Abstract
Hemoglobin (Hb) is well protected inside the red blood cells (RBCs). Upon hemolysis and when free in circulation, Hb can be involved in a range of radical generating reactions and may thereby attack several different biomolecules. In this study, we have examined the potential damaging effects of cell-free Hb on plasmid DNA (pDNA). Hb induced cleavage of supercoiled pDNA (sc pDNA) which was proportional to the concentration of Hb applied. Almost 70% of sc pDNA was converted to open circular or linear DNA using 10 µM of Hb in 12 h. Hb can be present in several different forms. The oxy (HbO2) and met forms are most reactive, while the carboxy-protein shows only low hydrolytic activity. Hemoglobin A (HbA) could easily induce complete pDNA cleavage while fetal hemoglobin (HbF) was three-fold less reactive. By inserting, a redox active cysteine residue on the surface of the alpha chain of HbF by site-directed mutagenesis, the DNA cleavage reaction was enhanced by 82%. Reactive oxygen species were not directly involved in the reaction since addition of superoxide dismutase and catalase did not prevent pDNA cleavage. The reactivity of Hb with pDNA can rather be associated with the formation of protein based radicals.
Abbreviations: Hb, hemoglobin; metHb, ferric hemoglobin (Fe3+), HbA, adult hemoglobin; pDNA, plasmid DNA; sc pDNA, supercoiled plasmid DNA; ocDNA, open circular plasmid DNA; LDNA, linear plasmid DNA, CO, carbon monoxide; KCN, potassium cyanide; HbF, fetal hemoglobin; Hp, haptoglobin; SOD, superoxide dismutase; DMSO, dimethyl sulfoxide
Keywords: Adult hemoglobin, Fetal hemoglobin, Supercoiled plasmid DNA, DNA cleavage, Cysteine, Protein radicals
Highlights
-
•
Hemoglobin induced plasmid DNA cleavage in the absence of hydrogen peroxide.
-
•
Fetal hemoglobin was three-fold less reactive compared to the adult protein on plasmid DNA.
-
•
Insertion of a cysteine residue in the alpha chain enhanced the DNA cleavage reaction by 82%.
-
•
Protein based radicals are associated with the DNA cleavage activity of hemoglobin.
1. Introduction
Hemoglobin (Hb) is a tetrameric protein, composed of two alpha (141 residues) and two beta (146 residues) chains, forming an α2β2 heterotetramer of 64 kDa. Each subunit harbors a heme in the center pocket responsible for its oxygen binding capability [1]. Although often viewed solely as an oxygen transporting protein, Hb has a very rich chemistry related to the reactivity of the iron atom in the heme groups. Hb is thus involved in several redox reactions and shows e.g. peroxidase-like activity, in which ferric or metHb (Fe3+) can react with hydrogen peroxide (H2O2) to form a potent oxidant ferryl Hb (Fe4+) along with protein-based radicals. These oxidized forms of Hb are in turn highly reactive and are involved in oxidative damages to a variety of biological molecules [2], [3]. Modifications of particularly lipids and protein side chains caused by Hb have been examined in detail. Moreover, Hb can bind and react with nitric oxide (NO) to form nitrate and ferric heme. NO depletion through these reactions leads to vasoconstriction and platelet aggregation [4]. Hb is normally encapsulated in the protective environment of the erythrocytes, but when released upon hemolysis its redox reactions may cause serious damages to surrounding cells and tissues. This becomes especially pronounced at various hematological disorders or at blood transfusion events [5], [6], [7], when substantial amounts of cell-free or acellular Hb are released into the circulatory system. To prevent these harmful reactions, several defence proteins have evolved. The plasma protein haptoglobin (Hp), binds cell-free Hb rapidly and almost irreversibly, and transports the complex Hp-Hb to the CD163 receptors mainly located on the Kuppfer cells of the liver for enzymatic degradation and reuse [8], [9]. Hemopexin and alpha-1-microglobulin are also involved in the protective system against toxic acellular Hb [10], [11]. Cell-free Hb may thus insert a substantial burden to our body at a range of conditions and the use of Hb protective and degradative proteins have been proposed to soon also find clinical practice [12].
Despite its clinical manifestations, only limited knowledge of the interactions between Hb and nucleic acids are available. Hb can bind to calf thymus DNA and the protein also carries an endonuclease-like activity which has been examined by the conversion of supercoiled plasmid DNA (sc pDNA) to nicked circular DNA in the presence of H2O2 [13]. Similarly, myoglobin (Mb) showed a distinct pathway for cleavage of DNA upon chemically induced reductions in presence of oxygen or when Mb was present in its met form [14], [15]. Hb can also induce DNA damages directly at the cellular level by rapid uptake and cleavage of available nucleic acids. This has been demonstrated for primary colon cells [16], leukocytes [17] and lymphocytes [18]. The use of the comet assay has been instrumental for quantifying the genotoxic effects of Hb, which occur rapidly and already at low concentrations.
In the human population several different Hb variants have been identified [19]. In this study, wild-type and mutant Hb molecules have therefore been examined to characterize their reactivity to nucleic acids. For this we have studied plasmid DNA (pDNA) cleavage by Hb. Purified pUC18 plasmid was incubated with various Hb samples, adult and fetal Hb, i.e. HbA and HbF, respectively, at different experimental conditions. Hemin (free heme in the met form) was included in the study to evaluate the effects of globin polypeptides and heme separately. It was observed that metHb interacts with pDNA and exhibit high endonuclease activity, responsible for conversion of sc pDNA into open circular DNA (ocDNA) or linear DNA (LDNA). Interestingly, HbF, which shares the same alpha chains as HbA, but has unique gamma chains, exhibited no or only low measurable reactivity against nucleic acids. This may have several clinical implications for treatment of a range of hematological conditions, like sickle cell anemia (SCA) and different forms of thalassemia.
2. Material and methods
2.1. Plasmid purification
Plasmid was prepared and isolated as described in our previous work [20]. The E. coli strain TG1 (GE Healthcare, Uppsala, Sweden) was used as host in all experiments for production of plasmid pUC18 of 2686 base pairs (GE Healthcare, Uppsala, Sweden). Cells were grown in Luria-Bertani (LB) media at 37 °C overnight, harvested by centrifugation and plasmid DNA was isolated using a dedicated purification kit (NucleoSpin Plasmid, MACHEREY-NAGEL, Germany). The DNA was eluted with 20 mM sodium phosphate buffer pH 7.2 and purified plasmid was stored at −80 °C until further use. The concentration of pDNA was determined by an Implen Nanophotometer (Labvision AB, Sweden).
