Abstract
Targeted delivery is a key element for improving the efficiency and safety of non-viral vectors for gene therapy. We have recently developed a CD44 receptor targeted, hyaluronic acid-adamantamethamidyl based pendant polymer system (HA-Ad), capable of forming complexes with cationic β-cyclodextrins (CD-PEI+) and pDNA. Complexes formed using these compounds (HA-Ad:CD-PEI+:pDNA) display high water solubility, good transfection efficiency, and low cytotoxicity. Spatial and dynamic tracking of the transfection complexes by confocal microscopy and multicolor flow cytometry techniques was used to evaluate the target specificity, subcellular localization, and endosomal escape process. Our data shows that cells expressing the CD44 receptor undergo enhanced cellular uptake and transfection efficiency with HA-Ad:CD PEI+:pDNA complexes. This transfection system, comprised of non-covalent assembly of cyclodextrin:adamantamethamidyl-modified hyaluronic acid via host:guest interactions to condense pDNA, is a potentially useful tool for targeted delivery of nucleic acid therapeutics.
Keywords: CD44 receptor, Hyaluronic acid conjugate, pDNA transfection, cationic β-cyclodextrin
Graphical abstract
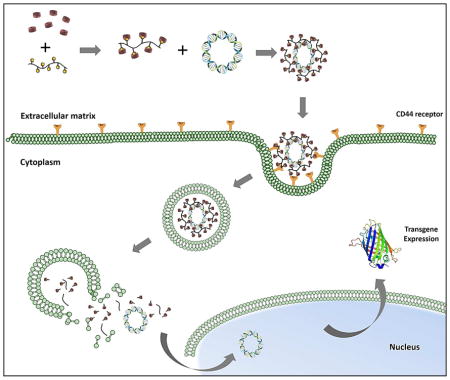
Conceptual diagram HA-Ad:CD-PEI+:pDNA assembly, CD44 receptor-mediated cellular binding, internalization, trafficking, and disassembly within the endosomal compartments of target cells, with subsequent transgene expression of GFP.
INTRODUCTION
Nucleic acid delivery using non-viral vectors against various cancer targets is an attractive therapeutic approach due to its low pathogenicity and industrial scale manufacturability. Unfortunately, low efficacy and off target effects still limit the widespread clinical application of this strategy 1, 2. Targeted nucleic acid delivery is an additional challenge to be met by non-viral vectors 3–5. Although various targeting strategies have been explored, low efficacy is most often due to inefficient internalization, poor protection of the nucleic acid cargo and/or inefficient release of the nucleic acids into the cell 6.
Among various targeting elements, hyaluronic acid (HA) is reported to be a highly effective ligand 7–9. Hyaluronan is a high molecular weight (106 Da) glycosaminoglycan that is a major component of extracellular matrix. It is naturally synthesized by a class of integral membrane proteins called hyaluronan synthases and degraded by a family of mammalian enzymes called hyaluronidases that are found in various forms throughout the body 10. HA plays essential role in proper cell growth, structural stability of organs, and tissue organization. Many studies have reported high concentrations of hyaluronan in regions of high cell division and invasion (wound repair, embryonic morphogenesis, inflammation, and cancer), thus suggesting it plays an important role in tumorigenesis. 7, 8, 11, 12
One of the HA receptors, CD44, belongs to a family of cell adhesion molecules (CAMs)13. It is a widely-distributed transmembrane glycoprotein that plays a critical role in malignant cell activities, including adhesion, migration, invasion, and survival; it is also strongly implicated in the cell signaling cascades associated with cancer initiation and progression. CD44 is a key component in the internalization and metabolism of HA and is endogenously expressed at low levels on various cell types in normal tissues 14. Tumor-derived cells, however, express CD44 in a high-affinity state that is capable of promoting the binding and internalization of HA. Mechanistic studies reveal that CD44 regulates lymphocyte adhesion to cells of the endothelial venules during lymphocyte migration, a process that displays many similarities to the metastatic distribution of solid tumors 15. Recent reports have suggested that CD44 is a breast cancer stem cell marker 16–18. Owing to its overexpression on a variety of cancer cells and macrophages and its interaction with HA, the presence of CD44 receptors on rapidly dividing cells makes HA an attractive choice for active targeting of nucleic acid therapeutics to CD44+ tumor cells 9, 16.
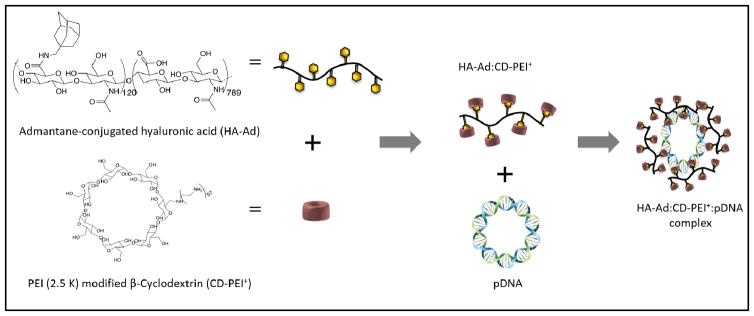
HA is an intriguing building block for development of novel delivery systems because it is biodegradable, nontoxic, hydrophilic, and non-immunogenic biopolymer that only requires a small block of 6–8 saccharide units to interact with CD44. These features also suggest that the intrinsic affinity of HA for CD44 may make it a useful interaction for targeting HA-derived nanoparticles to CD44+ tumor cells 19, 20. Based on these attributes and prior efforts to create HA-based carrier systems 21, we developed a new hyaluronic acid (HA)-based pendant polymer system capable of forming complexes with cationic β-cyclodextrins and pDNA. HA (350 kDa) conjugated with aminomethyladamantane (HA-Ad) forms a pendant polymer system, wherein the hydrophobic guest ligand, adamantane, interacts with the hydrophobic cavity of cationic β-CD to form a strong guest:host interaction that promotes self–assembly of the non-viral pendant polymer vector (Figure 1). We believe this design provides advantages such as biocleavable, biodegradability and high solubility due to use of HA as polymer backbone. Additionally, β-CD complexation provides additional flexibility to electrostatically interact with pDNA, resulting in stable nanoparticles.
Figure 1.
Chemical structures of (left) HA-Ad and (right) CD-PEI
Modification of β-CD with 2.5kD poly(ethyleneimine) (CD-PEI+) on the 1° rim of the cyclodextrin unit enables the formation of stable transfection complexes with negatively charged nucleic acid cargo. HA-Ad:CD-PEI+:pDNA complexes display high water solubility, efficient transfection, and good cell viability; however, the target specificity and mechanistic details of the transfection process with these polyplexes has not been reported22, 23. This study describes the use of confocal microscopy and multicolor flow cytometry tools to provide new spatial and dynamic tracking information that may aid the design of transfection complexes targeted to CD44+ cells.
