Abstract
Single cell sequencing (SCS) has become a new approach to study biological heterogeneity. The advancement in technologies for single cell isolation, amplification of genome/transcriptome and next-generation sequencing enables SCS to reveal the inherent properties of a single cell from the large scale of the genome, transcriptome or epigenome at high resolution. Recently, SCS has been widely applied in various clinical and research fields, such as cancer biology and oncology, immunology, microbiology, neurobiology and prenatal diagnosis. In this review, we will discuss the development of SCS methods and focus on the latest clinical and research applications of SCS.
Introduction
The majority of experimental and clinical results from cell culture or tissues are based on the assumption that all of the cells in a culture or tissue are homogeneous. The thriving omics fields of study (genomics, proteomics, transcriptomics, etc.) analyze and mine biomarkers mainly based on the bulk of cells or tissue samples. However, this averaging of messages always misses the critical information because the heterogeneity of the samples is ignored, while the nature of biology is diverse. Heterogeneity is generally explained at three different levels in the biological universe: first, there is heterogeneity in different organisms; second, there is heterogeneity in different organs or tissues from an organism; third, cellular heterogeneity exists in the same organ or tissue. In fact, the concept of cellular heterogeneity was proposed as early as 1957 [1]. Each cell was considered as a unique unit with molecular coding across the DNA, RNA, and protein conversions [2]. Thus, it is necessary to conduct studies, especially omics studies, at the single cell level.
A single cell is the smallest structural and functional unit of an organism. The estimated number of single cells in the human body is 3.72 × 1013 [3]. The size or weight of a cell varies from different tissue backgrounds. The major components of a cell include water, inorganic ions, small organic molecules, proteins, RNA and DNA. However, the minute numbers of copies of a gene (10–12 M) in a single cell are more than enough for conventional genomic analysis [2, 4]. In 2009, the first single cell whole transcriptome sequencing (WTA) protocol was applied to analyze transcriptome complexity in individual cells [5]. Subsequently, single cell whole genome sequencing (WGS) was created in 2011 [6], single cell whole exome sequencing (WES) was developed in 2012 [7, 8], and single cell epigenomic sequencing was developed in 2013 [9]. Currently, single cell sequencing (SCS) has been applied in various research and clinical fields, and the top five areas of SCS studies in order are cancer, embryonic development, microbiology, neurobiology and immunology, according to the reported statistics [10]. The number of SCS publications in these five areas has been increasing every year. Thus, this article will enable us to have a deep and broad view of SCS methods and to focus on the latest application of SCS in basic and clinical research.
Single cell isolation methods
Isolating single cells from a tissue mass or from cell culture is the first key step prior to SCS. Currently, the alternative methods used to isolate single cells from abundant populations include serial dilution, mechanical micromanipulation, laser capture microdissection (LCM), fluorescence activated cell sorting (FACS), and microfluidics [11, 12]. Although serial dilution is the simplest method to obtain a single cell in a single well via serial double dilution, it is a coarse and imprecise method that is rarely used in SCS (Fig. 1a). Our team has tried to use this method to isolate a single cell from primary lung cancer cells in cell suspension and found that it was hard to control the quality and quantity [13].
Fig. 1.

The current methods for single cell isolation. a Serial dilution. b Mechanical micromanipulation. c Laser capture microdissection (LCM). d Fluorescence activated cell sorting (FACS). e Microfluidics. f The representative platform for circulating tumor cells (CTCs) isolation: CellSearch
Mechanical micromanipulation is a classic method to isolate uncultivated microorganisms or early embryos, and it involves using a capillary pipette to suck up a single cell from a cell suspension with visual inspection of cellular morphology and coloring characteristics under a microscope [13, 14] (Fig. 1b). The drawback of mechanical micromanipulation is that it is low-throughput and time-consuming and can cause cellular injury from mechanical shearing during manipulation [15]. Additionally, it often leads to a failure for an unskilled manipulator or misidentification of the cellular morphology under the microscope.
FACS is the most efficient and economical method to isolate hundreds of thousands of individual cells per minute based on their size, granularity and fluorescence properties [4] (Fig. 1c). The high-throughput, time-saving and automatic properties are the main advantage of FACS. Additionally, it allows researchers to isolate specific individual cells from heterogeneous cell samples by labeling the targeted cells with specific fluorescent antibodies [16], and it allows researchers to sort a single viral particle from a mixed viral assemblage for single viral genome sequencing [17]. BD Aria II/III (BD Biosciences, San Jose, CA, USA) and Beckman Coulter MO-FLO XDP cell sorter (Beckman Coulter, Brea, CA, USA) are two widely used commercial instruments for flow cytometry [11]. Our team has used the BD Aria III to sort individual living cells from lung cancer tissue single cell suspensions that were stained with carboxyfluorescein diacetate succinimidyl ester (CFSE) and sorted into 96- well plates [18]. However, a bulk population of the cells (at least 5 × 105–1 × 106/ml) should be prepared as sorting material, which is greatly limited in accommodating low-abundance cell subpopulations. The high-speed fluid and fluorescent dye can damage the viability of cells.
Microfluidics is a newly developed and highly integrated system that sequentially processes or manipulates small volumes of fluids (10−9–10−18 l) in channels with dimensions of tens to hundreds of micrometers to achieve single cell culture and sequencing, that has been applied to single cell experiments [19, 20] (Fig. 1e). Recently, various microfluidics platforms have emerged for single cell whole-genome, whole-transcriptome or epigenomics sequencing [21–23]. The advancement of microfluidics research has extended to separate nanoparticles, such as DNA isolation [24]. The advantages for microfluidics are the ability to input nanoliter-to-picoliter volumes of samples and to output accurate results with high resolution and sensitivity [19]. Additionally, microfluidics can provide parallel and timely analyses to make studies more efficient.
The main limitation of the above-mentioned methods is that the sample must be prepared in suspension and thus have lost the spatial location of the cells in the tissue. LCM overcomes this limitation and directly isolates single cells from tissue sections based on the cellular morphology (Fig. 1d). The targeted single cell can be stained with fluorescent or chromogenic antibodies for LCM [11]. The main drawbacks include low-throughput, slicing the cells during the course of tissue sectioning, and UV damage to nuclei from the laser [12].