2.2. Hb preparation
Recombinant HbA was constructed, expressed and purified as described earlier [3]. In brief, the alpha and beta chains of Hb were cloned in the pETDuet-1 vector and transformed into E. coli strain BL21 (DE3). Transformed cells were grown in Terrific Broth (TB) media with vigorous shaking (150 rpm) in baffled 2 L flasks at 37 °C until OD620≥2. The cultures were then induced with 0.1 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG, Saveen Werner AB, Sweden) and supplemented with 0.3 mM δ-aminolevulinic acid (Sigma Aldrich), and allowed to grow overnight at 22 °C with reduced aeration (60 rpm). Cells were collected and sonicated in 10 mM sodium phosphate buffer pH 6.0. Protein was purified using a weak (CM Sepharose, GE Healthcare, Sweden) followed by strong (Q Sepharose HP, GE Healthcare, Sweden) ion exchange chromatography. The purified recombinant Hb samples were identical with the native protein and stored at −80 °C until further use. To keep the Hb samples in a stable form, carbon monoxide (CO) gas was bubbled through all solutions used at induction and each step of the purification.
The ferric form of Hb (metHb) was obtained by incubating Hb with 10 mM excess of potassium ferricyanide (Sigma Aldrich). This solution was passed through a gel filtration column Sephadex G-25 (5×0.5 cm, GE Healthcare, Sweden) to remove excess of ferricyanide. The concentration of Hb was determined by reducing an aliquot of the ferric Hb with sodium dithionite (Sigma Aldrich) to the deoxy form (430 nm=133 mM−1 cm−1). The Hb concentration used in all experiments is given on the basis of molar heme. To obtain the oxy form of Hb, the CO-Hb was exposed to a continuous stream of oxygen gas in presence of light. All the experiments were carried out in 20 mM sodium phosphate buffer pH 7.2 at 37 °C using the ferric form of Hb or as otherwise specified.
2.3. DNA cleavage assay
The pDNA (25 µg/ml) was incubated with different concentrations (1–100 µM) of the ferric Hb at 37 °C using PCR tubes (total reaction volume of 20 µl). Sample was removed after every 2 h and analysed by agarose gel (1%) electrophoresis carried out for 60 min at 100 V using TAE buffer (Tris-acetate-EDTA, pH 8.0). A control sample was included in all experiments containing only pDNA. The different forms of DNA (supercoiled, open circular and linear) were quantified by densitometric analysis using the ‘Quantity one’ software from Biorad. To evaluate the temperature dependent degradation of pDNA, Hb (20 µM) was incubated with pDNA at different temperatures (10–40 °C). The DNA damage effects were also evaluated in different concentrations of sodium phosphate buffer (20, 50 and 100 mM, pH 7.2). Similarly, oxy and CO adducts of Hb were examined. The absorption spectra of Hb (5 µM) with or without pDNA was taken every 30 min at 37 °C for 12 h using a Cary60 UV–Vis spectrophotometer (Agilent Technologies). All data were analysed from minimum three independent experiments.
2.4. Preparation of HbF mutants
Site-directed mutagenesis was employed for removing a cysteine residue in the gamma chain (γ-C93A) close to the heme group. Similarly, an alanine residue located on the surface of the alpha chain was substituted to cysteine (α-A19C). Primers used were purchased from integrated DNA technologies (IDT, Germany). Forward sequence of primers employed are as follows (reverse primers were complementary to forward primers): bold sequence represents mutation, 5′-GGGGTAAAGTTGGTTGCCATGCCGGTGAA-3′ (α-A19C) and 5′-GAGTGAACTGCACGCCGATAAACTGCAC −3′ (γ-C93A). A double mutant was prepared using similar primers. After PCR, the methylated DNA was removed using DpnI (Thermo Scientific) digestion. These mixtures were transformed into E. coli strain BL21 (DE3). Sequences of the transformed clones were confirmed (GATC Biotech, Germany). The expression conditions were slightly different than for HbA. In brief, the starter culture was inoculated into 500 ml TB media and was induced immediately. This culture was allowed to grow overnight at 30 °C and 150 rpm. These mutants and wildtype HbF variants were purified as described elsewhere [21]. HbF and HbF mutants (25 µM) were incubated with supercoiled plasmid pUC 18 (10 µg/ml) at 37 °C. Sample were removed after 1, 2 and 3 h respectively, and analysed by agarose gel electrophoresis.
2.5. Inhibition studies of Hb reactivity
A potassium cyanide (KCN) solution (100 µM, Sigma Aldrich) was mixed with Hb (50 µM) prior to the experiment for 1 h to allow complete complexation of the heme iron to prevent any oxidation reaction. The complex was subsequently incubated with pDNA (25 µg/ml) at 37 °C for 12 h. In another set of experiment, Hb samples were incubated together with pDNA containing sodium chloride (100 mM and 500 mM). Similarly, metHb (20 µM) and pDNA was incubated with superoxide dismutase (SOD, 20 units, Sigma Aldrich), catalase (20 units, Roche), ascorbate (20 µM, Sigma Aldrich) and dimethyl sulfoxide (DMSO, 100 µM).
2.6. Protection by haptoglobin
A haptoglobin (Hp) sample containing primarily dimers (Hp 1-1) and to a lesser extent polymer (Hp 2-1, and Hp 2-2) was kindly provided by Bio Products Laboratory (BPL, Hertfordshire, UK). Hp: Hb in ratios (1:1.25, 1:0.625, 1:0.25, 1:0.125) were incubated for 10 min. The Hb-Hp complex formed was subsequently incubated with pDNA (25 µg/ml) at 37 °C and analysed as described earlier.
3. Results
Hemoglobins have in general a rich redox chemistry, which results in oxidation or reduction of the heme iron frequently followed by formation of protein radicals. Several alternative approaches have been used to monitor these reactions, both in vitro and in vivo.
3.1. DNA cleavage activity of HbA and factors responsible
In this study, we have used sc pDNA as screening nucleic acid molecule to characterize the DNA cutting activity of different Hb samples. Upon cleavage of one of the strands, the sc pDNA form is converted into ocDNA. If such a cleavage reaction is allowed to proceed, a linear DNA molecule is generated (Fig. 1A). These modifications of pDNA in the presence of Hb, can easily be monitored over time using agarose gel electrophoresis. Different concentrations of ferric Hb were first incubated with pDNA at 37 °C in 20 mM sodium phosphate buffer pH 7.2. Hb (10 µM) induced significant cleavage of pDNA and the reaction could be quantified over time. As shown in Fig. 1B, under the conditions used, only 30% sc pDNA remained after 12 h, while ocDNA and LDNA increased to 46% and 23%, respectively, of the total DNA composition. These data thus suggest that sc pDNA can be rapidly degraded into ocDNA and/or LDNA in the presence of ferric Hb alone.