MATERIALS AND METHODS
Reagents
All solvents were reagent grade, purchased from commercial sources and used without further purification, except DMF and toluene, which were dried over CaH2 under N2, filtered, and distilled under reduced pressure. β-CD and rabbit anti-HYAL1 (ab1) antibody was obtained from Sigma-Aldrich. PEI (2.5kD) was purchased from Polysciences, Inc. Dialysis membranes (MWCO: 6000–8000) were purchased from Fisher Scientific. 1H NMR spectra were recorded on a 300 MHz Varian INOVA 300 spectrometer at 30 °C. Chemical shifts were referenced to the residual protonated solvent peak. Ultra-pure H2O (resistivity ≈ 18.0 MΩ/cm−1) was generated using a NANOpure Ultrapure H2O system. AcGFP1 plasmid vector and MTS reagent were purchased from PromegaR. pDNA was amplified in E. coli and purified according to the supplier’s protocol (Qiagen, Hilden, Germany). HCAM siRNA and HYAL1 siRNA (h) was purchased from Santa Cruz Biotechnology. Secondary antibody IRDye® 680 RD anti-Rabbit IgG (H+L) and IRDye® 800 RD anti-mouse IgG (H+L) were purchased from LI-COR. Anti-β-actin mouse antibody was purchased from Cell Signaling. Mouse anti-human CD44 antibody-BV421 (anti-CD44-BV421) was purchased from BD Biosciences. The concentration and purity of the pDNA was determined by absorption at 260 and 280 nm and by agarose gel electrophoresis, respectively. The purified pDNA was re-suspended in TE buffer (10 mM Tris- Cl, pH 7.5, 1 mM EDTA) and kept frozen in aliquots at a concentration of 0.5 mg/mL. LipofectamineR2000 (L2K) transfection reagent, LysoTrackerRRed DND-99, and Alexa 680- Wheat Germ Agglutinin conjugate (Alexa680-WGA) were obtained from Life Technologies. Fluorescein Label ITR nucleic acid labeling kit was obtained from Mirus. Cell culture reagents such as DMEM and RPMI media, FBS, sodium pyruvate, trypsin, and PBS were obtained from Atlanta Biologicals.
Synthesis of 1-Methaminoadamantane-modified Hyaluronic Acid Pendant Polymer (HA-Ad)
Hyaluronic acid-based pendant polymer was synthesized via EDC-mediated coupling of 350 kD HA and adamantane methylamine as previously reported 21. (ESI†)
Synthesis of CD-PEI+
Poly(ethyleneimine) (2.5 kDa) was used to introduce a single modification on the β-cyclodextrin as previously described 21, 24.
Formulation of HA-Ad:CD-PEI+:pDNA Complexes
HA-Ad pendant polymers solution (1 mM equivalent of Ad groups) were vortex mixed with an equal volume of β-CD-PEI+ (1mM equivalent of PEI) solution in water to form HA-Ad:β-CD-PEI+ complexes. Transfection complexes were formed by adding the appropriate amount of HA-Ad:β-CD-PEI+ complexes to 1 μg of pDNA (unmodified or FITC-labelled) dissolved in 30 μL of TE buffer in 1.5 mL centrifuge tubes. The amount of HA-Ad:β-CD-PEI+ added to DNA varied from 1–3 μL so as to achieve final N/P ratios of 5, 10, 20 & 30. The above solution was incubated at 4 °C for 1 h before use in transfection experiments.
Particle Size and Zeta (ζ) Potential Measurements
The diameters, size distributions, and ζ-potentials of the materials were evaluated by dynamic light scattering (DLS) using a Zetasizer Nano S (Malvern Instruments Ltd.) at 20 °C with a scattering angle of 900. At least 40 measurements were made and averaged for each sample. The particles were diluted to 1 mL with HEPES buffer prior to analysis.
Gel Retardation Assay
The complexation ability of HA-Ad:CD-PEI+ with pDNA was determined by low melting point 1% agarose gel electrophoresis. The agarose gels were precast in TBE buffer with GelRed® dye at 1:10,000 dilution. HA-Ad:CD-PEI+:pDNA complexes containing 0.2 μg of pDNA at different N/P ratios were loaded onto the gel before addition of loading dye (1:5 dilution) to each well and electrophoresis at a constant voltage of 55 V for 2 h in TBE buffer. The pDNA bands were then visualized under a UV transilluminator at λ = 365 nm.
Cellular Uptake Studies
CD44+ HeLa, CD44− HeLa, and NIH 3T3 cells were used to study the uptake of the complexes by plating 75,000 cells per well in 24-well plates and incubating for 24 h before the experiment. HA-Ad:CD-PEI+:pDNA-FITC (1 μg per well) complexes at different N/P ratios were incubated with cells for 4 h at 37 °C in serum free DMEM media. After 4 h, the cells were stained with anti-CD44-BV421 at a concentration of 1 μg/mL for 30 min. After incubation, the spent media was removed, the cells washed with PBS (3X), and trypsinized. The cells were then collected and analyzed for cellular localization of HA-Ad:CD-PEI+:pDNA-FITC and CD44 expression level using anti-CD44-BV421 by flow cytometry using a BD FACS Aria III. Statistical analysis for the biological studies was conducted using R software (http://cran.r-project. org). Data are reported as absolute values or as quantities relative to control values. Normality was evaluated for each variable using the Shapiro-Wilk test. Data with normal distribution were compared using ANOVA. Bonferroni was used as post-hoc analysis.
Free HA Competition Assay
To further validate CD44 receptor-mediated cellular uptake of HA-Ad:CD-PEI+:pDNA in CD44+ HeLa, a cellular uptake experiment in presence of free HA was carried out. HA (5 mg/mL) was added to the media 2 h before addition of the particles. The particles were added to the HA containing transfection media and were incubated for an additional 4 h25. After incubation, the spent media was removed, the cells washed with PBS (3X), trypsinized and analyzed using a FC 500.
In Vitro Transfection Assay
CD44+ HeLa, CD44− HeLa and NIH 3T3 cells were cultured in complete DMEM medium with 10% FBS. All cells were incubated at 37 °C, 5% CO2, and 95% relative humidity, respectively, at a cell density of 75,000 cells/well in 24-well plates. After 24 h, the culture media was replaced with serum free media and HA-Ad:CD-PEI+:pDNA complexes containing 1 μg of AcGFP added at N/P ratios of 5, 10, 20 and 30. The cells were incubated with the complexes for 4 h, after which the spent media was aspirated and fresh serum-supplemented media was added. After a total of 36 h incubation, the media was aspirated and the cells were washed with PBS, trypsinized, and analyzed by flow cytometry using a FC500. The % GFP mean fluorescence intensity was calculated relative to L2k controls, considered as 100%.