The increasing number of studies on rare single cells (<1%) poses a challenge on the current methods for single cell isolation. Now, several new technologies have been developed to cover the shortcomings of the above-mentioned methods in rare single cancer cell isolation, such as CellSearch (Johnson & Johnson), MagSweeper (Illumina Inc.), DEP-Array (Silicon Biosciences), CellCelector (Automated Lab Solutions), and nanofabricated filters (CellSieve) [25]. The FDA-cleared CellSearch system is the most-advanced commercially available technology using anti-EpCAM ferrofluid and has been applied to the monitoring of patients with metastatic prostate, breast, or colorectal cancer in hospitals [26, 27] (Fig. 1f). MagSweeper is an automated immunomagnetic separation technology for enrichment of rare cells in mixed populations with high purity [28]. DEP-Array uses a microfluidics chip with dielectrophoretic cages to isolate single cells by charge [12, 29]. The CellCelector uses a robotic arm carrying a module to retrieve single cells from microwells for micromanipulation [30]. The CellSieve system can capture a variety of circulating tumor cells based on size discrimination instead of specific cell surface markers [31].
Single cell sequencing methods
The advance in the next-generation sequencing (NGS) technologies has promoted the rapid development of SCS, including single cell whole-genome sequencing, single cell whole-exome sequencing, single cell whole-transcriptomic sequencing and single cell epigenomic sequencing [32–34].
Single cell whole-genome/whole-exome sequencing
The amount of DNA (approximately 6 pg) in a single cell is insufficient to meet the demand for next-generation sequencing, and thus whole genome amplification (WGA) was developed to amplify the DNA by the hundreds of thousands [25, 35]. Recently, the alternative WGA technologies have polymerase chain reaction (PCR), multiple displacement amplification (MDA), or a combination of displacement pre-amplification and PCR amplification [36]. In PCR-based WGA methods, degenerate oligonucleotide-primed PCR (DOP-PCR) is the most widely used method to amplify the entire genome [37, 38]. The principle of DOP-PCR is to perform a low annealing degenerate primer extension on the DNA template and then to amplify the tagged sequences at a high annealing temperature [37, 39] (Fig. 2a). The main shortcoming for DOP-PCR is the low physical coverage (approximately 10%) of a single cell genome, which is prone to miss single-nucleotide polymorphisms (SNPs), but DOP-PCR is the optimal method for copy-number variations (CNVs) or aneuploidy detection because of the uniformity of amplification during WGS [12, 40, 41]. The established MDA technologies are based on the discovery of two specific DNA polymerases: Phi29 DNA polymerase isolated from the Bacillus subtilis, and Bst DNA polymerase isolated from Bacillus stearothermophilus [42–44]. The mechanism of MDA is to yield continuous strand displacement DNA amplification using Phi29 or Bst DNA polymerase and random primers under isothermal conditions [45] (Fig. 2b). Phi29 DNA polymerase has been considered the optimal choice for MDA because it shows higher efficiency, higher fidelity and a lower error rate compared with Bst DNA polymerase which has no proofreading activity [10, 46]. The advantages of MDA are that it has high single cell genome or exome coverage (>90%), which can accurately measure mutations at base-pair resolution and that it yields adequate quantities of product (average length >10 kb) from single cell genomic DNA in a short time with high fidelity [47]. However, the main drawbacks of MDA are uneven genome coverage, chimeric sequences, and contamination issues [15]. Multiple annealing and looping based amplification cycles (MALBAC) is the newly applied WGS method that combines quasi-linear strand displacement pre-amplification by a polymerase and exponential amplification by PCR [33] (Fig. 2c). Remarkably, MALBAC has low amplification bias and can achieve 93% genome coverage ≥1× and 25× mean sequencing depth in a single cell during WGS. Moreover, MALBAC shows higher efficiency to detect CNVs and SNPs for its improved uniformity and a lower allele dropout rate, compared with MDA [36]. The pitfall of MALBAC is the extremely high false positive rate for SNV detection because of the low fidelity of Bst DNA polymerase, and the loss of underamplified regions of the genome [48]. Another improved method, nuc-seq or single nucleus exome sequencing (SNES), has been developed to reduce the bias, this method combines flow-sorting of single G1/0 or G2/M nuclei, time-limited isothermal MDA, exome capture, and NGS [49, 50]. The main advantage of this method is the high detection efficiencies for single-nucleotide variations (SNVs) and indels benefiting from the high physical coverage (96%) of the single cell genome and exome [50].
Fig. 2.
