanation is that despite the severity of hepatocellular damage in these models, the surviving hepatocytes are sufficient to regenerate the organ. In Mdm2 mutant mice in which all hepatocytes undergo apoptosis, necrosis and senescence, Sox9+ HPCs contribute to liver regeneration [44]. In zebrafish, when all hepatocytes are killed by the nitroreductase-mediated cell ablation system, Notch-active biliary cells convert into hepatocytes to regenerate the liver [45–47]. The conversion of Notch-active cells to hepatocytes is blocked in sox9b mutants and liver regeneration is perturbed [46]. It is noteworthy that these zebrafish studies were conducted at larval stage when the liver is still developing. It is plausible that some Notch-active cells are hepatoblasts rather than differentiated biliary cells and that they are the main supplier for new hepatocytes. Lineage tracing using biliary and hepatoblast-specific Cre lines will allow one to assess the contribution of either cell type to hepatocyte regeneration. Similar extreme hepatocyte ablation experiment should also be repeated in adult zebrafish liver.
Can hepatocytes also convert into biliary cells in liver injury? Two elegant lineage tracing studies showed that following 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) injury, hepatocytes gave rise to the intermediate cells that express both SOX9 and hepatocyte marker HNF4α (Fig. 1C). Whether they eventually form functional biliary cells is still in debate [48, 49]. Hepatocyte-to-biliary cell conversion has been suggested to contribute to intrahepatic bile duct regeneration in a mouse model of intrahepatic bile duct paucity. Albumin-Cre-mediated liver-specific double knockout of Hnf6 and Rbpjκ blocks the initial development of peripheral intrahepatic bile ducts [50]. With age, the double mutant liver exhibits ductular reactions and subsequently regenerates communicating peripheral intrahepatic bile ducts. The biliary cells in the ductular reactions and the regenerated peripheral intrahepatic bile ducts are derived from parenchymal cells as they do not express Hnf6 or Rbpjκ. They do not express marker for the hilar intrahepatic bile ducts at all stages, suggesting that they may be derived from hepatocytes. It has been shown that ectopic activation of Notch signaling is sufficient to induce expression of Sox9 and other biliary markers in hepatocytes without any injury [49]. Rbpjκ mutant liver contains fewer Sox9-expressing cells upon DDC injury [49] and shows impaired biliary regeneration following partial hepatectomy [51], suggesting that Notch promotes Sox9 expression and hepatocyte-to-biliary conversion. Meanwhile the few Sox9+ intermediate cells present in the Hnf6;Rbpjκ double mutant liver indicate that Sox9 expression can also be triggered by a Notch-independent mechanism [50]. Hypoxia may induce Sox9 expression as the Sox9+ intermediate cells are often located near the focal necrotic regions in Hnf6;Rbpjκ double knockout mice [50]. Chromatin immunoprecipitation demonstrated direct binding of hypoxia-inducible factor 1 alpha (HIF1α) to the Sox9 promoter [52]. As described in 2.3, Sox4/Sox9 double knockout mice exhibit intrahepatic bile duct paucity [27]. It will be interesting to follow these double mutants to determine whether SOX4 and SOX9 are required for intrahepatic bile duct regeneration.
SOX9 expression has also been observed in activated hepatic stellate cells that generate scarring tissues to cause liver fibrosis (Fig. 1C) [53, 54]. Whereas SOX9 is not detected in quiescent hepatic stellate cells, its expression is drastically induced in culture-activated hepatic stellate cells [53]. Inhibition of SOX9 in culture-activated hepatic stellate cells by siRNA significantly reduces the expression of Collagen I. In vivo, SOX9 is colocalized with the activated hepatic stellate cell marker α-smooth muscle actin (aSMA) in the fibrotic area following chronic CCl4 treatment or MCDE diet [54]. Validating the regulation of collagen deposition by SOX9 in vivo will be necessary to determine whether SOX9 can be used as a molecular target for fibrosis therapy. Notably, the Sox9-expressing reactive ductules also exhibit some mesenchymal features [55]. Thus it is important to determine the origin of these Sox9+;aSMA+ cells to confirm the involvement of SOX9 in stellate cell activation.
3. SOX transcription factors in pancreas development, homeostasis, and β-cell regeneration
The pancreas is composed of two morphologically and functionally distinct tissues [56]. The exocrine compartment contains acinar cells that secrete digestive enzymes and pancreatic ducts that transport digestive fluid. The endocrine pancreas is arranged as Islets of Langerhans that are populated by five hormone-producing cell types, including glucagon-secreting α-cells, insulin-secreting β-cells, somatostatin-secreting δ-cells, ghrelin-secreting ε-cells, and pancreatic polypepetide-generating Y (PP) cells. From specification of pancreatic domain in the foregut endoderm to formation of functional exocrine and endocrine cells, pancreas development is a precisely orchestrated multi-stage process that is governed by hierarchies of transcription factors [56–60]. Expression of many Sox transcription factors has been detected in murine fetal pancreas and adult islets (Table 1) [61, 62]. In this review, we will focus on the two most well characterized SOX factors SOX9 and SOX4.
Table 1.
Expression and function of SOX transcription factors in fetal and adult pancreas.
| Subgroup | Name | Stage expressed | Cell type expressed | Function | Reference |
|---|---|---|---|---|---|
| SOXB1 | Sox2 | Fetal (m*) Adult (h) |
Stem cell-like progenitors in the adult human pancreas | May regulate cell proliferation and stemness; dysregulated in pancreatic cancer. | [100, 101] |
| SOXB2 | Sox21 | Fetal (m) | ND | ND | [62] |
| SOXC | Sox4 | Fetal (m, h) Adult (m, h) |
Pancreas epithelium during embryonic development and insulin-producing β-cells and a subset of acinar cells in adult | Regulates endocrine differentiation and islet organization during embryogenesis; insulin secretion and glucose tolerance in adult. | [27, 62, 91–93] |
| Sox11 | Fetal (m, h) | Mesenchyme surrounding pancreatic buds at E9.5 to E10.5, and in endocrine cells during later development. | Sox11-deficient embryos exhibit pancreatic hypoplasia. | [99] | |
| Sox12 | Fetal (m, h) Adult (m, h) |
Pancreas epithelium during development. | Mutant does not have pancreatic phenotype. | [98] | |
| SOXD | Sox5 | Fetal (m, h) Adult (m, h) |
A subset of pancreas epithelium during embryogenesis; α- and β-cells in adult islet. |
Attenuates glucose-stimulated insulin secretion in adult. | [102] |
| Sox6 | Fetal (m) Adult (m) |
A subset of pancreas epithelium during embryogenesis; α- and β-cells in adult islet. |
Suppresses glucose-stimulated insulin secretion and β-cell proliferation in adult; associated with hyperinsullinemia. | [102, 103] | |
| Sox13 | Fetal (m, h) Adult (m, h) |
A subset of pancreas epithelium during embryogenesis; α- and β-cells in adult islet. | Attenuates glucose-stimulated insulin secretion in adult. Diabetes autoantigen. | [104, 105] | |
| SOXE | Sox8 | Fetal (m) | Scattered cells at the epithelial/mesenchymal boundary at E10.5 to E12.5, and glial cells at the periphery of islet during late embryogenesis and adult. | Mutant does not have pancreatic phenotype. | [61] |
| Sox9 | Fetal (m, h) Adult (m, h) |
Pancreatic epithelium during early embryogenesis; islets, a subset of ductal epithelial cells, and few exocrine acinar cells in later stage. | Regulates pancreatic specification, differentiation, and duct morphology. | [65, 70, 72, 73, 75] | |
| Sox10 | Fetal (m, h) Adult (m, h) |
Scattered cells at the epithelial/mesenchymal boundary at E10.5 to E12.5, and glial cells at the periphery of islet during late embryogenesis and adult. | Mutant does not have pancreatic phenotype. | [61] | |
| SOXF | Sox7 | Fetal (m) | ND | ND | [61, 62, 97] |
| Sox17 | Fetal (m, h) | ND | Control segregation of liver, biliary system, and pancreas; regulates insulin trafficking and secretion in β-cells. | [6, 7, 32, 106] | |
| Sox18 | Fetal (m) | ND | ND | [61, 62, 97] | |
| SOXG | Sox15 | Fetal (m) Adult (m) |
ND | ND | [61, 62, 97] |
h, human; m, mouse; ND, not determined.