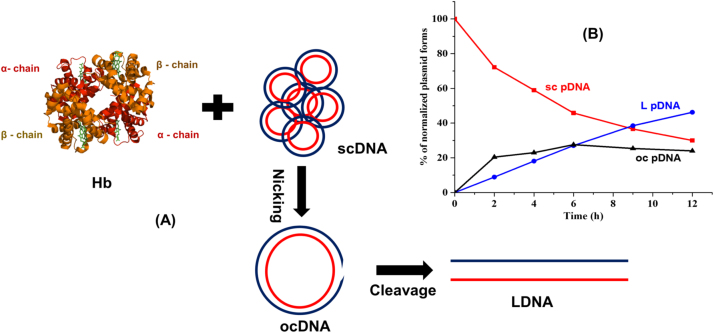
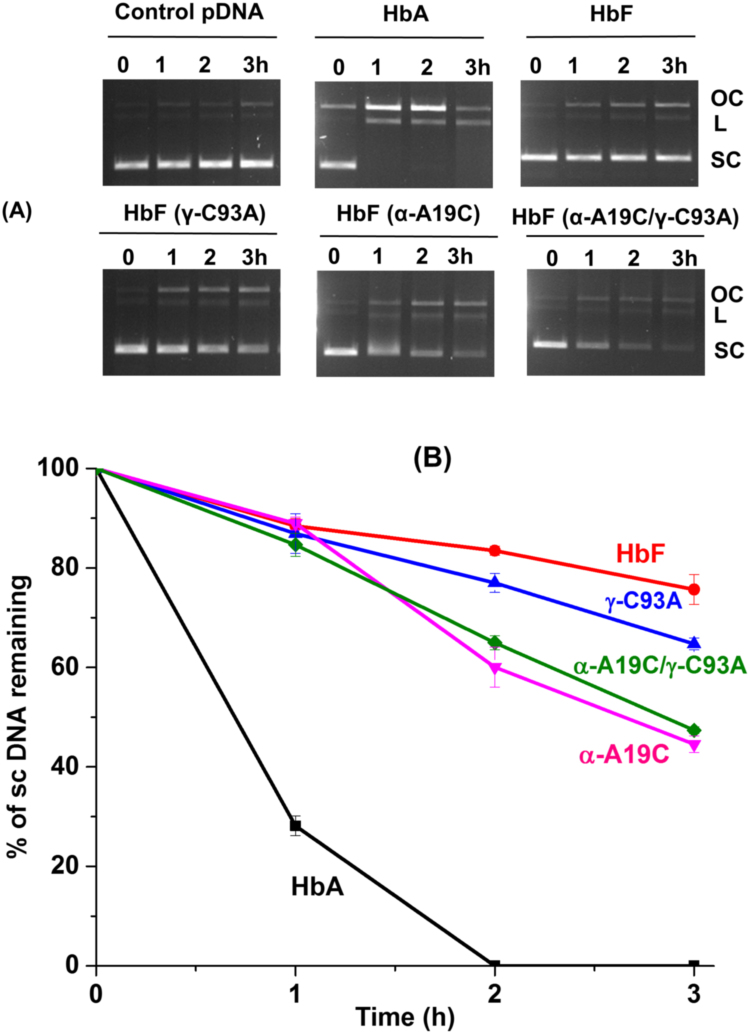
Fig. 1.
Schematic representation of pDNA cleavage activity by metHb. A) Hb tetramers (alpha and beta chains in red and orange color, respectively) interact with sc pDNA (red and blue strands) and convert it to open circular (oc) DNA by nicking. The ocDNA may be further cut into linear (L) DNA. B) Different isoforms of pDNA were observed after incubation with Hb (10 µM). The relative fraction of each isoform, as determined by agarose gel electrophoresis, was plotted against time and the gradual increase of ocDNA and LDNA , respectively, was monitored. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
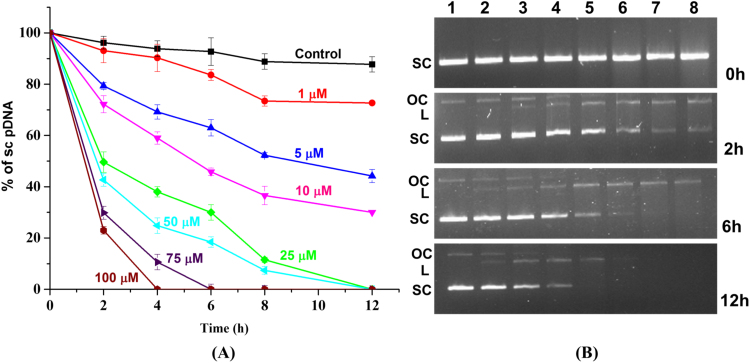
Agarose gel electrophoresis could be used to quantitatively follow the progressive cleavage of sc pDNA in the presence of ferric Hb. It was observed that even low Hb concentrations could initiate the unwinding of sc pDNA. After 12 h, 5 and 10 µM of Hb showed 50% and 70% conversion of sc pDNA, respectively. The degree of pDNA cleavage proved to be directly correlated with the concentration of Hb used in the experiment (Fig. 2A). When using higher concentration of Hb (75 µM and 100 µM) complete degradation of pDNA was observed over the time (Fig. 2B). When the remaining sc pDNA concentration was plotted against time, the degradation could be fitted to an exponential decay equation to obtain the reaction rate constant. The decay constant was found to be 3300 M−1 h−1, with R2 value of 0.99.
Fig. 2.
pDNA cleavage kinetics by ferric Hb as determined by agarose gel electrophoresis. (A) Characterization of the dose dependent cleavage of sc pDNA by Hb in the concentration range 0–100 µM at 37 °C (B) Agarose gel electrophoresis of DNA samples isolated at 0, 2, 6 and 12 h using pUC18 pDNA (25 µg/ml). Lane 1, pUC18 DNA without Hb addition; lane 2, 1 µM Hb+pDNA; lane 3, 5 µM Hb+pDNA; lane 4, 10 µM Hb+pDNA; lane 5, 25 µM Hb+pDNA; lane 6, 50 µM Hb+pDNA; lane 7, 75 µM Hb+pDNA; lane 8, 100 µM Hb+pDNA. Data present mean±S.D., n=3.
Most of the experiments were carried out at physiological temperature, i.e. at 37 °C. The GC content of pUC 18 plasmid is 50.6% and the melting temp (Tm) is approximately 75 °C (online tool), while for the human hemoglobin it is 64 °C. In order to analyse if any small structural rearrangements in the pDNA or Hb molecules could influence the degradation, the reaction was also monitored in the temperature range (10–40 °C). There was a clear temperature dependence and pDNA cleavage was more rapid at the higher temperature analysed. The sc pDNA was completely cleaved at 40 °C in 12 h. However, at the lower temperature (10 °C), 50% of sc pDNA remained after 12 h. Similarly, at 20 °C and 30 °C, almost 70% and 90% of sc pDNA were damaged after 12 h, respectively (data not shown). These data clearly suggest that increasing temperature also influence the pDNA cleavage action of Hb. The buffer component also influences the DNA cleavage activity of iron, Saran et al. [22] explained ‘crypto OH radicals’, as reactive species responsible for interaction of phosphate buffer and iron (Fe2+). In our studies, low buffer concentrations (20 mM) initiate the pDNA cleavage activity at slower rate. Notably, at 100 mM buffer concentration showed the highest pDNA cleavage, almost complete degradation of sc pDNA was observed. However, 43% and 9% of sc pDNA remained using 20 mM and 50 mM buffer concentration, respectively .