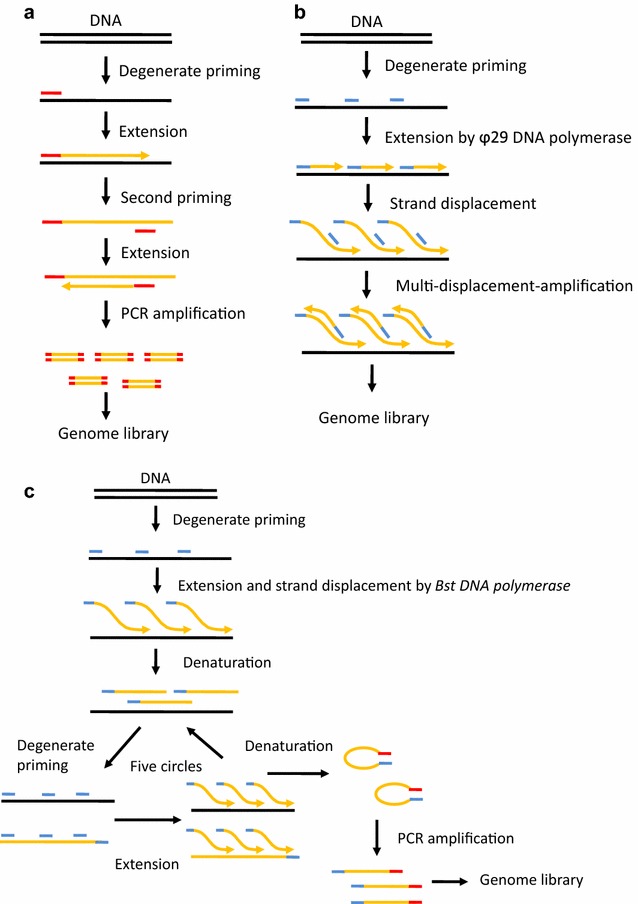
Schematic of the whole genome amplification methods for single cell sequencing. a Degenerate oligonucleotide-primed PCR (DOP-PCR). b Multiple displacement amplification (MDA). c Multiple annealing and looping based amplification cycles (MALBAC)
Single cell whole-transcriptomic sequencing
It is estimated that the amount of total RNA or mRNA is only approximately 10 pg or approximately 0.1 pg, respectively, in a single cell [10]. Thus, WTA is a necessary step to construct a cDNA library for single cell transcriptomic sequencing. WTA has been applied to amplify RNA from a single cell to obtain the gene expression profile in microarray prior to the advent of NGS [51, 52]. Tang et al. [5] improved the single cell whole-transcriptome amplification method and used NGS instead of microarray to identify more genes and previously unknown splice junctions in single cells. The principle of this method is to use oligo dT primers conjugated to adapter sequences for reverse transcription and selective amplification of polyadenylated mRNA by PCR [5, 10, 53] (Fig. 3a). However, this method generates 3′-end skew bias during reverse-transcription to miss proximal splicing events [34]. Another WTA method, called SMART-seq, was developed to use Moloney murine leukemia virus (MMLV) reverse transcriptase to construct full-length cDNA libraries [54]. The two key features, template-switching and terminal transferase activity, of the enzyme can lead to adding a few non-templated C nucleotides to the cDNA and switching templates to transcribe the other strand [55] (Fig. 3b). The advantage of SMART-seq is to generate and amplify full-length cDNA from single cell RNA, leading to the detection of alternatively spliced exons [56]. The low sensitivity of SMART is the main shortcoming that was improved in a subsequently developed method, called SMART-seq2 [57]. Similarly, single cell tagged reverse-transcription (STRT) is based on the template-switching property of the reverse transcriptase to tag the 5′-end of cDNA [58]. This method enables researchers to compare gene expression profile differences without bias in multiple samples, but it yields a strong 5′-end bias. Cell expression by linear amplification and sequencing (Cel-seq) labels cDNA with a barcode and pools these cDNA from multiple single cells for in vitro transcription (IVT) to linearly amplify cDNA [59]. The CEL-Seq generates more reproducible, linear, and sensitive results in comparison with the PCR-based amplification method, but it yields a high 3′-end skew bias and loses the full spectrum of transcript variant detection [60]. Additionally, the unique molecular identifiers (UMIs) labeling technique is applied in single cell WTA to achieve quantitative single cell RNA sequencing [61] (Fig. 3c). This method obviously increases the accuracy in single cell whole-transcriptome sequencing by eliminating amplification bias. Recently, two new droplet-based RNA-seq technologies, named as Drop-seq and inDrop (indexing droplets), has been exploited to sequence in parallel thousands of single cells from a tissue [62, 63]. Each nanoliter-scale aqueous droplet is a tiny reaction chamber that contains a single cell, barcoded and UMI-labeled primers, and reaction buffer. STAMPs (Single-cell Transcriptomes Attached to Microparticles) is PCR amplified for sequencing in Drop-seq, while Cell-seq is used by inDrop for sequencing. The advantages of these methods are to differentiate the cell-of-origin of each mRNA which helps to develop single cell analysis in a complicated tissue, and the low technical noise that allows the analysis of thousands of different cells in parallel. The latest commercial platform—Chromium™ System from 10× Genomics—integrates the Gemcode platform, which separates long pieces of DNA into droplets to create barcoded sequencing libraries [64, 65]. The high efficiency and flexible throughput of this method allows researchers to dynamically detect transcriptional profiles of single cells at scale [66].
Fig. 3.

Schematic of the whole transcriptome amplification methods for single cell sequencing. a oligo dT-Anchor approach. b Template-switching approach (including SMART-seq, SMART-seq2 and STRT). c Unique molecular identifiers (UMIs)
Single cell epigenomic sequencing
Epigenomics is defined as a phenomenon that changes the final outcome of a chromosome without changing the underlying DNA sequence, including DNA methylation, histone modifications, chromatin packaging, small RNA, etc. [67]. Recently, single cell epigenomic sequencing studies are on the rise with the application of new single cell epigenomic sequencing methods. Single cell reduced representation bisulfite sequencing (scRRBS) integrates all the experimental steps before PCR amplification into a single-tube reaction to avoid unnecessary DNA loss and enables the detection of approximately 40% of the CpG sites in comparison with standard RRBS using thousands of cells [68, 69]. Another method, single cell bisulfite sequencing (scBS-seq), modifies the Post-Bisulfite Adaptor Tagging (PBAT) to perform bisulfite conversion prior to successive primer extension with oligo1 and oligo2 tagged random primers to generate amplicons [70]. The drawbacks of these methods are DNA loss, purification, and disability to discriminate 5mC from 5hmC for bisulfite conversion [34]. Moreover, single cell chromatin immunoprecipitation followed by sequencing (scChIP-seq) combines microfluidics, DNA barcoding and sequencing to collect low coverage maps of the chromatin state at single cell resolution [71]. Additionally, other methods have been developed for single cell epigenomic sequencing, such as Hi-C methods that characterize chromatin interactions in the genome of single cells [9], and single chromatin molecule analysis at the nanoscale (SCAN) that extracts single chromatin with fluorescent antibodies through fluidic channels [72].
Application of single cell sequencing
Cancer
Cancer heterogeneity comes from clone diversity and mutational evolution, which promote cancer cell survival and metastasis and confound the cancer diagnosis and treatment [10, 73]. A deep understanding of cancer heterogeneity can contribute to therapeutic decisions. Thus, SCS as an ideal tool has been increasingly applied to reveal intratumor heterogeneity in various primary tumors, such as breast cancer [6, 49, 74], lung cancer [75], brain cancer [76], colon cancer [33, 77], bladder cancer [78], acute myeloid leukemia [79, 80] and melanoma [81].