The expression of Sox2 in fetal murine pancreas has only been reported by Wilson et al.[62].
The number of Sox2+ cells in the pancreas is extremely low. There was also concern about contamination of intestinal cells in the sample.
3.1 Overview of pancreas development
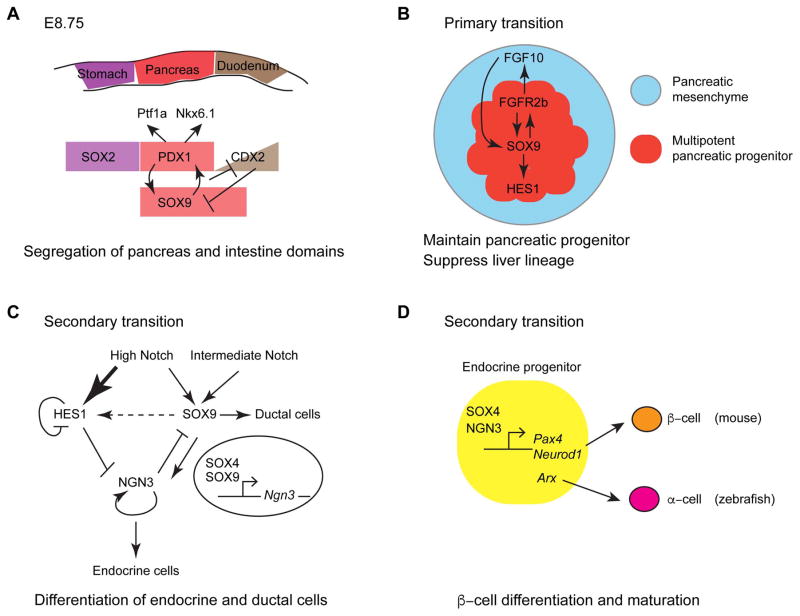
Our understanding of pancreas development relies heavily on the studies of murine models. Pancreas development starts at around E9 in mouse as two epithelial buds emerge on either side of the foregut endoderm at the boundary between the stomach and duodenum [63–65]. Extensive proliferation of pancreatic progenitors rapidly increases the size of the dorsal and ventral buds, which at the same time undergo active tubulogenesis and patterning events [66]. At the end of this first wave of pancreas development, referred to as primary transition, the dorsal and ventral buds fuse by E12.5 to form a unified organ. The largely undifferentiated pancreatic epithelium is segregated into two distinct domains [56]: the tip domain is marked by Ptf1a expression and contains multipotent progenitor cells that give rise to endocrine, duct, and acinar cells. The trunk domain expresses Nkx6.1 and only gives rise to duct and endocrine cells [67]. The second period of pancreas development, namely secondary transition, starts at E13.5 and is characterized by massive waves of organ morphogenesis and endocrine and exocrine differentiation. The topological organization of pancreas is set by the end of secondary transition, but the functional maturation of exocrine and endocrine cells continues postnatally [68].
3.2 SOX9 is a master regulator of pancreas development
The discovery that CD patients often show islet hypoplasia and reduction of endocrine hormone expression provides prevailing evidence for the requirement of SOX9 in pancreas development [69]. Studies of conditional Sox9 knockout mice revealed critical roles of SOX9 in every step of pancreas development (summarized in Fig. 2). Sox9 expression is first detected in the presumptive pancreas and proximal duodenum between anterior foregut and mid/hindgut at E8.75, prior to the emergence of pancreatic buds (Fig. 2A) [70]. Its expression in the pre-pancreatic domain overlaps with Pdx1. SOX9 and PDX1 reinforces each other’s expression as PDX1 occupies Sox9 regulatory sequences and vice versa [71, 72]. Whereas pancreas remnants still arise in Sox9 and Pdx1 individual mutant mice, compound mutants carrying different combinations of the Pdx1 null and gut-specific Sox9 mutant alleles exhibit a spectrum of defects in pancreatic buds, ranging from a smaller ventral bud in the compound heterozygous mutants to the absence of both dorsal and ventral buds in the compound homozygous mutants. In Sox9;Pdx1 compound homozygous mutants but not in the individual mutants, the intestinal marker Cdx2 becomes ectopically expressed in the pre-pancreatic domain. Conversely ectopic expression of Sox9 is sufficient to induce Pdx1 expression in intestinal progenitor cells. Thus PDX1 and SOX9 cooperatively induce pancreatic lineage and repress intestinal fate. Genomics analysis reveals that PDX1 and SOX9 function as direct activators of pancreatic genes and repressors of intestinal genes by binding to lineage-specific promoters and enhancers [70].
Figure 2. Overview of SOX4 and SOX9 function in pancreas development.
(A) A schematic showing a positive feedback loop between PDX1 and SOX9 that promotes pancreatic specification while repressing intestinal fate at E8.75 [modified from 70]. (B) During primary transition, FGF10 from the pancreatic mesenchyme is transduced by FGFR2b to maintain SOX9 expression in the multipotent pancreatic progenitors. SOX9 in turn maintains FGFR2b expression in the same cell. SOX9 also regulates HES1 expression. The interplay between SOX9, FGF10, FGFR2b, and HES1 preserves the pancreatic cell fate [modified from 72]. (C) Different levels of Notch signaling control endocrine versus ductal cell fate through regulating expression of SOX9 and HES1 during secondary transition [modified from 65]. (D) A proposed model showing that SOX4 and NGN3 co-bind the promoters of downstream transcription factors to regulate differentiation and maturation of β-cells in mouse and α-cells in zebrafish.
Sox9 continues being co-expressed with Pdx1 in uncommitted and pluripotent pancreatic progenitors during primary transition [73]. Sanders group generated Pdx1-Cre-mediated Sox9 knockout mice that abolish Sox9 expression in pancreatic progenitors from E10.5 onwards [72, 73]. The pancreas in Sox9fl/fl;Pdx1-Cre mutants is hypoplastic due to impaired proliferation and survival of pancreatic progenitors. The remnant progenitors exit cell cycle precociously to differentiate into glucagon-expressing cells. Some of the surviving pancreatic progenitors in the mutant undergo a pancreas-to-liver cell fate conversion [72]. Consistently, genome wide DNA occupancy study suggests that SOX9 and PDX1 co-bind hepatic genes to repress their expression [70].
Several lines of evidence indicate that SOX9 maintains pancreatic progenitor pool through modulating Notch signal transduction: SOX9 and HES1 are co-localized in pancreatic progenitors; Hes1 mutant exhibits pancreas hypoplasia due to depletion of pancreatic progenitors similar to Sox9 mutant; Sox9-deficient pancreas contains fewer Hes1+ cells [73]. An Fgf10/Fgfr2b/Sox9 feed-forward loop that acts between pancreatic progenitors and neighboring mesenchyme is also essential for maintaining pancreatic progenitors [72]. Specifically, SOX9 regulates the expression of Fgfr2b in pancreatic progenitors, and FGFR2b transduces Fgf10 signal from the mesenchyme. FGF10 in turn maintains Sox9 and Fgfr2b expression in pancreatic progenitors (Fig. 2B).
During secondary transition, Sox9 is expressed in multipotent pancreatic progenitors in the trunk epithelial cords that give rise to endocrine progenitors, differentiated endocrine cells, ductal cells, and exocrine acinar cells [36, 71, 73–75]. Its expression is eliminated from the lineage-committed progenitors and differentiated cells. Sox9fl/+;Pdx1-Cre heterozygous mutants show a 50% reduction of mature endocrine cells due to a decrease in endocrine progenitors, but no defect in exocrine differentiation [75]. This result implicates that differentiation of endocrine lineage is more sensitive to the reduction of Sox9 dosage.