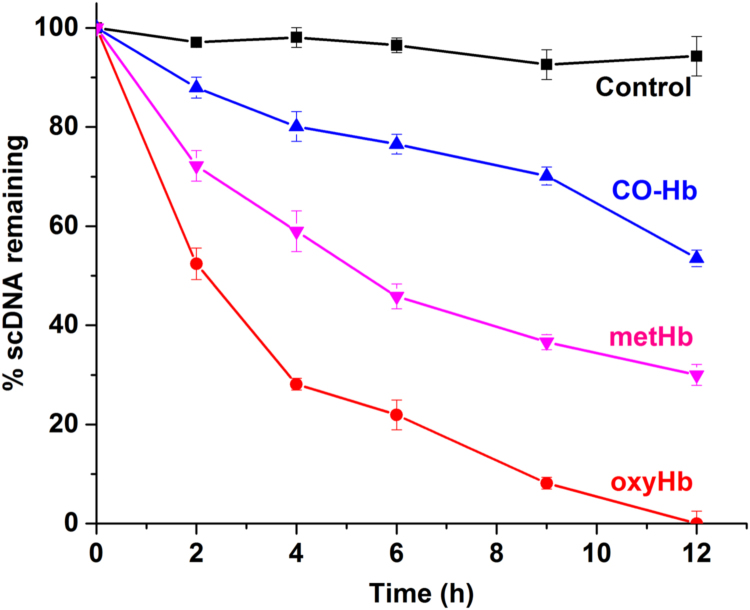
Besides the met form, Hb can also be present in several other different redox states. In addition, different ligands may be bound to the heme iron, including oxygen and CO. As shown in Fig. 3, the carboxy-Hb form (CO-Hb) reduced the pDNA cleavage activity compared to the oxy and ferric forms of Hb. CO binds strongly to the ferrous form of Hb and block further redox activities.
Fig. 3.
Dependence of Hb redox state and ligand binding on pDNA degradation. The different forms of Hb were incubated in presence of pDNA at 37 °C for 12 h and the percentage of remaining sc pDNA was determined. Control (black), CO (blue), oxy (red) and ferric (pink) form of Hb (10 µM) was incubated with pDNA and samples were taken at intervals of 2 h. CO-Hb was less reactive compared to oxy and metHb. Data present mean±S.D., n=3. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
The CO-Hb molecule is also stable and was therefore used during purification of the protein [23]. CO-Hb thus showed 40% damage to sc pDNA compared to 70% damage by metHb. Autoxidation of oxy-Hb (HbO2) easily produces metHb and superoxide radicals (O2•−) which can dismutate to O2 and H2O2, followed by formation of ferryl (Fe4+) species. This can in turn oxidize surrounding biological molecules i.e. DNA, lipids and proteins [24]. This could also be observed in our pDNA assay showing that oxyHb was six-fold faster compared to CO-Hb.
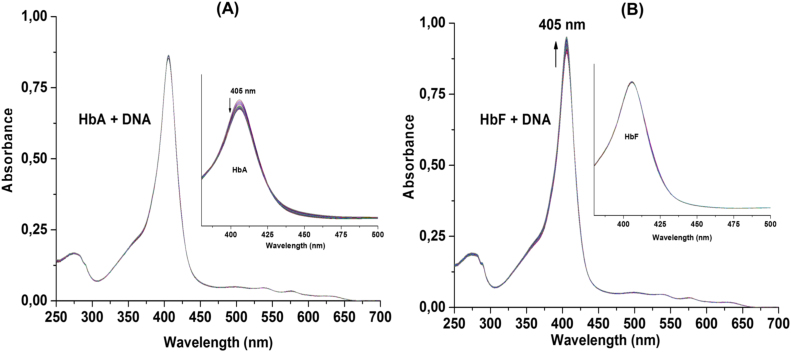
The physical interactions between DNA and Hb have been studied earlier, and it has been demonstrated that protein conformational changes increase the absorbance. Since HbA and HbF differ in their DNA degradation activities, absorption spectra were recorded for Hb in presence of pDNA for 12 h and scanned every 30 min at 37 °C. After 12 h, no shift in the soret region peak (405 nm) was observed. However, the absorbance intensity of HbA alone decreased by 15% (Fig. 4A). In the case of HbF, no such changes were detected. Interestingly, an addition of DNA increased the stability of HbA, while HbF showed gradual increase in peak intensity at soret region (Fig. 4B). In another set of experiment, protein unfolding was monitored at increasing temperatures, intrinsic tryptophan and tyrosine fluorescence was analysed to evaluate the Tm of the Hb variants examined. It was observed that the Tm values of the proteins were decreased by 2–3 °C in presence of pDNA. This suggests that physical interactions between pDNA and Hb occur before the cleavage activity. The pDNA alone did not show any significant differences in its spectral properties (data not shown).
Fig. 4.
The absorption spectras of metHb (5 µM) and pDNA (25 µg/ml) were recorded every 30 min for 12 h at 37 °C. A) The absorption spectra of HbA was not altered in presence of pDNA, however, gradual decrease in peak intensity of HbA (without pDNA) was observed (A, inset). B) The peak intensity of HbF, in presence of pDNA was increased over the time. HbF alone did not showed any change in absorbance during an incubation period (B, inset).
3.2. Protective nature of HbF and mutants
Several different Hb variants may be present in the human body. Of particular interest is the fetal Hb (HbF) which has been shown to harbor a different redox activity compared with HbA. This has been shown to be due to the absence of a redox active amino acid network in HbF [25]. It was found that HbF cleaved pDNA at a much slower rate. For instance, while HbA under the same conditions initiated pDNA cleavage after 1 h incubation and complete degradation of sc pDNA was observed in 2 h, HbF initiated unwinding of sc pDNA first after 3 h and 76% of sc pDNA was retained (Fig. 5A). Besides the wildtype, different HbF mutants carrying redox active side chains were examined. The cysteine residue present on the gamma chain of HbF, γ-C93, is the first primary location of protein radical formation, and is known to be redox active in Hb introduced on the protein surface [25], [26]. When this cysteine residue was replaced with alanine (γ-C93A), prominent difference in DNA cleavage was not observed, 36% of sc pDNA was cleaved compared to 24% for the native protein. However, introduction of cysteine on the surface of the alpha chain (α-A19C) promoted the reaction substantially, resulting in conversion of 55% of the sc pDNA after 3 h, an 82% escalation of the reaction rate. The double mutant (αA19C/γC93A) showed similar pDNA activity as α-A19C (Fig. 5B). These data clearly suggest the role of redox active residues on the protein surface in the pDNA cleavage activity of Hb.