Navin et al. [6] first applied single nucleus sequencing (SNS) to study tumor population structure and evolution in two breast cancer cases by analysis of genome copy number variation. The results found punctuated clonal evolution in tumors and confirmed that metastatic cells emerged from a main advanced expansion. Another study used nuc-seq to find a difference in the pattern of occurrence for aneuploid rearrangements and point mutations in breast tumor evolution [49]. Furthermore, Eirew et al. [74] studied the dynamics of genomic clones in breast cancer patient xenografts at single cell resolution to indicate that genomic aberrations can be reproducible determinants of evolutionary trajectories. Interestingly, a single cell whole genome sequencing study for colon cancer identified an abundant amount of mutated gene SLC12A5 at the individual level, which was sparse at the bulk cells level, and discovered that colon cancer had a biclonal origin [77]. However, another study using single cell exome sequencing to reveal the evolutionary process in bladder cancer indicated that 66 individual bladder cancer cells were derived from a single ancestral cell, but they developed into two distinct tumor cell subpopulations with subsequent evolution [78]. Single cell exome sequencing was also applied to elucidate the intratumoral genetic characteristics at a single cell level in a kidney cancer [8]. Additionally, the clonal evolution has been studied in hematopoietic tumors. Hughes et al. [79]. sequenced single cells from three myelodysplastic syndrome (MDS)-derived secondary acute myeloid leukemias (sAMLs) to confirm the clonal architecture that was identified from the bulk sample analysis. Single cell exome sequencing revealed a monoclonal evolution in a JAK2-negative myeloproliferative neoplasm and further identified candidate gene mutations for neoplasm progression [7].
In addition to single cell DNA and exome sequencing applications, single cell RNA-seq has also been widely used to study clonal evolution in different cancers. Single cell RNA-seq demonstrated subclonal heterogeneity in xenograft tumor cells and found a candidate tumor cell subpopulation associated with anti-cancer drug resistance in lung adenocarcinoma (ADC) [75]. Equally, intertumor and intratumor heterogeneity was elucidated in melanoma by single cell RNA-seq [81]. Tirosh et al. [76] used single cell RNA-seq to find a new subpopulation marked with stem or progenitor cell-like characteristics, which supported developmental programs in oligodendroglioma. Furthermore, another study identified several rare tumor-related genes in squamous cell carcinoma of urinary bladder using single cell RNA-seq [81].
Recently, an array of studies that used SCS to understand the necessary knowledge of different rare circulating cancer cells have been published. Ni et al. [82] combined MALBAC with NGS to elucidate the CNV patterns for metastasis of cancer in circulating tumor cells (CTCs) from lung ADC. Lohr et al. [83] sequenced entire exomes of CTCs from two prostate cancer patients and observed 73% CTC mutations that were identified in bulk tissue. The results were consistent with another study that compared CTCs with tissue using WGS in prostate cancer [84]. Additionally, one study built a new system to assess the genomic heterogeneity of single CTCs from metastatic breast cancer patients and found a cell subpopulation related to drug resistance [85]. However, a recent study indicated that a targeted mutation detection rate is approximately 27.7% in CTCs from pancreatic cancer compared with bulk cells but is negative in white blood cells [86]. Furthermore, single CTCs studies based on whole RNA-seq have also been published. Lohr et al. [87] classified multiple myeloma (MM) and quantitatively assessed prognosis related genes using single CTC RNA-seq. Another CTC RNA-seq study revealed that noncanonical Wnt signaling took part in antiandrogen resistance in prostate cancer [88].
Immunology
The heterogeneity of the immune system contributes to an efficient defense against a multitude of different pathogens [89]. The SCS technologies can help to define new classifications and differentiation trajectories of immune cells. CD4+ T helper cell, which play a key role in adaptive immune responses, are further investigated to unravel the heterogeneity of this celluar population at the single cell level. Mahata et al. [90] used single cell RNA-seq to reveal the extensive heterogeneity within the Th2 population and to identify a new Th2 cell subpopulation marked with Cyp11a1 that modulated the steroid synthesis pathway. Additionally, functional and structural studies of the T cell receptor repertoire have also benefited from SCS approaches. Dash et al. [91] developed a new method to sequence the TCRα and TCRβ chains from single CD8+ T cells. The data showed a characterized expression of TCRα for an influenza epitope. Another study combined TCRα and TCRβ sequencing with phenotypic analysis to reveal the clonal structure of T cells at the single cell level [92]. In addition to T cells, Shalek et al. [93] examined the mouse bone-marrow-derived dendritic cells (BMDCs), which is an important antigen-presenting cell subpopulation in the adaptive immune system, using single cell RNA-seq. The results indicated that hundreds of immune related genes displayed bimodal expression in single cells. Further study demonstrated that paracrine signaling from early-induced dendritic cells plays an important role in inflammatory program [94]. Although the application of SCS to study the immune system is limited at present, SCS has shown robust potential for defining immune cell subpopulations and for examining gene expression variability, differential splicing and gene-regulatory networks [89].
Microbiology
The vast majority of microorganisms are uncultivated with current culturing methods which has extremely limited our ability to understand the biological diversity of the microbiome [95]. Recently, the difficulty in microbial research has been overcome with the development of SCS. The first study combined FACS with MDA to sequence single TM7 bacterial cells from the soil and gained a deep insight into the evolution and metabolism of these cells [96]. The member of TM7 phylum from the human mouth was also investigated in a similar method [97]. The subsequent study conducted the single cell genomic sequencing in other candidate uncultured phyla from different environments, including anoxic spring-derived OP11 [98], human microbiota-derived SR1 [99], hospital sink biofilm-derived TM6 [100] and hot spring sediments-derived OP9 [101]. In addition to sequencing the genome of various bacterial phyla, SCS can reveal the lifestyle and metabolism of uncultivated microorganisms, supporting the potential development of cultivation approaches and commercial applications. Marc et al. [102] sequenced over 70% of the genome of Beggiatoa from the surface of marine sediment and confirmed the chemolithoautotrophic physiology via investigating the pathway for sulfur oxidation, oxygen and nitrate respiration, and carbon metabolism. The findings supported the establishment of a particulate cultivating environment in which there was coexistence of different members of the microbial community and some missing supplementary materials [95]. In another study, Mason et al. [103] used MDA to sequence and assemble the single cell genome of Oceanospirillales from seawater after the Deepwater Horizon oil spill and identified enzymes that can degrade crude oil.