Endocrine differentiation is governed by a molecular network involving SOX9, Notch signaling components, and neurogenin 3 (NGN3) (Fig. 2C) [65]. Notch signaling activates Sox9 expression in multipotent pancreatic progenitors, and Sox9 cell-autonomously induces Ngn3 expression either by direct binding or possibly by mediating Pdx1 expression [65, 71]. Once Ngn3 expression is initiated in a subset of pancreatic progenitors, it represses Sox9 expression in the same cell so that Ngn3+ cells are seen intercalating with Sox9+ cells [73, 75]. NGN3 positively enhances its own expression and is necessary and sufficient for endocrine differentiation. The remaining Sox9+ cells that do not express Ngn3 adopt a ductal fate. Notch signaling also induces expression of HES1, which acts as a repressor of NGN3. SOX9 and HES1 respond to Notch activity at different thresholds [65]. At high Notch activity, the repressive effect of HES1 on NGN3 is dominant and endocrine differentiation is blocked. Pancreatic progenitors instead turn on ductal markers. At intermediate Notch activity, HES1 expression is reduced and Sox9 expression is maintained, which derepresses NGN3 expression and allows endocrine differentiation to occur [65].
The islets from pancreatic Sox9-haploinsufficient mice, albeit small, show normal insulin secretion, indicating that SOX9 is not required for β-cell function [71]. These animals only develop mild glucose intolerance as the mutant β-cells undergo extensive proliferation between E18.5 and six weeks to partially compensate for the initial reduction in β-cell mass [71]. It has been reported that human β-cells have a much lower proliferative capacity than mouse β-cells [76]. Therefore reduced SOX9 expression may have a more confound impact on glucose homeostasis in human than in mice due to a lack of compensatory proliferation mechanism to restore the β-cell mass.
Sox9+ progenitors also give rise to exocrine lineages including acinar cells and pancreatic ductal cells, yet how it contributes to exocrine development is less understood. Notch over-activation fails to induce ductal cells in Sox9-deficient mice, suggesting that Sox9 is an obligatory effector of Notch-dependent ductal cell differentiation [65]. In Sox9fl/fl;Rosa26CreER mice in which Sox9 is ablated at E12.5 by tamoxifen administration, the ductal network is disorganized and forms cysts postnatally. Cystic phenotype is observed when Sox9 is ablated in adult ducts, indicating that Sox9 directly mediates pancreatic ductal cell differentiation and maintenance. Gene expression profiling analyses show that SOX9 controls the expression of putative ductal markers. It is required for primary cilia formation in the ductal epithelial cells likely through regulating the expression of polycystin 2 (PKD2). The requirement of SOX9 in pancreatic duct differentiation and morphogenesis is conserved in zebrafish. In sox9b mutant, ductal cells are reduced in number and fail to undergo branching morphogenesis [19, 20]. The ductal network remains rudimental with clusters of cells along the main duct [19].
The involvement of Sox9 in exocrine acinar cell development is illusive. During early secondary transition, Ptf1a+Sox9+ cells are located at tip-trunk interface and inside the tip region, intermingled with Ptf1ahiSox9lo cells [77]. Whereas Ptf1ahiSox9lo cells are proacinar progenitors, Ptf1a+Sox9+ cells are multipotent progenitors that give rise to acinar, ductal and endocrine cells. Ptf1a+Sox9+ cells rapidly decreases between E12.5 and E15.5, with the exception of those at tip-trunk interface that remain multipotent throughout pancreogenesis [36, 75]. Given that HES1 also represses Ptf1a [78], it is plausible that a SOX9/NOTCH/HES1/PTF1A network may control acinar cell development in a similar fashion as the SOX9/NOTCH/HES1/NGN3 network that regulates endocrine differentiation in the trunk region. However, pancreatic Sox9-haploinsufficiency mice and zebrafish sox9b mutants still form normal number of acinar cells [20, 75], thus SOX9 is not absolutely essential for the differentiation of acinar cells. Instead, SOX9 dosage may mediate the balance between proacinar and endocrine progenitors as Sox9 heterozygous mutants have more proacinar progenitors at the expense of endocrine progenitors [75]. Ablation of SOX9 at different embryonic stages using genetic mouse models may provide additional insights into the roles of SOX9 in exocrine development.
3.3 SOX9 in pancreas homeostasis and injury
Following secondary transition, SOX9 expression becomes restricted to ductal cells and centroacinar cells at trunk-tip interface [36, 75]. Two groups have performed lineage-tracing studies to evaluate the multipotency of Sox9+ cells during prenatal and adult stages. Kopp et al showed that when tamoxifen was administrated to the BAC Sox9CreERT2 transgenic mice at postnatal day 5, Sox9+ cells gave rise to predominantly ductal cells and few endocrine cells [36]. When tamoxifen administration was conducted at 3 weeks, Sox9+ cells exclusively formed ductal cells. These results suggest that Sox9+ cells are bipotent during early neonatal stage but become confined to ductal fate in adult. No acinar cells were labeled in either experiment, consistent with a previous report that new acinar cells are supplied by preexisting acinar cells [79]. Furuyama and colleagues performed lineage tracing using the Sox9IRES-CreERT2 knockin mice [26], and showed that in adult pancreas, Sox9+ cells gave rise to both ductal and acinar cells. The discrepancy between the two studies has been discussed earlier. Moreover, Sox9IRES-CreERT2 knockin mice, but not BAC Sox9CreERT2 mice, show decreased endogenous pancreatic Sox9 expression at adult stage [80, 81]. This observation suggests that reduced SOX9 dosage may increase the plasticity of SOX9+ cells to allow “ductal-to-acinar” transition.
As SOX9 maintains multipotent progenitors during pancreas development, it has been proposed that Sox9+ pancreatic ducts contain facultative progenitors that form both exocrine and endocrine cells in both healthy and injured adult pancreas. In support of this notion, Jin et al showed that a subpopulation of Sox9+ ductal cells expressed progenitor marker CD133 and formed insulin-producing β-cells, ductal cells, and acinar cells in vitro [82, 83]. However, neither of the two SOX9 lineage-tracing studies detected endocrine neogenesis from Sox9+ cells in healthy adult pancreas [26, 71]. Melton group lineage traced β-cells and demonstrated that adult β-cells are formed by self-duplication rather than stem-cell differentiation [84].
The contribution of ductal cells to β-cell neogenesis during pancreatic injury is highly controversial. Heimberg group and Bonner-Weir group reported that partial duct ligation triggered ductal-lining cells to transdifferentiate into new β-cells in adult pancreas [85–87]. In contrast, Sox9 lineage-tracing studies found no evidence of β-cell neogenesis from Sox9+ cells after partial ductal ligation [26, 71]. Furuyama and colleagues examined additional pancreas injury models, including partial pancreatectomy, cerulein-induced pancreatitis, and streptozotocin diabetes, and did not detect Sox9+ derived β-cells following any of these injuries [26]. A recent study reported that a combination of medium hyperglycemia and long-term administration of low dose gastrin and epidermal growth factor induced Sox9+ ductal cells to transdifferentiate into β-cells in nonautoimmune diabetic mice [88]. The conflicting results from different studies implicate that the type of pancreatic injury as well as its primary cell target have critical influence on whether ductal cells contribute to β-cell neogenesis [89]. Zebrafish larvae regenerate primary islet after nitroreductase-mediated β-cell ablation, and β-cell regeneration is blocked in sox9b mutant larvae [20]. However, these β-cell ablation experiments were conducted during the course of zebrafish pancreas development. The fact that sox9b mutants do not regenerate β-cells at this stage may reflect the requirement of Sox9b in endocrine differentiation rather than in β-cell neogenesis. sox9b adult mutant is not informative for studying β-cell neogenesis as it exhibits extensive necrosis and fibrosis in both exocrine and endocrine compartments [19].