Fig. 5.
HbF mutants (25 µM) were incubated with pUC18 (10 µg/ml), and agarose gel electrophoresis was carried out to determine the levels of remaining sc pDNA. A) Agarose gel electrophoresis of pDNA after treatment with HbF mutants. HbF did not induce pDNA cleavage in 3 h, while complete cleavage was observed in presence of HbA. An insertion of Cys residue in the alpha chain escalate the pDNA cleavage. OC, L and SC represent open circular, linear and supercoiled DNA, respectively. B) The percentage of remaining sc pDNA was plotted against the time interval. Data present mean±S.D., n=3.
3.3. Inhibition of DNA cleavage induced by Hb
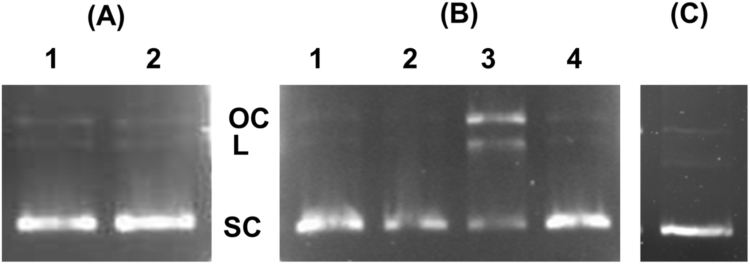
KCN has very high affinity for heme iron and effectively displaces other ligands from the protein and prevents further redox activity [23]. When the pDNA cleavage activity was monitored in the presence of KCN treated Hb, no reactivity was observed (Fig. 6A, lane 1). It suggests that involvement of the central heme, which initiates oxidative or radical formation reactions, is critical for pDNA cleavage. The absorption spectra also suggest physical interactions of Hb and pDNA, which can be based on both hydrophobic and ionic interactions. When the reaction was followed in the presence of high sodium chloride concentrations (500 mM), the pDNA cleavage activity of ferric Hb could be inhibited (Fig. 6B, lane 1). Also at lower (100 mM) NaCl levels (Fig. 6B, lane 3), the DNA cleavage was reduced, but did not completely inhibit the reaction. These results suggest that electrostatic interactions are needed between Hb and pDNA to generate DNA cleavage. Free hemin (10 µM), the reactive ferric protoporphyrin-IX group potentially released from cell-free Hb, was also examined. When added alone to the reaction mixture and incubated for 6 h, it was observed that hemin showed no pDNA cleavage activity (Fig. 6C). It clearly further supports that, not heme alone, but rather amino acid residues oxidized by the heme groups are responsible for pDNA cleavage.
Fig. 6.
Inhibition of the Hb cleavage activity by KCN and NaCl, evaluated by agarose gel electrophoresis of pDNA (25 µg/ml) and Hb (50 µM) after 12 h at 37 °C. (A) Inhibition was studied by using KCN (100 µM), which abolished the pDNA cleavage activity; Lane1, Hb and pDNA in presence of KCN; lane 2, pDNA incubated with KCN (B) Hb and pDNA were incubated with NaCl: lane1, Hb and pDNA with 500 mM NaCl; lane 2, pDNA with 500 mM NaCl; Lane 3, Hb and pDNA with 100 mM NaCl; lane 4, pDNA with 100 mM NaCl. (C) Effect of hemin (10 µM) was analysed and it did not induce any pDNA cleavage when observed on agarose gel electrophoresis after 6 h.
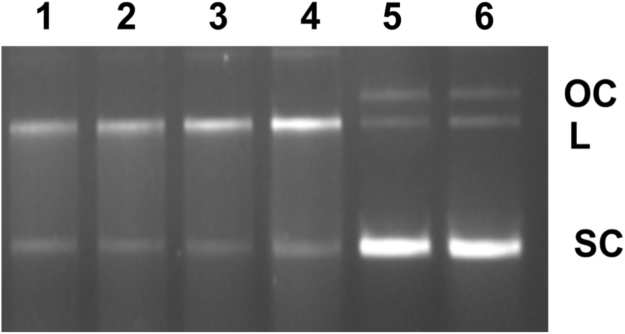
Hp is a plasma protein that binds and captures Hb released from red blood cells (RBCs), thereby minimizing the toxic effects of cell-free Hb [24], [27]. The Hb (10 µM) and pDNA were incubated in presence of Hp in different ratios (Hp: Hb-1:1.25, 1:0.625, 1:0.25, 1:0.125). It was observed that Hp could not inhibit the pDNA cleavage activity of Hb (Fig. 7).
Fig. 7.
Inhibition studies of DNA cleavage in presence of haptoglobin (Hp) as determined by agarose gel electrophoresis. The HbA was incubated with Hp, followed by addition of pDNA, agarose gel electrophoresis of different ratios of Hp:Hb. lane 1, Hb+pDNA+Hp (0.5 µM, 1:1.25); lane 2, Hb+pDNA+Hp (1 µM, 1: 0.625); lane 3, Hb+pDNA+Hp (2.5 µM, 1:0.25); lane 4, Hb+pDNA+Hp (5 µM, 1: 0.125); lane 5, pDNA+Hp (2.5 µM), lane 6, pDNA+Hp (5 µM).
4. Discussion
Hb is one of our most abundant proteins, normally densely packed inside the erythrocytes. The protein is well protected by an antioxidant network in the RBCs [28], but when released during hemolysis, acellular Hb can promote a cascade of deteriorating reactions [29]. Early studies have thus shown that stroma free Hb in circulation can induce a number of unwanted side effects including inflammation and hypertension [30], [31]. Several reactions regarding Hb toxicity have been elucidated in detail, including effects of heme loss, quenching of nitric oxide and redox activity of the protein. Particularly oxidative Hb reactions have been identified as a key factor contributing to the known pathophysiology of some Hb inherited disorders [4], [29], [31]. When bound to the globin chains, the redox active heme iron shows reduced oxidation of the ferrous iron. Nevertheless, it is well known that Hb undergoes continuous autoxidation in presence of oxygen with formation of the met form and production of superoxide anion as in Eq. (1) [5], [24].