Prenatal diagnosis
The application of SCS to prenatal diagnosis, including pre-implantation genetic diagnosis (PGD) and non-invasive prenatal diagnosis (NIPD) has greatly increased the opportunities for healthy birth [32]. Recently, SCS has been widely used to detect aneuploidy and SNPs in prenatal diagnosis. Well et al. [104] used a rapid WDA protocol to diagnosis of aneuploidy in embryo biopsy with high accuracy and cost-efficiency. In another study, Fiorentino et al. [105] confirmed the validation and accuracy of a single cell NGS-based method for aneuploidy screening in single blastomeres. In the subsequent study, they compared this protocol with array comparative genomic hybridization (array-CGH) and demonstrated that a single cell NGS-based method improved the aneuploidy detection with high-throughput, automation and reliability [106]. Furthermore, Vera-Rodríguez et al. [107] used single cell NGS to investigate the distribution patterns of segmental aneuploidies in trophectoderm biopsy. The efficiency of NGS in the detection of pure and mosaic segmental aneuploidies equated with that of CGH. Lu et al. [108] used MALBAC to sequence 99 sperm from an Asian male to detect aneuploidy and single nucleotide polymorphisms. The same method was used to accurately detect aneuploidy and SNPs in a single oocyte [109]. Additionally, using NIPD as a safe and reliable method to identify affected fetuses before birth is becoming increasingly popular for clinical and research applications in combination with NGS technologies. Zhang et al. [110] used low-coverage massively parallel sequencing to detect CNVs in four single cells from peripheral blood. The sensitivity and specificity for CNVs and aneuploidies were 99.63 and 97.71%, respectively. Hua et al. [111] used WGA and Illumina MiSeq to sequence single fetal nucleated red blood cells from placental villi and to diagnose aneuploidy in 5 cases in 10 single cells.
Neurobiology
Defining neuronal heterogeneity is an enormous task in nervous research [112]. SCS has been increasingly used to understand neural cell diversity and to classify neurons. Many studies have been reported to use single cell RNA-seq to classify the type of neurons in various regions of the mouse nervous system, including monoaminergic systems, dorsal root ganglia, cortex and retina [112]. Okaty et al. [113] used single cell RNA-seq to distinguish serotonergic neurons from five hindbrain rhombomeres and confirmed the subpopulation grouped from the population-scale transcriptomes. Additionally, the subtype-specific behavioral function-related genes were identified at the single cell level. In another study, Zeisel et al. [114] used large-scale single cell RNA-seq to sequence the neuronal cells from the somatosensory cortex and hippocampal CA1 region, and they identified an interneuron and a postmitotic oligodendrocyte labeled with Pax6 and ltpr2, respectively. Similarly, Tasic et al. [115] defined 49 transcriptomic cortical cell types, including 23 GABAergic, 19 glutamatergic and 7 non-neuronal types based on single cell RNA sequencing. In humans, the structure and function of brain is more complex. Johnson et al. [116] combined FACS and single cell RNA-seq to detect the heterogeneity in evolution of human outer radial glia (ORG). In another study, single cell RNA-seq was used to identify the transcriptome diversity in adult and fetal brains. The results indicated that there was differential gene expression between adult and fetal neurons, and the gradient patterns of gene expression contributed to the understanding of the evolution of neurons in the brain [117]. The latest study used single nucleus RNA-seq to sequence the single neuron from six distinct regions of the human cerebral cortex and identified 16 neuronal subtypes with subtype-specific transcriptome profiles [118]. Additionally, single cell DNA-seq was used for CNV detection in brain diseases. Using single cell DNA-seq, McConnell et al. [119] demonstrated that there were abundant mosaic CNVs in human neurons, especially in hiPSC-derived neurons. In another study, Cai et al. [120] used WGS to find a somatic CNV of chromosome 1q in more than 20% of neurons in a hemimegalencephaly (HMG) patient.
Conclusions
Biological heterogeneity must be considered in clinical and basic studies. With the advancement of next-generation sequencing, SCS, including single cell genomic, transcriptomic and epigenomic sequencing, has been become the major tool to unlock the secrets of biological diversity [41]. Recently, the application of SCS has been widespread in various research fields, such as cancer, immunology, microbiology, neurobiology and embryogenesis, and many successful commercial kits have emerged in the market [34]. Most exciting is the transformation of the use of SCS from bench to bedside. For example, SCS has been applied to the assessment of human embryos prior to implantation, non-invasive prenatal diagnosis and cancer diagnosis and prognosis [32]. However, there are still several shortcomings of SCS [10, 25]. It is hard to comprehensively and simultaneously sequence the genome, transcriptome and epigenome in a single cell. The high cost of SCS impedes its clinical application, and thus it will be a great challenge for researchers and engineers to provide highly efficient and low-cost technologies in SCS. Furthermore, in situ, real-time and in vivo sequencing and analysis of the DNA and RNA from single cells will be a new field that obtains deep insight into the spatial and temporal measurement of the molecular profiles of single cells. Lastly, new analysis models for the enormous data obtained from SCS should be built to unbiasedly mine the inherent properties of a single cell. Although still evolving, new SCS technologies have become powerful approaches for us to unravel the complexities of nature.
Authors’ contributions
JW was involved in the conception, design and drafting of the manuscript. YLS was involved in the revision and final acceptance of the manuscript. Both authors read and approved the final manuscript.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (81500026,81490533,81570028,81400018) and the Shanghai Science and Technology Committee (15DZ1930602). Dr. YL Song was supported by the State Key Basic Research Program (973) project (2015CB553404) and by the Doctoral Fund of the Ministry of Education of China (20130071110044).
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Jian Wang, Email: 251328610@qq.com.
Yuanlin Song, Email: song.yuanlin@zs-hospital.sh.cn.