Western Blot Analysis of Cell Lysate
HeLa- Hyal1 knockdown experiments were performed using HYAL1 siRNA (h) (Santa Cruz biotechnology) and Lipofectamine 2000 following the standard protocol suggested by the vendor. For Western blot, untreated HeLa and HeLa- Hyal1 knockdown cells were grown in 6 well plates to a density of 1.5–2 × 106 cells/mL before washing in ice-cold PBS and lysing by resuspension in a 10× pellet volume of RIPA lysis buffer containing 50 mM Tris-HCl, 150 mM NaCl, 0.25% deoxycholate, 1% NP-40, and 1mM EDTA, plus 0.1% SDS and protease inhibitor tablets. Lysates were cleared by centrifugation at 4 °C using a bench top centrifuge at 11,000 rpm for 10 min. Protein concentration was determined using a NanoDrop2000 at 280 nm. Aliquots (4 μg) of each lysate was resolved using SDS gels (10 % Acrylamide) transferred to PVDF and probed with the following antibodies in Odyssey blocking buffer: rabbit anti-HYAL1 (ab1) antibody (1:1000) and mouse anti-β-actin antibody (1:15000). Following primary Ab incubation, membranes were probed with IRDyeR 680RD Goat anti-Rabbit IgG (1: 15000) or IRDyeR 800 RD Goat anti-mouse IgG (1:15000) and imaged using the LiCOR Odyssey system.
In vitro Cell Viability Assay
Cell viability of HA-Ad:CD-PEI+:pDNA complexes in HeLa cells relative to Lipofectamine 2000 and untreated cells as positive and negative controls, respectively, was performed using the MTS assay. Cell viabilities were measured as a function of HA-Ad:CD-PEI +:pDNA complex N/P ratio. For the assay, HeLa cells were cultured overnight at a density of 10,000 cells per well in complete DMEM medium supplemented with 10% FBS at 37 °C, 5% CO2, and 95% relative humidity in 96 well microtiter plates (Greiner, 96-well multiwall plates-flat bottom). After 24 h, the cells were washed with PBS and the HA-Ad:CD-PEI+:pDNA complexes in serum free DMEM media were added for 24 h before addition of MTS reagent (15μL) and incubation for 2 h. The absorbance was measured at a wavelength of 492 nm using a multimode plate reader (Beckman Coulter, DTX880). The cell viability (%) relative to control cells cultured in media without any transfection agent was calculated as [A]test/[A]control × 100%, where [A]test was the absorbance of the wells with PR+ and [A]control was the absorbance of the control wells. All cytotoxicity values were measured in sextuplicate and averaged.
Intracellular Trafficking Analysis by Confocal Laser Scanning Microscopy (CLSM)
Confocal microscopy was used to study the intracellular traffcking of HA-Ad:CD-PEI+:pDNA complexes. CD44+ HeLa cells were cultured in complete DMEM medium containing 10% FBS. All cells were incubated at 37 °C, 5% CO2, and 95% relative humidity at a cell density of 40,000 cells/well in 4-chamber confocal slides. After 24 h, the culture media was replaced with DMEM media containing HA-Ad:CD-PEI+:pDNA-FITC complexes at N/P ratios of 20. The cells were incubated with the complexes for a maximum of 4 h before replacing the medium with serum-supplemented DMEM media. At specified time points, the media was aspirated and cells were washed with PBS (3×), after which they were stained with Alexa680-WGA and Lysotracker Red DND. After the last wash, 500 μL of PBS containing 5 μL of Alexa680-WGA stock solution was added to cells and incubated at 37 °C for 10 min. After incubation, the dye was removed by PBS washing (3X), followed by addition of 500 μL of PBS containing 5 μL of LysoTracker Red DND stock solution and again incubated at 37 °C for 10 min. After cell staining was complete, the cells were washed with PBS (3×) before addition of fresh PBS (500 μL) and collection of confocal microscope images using a Nikon A1R MP Multiphoton microscope. Images were recorded at 60× magnification using an oil immersion objective.
RESULTS AND DISCUSSION
HA-Ad:CD-PEI+:pDNA complexes prepared by flow mixing have been reported as effective transfection agents 21; however, their use in a receptor-mediated endocytosis approach for targeted delivery to CD44+ cells has not been described. In this study, we have compared the differences in cellular uptake, localization, and transfection of CD44+ and CD44− cells lines treated with HA-Ad:CD-PEI+:pDNA complexes. Three different cell lines, CD44+ HeLa, CD44− HeLa, and NIH 3T3 fibroblasts lacking functional CD44 hyaluronan receptors 26–28, were chosen for comparison. We hypothesized that HA-Ad:CD-PEI+:pDNA complexes would be internalized and efficiently transfect only the CD44+ cancer cell line. Comparison of the uptake and trafficking of the complexes in CD44+ versus CD44− cells would also aid in understanding the target specific internalization of these particles.
Physical Characterization of HA-Ad:CD-PEI+:pDNA Complexes
The abilities of the HA-Ad: CD-PEI+ pendant polymer assembly to form pDNA complexes was initially evaluated by analyzing their particle diameters and surface charges at different N/P ratios. Dynamic light scattering (DLS) was used to determine the number-averaged diameters produced using the different HA-Ad:CD-PEI+:pDNA complexes. Our data shows that the HA-Ad:CD-PEI+ pendant polymers formed complexes with diameters between 180 – 350 nm. At low N/P ratios (e.g., 5), complexes in the 300 – 400 nm size regime were produced; however, the observed diameters dropped gradually, below 200 nm when N/P ≥ 30, showing no appreciable change in diameter as the N/P ratio increased further (Figure S1-A). This is in agreement with similar observations of bPEI:pDNA complexes wherein the particle diameters become smaller when N/P ≥ 5. The PDIs for all the complexes were in the range of 0.3–0.4.
The ζ-potentials of the HA-Ad:CD-PEI+:pDNA vectors were measured to determine the extent of charge development on the HA-Ad:CD-PEI+:pDNA transfection complexes (Figure S2-B). We found that all the complexes produced an increase in the observed ζ-potential as the N/P ratio increased, with ζ-potentials observed in the range of + 20 to 40 mV. It was also observed that increasing the N/P ratio affected the ζ-potential significantly, such that the HA-Ad: CD-PEI+ complexes displayed ζ-potentials between 30 – 35 mV at N/P = 30. In general, ζ-potentials above 25 mV can be considered as an indication of a stable nanoparticle dispersion, since higher charge causes repulsive interactions between the particles, thereby preventing their aggregation 29, 30. For biological studies, a positive nanoparticle complex ζ-potential typically promotes efficient in vitro cellular uptake; however, it also adversely affects their in vivo performance due to non-specific uptake, macrophage clearance, and mononuclear phagocytic system opsonization. Consequently, the formation of transfection complexes with modest postive ζ-potentials may contribute to their improved in vivo performance.