| Hb (Fe2+O2)4 —› metHb (Fe3+) + 2O2•− | (1) |
| 2O2•− + 2H+ —› H2O2 + O2 | (2) |
| Hb (Fe3+) + H2O2 —› •Hb (Fe4+) + H2O | (3) |
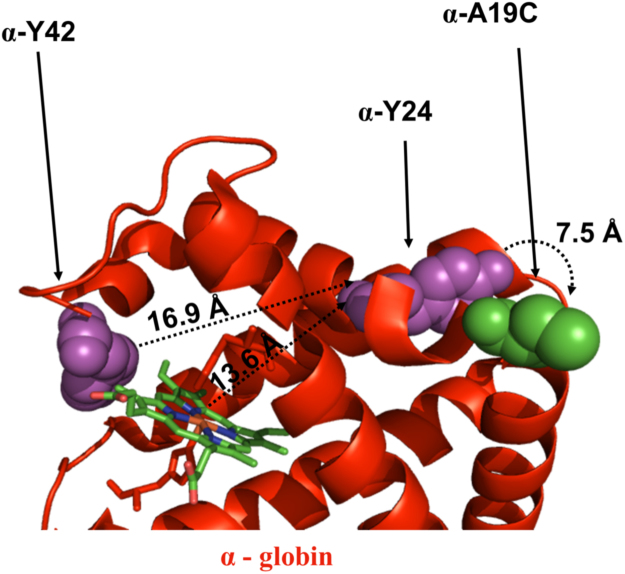
Superoxide dismutase acts on the superoxide (O2•−) anions, forming molecular oxygen and H2O2 (Eq. (2)). Hydrogen peroxide can in turn be decomposed into water in the presence of catalase but can also further oxidize metHb to the ferryl form (Eq. (3)). The protein based radicals are generated within the Hb molecule by the redox active heme molecule when the ferryl protein is produced. The radicals located within the Hb molecules, can migrate to individual amino acid residues within the protein and/or to surrounding biological molecules. Previous studies have demonstrated that the protein based radicals are unstable and easily can migrate far away from the heme moiety [32]. Tyrosine residues have been proposed to be one of the initial target locations [33]. Particularly β–Y145 is known to be one of the radical sites in the Hb molecule. Natural mutations have also been identified at this location [34]. In addition, site-directed mutagenesis to β-Y145F has been made to characterize the redox property of the mutant. Replacement of this active tyrosine reduced the DNA cleavage activity [18]. Cysteine residues are also highly susceptible to redox modifications and particularly the irreversible oxidation of β-Cys93 have been emphasised [25], [26]. This cysteine residue is conserved amongst all vertebrates and was evolved during transition of vertebrates from aquatic to terrestrial life. The antioxidant function of β-C93 has also been examined in a mouse model expressing human Hb [28]. The γ-C93A mutant slightly enhanced DNA cleavage compare to the native Hb, most likely due to migration of the protein radicals to other residues on the proteins. However, insertion of a cysteine (α-A19C) residue on the surface of the alpha chain had a major impact on the pDNA damage, which was almost 82% higher than the native HbF. As shown in Fig. 8, a plausible pathway for protein based radicals is the transfer of electrons from the α-heme iron to α-Tyr24 (13.6 Å), and subsequently to α-Cys19 (7.5 Å). Additionally, α-Tyr42 (5.3 Å) can receive electrons from the heme iron, and transfer them to α-Cys19 via α-Tyr24 (16.9 Å).
Fig. 8.
Schematic model of a primary radical formation sites in the alpha chain of HbF. The alpha (red) chain (PDB code 4MQJ), amino acids prone to oxidation are shown (magenta) and cysteine mutant (green) as a spheres. The generated radical transfer from α-Tyr42/α-Tyr24 to an engineered α-Cys19 residue, consequently damages the pDNA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
The involvement of protein radicals is further supported by the fact that superoxide radicals were not responsible for DNA cleavage. The pDNA cleavage activity thus remained largely unchanged in the presence of SOD and catalase. In addition, formation of CO-Hb or CN-Hb blocks the oxidative conversion of iron in the heme pocket [23] and thereby slows down the radical reactions. Furthermore, DMSO, did not inhibit the pDNA cleavage activity, which strengthens the hypothesis that Hb produces protein based radicals which are responsible for pDNA cleavage.
The binding reaction between DNA and Hb has been studied previously, resulting in a high affinity constant (10−5 M−1) [13]. The binding of Hb to DNA can be followed by a small change in the spectroscopic absorbance of the protein. This has been observed for Hb in presence of salmon [35] and calf thymus DNA [13]. When using pDNA, this interaction did not alter the heme environment since no change in the location of the soret peak was observed. However, there was a small increase in peak absorbance. This elevation of absorbance in the presence of pDNA further supports that one or more Hb molecules bind to pDNA. Taken together, this suggests that Hb firstly interacts with DNA, and electrons from the heme iron distributed to different protein locations subsequently contribute to the pDNA cleavage activity of Hb.
Erythrocytes do not normally contain a nucleus and are unable to proliferate. Enucleated RBCs are present in the blood of all mammals, suggesting that nuclear removal provides an evolutionary advantage. One of the reasons for the absence of nucleus can be related to the aggressive character of HbA on DNA as demonstrated here. This is also supported by the fact that Hb easily can penetrate the nuclear membrane and thereby generate fragmentation of the chromosomes [36], [37]. The precursor cells of RBCs, the pro-erythroblasts, pass a series of cell divisions where Hb is gradually accumulated while the size of the nuclease is reduced [38]. During the gestational period (fetal life), HbF is produced which is followed by a switch to HbA only after birth. The tetrameric structure of these Hbs consists of two alpha chains and either two beta chains in HbA or two gamma chains in HbF [39]. The 39 amino acids difference in the gamma compared to the beta subunit has modified the properties of HbF in terms of enhanced solubility and oxygen affinity. Recent studies have also shown that HbF is oxidatively more stable compared to HbA [25], A lack of a cysteine residue at position 112 in the gamma chain, has been proposed to be responsible for this enhanced oxidative stability. The present investigation also indicates that HbF is less prone to DNA cleavage activity compared to HbA which may be an efficient way of protecting the nucleus in the first development phases of the embryo.
Acellular Hb molecules are normally captured by the plasma protein Hp, which binds almost irreversibly to Hb dimers. This Hb-Hp complex is cleared by CD163 present on the macrophages [8], [9]. Hp not only removes acellular Hb from circulation, but is also responsible for attenuation of the oxidative reactions raised by Hb [34], [40]. However, in this study it was shown that the Hb-Hp complex did not mitigate the pDNA cleavage activity. It may be explained by the large freely accessible surface of Hb which can interact with DNA even after Hp complex formation [41].
Besides, developing a basic understanding of the interactions between Hb and nucleic acids, it is important to stress that such knowledge is highly relevant also in clinical settings. Since the 1980s, hemoglobin based oxygen carriers (HBOCs) have been extensively evaluated as a blood substitutes. Initial safety tests for HBOCs development include peroxidase [27], [42], mass spectrometry [26], [31] and electron paramagnetic resonance measurements [24], [32]. The present approach may be explored as a simple and rapid technique for quantitative and safety characterization of HBOCs requiring minimal quantities of samples. It is also supports the use of HbF instead of HbA for such applications [21].