While it is still under debate whether Sox9+ ductal cells in the pancreas participate in βcell neogenesis, Sox9+ cells in the liver can be reprogrammed into insulin-secreting ducts in vivo by introduction of three transcription factors PDX1, NGN3, and MAFa [90]. This is not surprising as the pancreas and liver arise from adjacent regions of the foregut endoderm. In adult liver, Sox9 is expressed in the periportal small intrahepatic ducts [16, 26], periportal hepatocytes [31], and peribilliary glands lining the large bile ducts [32, 33]. Which of these three cell types are capable of forming insulin-secreting ducts is yet to be determined. It will also be interesting to investigate whether Sox9-deficient liver cells can still be reprogrammed into insulin-producing ducts. Intriguingly, the insulin-producing ducts in the liver relieved diabetes in adult animal, thus reprogramming liver cells could potentially be used as a novel strategy for diabetes therapy.
3.4 Sox4 in endocrine cell differentiation and maturation
SOX4 is also expressed throughout pancreas development [61, 62]. During primary transition, Sox4 is expressed in both the tip and trunk progenitors [91]. During secondary transition, Sox4 expression gradually decreases from the pancreas epithelium but remains at high levels in the Ngn3-expressing endocrine progenitors and newly formed endocrine cells. At the end of gestation, Sox4 becomes restricted to the endocrine cells and this expression pattern maintains in adult. Sox4 global knockout mice show no morphological defects in the pancreas at stages up to E12.5 [62], suggesting that SOX4 is dispensable for the formation of pancreatic progenitors during primary transition. The mutants die at E14.5 due to cardiac defects, preventing characterization of SOX4 function during later pancreas development. To overcome this issue, Wilson and colleagues isolated explants of pancreas from Sox4 mutants at E11.5 and showed that they continued to develop ex vivo [62]. During secondary transition, Sox4−/− pancreatic explants form fewer endocrine cells. The islets in these mutants lose the stereotypical organization with a core of β-cells surrounded by α-cells, indicating that Sox4 also mediates islet morphogenesis. Conditional knockout mice in which Sox4 is deleted from either pancreatic epithelium or endocrine progenitors exhibit reduction of all endocrine cells except for ε-cells [91]. It is not due to dysregulated cell division or cell death [62, 91]. Instead, depletion of Sox4 in endocrine progenitors keeps these cells in a “de-differentiated” state [91]. SOX4 also mediates adult β-cell function as two N-ethyl-N-nitrosourea (ENU)-induced Sox4 missense mutations cause impaired glucose tolerance and reduced insulin secretion [92]. Whereas loss of SOX4 function in mouse causes a more drastic reduction of β-cells than α-cells [62, 91], knockdown of sox4b in zebrafish by injecting either an anti-sense morpholino oligonucleotide or a sox4b mutant mRNA lacking the transactivation domain causes severe reduction in α-cells without affecting β-cells [93]. Such a discrepancy may reflect a fundamental difference in endocrine pancreas development between the two species. In zebrafish, dorsal bud-derived β-cells are quiescent and express low level of insulin after embryogenesis [94]. Ventral bud-derived β-cells that emerge late in development are proliferative and maintain high expression of insulin postembryonically, thus are fully functional β cells. The study of sox4b mutants focused on the endocrine cells in the dorsal bud [93]. Whether loss of Sox4b function affects ventral bud-derived β cells remains to be determined. The difference between the fish and mouse data could also be attributed to the limitation of the knockdown strategy used in the fish study: the efficacy of both morpholino and mRNA decreases as the embryo grows older [95] and may not completely block Sox4b function during pancreas development. Furthermore, morpholino-induced phenotypes do not necessarily always correlate with mutant phenotypes [96]. Therefore, it will be necessary to validate the endocrine phenotypes in a genetic mutant of sox4b.
Where does SOX4 fit within the cascade of transcription factors that control endocrine differentiation (Fig. 2D)? SOX4 is not required for maintaining the expression of Sox9, Nkx2.2, and Nkx6.1 in multipotent pancreatic progenitors [62]. Meanwhile, SOX4 can activate the Ngn3 promoter by direct binding. Thus it facilitates endocrine progenitor formation by enhancing Ngn3 expression in a subset of bipotent trunk cells [91]. Following secondary transition, SOX4 regulates the maturation of β-cells by binding to and activating the promoters of NeuroD1 and Pax4 transcription factors, both are NGN3 targets [91]. In zebrafish Sox4b is required for the expression of transcription factor Arx in α-cell differentiation, consistent with the fact that only α-cells are greatly reduced in sox4b mutants [93].
Although Sox4 is abundantly expressed throughout pancreas development, inactivation of SOX4 does not affect the formation of pancreatic progenitors [62, 91]. Ngn3+ endocrine progenitors and differentiated endocrine cell types are not completely absent in Sox4 mutant pancreas, suggesting that other transcription factors may compensate for the loss of SOX4 function. Indeed, the two other SOXC family members, Sox11 and Sox12, are both expressed in the fetal pancreas [61, 91, 97]. Whereas Sox12 mutants do not have obvious gross morphologic defects [98], Sox11 mutants exhibit hypoplasia of the pancreas [99]. More importantly, Sox11 expression is elevated in the Sox4 mutant pancreas, thus may compensate for the absence of Sox4 in these animals [62]. Moreover, SOX4 may partner with SOX9 to promote Ngn3 expression in the endocrine progenitors as both of them bind to and activate the Ngn3 promoter. Within the endocrine progenitors, SOX4 may cooperate with NGN3 to regulate the formation of mature β-cells by activating the NeuoD1 and Pax4 promoters. Generation of pancreas-specific double knockout mice will allow one to confirm these interactions.
4. Conclusions
The liver and pancreas, two organs with distinct morphology and function but originated from close foregut endoderm domains, are the best examples to demonstrate the versatile function of SOX transcription factors in development. Prior to organ formation, SOX17 interacts with HHEX and PDX1 to segregate the liver, biliary system, and ventral pancreas domains within the foregut endoderm. SOX9 and PDX1 function in a positive feedback loop to promote pancreas specification while suppressing intestine fate. During liver development, SOX4 and SOX9 are required for the differentiation and morphogenesis of intrahepatic biliary cells. In the pancreas, SOX9 interacts with FGF10 and Notch signaling to maintain pancreatic progenitor during primary transition. During secondary transition, SOX9 regulates the differentiation of endocrine and ductal cells in response to different levels of Notch activity. SOX4 mediates β-cell differentiation and maturation. SOX factors continue being expressed in the liver and pancreas beyond embryonic stage and play critical roles in adult organ homeostasis.
There are still many important questions that remain unanswered. The upstream signals that regulate the temporal and spatial expression of SOX factors are not well understood. SOX proteins undergo posttranslational modification, including phosphorylation, acetylation, SUMOylation and ubiquitination [1]. Given that SOX factors often act in a dose-dependent fashion, it will be interesting to study whether posttranslational modification mediates the dosage of their expression during liver and pancreas development. A systematic survey of SOX factor expression and function in the liver has not been reported. The pancreas expresses many other SOX factors besides SOX4 and SOX9, yet their function remains largely unknown.
No mutations in SOX genes have yet been associated with liver diseases or diabetes. However, the relevance of SOX17 in biliary atresia and SOX9 in liver fibrosis suggest that SOX factors may represent as important disease modifiers and thus are potential molecular targets for gene therapies. Directed differentiation of human pluripotent stem cells into liver cells and β-cells in culture could lead to new cell therapies for liver diseases and diabetes, respectively. The main challenges are the efficiency of directed differentiation and to what extend the stem cell-derived cells are able to carry out the physiological function of bona fida cells. In this regard, SOX factors could be the crucial missing pieces in the current methodology. Expressing the SOX factors that are key regulators of liver and pancreas development in stem cells may increase the efficiency of their differentiation. Given the facts that SOX4 and SOX17 are required for making functional β-cells and SOX9 maintains pancreatic ductal epithelium, introducing SOX factors may improve the maturation and function of stem cell-derived cells. Lastly, although highly controversial, SOX9-expressing cells in the liver and pancreas have been proven to be able to contribute to hepatocyte regeneration and β-cell neogenesis in certain types of injury. Liver regeneration and β-cell neogenesis are perturbed in zebrafish sox9b mutants. These observations implicate that SOX factors may be attractive molecular targets in regenerative medicine for treating liver diseases and diabetes. To incorporate SOX factors in clinical therapies requires further characterization of their upstream regulators, binding partners, and the downstream molecular and cellular events.