Complexation Properties and Colloidal Stability of HA-Ad:CD-PEI+:pDNA Complexes
The complexation properties and colloidal stabilities of the HA-Ad:CD-PEI+:pDNA complexes were determined by gel retardation assays31. Our data show that the complexation ability of HA-Ad: CD-PEI+ with pDNA increases with increasing N/P ratio. Although the complexes displayed modest condensation ability at N/P = 5, it improved significantly at N/P ratios of 10, 20 and 30, where pDNA release was almost insignificant after ≥10 (Figure S2).
Analysis of CD44 Expression
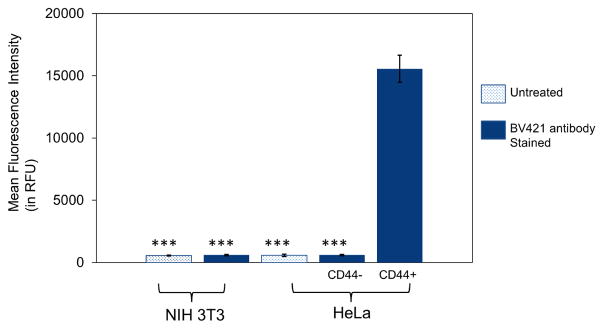
In order to determine the CD44 status of the cells, the HeLa and NIH3T3 cell lines were stained with anti-CD44-BV421 and analyzed using flow cytometry. Significant differences in CD44 expression levels were observed, with ~16,000 times higher mean fluorescence intensity (MFI) found for CD44+ HeLa relative to the MFI levels observed for CD44− HeLa and NIH 3T3 cells (Figure 2 & S3). These results were in accordance with literature precedent suggesting that there was an overexpression of CD44 receptors in CD44+ HeLa and very low expression levels in CD44− HeLa and NIH 3T3 cells28,32–34.
Figure 2.
Flow cytometry analysis using anti-CD44-BV421 antibody labeling to determine the CD44 receptor expression levels in CD44+ HeLa cells, CD44− HeLa cells, and CD44− NIH 3T3 fibroblasts. The CD44 receptor expression levels were determined by plating 75,000 cells per well in 24-well plates and incubating for 24 h before the experiment. Cells were then stained with CD44 antibody (mouse anti-human-CD44 antibody labeled with BV421) at a concentration of 1μg/mL in DMEM media for 30 min. After incubation, the spent media was removed and the cells were washed 3 times with PBS before trypsinization. The cells were then collected and analyzed using a BD FACS Aria III flow cytometer. Data points represent group mean ± SD (n = 6; ***P < 0.005; ANOVA).
Cellular Uptake
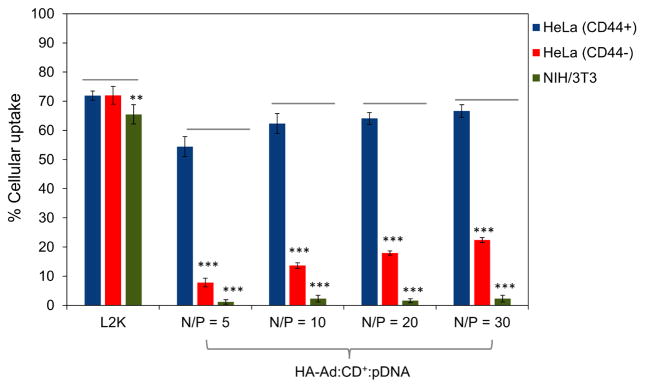
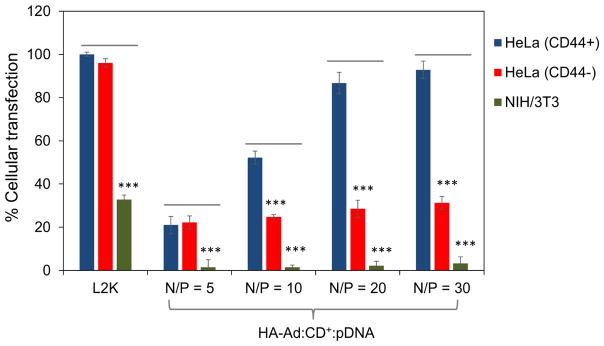
Cellular uptake of polyplexes was performed simultaneously in all three cell lines and analyzed by flow cytometry using FITC-labeled pDNA (HA-Ad:CD-PEI+:pDNA-FITC) and anti-CD44-BV421 to probe for the presence of CD44 on the cell surface. Untreated, non-labeled cells were used as a negative control and L2K:pDNA-treated cells were used as a positive, untargeted control. L2K complexes showed cellular uptake equally well in all three cell lines, regardless of CD44 expression. In the case of HA-Ad:CD-PEI+:pDNA-FITC particles; however, significantly higher levels of transfection complex uptake were observed for CD44+ HeLa cells relative to CD44− HeLa cells (3 – 7 fold increase, depending on N/P ratio). Moreover, CD44+ HeLa cells exhibited a 40-fold increase in uptake relative to NIH 3T3 fibroblasts (Figures 3 and S4). Overall, HA complexes showed good cellular association with HeLa CD44+, where the maximum uptake efficiency (at N/P=30) was comparable to L2K complexes (~70%). It is interesting to note that cellular uptake was found to gradually increase with increasing N/P ratios for both HeLa CD44+ and HeLa CD44−; however a saturation effect was observed beyond N/P of 20. We attribute the gradual increase in cellular uptake to two factors in the case of CD44+ HeLa. The higher N/P ratio produces a higher polymer loading in the transfection complexes, leading to the more efficient binding of pDNA complexes to CD44+ HeLa cells at high N/P due to increased receptor-specific binding of the HA-Ad:CD-PEI+:pDNA particles. Furthermore, the higher positive charge resulting from higher N/P ratios (Figure S1) likely produces a greater number of particles that associate with the cells via non-specific electrostatic interactions with the negatively-charged plasma membrane, thereby contributing to increased pDNA uptake (Figure S2).
Figure 3.
Cellular uptake of HA-Ad:CD-PEI+:pDNA and L2K:pDNA complexes after incubation for 4 h with HeLa and NIH 3T3 cells. CD44+ HeLa, CD44− HeLa, and NIH 3T3 cells were used to study the uptake of the complexes by plating 75,000 cells per well in 24-well plates and incubating for 24 h before the experiment. HA-Ad:CD-PEI+:pDNA-FITC (1 μg per well) complexes at different N/P ratios were incubated with cells for 4 h at 37 °C in serum free DMEM media. After 4 h, the cells were stained with anti-CD44-BV421 (BD Biosciences) at a concentration of 1 μg/mL for 30 min. After incubation, the spent media was removed, the cells washed with PBS (3X), and trypsinized. The cells were then collected and analyzed by flow cytometry for the expression of CD44 and FITC-labelled complexes. Data points are expressed as a percentage of total parent population of cells and represent group mean ± SD (n = 6; ***P < 0.005; ANOVA).