5. Conclusion
To conclude, it was observed for the first time that Hb can initiate DNA cleavage in the absence of hydrogen peroxide. The reaction mechanism has not been elucidated but certainly involves a redox reaction initiated by Hb. Importantly, DNA cleavage was not observed for HbF.
Acknowledgement
The authors would like to thanks Dr. Michael T. Wilson (University of Essex, UK) and Dr. Abdu Alayash (FDA, USA) for valuable discussion and comments. This work was supported by the Erasmus Mundus External Cooperation Window Programme [Lot 13]; the Swedish Research council [VR 5607,2014] and the Swedish Fund for Strategic Research [RBP14-0055].
References
- 1.Nagatomo S., Nagai Y., Aki Y., Sakurai H., Imai K., Mizusawa N., Ogura T., Kitagawa T., Nagai M. An origin of cooperative oxygen binding of human adult hemoglobin: different roles of the α and β subunits in the α2β2 Tetramer. PLoS One. 2015;10:e0135080. doi: 10.1371/journal.pone.0135080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alayash A.I., Patel R.P., Cashon R.E. Redox reactions of hemoglobin and myoglobin: biological and toxicological implications. Antioxid. Redox Signal. 2001;3:313–327. doi: 10.1089/152308601300185250. [DOI] [PubMed] [Google Scholar]
- 3.Reeder B.J., Grey M., Silaghi-Dumitrescu R.-L., Svistunenko D.A., Bülow L., Cooper C.E., Wilson M.T. Tyrosine residues as redox cofactors in human hemoglobin implications for engineering nontoxic blood substitutes. J. Biol. Chem. 2008;283:30780–30787. doi: 10.1074/jbc.M804709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olson J.S., Foley E.W., Rogge C., Tsai A.-L., Doyle M.P., Lemon D.D. NO scavenging and the hypertensive effect of hemoglobin-based blood substitutes. Free Radic. Biol. Med. 2004;36:685–697. doi: 10.1016/j.freeradbiomed.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 5.Rifkind J.M., Mohanty J.G., Nagababu E. The pathophysiology of extracellular hemoglobin associated with enhanced oxidative reactions. Front. Physiol. 2014;5:500. doi: 10.3389/fphys.2014.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rother R.P., Bell L., Hillmen P., Gladwin M.T. The clinical sequelae of intravascular hemolysis and extracellular plasma hemoglobin: a novel mechanism of human disease. JAMA. 2005;293:1653–1662. doi: 10.1001/jama.293.13.1653. [DOI] [PubMed] [Google Scholar]
- 7.Koch C.G., Li L., Sessler D.I., Figueroa P., Hoeltge G.A., Mihaljevic T., Blackstone E.H. Duration of red-cell storage and complications after cardiac surgery. N. Engl. J. Med. 2008;358:1229–1239. doi: 10.1056/NEJMoa070403. [DOI] [PubMed] [Google Scholar]
- 8.Kristiansen M., Graversen J.H., Jacobsen C., Sonne O., Hoffman H.-J., Law S.K.., Moestrup S.K. Identification of the haemoglobin scavenger receptor. Nature. 2001;409:198–201. doi: 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen M.J., Andersen C.B., Moestrup S.K. CD163 binding to haptoglobin-hemoglobin complexes involves a dual-point electrostatic receptor-ligand pairing. J. Biol. Chem. 2013;288:18834–18841. doi: 10.1074/jbc.M113.471060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Åkerström B., Maghzal G.J., Winterbourn C.C., Kettle A.J. The lipocalin α1-microglobulin has radical scavenging activity. J. Biol. Chem. 2007;282:31493–31503. doi: 10.1074/jbc.M702624200. [DOI] [PubMed] [Google Scholar]
- 11.Anderson U.D., Gram M., Ranstam J., Thilaganathan B., Åkerström B., Hansson S.R. Fetal hemoglobin, α 1-microglobulin and hemopexin are potential predictive first trimester biomarkers for preeclampsia. Pregnancy Hypertens. 2016;6:103–109. doi: 10.1016/j.preghy.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Schaer D.J., Buehler P.W., Alayash A.I., Belcher J.D., Vercellotti G.M. Hemolysis and free hemoglobin revisited: exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins. Blood. 2013;121:1276–1284. doi: 10.1182/blood-2012-11-451229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan W.B., Cheng W., Webber A., Bhambhani A., Duff M.R., Kumar C.V., McLendon G.L. Endonuclease-like activity of heme proteins. J. Biol. Inorg. Chem. 2005;10:790–799. doi: 10.1007/s00775-005-0028-y. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y., Du K.J., Gao S.Q., He B., Wen G.B., Tan X., Lin Y.-W. Distinct mechanisms for DNA cleavage by myoglobin with a designed heme active center. J. Inorg. Biochem. 2016;156:113–121. doi: 10.1016/j.jinorgbio.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Deshpande M.S., Junedi S., Prakash H., Nagao S., Yamanaka M., Hirota S. DNA cleavage by oxymyoglobin and cysteine-introduced metmyoglobin. Chem. Commun. 2014;50:15034–15036. doi: 10.1039/c4cc06617k. [DOI] [PubMed] [Google Scholar]
- 16.Glei M., Klenow S., Sauer J., Wegewitz U., Richter K., Pool-Zobel B.L. Hemoglobin and hemin induce DNA damage in human colon tumor cells HT29 clone 19A and in primary human colonocytes. Mutat. Res-Fundam. Mol. M. 2006;594:162–171. doi: 10.1016/j.mrfmmm.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Park J.H., Park E. Influence of iron-overload on DNA damage and its repair in human leukocytes in vitro. Mutat. Res-Gen. Toxicol. Environ. 2011;718:56–61. doi: 10.1016/j.mrgentox.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Chakane S., Markad V., Kodam K., Bulow L. The Penultimate tyrosine residues are critical for the genotoxic effect of human hemoglobin. Adv. Exp. Med. Biol. 2017;977 doi: 10.1007/978-3-319-55231-6_46. [DOI] [PubMed] [Google Scholar]
- 19.Patrinos G.P., Giardine B., Riemer C., Miller W., Chui D.H., Anagnou N.P., Wajcman H., Hardison R.C. Improvements in the HbVar database of human hemoglobin variants and thalassemia mutations for population and sequence variation studies. Nucleic Acids Res. 2004;32:D537–D541. doi: 10.1093/nar/gkh006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansson H.O., Matos T., Luz J.S., Feitosa E., Oliveira C.C., Pessoa A., Bülow L., Tjerneld F., Plasmid DNA partitioning and separation using poly (ethylene glycol)/poly (acrylate)/salt aqueous two-phase systems. J. Chromatogr. A. 2012;1233:30–35. doi: 10.1016/j.chroma.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 21.Ratanasopa K., Cedervall T., Bülow L. Possibilities of using fetal hemoglobin as a platform for producing hemoglobin-based oxygen carriers (HBOCs) Adv. Exp. Med. Biol. 2016;876:445–453. doi: 10.1007/978-1-4939-3023-4_56. [DOI] [PubMed] [Google Scholar]