References
- 1.Novick A, Weiner M. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci USA. 1957;43(7):553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coskun AF, Eser U, Islam S. Cellular identity at the single-cell level. Mol Bio Syst. 2016;12(10):2965–2979. doi: 10.1039/c6mb00388e. [DOI] [PubMed] [Google Scholar]
- 3.Bianconi E, Piovesan A, Facchin F, et al. An estimation of the number of cells in the human body. Ann Hum Biol. 2013;40(6):463–471. doi: 10.3109/03014460.2013.807878. [DOI] [PubMed] [Google Scholar]
- 4.Lindstrom S, Andersson-Svahn H. Overview of single-cell analyses: microdevices and applications. Lab Chip. 2010;10(24):3363–3372. doi: 10.1039/c0lc00150c. [DOI] [PubMed] [Google Scholar]
- 5.Tang F, Barbacioru C, Wang Y, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 6.Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou Y, Song L, Zhu P, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148(5):873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 8.Xu X, Hou Y, Yin X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148(5):886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagano T, Lubling Y, Stevens TJ, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502(7469):59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol Cell. 2015;58(4):598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navin N, Hicks J. Future medical applications of single-cell sequencing in cancer. Genome Med. 2011;3(5):31. doi: 10.1186/gm247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navin NE. Cancer genomics: one cell at a time. Genome Biol. 2014;15(8):452. doi: 10.1186/s13059-014-0452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang D, Wang X (2015) A simple protocol for single lung cancer cell isolation-making the single cell based lung cancer research feasible for individual investigator. In Single cell sequencing and systems immunology. Springer, Berlin
- 14.Brehm-Stecher BF, Johnson EA. Single-cell microbiology: tools, technologies, and applications. Microbiol Mol Biol Rev. 2004;68(3):538–559. doi: 10.1128/MMBR.68.3.538-559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yilmaz S, Singh AK. Single cell genome sequencing. Curr Opin Biotechnol. 2012;23(3):437–443. doi: 10.1016/j.copbio.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Boehmer L, Liu C, Ackerman S, et al. Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat Protoc. 2016;11(10):1908–1923. doi: 10.1038/nprot.2016.102. [DOI] [PubMed] [Google Scholar]
- 17.Allen LZ, Ishoey T, Novotny MA, et al. Single virus genomics: a new tool for virus discovery. PLoS ONE. 2011;6(3):e17722. doi: 10.1371/journal.pone.0017722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Min Z, Jin M, et al (2015) Protocol for single cell isolation by flow cytometry. In Single cell sequencing and systems immunology. Springer, Berlin
- 19.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442(7101):368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 20.Le Gac S, Nordhoff V. Microfluidics for mammalian embryo culture and selection: where do we stand now? Mol Hum Reprod. 2016;27:61. doi: 10.1093/molehr/gaw061. [DOI] [PubMed] [Google Scholar]
- 21.Streets AM, Zhang X, Cao C, et al. Microfluidic single-cell whole-transcriptome sequencing. Proc Natl Acad Sci USA. 2014;111(19):7048–7053. doi: 10.1073/pnas.1402030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han L, Zi X, Garmire LX, et al. Co-detection and sequencing of genes and transcripts from the same single cells facilitated by a microfluidics platform. Sci Rep. 2014;4:6485. doi: 10.1038/srep06485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu AR, Kawahara TL, Rapicavoli NA, et al. High throughput automated chromatin immunoprecipitation as a platform for drug screening and antibody validation. Lab Chip. 2012;12(12):2190–2198. doi: 10.1039/c2lc21290k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salafi T, Zeming KK, Zhang Y. Advancements in microfluidics for nanoparticle separation. Lab Chip. 2016;17:11–33. doi: 10.1039/C6LC01045H. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Marjani SL, Hu Z, et al. Single-cell sequencing for precise cancer research: progress and prospects. Cancer Res. 2016;76(6):1305–1312. doi: 10.1158/0008-5472.CAN-15-1907. [DOI] [PubMed] [Google Scholar]
- 26.Swennenhuis JF, van Dalum G, Zeune LL, et al. Improving the cell search(R) system. Expert Rev Mol Diagn. 2016;16(12):1291–1305. doi: 10.1080/14737159.2016.1255144. [DOI] [PubMed] [Google Scholar]
- 27.Pantel K, Alix-Panabieres C, Riethdorf S. Cancer micrometastases. Nat Rev Clin Oncol. 2009;6(6):339–351. doi: 10.1038/nrclinonc.2009.44. [DOI] [PubMed] [Google Scholar]
- 28.Talasaz AH, Powell AA, Huber DE, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci USA. 2009;106(10):3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altomare L, Borgatti M, Medoro G, et al. Levitation and movement of human tumor cells using a printed circuit board device based on software-controlled dielectrophoresis. Biotechnol Bioeng. 2003;82(4):474–479. doi: 10.1002/bit.10590. [DOI] [PubMed] [Google Scholar]
- 30.Choi JH, Ogunniyi AO, Du M, et al. Development and optimization of a process for automated recovery of single cells identified by microengraving. Biotechnol Prog. 2010;26(3):888–895. doi: 10.1002/btpr.374. [DOI] [PubMed] [Google Scholar]
- 31.Adams DL, Adams DK, Alpaugh RK, et al. Circulating cancer-associated macrophage-like cells differentiate malignant breast cancer and benign breast conditions. Cancer Epidemiol Biomark Prev. 2016;25(7):1037–1042. doi: 10.1158/1055-9965.EPI-15-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu W, Zhang XY, Marjani SL, et al. Next-generation molecular diagnosis: single-cell sequencing from bench to bedside. Cell Mol Life Sci. 2016;13:1. doi: 10.1007/s00018-016-2368-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zong C, Lu S, Chapman AR, et al. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338(6114):1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang J, Cai W, Sun Z. Single-cell sequencing technologies: current and future. J Genet Genom. 2014;41(10):513–528. doi: 10.1016/j.jgg.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Van Loo P, Voet T. Single cell analysis of cancer genomes. Curr Opin Genet Dev. 2014;24:82–91. doi: 10.1016/j.gde.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 36.Grun D, van Oudenaarden A. Design and analysis of single-cell sequencing experiments. Cell. 2015;163(4):799–810. doi: 10.1016/j.cell.2015.10.039. [DOI] [PubMed] [Google Scholar]
- 37.Telenius H, Carter NP, Bebb CE, et al. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13(3):718–725. doi: 10.1016/0888-7543(92)90147-K. [DOI] [PubMed] [Google Scholar]
- 38.Huang L, Ma F, Chapman A, et al. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genom Hum Genet. 2015;16:79–102. doi: 10.1146/annurev-genom-090413-025352. [DOI] [PubMed] [Google Scholar]
- 39.Arneson N, Hughes S, Houlston R, et al. Whole-genome amplification by degenerate oligonucleotide primed PCR (DOP-PCR) CSH Protoc. 2008;2008:t4919. doi: 10.1101/pdb.prot4919. [DOI] [PubMed] [Google Scholar]