Acknowledgments
We would like to thank Drs Frederic Lemaigre, James Wells, and Stacey Huppert for insightful discussion during the preparation of this review. This work was supported by the National Institutes of Health [R00AA020514, P30 DK078392], and a pilot grant from Center of Pediatric Genomics in Cincinnati Childern’s Hospital.
Abbreviations
- AAV
adeno-associated virus
- Arx
aristaless-related homeobox
- BA
biliary atresia
- BAC
bacterial artificial chromosome
- BDL
bile duct ligation
- CD
campomelic dysplasia
- CDE
choline-deficient, ethionine-supplemented diet
- Cdx2
caudal type homeobox 2
- DDC
3,5-diethoxycarbonyl-1,4-dihydrocollidine
- ENU
N-ethyl-N-nitrosourea
- Fgf
fibroblast growth factor
- Foxa3
forkhead box A3
- Hes1
hairy and enhancer of split 1
- HHEX
hematopoietically expressed homeobox
- Hif1α
hypoxia-inducible factor 1 alpha
- Hnf6
hepatocyte nuclear factor 6
- HMG
high mobility-group
- HPC
hepatic progenitor cell
- HPD
hepatopancreatic duct
- HybHP
hybrid periportal hepatocyte
- IRES
internal ribosome entry site
- MAFA
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a
- MCDE
methionine choline-deficient, ethionine-supplemented diet
- Mdm2
mouse double minute 2 homolog
- NeuroD
neuronal differentiation 1
- Ngn3
neurogenin 3
- Nkx6.1
Nk6 homeobox protein 1
- Pdx1
pancreatic and duodenal homeobox 1
- Pkd2
polycystin 2
- PP
pancreatic polypeptide
- Ptf1a
pancreas transcription factor 1 subunit alpha
- Rbpjκ
recombination signal binding protein for immunoglobulin kappa J region
- Sox
sex-determining region of Y chromosome-related high mobility-group box
- TGFβ
transforming growth factor beta;
- TβRII
transforming growth factor β receptor II.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.She ZY, Yang WX. SOX family transcription factors involved in diverse cellular events during development. Eur J Cell Biol. 2015;94(12):547–63. doi: 10.1016/j.ejcb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 2010;18(2):175–89. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Alexander J, Stainier DY. A molecular pathway leading to endoderm formation in zebrafish. Curr Biol. 1999;9(20):1147–57. doi: 10.1016/S0960-9822(00)80016-0. [DOI] [PubMed] [Google Scholar]
- 4.Clements D, Woodland HR. Changes in embryonic cell fate produced by expression of an endodermal transcription factor, Xsox17. Mech Dev. 2000;99(1–2):65–70. doi: 10.1016/s0925-4773(00)00476-7. [DOI] [PubMed] [Google Scholar]
- 5.Hudson C, Clements D, Friday RV, Stott D, Woodland HR. Xsox17alpha and -beta mediate endoderm formation in Xenopus. Cell. 1997;91(3):397–405. doi: 10.1016/s0092-8674(00)80423-7. [DOI] [PubMed] [Google Scholar]
- 6.Kanai-Azuma M, Kanai Y, Gad JM, Tajima Y, Taya C, Kurohmaru M, Sanai Y, Yonekawa H, Yazaki K, Tam PP, Hayashi Y. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129(10):2367–79. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- 7.Spence JR, Lange AW, Lin SC, Kaestner KH, Lowy AM, Kim I, Whitsett JA, Wells JM. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell. 2009;17(1):62–74. doi: 10.1016/j.devcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uemura M, Hara K, Shitara H, Ishii R, Tsunekawa N, Miura Y, Kurohmaru M, Taya C, Yonekawa H, Kanai-Azuma M, Kanai Y. Expression and function of mouse Sox17 gene in the specification of gallbladder/bile-duct progenitors during early foregut morphogenesis. Biochem Biophys Res Commun. 2010;391(1):357–63. doi: 10.1016/j.bbrc.2009.11.063. [DOI] [PubMed] [Google Scholar]
- 9.Matsui T, Kanai-Azuma M, Hara K, Matoba S, Hiramatsu R, Kawakami H, Kurohmaru M, Koopman P, Kanai Y. Redundant roles of Sox17 and Sox18 in postnatal angiogenesis in mice. J Cell Sci. 2006;119(Pt 17):3513–26. doi: 10.1242/jcs.03081. [DOI] [PubMed] [Google Scholar]
- 10.Shin D, Weidinger G, Moon RT, Stainier DY. Intrinsic and extrinsic modifiers of the regulative capacity of the developing liver. Mech Dev. 2012;128(11–12):525–35. doi: 10.1016/j.mod.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uemura M, Ozawa A, Nagata T, Kurasawa K, Tsunekawa N, Nobuhisa I, Taga T, Hara K, Kudo A, Kawakami H, Saijoh Y, Kurohmaru M, Kanai-Azuma M, Kanai Y. Sox17 haploinsufficiency results in perinatal biliary atresia and hepatitis in C57BL/6 background mice. Development. 2013;140(3):639–48. doi: 10.1242/dev.086702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorent K, Gong W, Koo KA, Waisbourd-Zinman O, Karjoo S, Zhao X, Sealy I, Kettleborough RN, Stemple DL, Windsor PA, Whittaker SJ, Porter JR, Wells RG, Pack M. Identification of a plant isoflavonoid that causes biliary atresia. Sci Transl Med. 2015;7(286):286ra67. doi: 10.1126/scitranslmed.aaa1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waisbourd-Zinman O, Koh H, Tsai S, Lavrut PM, Dang C, Zhao X, Pack M, Cave J, Hawes M, Koo KA, Porter JR, Wells RG. The toxin biliatresone causes mouse extrahepatic cholangiocyte damage and fibrosis via decreased glutathione and SOX17. Hepatology. 2016 doi: 10.1002/hep.28599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster JW, Dominguez-Steglich MA, Guioli S, Kwok C, Weller PA, Stevanovic M, Weissenbach J, Mansour S, Young ID, Goodfellow PN, et al. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994;372(6506):525–30. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 15.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, Wolf U, Tommerup N, Schempp W, Scherer G. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79(6):1111–20. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 16.Antoniou A, Raynaud P, Cordi S, Zong Y, Tronche F, Stanger BZ, Jacquemin P, Pierreux CE, Clotman F, Lemaigre FP. Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology. 2009;136(7):2325–33. doi: 10.1053/j.gastro.2009.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorent K, Moore JC, Siekmann AF, Lawson N, Pack M. Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Dev Dyn. 2010;239(3):855–64. doi: 10.1002/dvdy.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan YL, Willoughby J, Liu D, Crump JG, Wilson C, Miller CT, Singer A, Kimmel C, Westerfield M, Postlethwait JH. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132(5):1069–83. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- 19.Delous M, Yin C, Shin D, Ninov N, Debrito Carten J, Pan L, Ma TP, Farber SA, Moens CB, Stainier DY. Sox9b is a key regulator of pancreaticobiliary ductal system development. PLoS Genet. 2012;8(6):e1002754. doi: 10.1371/journal.pgen.1002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manfroid I, Ghaye A, Naye F, Detry N, Palm S, Pan L, Ma TP, Huang W, Rovira M, Martial JA, Parsons MJ, Moens CB, Voz ML, Peers B. Zebrafish sox9b is crucial for hepatopancreatic duct development and pancreatic endocrine cell regeneration. Dev Biol. 2012;366(2):268–78. doi: 10.1016/j.ydbio.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, Costa T, Pierpont ME, Rand EB, Piccoli DA, Hood L, Spinner NB. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16(3):243–51. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 22.McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet. 2006;79(1):169–73. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann JJ, Zovein AC, Koh H, Radtke F, Weinmaster G, Iruela-Arispe ML. Jagged1 in the portal vein mesenchyme regulates intrahepatic bile duct development: insights into Alagille syndrome. Development. 2010;137(23):4061–72. doi: 10.1242/dev.052118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kodama Y, Hijikata M, Kageyama R, Shimotohno K, Chiba T. The role of notch signaling in the development of intrahepatic bile ducts. Gastroenterology. 2004;127(6):1775–86. doi: 10.1053/j.gastro.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, Stanger BZ. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136(10):1727–39. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, Masui T, Kawaguchi M, Takaori K, Doi R, Nishi E, Kakinoki R, Deng JM, Behringer RR, Nakamura T, Uemoto S. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43(1):34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- 27.Poncy A, Antoniou A, Cordi S, Pierreux CE, Jacquemin P, Lemaigre FP. Transcription factors SOX4 and SOX9 cooperatively control development of bile ducts. Dev Biol. 2015;404(2):136–48. doi: 10.1016/j.ydbio.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 28.Zong Y, Stanger BZ. Molecular mechanisms of bile duct development. Int J Biochem Cell Biol. 2011;43(2):257–64. doi: 10.1016/j.biocel.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clotman F, Jacquemin P, Plumb-Rudewiez N, Pierreux CE, Van der Smissen P, Dietz HC, Courtoy PJ, Rousseau GG, Lemaigre FP. Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev. 2005;19(16):1849–54. doi: 10.1101/gad.340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plumb-Rudewiez N, Clotman F, Strick-Marchand H, Pierreux CE, Weiss MC, Rousseau GG, Lemaigre FP. Transcription factor HNF-6/OC-1 inhibits the stimulation of the HNF-3alpha/Foxa1 gene by TGF-beta in mouse liver. Hepatology. 2004;40(6):1266–74. doi: 10.1002/hep.20459. [DOI] [PubMed] [Google Scholar]
- 31.Font-Burgada J, Shalapour S, Ramaswamy S, Hsueh B, Rossell D, Umemura A, Taniguchi K, Nakagawa H, Valasek MA, Ye L, Kopp JL, Sander M, Carter H, Deisseroth K, Verma IM, Karin M. Hybrid Periportal Hepatocytes Regenerate the Injured Liver without Giving Rise to Cancer. Cell. 2015;162(4):766–79. doi: 10.1016/j.cell.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardinale V, Wang Y, Carpino G, Cui CB, Gatto M, Rossi M, Berloco PB, Cantafora A, Wauthier E, Furth ME, Inverardi L, Dominguez-Bendala J, Ricordi C, Gerber D, Gaudio E, Alvaro D, Reid L. Multipotent stem/progenitor cells in human biliary tree give rise to hepatocytes, cholangiocytes, and pancreatic islets. Hepatology. 2011;54(6):2159–72. doi: 10.1002/hep.24590. [DOI] [PubMed] [Google Scholar]
- 33.Carpino G, Cardinale V, Onori P, Franchitto A, Berloco PB, Rossi M, Wang Y, Semeraro R, Anceschi M, Brunelli R, Alvaro D, Reid LM, Gaudio E. Biliary tree stem/progenitor cells in glands of extrahepatic and intraheptic bile ducts: an anatomical in situ study yielding evidence of maturational lineages. J Anat. 2012;220(2):186–99. doi: 10.1111/j.1469-7580.2011.01462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpentier R, Suner RE, van Hul N, Kopp JL, Beaudry JB, Cordi S, Antoniou A, Raynaud P, Lepreux S, Jacquemin P, Leclercq IA, Sander M, Lemaigre FP. Embryonic ductal plate cells give rise to cholangiocytes, periportal hepatocytes, and adult liver progenitor cells. Gastroenterology. 2011;141(4):1432–8. 1438 e1–4. doi: 10.1053/j.gastro.2011.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorrell C, Erker L, Schug J, Kopp JL, Canaday PS, Fox AJ, Smirnova O, Duncan AW, Finegold MJ, Sander M, Kaestner KH, Grompe M. Prospective isolation of a bipotential clonogenic liver progenitor cell in adult mice. Genes Dev. 2011;25(11):1193–203. doi: 10.1101/gad.2029411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopp JL, Dubois CL, Schaffer AE, Hao E, Shih HP, Seymour PA, Ma J, Sander M. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development. 2011;138(4):653–65. doi: 10.1242/dev.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarlow BD, Finegold MJ, Grompe M. Clonal tracing of Sox9+ liver progenitors in mouse oval cell injury. Hepatology. 2014;60(1):278–89. doi: 10.1002/hep.27084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malato Y, Naqvi S, Schurmann N, Ng R, Wang B, Zape J, Kay MA, Grimm D, Willenbring H. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J Clin Invest. 2011;121(12):4850–60. doi: 10.1172/JCI59261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanger K, Knigin D, Zong Y, Maggs L, Gu G, Akiyama H, Pikarsky E, Stanger BZ. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell. 2014;15(3):340–9. doi: 10.1016/j.stem.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alison MR, Lin WR. Diverse routes to liver regeneration. J Pathol. 2016;238(3):371–4. doi: 10.1002/path.4667. [DOI] [PubMed] [Google Scholar]
- 41.Carpino G, Renzi A, Franchitto A, Cardinale V, Onori P, Reid L, Alvaro D, Gaudio E. Stem/Progenitor Cell Niches Involved in Hepatic and Biliary Regeneration. Stem Cells Int. 2016;2016:3658013. doi: 10.1155/2016/3658013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyajima A, Tanaka M, Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell. 2014;14(5):561–74. doi: 10.1016/j.stem.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 43.Schaub JR, Malato Y, Gormond C, Willenbring H. Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep. 2014;8(4):933–9. doi: 10.1016/j.celrep.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu WY, Bird TG, Boulter L, Tsuchiya A, Cole AM, Hay T, Guest RV, Wojtacha D, Man TY, Mackinnon A, Ridgway RA, Kendall T, Williams MJ, Jamieson T, Raven A, Hay DC, Iredale JP, Clarke AR, Sansom OJ, Forbes SJ. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol. 2015;17(8):971–83. doi: 10.1038/ncb3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi TY, Ninov N, Stainier DY, Shin D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology. 2014;146(3):776–88. doi: 10.1053/j.gastro.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He J, Lu H, Zou Q, Luo L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology. 2014;146(3):789–800 e8. doi: 10.1053/j.gastro.2013.11.045. [DOI] [PubMed] [Google Scholar]
- 47.Huang M, Chang A, Choi M, Zhou D, Anania FA, Shin CH. Antagonistic interaction between Wnt and Notch activity modulates the regenerative capacity of a zebrafish fibrotic liver model. Hepatology. 2014;60(5):1753–66. doi: 10.1002/hep.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarlow BD, Pelz C, Naugler WE, Wakefield L, Wilson EM, Finegold MJ, Grompe M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell. 2014;15(5):605–18. doi: 10.1016/j.stem.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, Wells RG, Greenbaum LE, Stanger BZ. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27(7):719–24. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walter TJ, Vanderpool C, Cast AE, Huppert SS. Intrahepatic bile duct regeneration in mice does not require Hnf6 or Notch signaling through Rbpj. Am J Pathol. 2014;184(5):1479–88. doi: 10.1016/j.ajpath.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu J, Zhou Y, Hu T, Zhang H, Shen M, Cheng P, Dai W, Wang F, Chen K, Zhang Y, Wang C, Li J, Zheng Y, Yang J, Zhu R, Wang J, Lu W, Zhang H, Wang J, Xia Y, De Assuncao TM, Jalan-Sakrikar N, Huebert RC, Bin Z, Guo C. Notch Signaling Coordinates Progenitor Cell-Mediated Biliary Regeneration Following Partial Hepatectomy. Sci Rep. 2016;6:22754. doi: 10.1038/srep22754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amarilio R, Viukov SV, Sharir A, Eshkar-Oren I, Johnson RS, Zelzer E. HIF1alpha regulation of Sox9 is necessary to maintain differentiation of hypoxic prechondrogenic cells during early skeletogenesis. Development. 2007;134(21):3917–28. doi: 10.1242/dev.008441. [DOI] [PubMed] [Google Scholar]