Nonetheless, both CD44+ and CD44− HeLa cells showed modest (~15%) increases in uptake with increased N/P ratio. We infer from these findings that N/P ratio played a minor role in guiding the cellular uptake of these particles. Furthermore, the recycling rate for CD44 is known to be slow (1–2 d), therefore, the modest increases in uptake observed at high N/P ratios are likely due to the slow internalization kinetics of these CD44 receptor-HA ligand complexes 35, 36. No appreciable localization of HA-Ad:CD-PEI+:pDNA-FITC was observed in case of NIH 3T3 fibroblasts, an observation that we attribute to their low abundance of CD44 receptors and the widely reported finding that fibroblast cells are difficult to transfect with polymeric transfection agents 37.
Hyaluronic Acid Competition Assay
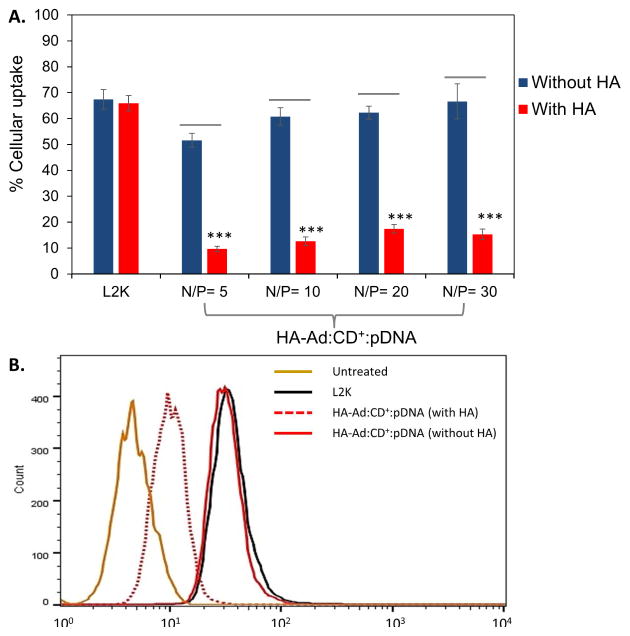
CD44 receptor-mediated binding and uptake of HA-Ad: CD-PEI+:pDNA complexes was further probed by competition assay using excess free HA ligand in the culture media (Figure 4). The uptake efficiency of HA-Ad:CD-PEI+:pDNA was reduced by almost 75% in CD44+ HeLa cell cultures containing 5 mg/mL additional free HA relative to cells incubated in the absence of free HA. Uptake of L2K:pDNA complexes was not affected by the presence of excess HA. Receptor-mediated endocytosis is reported to be a major mechanism of cellular uptake for HA-based nanoparticles 38, 39, therefore, the enhanced cellular uptake of HA-Ad:CD-PEI+:pDNA complexes, and significant reduction in presence of free HA, is consistent with the specificity and competition for CD44 receptors on these cells 25.
Figure 4.
A: Cellular uptake of HA-Ad:CD-PEI+:pDNA and L2K:pDNA complexes in the absence and presence of free HA at different N/P ratios. B: Flow cytometry data for cellular uptake of L2K:pDNA and HA-Ad:CD-PEI+:pDNA complexes at N/P = 30 in the presence and absence of free HA. To further validate CD44 receptor-mediated cellular uptake of HA-Ad: CD-PEI+:pDNA in HeLa (CD44+), a cellular uptake experiment in the presence of free HA was carried out. HA (10 mg/mL) was added to the media 2 h before addition of the particles. The particles were added to the HA containing transfection media and were incubated for an additional 4 h. After incubation, the spent media was removed, the cells washed with PBS (3X), trypsinized and analyzed for using a FC500 (Beckman Coulter). Data points are expressed as a percentage of total parent population of cells and represent group mean ± SD (n = 6; ***P < 0.005; ANOVA).
Transfection Efficiency
GFP expression in HeLa cells transfected with L2K complexes was not dependent on CD44 expression. Conversely, HA particles displayed a transfection performance that was strongly influenced by the CD44 status of the cells (Figures 5 & S5). High efficiencies were observed in CD44+ HeLa cells treated at higher N/P ratios (e.g., ~90% at N/P = 30). The complexes formed at N/P of 5 showed very poor transfection (~ 20 %), which can be attributed to poor stability as shown by their relative agarose gel shift assay behavior (Figure S2). Even though the complexes formed at N/P = 5 localize on the cell membrane surface, their lower colloidal stability during binding and uptake may result in the loss of particle morphology and pDNA retention, such that they are not internalized efficiently. It is noteworthy that cells treated at N/P of 20 or 30 were about 40% more efficient than treatment with N/P = 10 complexes, even though their cellular association levels were very similar at all three N/P ratios (Figure 3). It has been reported that once the polyplexes are internalized, the transfection efficiency is typically dependent on the cargo release rate, i.e., the disassembly/degradation rate of the complexes within the endo/lysosomal compartment 40, 41. For the HA-Ad:CD-PEI+:pDNA complexes, we infer from our findings that disassembly occurs due to catalytic HA degradation by the hyaluronidase-1 (Hyal1) enzyme present in the endosomal and lysosomal compartments.42, 43 Since all the HA-Ad:CD-PEI+:pDNA complexes have HA as the core main chain polymer element, they can all be readily degraded by the Hyal1, regardless of N/P ratios at which they were formulated. Consequently, complexes formed at higher N/P showed higher transfection efficiencies due to their higher degrees of pDNA association with the CD44+ cells.
Figure 5.
Flow cytometry analysis of in vitro transfection efficiencies in HeLa and NIH 3T3 cells treated with HA-Ad:CD-PEI+:pDNA complexes and L2K controls (4 h incubation, followed by readout after 36 h). CD44+ HeLa, CD44− HeLa, and NIH 3T3 cells were cultured in complete DMEM medium with 10% FBS. All cells were incubated at 37 °C, 5% CO2, and 95% relative humidity, respectively, at a cell density of 75,000 cells/well in 24-well plates. After 24 h, the culture media was replaced with serum free media, with addition of HA-Ad: CD-PEI+:pDNA complexes containing 1 μg of AcGFP, at N/P ratios of 5, 10, 20 and 30. The cells were incubated with the complexes for 4 h, after which the spent media was aspirated and fresh serum-supplemented media was added. After a total of 36 h incubation, the media was aspirated and the cells were washed with PBS, trypsinized and analyzed by flow cytometry. The % GFP mean fluorescence intensity was calculated relative to L2k controls, considered as 100%. Data points represent group mean ± SD (n = 6; ***P < 0.005; ANOVA).