- 40.Hou Y, Wu K, Shi X, et al. Comparison of variations detection between whole-genome amplification methods used in single-cell resequencing. Gigascience. 2015;4:37. doi: 10.1186/s13742-015-0068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baslan T, Hicks J. Single cell sequencing approaches for complex biological systems. Curr Opin Genet Dev. 2014;26:59–65. doi: 10.1016/j.gde.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 42.Zhang DY, Zhang W, Li X, et al. Detection of rare DNA targets by isothermal ramification amplification. Gene. 2001;274(1–2):209–216. doi: 10.1016/S0378-1119(01)00607-2. [DOI] [PubMed] [Google Scholar]
- 43.Aliotta JM, Pelletier JJ, Ware JL, et al. Thermostable Bst DNA polymerase I lacks a 3′ –> 5′ proofreading exonuclease activity. Genet Anal. 1996;12(5–6):185–195. doi: 10.1016/S1050-3862(96)80005-2. [DOI] [PubMed] [Google Scholar]
- 44.Baner J, Nilsson M, Mendel-Hartvig M, et al. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res. 1998;26(22):5073–5078. doi: 10.1093/nar/26.22.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dean FB, Nelson JR, Giesler TL, et al. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11(6):1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spits C, Le Caignec C, De Rycke M, et al. Optimization and evaluation of single-cell whole-genome multiple displacement amplification. Hum Mutat. 2006;27(5):496–503. doi: 10.1002/humu.20324. [DOI] [PubMed] [Google Scholar]
- 47.Dean FB, Hosono S, Fang L, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99(8):5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lasken RS. Single-cell sequencing in its prime. Nat Biotechnol. 2013;31(3):211–212. doi: 10.1038/nbt.2523. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Waters J, Leung ML, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512(7513):155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leung ML, Wang Y, Waters J, et al. SNES: single nucleus exome sequencing. Genome Biol. 2015;16:55. doi: 10.1186/s13059-015-0616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurimoto K, Yabuta Y, Ohinata Y, et al. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res. 2006;34(5):e42. doi: 10.1093/nar/gkl050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iscove NN, Barbara M, Gu M, et al. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat Biotechnol. 2002;20(9):940–943. doi: 10.1038/nbt729. [DOI] [PubMed] [Google Scholar]
- 53.Tang F, Barbacioru C, Nordman E, et al. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc. 2010;5(3):516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramskold D, Luo S, Wang YC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30(8):777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu YY, Machleder EM, Chenchik A, et al. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques. 2001;30(4):892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
- 56.Goetz JJ, Trimarchi JM. Transcriptome sequencing of single cells with Smart-Seq. Nat Biotechnol. 2012;30(8):763–765. doi: 10.1038/nbt.2325. [DOI] [PubMed] [Google Scholar]
- 57.Picelli S, Bjorklund AK, Faridani OR, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10(11):1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 58.Islam S, Kjallquist U, Moliner A, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21(7):1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hashimshony T, Wagner F, Sher N, et al. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2(3):666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 60.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14(9):618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 61.Islam S, Zeisel A, Joost S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014;11(2):163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 62.Macosko EZ, Basu A, Satija R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klein AM, Mazutis L, Akartuna I, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eisenstein M. Startups use short-read data to expand long-read sequencing market. Nat Biotechnol. 2015;33(5):433–435. doi: 10.1038/nbt0515-433. [DOI] [PubMed] [Google Scholar]
- 65.Coombe L, Warren RL, Jackman SD, et al. Assembly of the complete Sitka Spruce chloroplast genome using 10× genomics’ GemCode sequencing data. PLoS ONE. 2016;11(9):e163059. doi: 10.1371/journal.pone.0163059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 68.Guo H, Zhu P, Guo F, et al. Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing. Nat Protoc. 2015;10(5):645–659. doi: 10.1038/nprot.2015.039. [DOI] [PubMed] [Google Scholar]
- 69.Guo H, Zhu P, Wu X, et al. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013;23(12):2126–2135. doi: 10.1101/gr.161679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smallwood SA, Lee HJ, Angermueller C, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11(8):817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rotem A, Ram O, Shoresh N, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33(11):1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cipriany BR, Zhao R, Murphy PJ, et al. Single molecule epigenetic analysis in a nanofluidic channel. Anal Chem. 2010;82(6):2480–2487. doi: 10.1021/ac9028642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park SY, Gonen M, Kim HJ, et al. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120(2):636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eirew P, Steif A, Khattra J, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015;518(7539):422–426. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim KT, Lee HW, Lee HO, et al. Single-cell mRNA sequencing identifies subclonal heterogeneity in anti-cancer drug responses of lung adenocarcinoma cells. Genome Biol. 2015;16:127. doi: 10.1186/s13059-015-0692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tirosh I, Venteicher AS, Hebert C, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539(7628):309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu C, Yu J, Yao X, et al. Discovery of biclonal origin and a novel oncogene SLC12A5 in colon cancer by single-cell sequencing. Cell Res. 2014;24(6):701–712. doi: 10.1038/cr.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y, Xu X, Song L, et al. Single-cell sequencing analysis characterizes common and cell-lineage-specific mutations in a muscle-invasive bladder cancer. Gigascience. 2012;1(1):12. doi: 10.1186/2047-217X-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hughes AE, Magrini V, Demeter R, et al. Clonal architecture of secondary acute myeloid leukemia defined by single-cell sequencing. PLoS Genet. 2014;10(7):e1004462. doi: 10.1371/journal.pgen.1004462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paguirigan AL, Smith J, Meshinchi S, et al. Single-cell genotyping demonstrates complex clonal diversity in acute myeloid leukemia. Sci Transl Med. 2015;7(281):281r–282r. doi: 10.1126/scitranslmed.aaa0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gerber T, Willscher E, Loeffler-Wirth H, et al. Mapping heterogeneity in patient-derived melanoma cultures by single-cell RNA-seq. Oncotarget. 2016;26:8. doi: 10.18632/oncotarget.13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ni X, Zhuo M, Su Z, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci USA. 2013;110(52):21083–21088. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lohr JG, Adalsteinsson VA, Cibulskis K, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32(5):479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang R, Lu YT, Ho H, et al. A comparison of isolated circulating tumor cells and tissue biopsies using whole-genome sequencing in prostate cancer. Oncotarget. 