- 22.Saran M., Michel C., Stettmaier K., Bors W. Arguments against the significance of the Fenton reaction contributing to signal pathways under in vivo conditions. Free Radic. Res. 2000;33:567–579. doi: 10.1080/10715760000301101. [DOI] [PubMed] [Google Scholar]
- 23.Antonini E., Brunori M. North-Holland Pub. Co.; Amsterdam: 1971. Hemoglobin and Myoglobin in Their Reactions with Ligands. [Google Scholar]
- 24.Mollan T.L., Jia Y., Banerjee S., Wu G., Kreulen R.T., Tsai A.-L., Olson J.S., Crumbliss A.L., Alayash A.I. Redox properties of human hemoglobin in complex with fractionated dimeric and polymeric human haptoglobin. Free Radic. Biol. Med. 2014;69:265–277. doi: 10.1016/j.freeradbiomed.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratanasopa K., Strader M.B., Alayash A.I., Bulow L. Dissection of the radical reactions linked to fetal hemoglobin reveals enhanced pseudoperoxidase activity. Front. Physiol. 2015;6 doi: 10.3389/fphys.2015.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kassa T., Jana S., Strader M.B., Meng F., Jia Y., Wilson M.T., Alayash A.I. Sickle cell hemoglobin in the ferryl state promotes βCys-93 oxidation and mitochondrial dysfunction in epithelial lung cells (E10) J. Biol. Chem. 2015;290:27939–27958. doi: 10.1074/jbc.M115.651257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapralov A., Vlasova I.I., Feng W., Maeda A., Walson K., Tyurin V.A., Huang Z., Aneja R.K., Carcillo J., Bayır H., Kagan V.E. Peroxidase activity of hemoglobin·haptoglobin complexes covalent aggregation and oxidative stress in plasma and macrophages. J. Biol. Chem. 2009;284:30395–30407. doi: 10.1074/jbc.M109.045567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vitturi D.A., Sun C.-W., Harper V.M., Thrash-Williams B., Cantu-Medellin N., Chacko B.K., Peng N., Dai Y., Wyss J.M., Townes T., Patel R.P. Antioxidant functions for the hemoglobin β93 cysteine residue in erythrocytes and in the vascular compartment in vivo. Free Radic. Biol. Med. 2013;55:119–129. doi: 10.1016/j.freeradbiomed.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reeder B.J. The redox activity of hemoglobins: from physiologic functions to pathologic mechanisms. Antioxid. Redox Signal. 2010;13:1087–1123. doi: 10.1089/ars.2009.2974. [DOI] [PubMed] [Google Scholar]
- 30.Moore E.E. Blood substitutes: the future is now. J. Am. Coll. Surg. 2003;196:1–17. doi: 10.1016/s1072-7515(02)01704-0. [DOI] [PubMed] [Google Scholar]
- 31.Strader M.B., Alayash A.I. Exploring oxidative reactions in hemoglobin variants using mass spectrometry: lessons for engineering oxidatively stable oxygen therapeutics. Antioxid. Redox Signal. 2016 doi: 10.1089/ars.2016.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svistunenko D.A., Dunne J., Fryer M., Nicholls P., Reeder B.J., Wilson M.T., Bigotti M.G., Cutruzzolà F., Cooper C.E. Comparative study of tyrosine radicals in hemoglobin and myoglobins treated with hydrogen peroxide. Biophys. J. 2002;83:2845–2855. doi: 10.1016/S0006-3495(02)75293-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H., Xu Y., Joseph J., Kalyanaraman B. Intramolecular electron transfer between tyrosyl radical and cysteine residue inhibits tyrosine nitration and induces thiyl radical formation in model peptides treated with myeloperoxidase, H2O2, and NO2-EPR spin trapping studies. J. Biol. Chem. 2005;280:40684–40698. doi: 10.1074/jbc.M504503200. [DOI] [PubMed] [Google Scholar]
- 34.Cooper C.E., Schaer D.J., Buehler P.W., Wilson M.T., Reeder B.J., Silkstone G., Svistunenko D.A., Bulow L., Alayash A.I. Haptoglobin binding stabilizes hemoglobin ferryl iron and the globin radical on tyrosine β145. Antioxid. Redox Signal. 2013;18:2264–2273. doi: 10.1089/ars.2012.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong L., Liu Z., Hu X., Liu S., Meng J. Spectroscopic study on interaction between bovine hemoglobin and salmon DNA and the analytical applications. J. Lumin. 2013;137:186–190. [Google Scholar]
- 36.Zhang Z.W., Cheng J., Xu F., Chen Y.E., Du J.B., Yuan M., Zhu F., Xu X.C., Yuan S. Red blood cell extrudes nucleus and mitochondria against oxidative stress. IUBMB Life. 2011;63:560–565. doi: 10.1002/iub.490. [DOI] [PubMed] [Google Scholar]
- 37.Lang K.S., Duranton C., Poehlmann H., Myssina S., Bauer C., Lang F., Wieder T., Huber S.M. Cation channels trigger apoptotic death of erythrocytes. Cell Death Differ. 2003;10:249–256. doi: 10.1038/sj.cdd.4401144. [DOI] [PubMed] [Google Scholar]
- 38.Hattangadi S.M., Wong P., Zhang L., Flygare J., Lodish H.F. From stem cell to red cell: regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood. 2011;118:6258–6268. doi: 10.1182/blood-2011-07-356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sankaran V.G., Xu J., Orkin S.H. Advances in the understanding of haemoglobin switching. Br. J. Haematol. 2010;149:181–194. doi: 10.1111/j.1365-2141.2010.08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vallelian F., Garcia-Rubio I., Puglia M., Kahraman A., Deuel J.W., Engelsberger W.R., Mason R.P., Buehler P.W., Schaer D.J. Spin trapping combined with quantitative mass spectrometry defines free radical redistribution within the oxidized hemoglobin: haptoglobin complex. Free Radic. Biol. Med. 2015;85:259–268. doi: 10.1016/j.freeradbiomed.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 41.Nantasenamat C., Prachayasittikul V., Bulow L. Molecular modeling of the human hemoglobin-haptoglobin complex sheds light on the protective mechanisms of haptoglobin. PLoS One. 2013;8:e62996. doi: 10.1371/journal.pone.0062996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaer D.J., Vinchi F., Ingoglia G., Tolosano E., Buehler P.W. Haptoglobin, hemopexin, and related defense pathways—basic science, clinical perspectives, and drug development. Front. Physiol. 2014;5:415. doi: 10.3389/fphys.2014.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]