In the case of CD44− HeLa cells, the maximum transfection efficiency is about 20%, in accordance with their modest extent of cellular association. Similarly, transfection levels were not significant for NIH 3T3 fibroblasts, since their degree of binding and uptake was low (i.e., less than 5% at all N/P ratios).
The relative cell viability profiles of the HA-Ad:CD-PEI+:pDNA complexes and L2K:pDNA complexes were evaluated by MTS assay. HA-Ad:CD-PEI+:pDNA complexes showed high cell viability (>90%) at all N/P ratios, which was comparable or higher than L2K (Figure S6).
Role of Hyal1 in Disassembly of HA-Ad:CD-PEI+:pDNA Complexes
To investigate if Hyal1 plays any role in the catabolism of the HA carrier backbone, thus contributing to disassembly of HA-Ad:CD-PEI+:pDNA complexes, we sought to knockdown the Hyal1 levels in CD44+ HeLa using Hyal1-siRNA, and to compare their transgene expression levels with unsilenced CD44+ HeLa cells treated with HA-Ad:CD-PEI+:pDNA complexes (Figure 6). The knockdown of Hyal1 was verified by Western blot assay, which clearly showed 62 % knockdown of Hyal1 relative to control Hyal1 expression levels. It was interesting to observe that while cellular uptake of complexes was not significantly different in both cell types, the transfection efficiency of of HA-Ad: CD-PEI+:pDNA was significantly decreased by ~39 % in the absence of Hyal1 relative to the transfection complex performance in the presence of Hyal1.
Figure 6.
Suppressed Hyal 1 expression led to decreased transfection performance of HA-Ad: CD-PEI+:pDNA complexes. (A) Transfection performance of naked pDNA, pDNA complexed with L2K and HA-Ad:CD-PEI+:pDNA at N/P ratio of 30 in HeLa vs. HeLa- Hyal1 knockdown cells. The transfection efficiency was analyzed using flow cytometry, with median intensity plotted as a relative percentage of L2K expression taken as 100%. (B) Treatment with Hyal1-siRNA results in reduced Hyal1 expression. HeLa cells were treated with 50 nM of siRNA for 36 h and Hyal expression was analyzed by Western blot. Analysis of β-actin was included as a loading control and (C) images were quantified using the Odyssey Infrared Imaging System software and plotted as the relative percentage of β-actin expression.
Intracellular Trafficking of HA Complexes Using CLSM
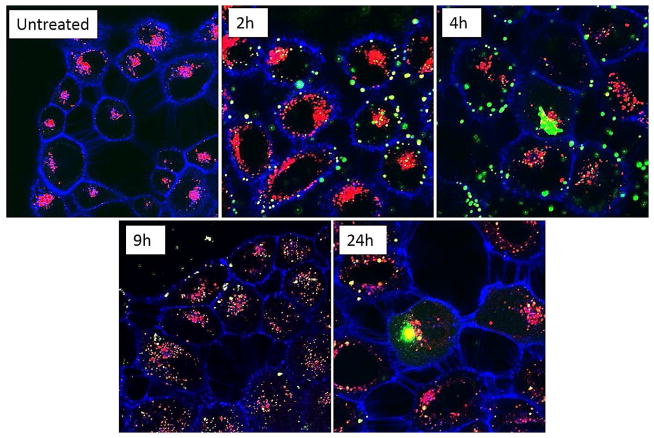
Intracellular trafficking of the HA-Ad: CD-PEI+:pDNA vehicles in CD44+ HeLa cells was monitored by confocal microscopy using Alexa680-WGA to label the cell membrane and LysotrackerR RED DND-99 to reveal the acidic endosomal and lysosomal compartments. HA-Ad:CD-PEI+:pDNA-FITC complexes were prepared to enable particle tracking within the cells at time points varying from 2 h to 24 h (Figures 7 & S6). Initial binding of the complexes occurred within the first 2 h, with subsequent internalization after 4 h and association of some HA-Ad:CD-PEI+:pDNA-FITC particles with acidic cellular compartments as described for other HA-based nanoparticle systems 39. Images collected 9 h after exposure to HA-Ad:CD-PEI+:pDNA-FITC complexes showed that the particles were fully internalized within acidic compartments that stained yellow due to colocalization of the green HA-Ad:CD-PEI+:pDNA-FITC complexes and the red Lysotracker signals. Intracellular localization of the particles was confirmed by z-stack analysis.
Figure 7.
Confocal microscopy images of HeLa cells treated with HA-Ad:CD-PEI+:pDNA complexes at N/P = 20 as a function of incubation time (indicated time, h). Top row: 60× magnification; bottom row: 100× magnification. Green: pDNA-FITC cargo in HA-Ad:CD-PEI +:pDNA complexes; Blue: plasma membrane labeled with Alexa680-WGA; Red: lysosomes labeled with Lysotracker DND-99.
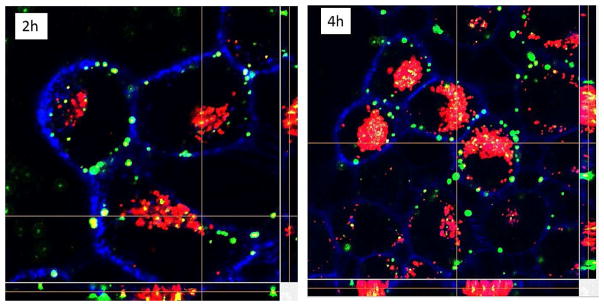
These images clearly showed that the particles were inside the cell rather than just bound to the surface and coincident with the acidic compartments (Figure 8). Images collected at later time points (e.g., 24 h in Figure S6) showed that internalized complexes present in the 9 h time frame had gradually disappeared by the 24 h time point and that no additional complexes were visible on the cell surface. We infer from these findings that the HA-Ad:CD-PEI+:pDNA particles disassemble on the 4 – 9 h time frame upon entry into the endosomal/lysosomal compartments to release the complexed pDNA throughout the cytosol, leading to dilution of the signal beyond the detection limit of CLSM as previously reported for internalized HA-based materials. 39
Figure 8.
Confocal microscopy z-stack images of HeLa cells treated with HA-Ad:CD-PEI +:pDNA complexes at N/P = 20 as a function of incubation time (indicated time, h). Green: pDNA-FITC cargo in HA-Ad:CD-PEI+:pDNA complexes; Blue: plasma membrane labeled with Alexa680-WGA; Red: lysosomes labeled with Lysotracker DND-99.