2015;6(42):44781–44793. doi: 10.18632/oncotarget.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Polzer B, Medoro G, Pasch S, et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol Med. 2014;6(11):1371–1386. doi: 10.15252/emmm.201404033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Court CM, Ankeny JS, Sho S, et al. Reality of single circulating tumor cell sequencing for molecular diagnostics in pancreatic cancer. J Mol Diagn. 2016;18(5):688–696. doi: 10.1016/j.jmoldx.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lohr JG, Kim S, Gould J, et al. Genetic interrogation of circulating multiple myeloma cells at single-cell resolution. Sci Transl Med. 2016;8(363):147r–363r. doi: 10.1126/scitranslmed.aac7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miyamoto DT, Zheng Y, Wittner BS, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349(6254):1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Proserpio V, Mahata B. Single-cell technologies to study the immune system. Immunology. 2016;147(2):133–140. doi: 10.1111/imm.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mahata B, Zhang X, Kolodziejczyk AA, et al. Single-cell RNA sequencing reveals T helper cells synthesizing steroids de novo to contribute to immune homeostasis. Cell Rep. 2014;7(4):1130–1142. doi: 10.1016/j.celrep.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Douek DC, Betts MR, Brenchley JM, et al. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol. 2002;168(6):3099–3104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- 92.Han A, Glanville J, Hansmann L, et al. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32(7):684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shalek AK, Satija R, Adiconis X, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498(7453):236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shalek AK, Satija R, Shuga J, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510(7505):363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lasken RS, McLean JS. Recent advances in genomic DNA sequencing of microbial species from single cells. Nat Rev Genet. 2014;15(9):577–584. doi: 10.1038/nrg3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Podar M, Abulencia CB, Walcher M, et al. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl Environ Microbiol. 2007;73(10):3205–3214. doi: 10.1128/AEM.02985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marcy Y, Ouverney C, Bik EM, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci USA. 2007;104(29):11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Youssef NH, Blainey PC, Quake SR, et al. Partial genome assembly for a candidate division OP11 single cell from an anoxic spring (Zodletone Spring, Oklahoma) Appl Environ Microbiol. 2011;77(21):7804–7814. doi: 10.1128/AEM.06059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Campbell JH, O’Donoghue P, Campbell AG, et al. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc Natl Acad Sci USA. 2013;110(14):5540–5545. doi: 10.1073/pnas.1303090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McLean JS, Lombardo MJ, Badger JH, et al. Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci USA. 2013;110(26):E2390–E2399. doi: 10.1073/pnas.1219809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dodsworth JA, Blainey PC, Murugapiran SK, et al. Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage. Nat Commun. 1854;2013:4. doi: 10.1038/ncomms2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mussmann M, Hu FZ, Richter M, et al. Insights into the genome of large sulfur bacteria revealed by analysis of single filaments. PLoS Biol. 2007;5(9):e230. doi: 10.1371/journal.pbio.0050230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mason OU, Hazen TC, Borglin S, et al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to deepwater horizon oil spill. ISME J. 2012;6(9):1715–1727. doi: 10.1038/ismej.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wells D, Kaur K, Grifo J, et al. Clinical utilisation of a rapid low-pass whole genome sequencing technique for the diagnosis of aneuploidy in human embryos prior to implantation. J Med Genet. 2014;51(8):553–562. doi: 10.1136/jmedgenet-2014-102497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fiorentino F, Biricik A, Bono S, et al. Development and validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of embryos. Fertil Steril. 2014;101(5):1375–1382. doi: 10.1016/j.fertnstert.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 106.Fiorentino F, Bono S, Biricik A, et al. Application of next-generation sequencing technology for comprehensive aneuploidy screening of blastocysts in clinical preimplantation genetic screening cycles. Hum Reprod. 2014;29(12):2802–2813. doi: 10.1093/humrep/deu277. [DOI] [PubMed] [Google Scholar]
- 107.Vera-Rodriguez M, Michel CE, Mercader A, et al. Distribution patterns of segmental aneuploidies in human blastocysts identified by next-generation sequencing. Fertil Steril. 2016;105(4):1047–1055. doi: 10.1016/j.fertnstert.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 108.Lu S, Zong C, Fan W, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338(6114):1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hou Y, Fan W, Yan L, et al. Genome analyses of single human oocytes. Cell. 2013;155(7):1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 110.Zhang C, Zhang C, Chen S, et al. A single cell level based method for copy number variation analysis by low coverage massively parallel sequencing. PLoS ONE. 2013;8(1):e54236. doi: 10.1371/journal.pone.0054236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hua R, Barrett AN, Tan TZ, et al. Detection of aneuploidy from single fetal nucleated red blood cells using whole genome sequencing. Prenat Diagn. 2015;35(7):637–644. doi: 10.1002/pd.4491. [DOI] [PubMed] [Google Scholar]
- 112.Poulin JF, Tasic B, Hjerling-Leffler J, et al. Disentangling neural cell diversity using single-cell transcriptomics. Nat Neurosci. 2016;19(9):1131–1141. doi: 10.1038/nn.4366. [DOI] [PubMed] [Google Scholar]
- 113.Okaty BW, Freret ME, Rood BD, et al. Multi-scale molecular deconstruction of the serotonin neuron system. Neuron. 2015;88(4):774–791. doi: 10.1016/j.neuron.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zeisel A, Munoz-Manchado AB, Codeluppi S, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347(6226):1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 115.Tasic B, Menon V, Nguyen TN, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19(2):335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Johnson MB, Walsh CA. Cerebral cortical neuron diversity and development at single-cell resolution. Curr Opin Neurobiol. 2016;42:9–16. doi: 10.1016/j.conb.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Darmanis S, Sloan SA, Zhang Y, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci USA. 2015;112(23):7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lake BB, Ai R, Kaeser GE, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352(6293):1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McConnell MJ, Lindberg MR, Brennand KJ, et al. Mosaic copy number variation in human neurons. Science. 2013;342(6158):632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cai X, Evrony GD, Lehmann HS, et al. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 2014;8(5):1280–1289. doi: 10.1016/j.celrep.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]