- 53.Hanley KP, Oakley F, Sugden S, Wilson DI, Mann DA, Hanley NA. Ectopic SOX9 mediates extracellular matrix deposition characteristic of organ fibrosis. J Biol Chem. 2008;283(20):14063–71. doi: 10.1074/jbc.M707390200. [DOI] [PubMed] [Google Scholar]
- 54.Pritchett J, Harvey E, Athwal V, Berry A, Rowe C, Oakley F, Moles A, Mann DA, Bobola N, Sharrocks AD, Thomson BJ, Zaitoun AM, Irving WL, Guha IN, Hanley NA, Hanley KP. Osteopontin is a novel downstream target of SOX9 with diagnostic implications for progression of liver fibrosis in humans. Hepatology. 2012;56(3):1108–16. doi: 10.1002/hep.25758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gouw AS, Clouston AD, Theise ND. Ductular reactions in human liver: diversity at the interface. Hepatology. 2011;54(5):1853–63. doi: 10.1002/hep.24613. [DOI] [PubMed] [Google Scholar]
- 56.Benitez CM, Goodyer WR, Kim SK. Deconstructing pancreas developmental biology. Cold Spring Harb Perspect Biol. 2012;4(6) doi: 10.1101/cshperspect.a012401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim SK, Hebrok M. Intercellular signals regulating pancreas development and function. Genes Dev. 2001;15(2):111–27. doi: 10.1101/gad.859401. [DOI] [PubMed] [Google Scholar]
- 58.Lyttle BM, Li J, Krishnamurthy M, Fellows F, Wheeler MB, Goodyer CG, Wang R. Transcription factor expression in the developing human fetal endocrine pancreas. Diabetologia. 2008;51(7):1169–80. doi: 10.1007/s00125-008-1006-z. [DOI] [PubMed] [Google Scholar]
- 59.Puri S, Hebrok M. Cellular plasticity within the pancreas--lessons learned from development. Dev Cell. 2010;18(3):342–56. doi: 10.1016/j.devcel.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sander M, German MS. The beta cell transcription factors and development of the pancreas. J Mol Med (Berl) 1997;75(5):327–40. doi: 10.1007/s001090050118. [DOI] [PubMed] [Google Scholar]
- 61.Lioubinski O, Muller M, Wegner M, Sander M. Expression of Sox transcription factors in the developing mouse pancreas. Dev Dyn. 2003;227(3):402–8. doi: 10.1002/dvdy.10311. [DOI] [PubMed] [Google Scholar]
- 62.Wilson ME, Yang KY, Kalousova A, Lau J, Kosaka Y, Lynn FC, Wang J, Mrejen C, Episkopou V, Clevers HC, German MS. The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes. 2005;54(12):3402–9. doi: 10.2337/diabetes.54.12.3402. [DOI] [PubMed] [Google Scholar]
- 63.Jennings RE, Berry AA, Strutt JP, Gerrard DT, Hanley NA. Human pancreas development. Development. 2015;142(18):3126–37. doi: 10.1242/dev.120063. [DOI] [PubMed] [Google Scholar]
- 64.Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240(3):530–65. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- 65.Shih HP, Kopp JL, Sandhu M, Dubois CL, Seymour PA, Grapin-Botton A, Sander M. A Notch-dependent molecular circuitry initiates pancreatic endocrine and ductal cell differentiation. Development. 2012;139(14):2488–99. doi: 10.1242/dev.078634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Villasenor A, Chong DC, Henkemeyer M, Cleaver O. Epithelial dynamics of pancreatic branching morphogenesis. Development. 2010;137(24):4295–305. doi: 10.1242/dev.052993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, Melton DA. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell. 2007;13(1):103–14. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 68.Shih HP, Wang A, Sander M. Pancreas organogenesis: from lineage determination to morphogenesis. Annu Rev Cell Dev Biol. 2013;29:81–105. doi: 10.1146/annurev-cellbio-101512-122405. [DOI] [PubMed] [Google Scholar]
- 69.Piper K, Ball SG, Keeling JW, Mansoor S, Wilson DI, Hanley NA. Novel SOX9 expression during human pancreas development correlates to abnormalities in Campomelic dysplasia. Mech Dev. 2002;116(1–2):223–6. doi: 10.1016/s0925-4773(02)00145-4. [DOI] [PubMed] [Google Scholar]
- 70.Shih HP, Seymour PA, Patel NA, Xie R, Wang A, Liu PP, Yeo GW, Magnuson MA, Sander M. A Gene Regulatory Network Cooperatively Controlled by Pdx1 and Sox9 Governs Lineage Allocation of Foregut Progenitor Cells. Cell Rep. 2015;13(2):326–36. doi: 10.1016/j.celrep.2015.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dubois CL, Shih HP, Seymour PA, Patel NA, Behrmann JM, Ngo V, Sander M. Sox9-haploinsufficiency causes glucose intolerance in mice. PLoS One. 2011;6(8):e23131. doi: 10.1371/journal.pone.0023131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seymour PA, Shih HP, Patel NA, Freude KK, Xie R, Lim CJ, Sander M. A Sox9/Fgf feed-forward loop maintains pancreatic organ identity. Development. 2012;139(18):3363–72. doi: 10.1242/dev.078733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seymour PA, Freude KK, Tran MN, Mayes EE, Jensen J, Kist R, Scherer G, Sander M. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci U S A. 2007;104(6):1865–70. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seymour PA. Sox9: a master regulator of the pancreatic program. Rev Diabet Stud. 2014;11(1):51–83. doi: 10.1900/RDS.2014.11.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seymour PA, Freude KK, Dubois CL, Shih HP, Patel NA, Sander M. A dosage-dependent requirement for Sox9 in pancreatic endocrine cell formation. Dev Biol. 2008;323(1):19–30. doi: 10.1016/j.ydbio.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52(1):102–10. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 77.Pan FC, Bankaitis ED, Boyer D, Xu X, Van de Casteele M, Magnuson MA, Heimberg H, Wright CV. Spatiotemporal patterns of multipotentiality in Ptf1a-expressing cells during pancreas organogenesis and injury-induced facultative restoration. Development. 2013;140(4):751–64. doi: 10.1242/dev.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fukuda A, Kawaguchi Y, Furuyama K, Kodama S, Horiguchi M, Kuhara T, Koizumi M, Boyer DF, Fujimoto K, Doi R, Kageyama R, Wright CV, Chiba T. Ectopic pancreas formation in Hes1 -knockout mice reveals plasticity of endodermal progenitors of the gut, bile duct, and pancreas. J Clin Invest. 2006;116(6):1484–93. doi: 10.1172/JCI27704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Desai BM, Oliver-Krasinski J, De Leon DD, Farzad C, Hong N, Leach SD, Stoffers DA. Preexisting pancreatic acinar cells contribute to acinar cell, but not islet beta cell, regeneration. J Clin Invest. 2007;117(4):971–7. doi: 10.1172/JCI29988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hosokawa S, Furuyama K, Horiguchi M, Aoyama Y, Tsuboi K, Sakikubo M, Goto T, Hirata K, Tanabe W, Nakano Y, Akiyama H, Kageyama R, Uemoto S, Kawaguchi Y. Impact of Sox9 dosage and Hes1-mediated Notch signaling in controlling the plasticity of adult pancreatic duct cells in mice. Sci Rep. 2015;5:8518. doi: 10.1038/srep08518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawaguchi Y. Sox9 and programming of liver and pancreatic progenitors. J Clin Invest. 