CONCLUSIONS
Our findings indicate that CD44 receptor expression has a major influence on cellular uptake and transgene expression levels of HA-Ad:CD-PEI+:pDNA transfection complexes. The HA-based complexes displayed significant target-specific cellular localization in CD44+ HeLa cells relative to CD44− HeLa and NIH 3T3 cells, regardless of N/P ratio. Transfection efficiencies of HA-Ad: CD-PEI+:pDNA complexes in CD44+ HeLa cells (>90 %) were comparable to the commercial standard, Lipofectamine2000; however, they were comparatively low in CD44− HeLa and NIH 3T3 fibroblasts. Binding of the complexes to the cell surface occurred within 2 – 4 h, with internalization within acidic endosome/lysosome compartments occurring between 4 – 9 h, leading to release of the pDNA cargo within 24 h. In conclusion, HA-based pendant polymer:CD-PEI+:pDNA complexes promote CD44 target-specific cellular uptake and efficient transgene expression in vitro. These materials may offer interesting opportunities for CD44- targeted transfections in vivo as well.
Supplementary Material
Acknowledgments
The support of NIH RO1 GM087016 and R21 CA15196 are gratefully acknowledged. The NMR, MS and flow cytometry data were acquired using facilities supported by a grant to Purdue University of Center for Cancer Research (P30 CA023168). We would like to thank Aaron Taylor and Jeffrey Woodliff for their assistance with the confocal microscopy and flow cytometry experiments, respectively.
ABBREVIATIONS
- HA
Hyaluronic acid
- Ad
Adamantane
- PEI
Polyethylenimine
- pDNA
Plasmid DNA
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Physical characterization and colloidal stability of HA-Ad:CD-PEI+:pDNA complexes as a function of N/P ratio, flow cytometry data for CD44 analysis using Brilliant Violet 421R antihuman CD44 antibody, cellular uptake of HA-Ad:CD-PEI+:pDNA and transfection analysis in HeLa and NIH 3T3 cells, confocal microscopy images of HA-Ad:CD-PEI+:pDNA complexes at N/P = 20 with incubation at various time points. The Supporting Information is available free of charge on the ACS Publications website at http://pubs.acs.org.
References
- 1.Jafari M, Soltani M, Naahidi S, Karunaratne DN, Chen P. Nonviral approach for targeted nucleic acid delivery. Curr Med Chem. 2012;19:197–208. doi: 10.2174/092986712803414141. [DOI] [PubMed] [Google Scholar]
- 2.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Cancer nanotechnology: The impact of passive and active targeting in the era of modern cancer biology. Adv Drug Del Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson DA, Juranek S, Lipps HJ. Designing nonviral vectors for efficient gene transfer and long-term gene expression. Mol Ther. 2006;14:613–626. doi: 10.1016/j.ymthe.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 4.Sundaram S, Trivedi R, Durairaj C, Ramesh R, Ambati BK, Kompella UB. Targeted drug and gene delivery systems for lung cancer therapy. Clin Cancer Res. 2009;15:7299–7308. doi: 10.1158/1078-0432.CCR-09-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aneja MK, Geiger JP, Himmel A, Rudolph C. Targeted gene delivery to the lung. Exp Op Drug Del. 2009;6:567–583. doi: 10.1517/17425240902927841. [DOI] [PubMed] [Google Scholar]
- 6.Elsabahy M, Nazarali A, Foldvari M. Non-viral nucleic acid delivery: key challenges and future directions. Curr Drug Del. 2011;8:235–244. doi: 10.2174/156720111795256174. [DOI] [PubMed] [Google Scholar]
- 7.Surace C, Arpicco S, Dufay-Wojcicki A, Marsaud V, Bouclier C, Clay D, Cattel L, Renoir JM, Fattal E. Lipoplexes targeting the CD44 hyaluronic acid receptor for efficient transfection of breast cancer cells. Mol Pharm. 2009;6:1062–1073. doi: 10.1021/mp800215d. [DOI] [PubMed] [Google Scholar]
- 8.Zoller M. CD44: physiological expression of distinct isoforms as evidence for organ-specific metastasis formation. J Mol Med. 1995;73:425–438. doi: 10.1007/BF00202261. [DOI] [PubMed] [Google Scholar]
- 9.Arpicco S, Milla P, Stella B, Dosio F. Hyaluronic acid conjugates as vectors for the active targeting of drugs, genes and nanocomposites in cancer treatment. Molecules. 2014;19:3193–3230. doi: 10.3390/molecules19033193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baier Leach J, Schmidt CE. HYALURONAN. Encyclopedia of Biomaterials and Biomedical Engineering. :1421–1431. [Google Scholar]
- 11.Ohene-Abuakwa Y, Pignatelli M. Adhesion molecules in cancer biology. Adv Exp Med Biol. 2000;465:115–126. doi: 10.1007/0-306-46817-4_11. [DOI] [PubMed] [Google Scholar]
- 12.Fattal E, Dufay A, Surace C, Marsaud V, Renoir M. Lipoplexes targeting the CD44 hyaluronic acid receptor for efficient transfection of breast cancer cell. Hum Gene Ther. 2009;20:682–682. doi: 10.1021/mp800215d. [DOI] [PubMed] [Google Scholar]
- 13.Culty M, Miyake K, Kincade PW, Silorski E, Butcher EC, Underhill C. The hyaluronate receptor is a member of the CD44 (H-Cam) family of cell-surface glycoproteins. J Cell Biol. 1990;111:2765–2774. doi: 10.1083/jcb.111.6.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh SC, Alpay SN, Klostergaard J. CD44: a validated target for improved delivery of cancer therapeutics. Exp Opin Ther Tar. 2012;16:635–650. doi: 10.1517/14728222.2012.687374. [DOI] [PubMed] [Google Scholar]
- 15.Gallatin M, Stjohn TP, Siegelman M, Reichert R, Butcher EC, Weissman IL. Lymphocyte homing receptors. Cell. 1986;44:673–680. doi: 10.1016/0092-8674(86)90832-9. [DOI] [PubMed] [Google Scholar]
- 16.Han L, Shi S, Gong T, Zhang Z, Sun X. Cancer stem cells: therapeutic implications and perspectives in cancer therapy. Acta Pharm Sinica B. 2013;3:65–75. [Google Scholar]
- 17.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 18.Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, Reilly JG, Chandra D, Zhou J, Claypool K, Coghlan L, Tang DG. Highly purified CD44(+) prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696–1708. doi: 10.1038/sj.onc.1209327. [DOI] [PubMed] [Google Scholar]
- 19.Pouyani T, Prestwich GD. Functionalized derivatives of hyaluronic-acid oligosaccharides - drug carriers and novel biomaterials. Bioconjugate Chem. 1994;5:339–347. doi: 10.1021/bc00028a010. [DOI] [PubMed] [Google Scholar]
- 20.Schante CE, Zuber G, Herlin C, Vandamme TF. Chemical modifications of hyaluronic acid for the synthesis of derivatives for a broad range of biomedical applications. Carbohyd Polym. 2011;85:469–489. [Google Scholar]
- 21.Kulkarni A, VerHeul R, DeFrees K, Collins CJ, Schuldt RA, Vlahu A, Thompson DH. Microfluidic assembly of cationic-β-cyclodextrin:hyaluronic acid-adamantane host:guest pDNA nanoparticles. Biomat Sci. 2013;1:1029–1033. doi: 10.1039/C3BM00189J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowenstein P, Kroeger K, Castro M. Immunology of neurological gene therapy: How T cells modulate viral vector-mediated therapeutic transgene expression through immunological synapses. Neurotherapeutics. 2007;4:715–724. doi: 10.1016/j.nurt.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang JS, Liu F, Conwell CC, Tan Y, Huang L. Mechanistic studies of sequential injection of cationic liposome and plasmid DNA. Mol Ther. 2006;13:429–437. doi: 10.1016/j.ymthe.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 24.Kulkarni A, Badwaik V, DeFrees K, Schuldt RA, Gunasekera DS, Powers C, Vlahu A, VerHeul R, Thompson DH. Effect of pendant group on pDNA delivery by cationic-β-cyclodextrin:alkyl-PVA-PEG pendant polymer complexes. Biomacromolecules. 2013;15:12–19. doi: 10.1021/bm401096v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee MS, Lee JE, Byun E, Kim NW, Lee K, Lee H, Sim SJ, Lee DS, Jeong JH. Target-specific delivery of siRNA by stabilized calcium phosphate nanoparticles using dopa–hyaluronic acid conjugate. J Control Release. 2014;192:122–130. doi: 10.1016/j.jconrel.2014.06.049. [DOI] [PubMed] [Google Scholar]
- 26.Cho HJ, Yoon IS, Yoon HY, Koo H, Jin YJ, Ko SH, Shim JS, Kim K, Kwon IC, Kim DD. Polyethylene glycol-conjugated hyaluronic acid-ceramide self-assembled nanoparticles for targeted delivery of doxorubicin. Biomaterials. 2012;33:1190–1200. doi: 10.1016/j.biomaterials.2011.10.064. [DOI] [PubMed] [Google Scholar]
- 27.Arpicco S, De Rosa G, Fattal E. Lipid-based nanovectors for targeting of CD44-overexpressing tumor cells. J Drug Del. 2013;2013:860780. doi: 10.1155/2013/860780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada Y, Hashida M, Harashima H. Hyaluronic acid controls the uptake pathway and intracellular trafficking of an octaarginine-modified gene vector in CD44 positive- and CD44 negative-cells. Biomaterials. 2015;52:189–198. doi: 10.1016/j.biomaterials.2015.02.027. [DOI] [PubMed] [Google Scholar]
- 29.Lyklema H. Fundamentals of Interface and Colloid Science Volume V Soft Colloids - preface to Volumes IV and V: Colloids. Fund Interface Coll. 2005;5:Vii–Ix. [Google Scholar]
- 30.Lyklema H. Fundamentals of Interface and Colloid Science Volume V Soft Colloids General Preface. Fund Interface Coll. 2005;5:V–Vi. [Google Scholar]
- 31.Buck CB, Pastrana DV, Lowy DR, Schiller JT. Efficient intracellular assembly of papillomaviral vectors. J Virol. 2004;78:751–757. doi: 10.1128/JVI.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu W, Yeo E, McMillan N, Yu C. Silencing oncogene expression in cervical cancer stem-like cells inhibits their cell growth and self-renewal ability. Cancer Gene Ther. 2011;18:897–905. doi: 10.1038/cgt.2011.58. [DOI] [PubMed] [Google Scholar]
- 33.Cho HJ, Yoon IS, Yoon HY, Koo H, Jin YJ, Ko SH, Shim JS, Kim K, Kwon IC, Kim DD. Polyethylene glycol-conjugated hyaluronic acid-ceramide self-assembled nanoparticles for targeted delivery of doxorubicin. Biomaterials. 2012;33:1190–1200. doi: 10.1016/j.biomaterials.2011.10.064. [DOI] [PubMed] [Google Scholar]
- 34.Arpicco S, De Rosa G, Fattal E. Lipid-based nanovectors for targeting of CD44-overexpressing tumor cells. J Drug Del. 2013;2013:8. doi: 10.1155/2013/860780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Almalik A, Day PJ, Tirelli N. HA-coated chitosan nanoparticles for CD44-mediated nucleic acid delivery. Macromol Biosci. 2013;13:1671–1680. doi: 10.1002/mabi.201300302. [DOI] [PubMed] [Google Scholar]
- 36.Almalik A, Karimi S, Ouasti S, Donno R, Wandrey C, Day PJ, Tirelli N. Hyaluronic acid (HA) presentation as a tool to modulate and control the receptor-mediated uptake of HA-coated nanoparticles. Biomaterials. 2013;34:5369–5380. doi: 10.1016/j.biomaterials.2013.03.065. [DOI] [PubMed] [Google Scholar]
- 37.Jainchill JL, Aaronson SA, Todaro GJ. Murine sarcoma and leukemia viruses: assay using clonal lines of contact-inhibited mouse cells. J Virol. 1969;4:549–553. doi: 10.1128/jvi.4.5.549-553.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eliaz RE, Nir S, Szoka FC. Interactions of hyaluronan-targeted liposomes with cultured cells: Modeling of binding and endocytosis. Method Enzymol. 2004;387:16–33. doi: 10.1016/S0076-6879(04)87002-2. [DOI] [PubMed] [Google Scholar]
- 39.Contreras-Ruiz L, de la Fuente M, Parraga JE, Lopez-Garcia A, Fernandez I, Seijo B, Sanchez A, Calonge M, Diebold Y. Intracellular trafficking of hyaluronic acid-chitosan oligomer-based nanoparticles in cultured human ocular surface cells. Mol Vis. 2011;17:279–290. [PMC free article] [PubMed] [Google Scholar]
- 40.Haisma H. Endosomal escape pathways for delivery of biologicals. Hum Gene Ther. 2011;22:A14–A14. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 41.Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 42.Gasingirwa MC, Thirion J, Mertens-Strijthagen J, Wattiaux-De Coninck S, Flamion B, Wattiaux R, Jadot M. Endocytosis of hyaluronidase-1 by the liver. Biochem J. 2010;430:305–313. doi: 10.1042/BJ20100711. [DOI] [PubMed] [Google Scholar]
- 43.Boonen M, Puissant E, Gills F, Flamion B, Jadot M. Mouse liver lysosomes contain enzymatically active processed forms of Hyal-1. Biochem Biophys Res Comm. 2014;446:1155–1160. doi: 10.1016/j.bbrc.2014.03.070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.