2013;123(5):1881–6. doi: 10.1172/JCI66022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jin L, Feng T, Shih HP, Zerda R, Luo A, Hsu J, Mahdavi A, Sander M, Tirrell DA, Riggs AD, Ku HT. Colony-forming cells in the adult mouse pancreas are expandable in Matrigel and form endocrine/acinar colonies in laminin hydrogel. Proc Natl Acad Sci U S A. 2013;110(10):3907–12. doi: 10.1073/pnas.1301889110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jin L, Feng T, Zerda R, Chen CC, Riggs AD, Ku HT. In vitro multilineage differentiation and self-renewal of single pancreatic colony-forming cells from adult C57BL/6 mice. Stem Cells Dev. 2014;23(8):899–909. doi: 10.1089/scd.2013.0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429(6987):41–6. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 85.Bonner-Weir S, Inada A, Yatoh S, Li WC, Aye T, Toschi E, Sharma A. Transdifferentiation of pancreatic ductal cells to endocrine beta-cells. Biochem Soc Trans. 2008;36(Pt 3):353–6. doi: 10.1042/BST0360353. [DOI] [PubMed] [Google Scholar]
- 86.Inada A, Nienaber C, Katsuta H, Fujitani Y, Levine J, Morita R, Sharma A, Bonner-Weir S. Carbonic anhydrase II-positive pancreatic cells are progenitors for both endocrine and exocrine pancreas after birth. Proc Natl Acad Sci U S A. 2008;105(50):19915–9. doi: 10.1073/pnas.0805803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu X, D’Hoker J, Stange G, Bonne S, De Leu N, Xiao X, Van de Casteele M, Mellitzer G, Ling Z, Pipeleers D, Bouwens L, Scharfmann R, Gradwohl G, Heimberg H. Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell. 2008;132(2):197–207. doi: 10.1016/j.cell.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 88.Zhang M, Lin Q, Qi T, Wang T, Chen CC, Riggs AD, Zeng D. Growth factors and medium hyperglycemia induce Sox9+ ductal cell differentiation into beta cells in mice with reversal of diabetes. Proc Natl Acad Sci U S A. 2016;113(3):650–5. doi: 10.1073/pnas.1524200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Criscimanna A, Speicher JA, Houshmand G, Shiota C, Prasadan K, Ji B, Logsdon CD, Gittes GK, Esni F. Duct cells contribute to regeneration of endocrine and acinar cells following pancreatic damage in adult mice. Gastroenterology. 2011;141(4):1451–62. 1462 e1–6. doi: 10.1053/j.gastro.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Banga A, Akinci E, Greder LV, Dutton JR, Slack JM. In vivo reprogramming of Sox9+ cells in the liver to insulin-secreting ducts. Proc Natl Acad Sci U S A. 2012;109(38):15336–41. doi: 10.1073/pnas.1201701109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu EE, Krentz NA, Tan S, Chow SZ, Tang M, Nian C, Lynn FC. SOX4 cooperates with neurogenin 3 to regulate endocrine pancreas formation in mouse models. Diabetologia. 2015;58(5):1013–23. doi: 10.1007/s00125-015-3507-x. [DOI] [PubMed] [Google Scholar]
- 92.Goldsworthy M, Hugill A, Freeman H, Horner E, Shimomura K, Bogani D, Pieles G, Mijat V, Arkell R, Bhattacharya S, Ashcroft FM, Cox RD. Role of the transcription factor sox4 in insulin secretion and impaired glucose tolerance. Diabetes. 2008;57(8):2234–44. doi: 10.2337/db07-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mavropoulos A, Devos N, Biemar F, Zecchin E, Argenton F, Edlund H, Motte P, Martial JA, Peers B. sox4b is a key player of pancreatic alpha cell differentiation in zebrafish. Dev Biol. 2005;285(1):211–23. doi: 10.1016/j.ydbio.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 94.Hesselson D, Anderson RM, Beinat M, Stainier DY. Distinct populations of quiescent and proliferative pancreatic beta-cells identified by HOTcre mediated labeling. Proc Natl Acad Sci U S A. 2009;106(35):14896–901. doi: 10.1073/pnas.0906348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. A primer for morpholino use in zebrafish. Zebrafish. 2009;6(1):69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, DeSantis DF, Sheppard-Tindell S, Ebarasi L, Betsholtz C, Schulte-Merker S, Wolfe SA, Lawson ND. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32(1):97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McDonald E, Krishnamurthy M, Goodyer CG, Wang R. The emerging role of SOX transcription factors in pancreatic endocrine cell development and function. Stem Cells Dev. 2009;18(10):1379–88. doi: 10.1089/scd.2009.0240. [DOI] [PubMed] [Google Scholar]
- 98.Hoser M, Potzner MR, Koch JM, Bosl MR, Wegner M, Sock E. Sox12 deletion in the mouse reveals nonreciprocal redundancy with the related Sox4 and Sox11 transcription factors. Mol Cell Biol. 2008;28(15):4675–87. doi: 10.1128/MCB.00338-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sock E, Rettig SD, Enderich J, Bosl MR, Tamm ER, Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol Cell Biol. 2004;24(15):6635–44. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.White MG, Al-Turaifi HR, Holliman GN, Aldibbiat A, Mahmoud A, Shaw JA. Pluripotency-associated stem cell marker expression in proliferative cell cultures derived from adult human pancreas. J Endocrinol. 2011;211(2):169–76. doi: 10.1530/JOE-11-0123. [DOI] [PubMed] [Google Scholar]
- 101.Zhao M, Amiel SA, Christie MR, Muiesan P, Srinivasan P, Littlejohn W, Rela M, Arno M, Heaton N, Huang GC. Evidence for the presence of stem cell-like progenitor cells in human adult pancreas. J Endocrinol. 2007;195(3):407–14. doi: 10.1677/JOE-07-0436. [DOI] [PubMed] [Google Scholar]
- 102.Iguchi H, Ikeda Y, Okamura M, Tanaka T, Urashima Y, Ohguchi H, Takayasu S, Kojima N, Iwasaki S, Ohashi R, Jiang S, Hasegawa G, Ioka RX, Magoori K, Sumi K, Maejima T, Uchida A, Naito M, Osborne TF, Yanagisawa M, Yamamoto TT, Kodama T, Sakai J. SOX6 attenuates glucose-stimulated insulin secretion by repressing PDX1 transcriptional activity and is down-regulated in hyperinsulinemic obese mice. J Biol Chem. 2005;280(45):37669–80. doi: 10.1074/jbc.M505392200. [DOI] [PubMed] [Google Scholar]
- 103.Iguchi H, Urashima Y, Inagaki Y, Ikeda Y, Okamura M, Tanaka T, Uchida A, Yamamoto TT, Kodama T, Sakai J. SOX6 suppresses cyclin D1 promoter activity by interacting with beta-catenin and histone deacetylase 1, and its down-regulation induces pancreatic beta-cell proliferation. J Biol Chem. 2007;282(26):19052–61. doi: 10.1074/jbc.M700460200. [DOI] [PubMed] [Google Scholar]
- 104.Kasimiotis H, Myers MA, Argentaro A, Mertin S, Fida S, Ferraro T, Olsson J, Rowley MJ, Harley VR. Sex-determining region Y-related protein SOX13 is a diabetes autoantigen expressed in pancreatic islets. Diabetes. 2000;49(4):555–61. doi: 10.2337/diabetes.49.4.555. [DOI] [PubMed] [Google Scholar]
- 105.Wang Y, Ristevski S, Harley VR. SOX13 exhibits a distinct spatial and temporal expression pattern during chondrogenesis, neurogenesis, and limb development. J Histochem Cytochem. 2006;54(12):1327–33. doi: 10.1369/jhc.6A6923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jonatan D, Spence JR, Method AM, Kofron M, Sinagoga K, Haataja L, Arvan P, Deutsch GH, Wells JM. Sox17 regulates insulin secretion in the normal and pathologic mouse beta cell. PLoS One. 2014;9(8):e104675. doi: 10.1371/journal.pone.0104675. [DOI] [PMC free article] [PubMed] [Google Scholar]