Abstract
Objective. This study was aimed at analyzing the expressions of long noncoding RNAs (lncRNAs) in Botulinum Toxin Type A (BoNTA) treated human dermal fibroblasts (HDFs) in vitro. Methods. We used RNA sequencing to characterize the lncRNAs and mRNAs transcriptome in the control and BoNTA treated group, in conjunction with application of GO (gene ontology) analysis and KEGG (kyoto encyclopedia of genes and genomes) analysis to delineate the alterations in gene expression. We also obtained quantitative real time polymerase chain reaction (qRT-PCR) to confirm some differentially expressed genes. Results. Numerous differentially expressed genes were observed by microarrays between the two groups. qRT-PCR confirmed the changes of six lncRNAs (RP11-517C16.2-001, FR271872, LOC283352, RP11-401E9.3, FGFR3P, and XXbac-BPG16N22.5) and nine mRNAs (NOS2, C13orf15, FOS, FCN2, SPINT1, PLAC8, BIRC5, NOS2, and COL19A1). Farther studies indicated that the downregulating effect of BoNTA on the expression of FGFR3P was time-related and the dosage of BoNTA at a range from 2.5 U/106 cells to 7.5 U/106 cells increased the expression of FGFR3P and COL19A1 in HDFs as well. Conclusion. The expression profiling of lncRNAs was visibly changed in BoNTA treated HDFs. Further studies should focus on several lncRNAs to investigate their functions in BoNTA treated HDFs and the underlying mechanisms.
1. Introduction
Botulinum Toxin Type A (BoNTA) is the most effective one among the seven neurotoxins secreted by Clostridium botulinum [1]. BoNTA causes muscle relaxation and this concept nowadays is widely being used in the cosmetic treatment of wrinkles [2, 3]. In 2005 and 2008, some researchers found a face-lifting effect after intradermal injection of BoNTA to the mid and lower face [4, 5]. However, some other researchers reported that needle pricks themselves without BoNTA can make the skin become smoother as well [6]. In order to find out whether BoNTA can affect the human dermal fibroblasts (HDFs) directly, in 2012, Oh et al. studied the in vitro effects of BoNTA on normal HDFs and found that BoNTA has a notable effect in increasing the level of collagen production and downregulating its degradation [7]. Collagen is the most abundant basic element of fibrous components in the dermis and is responsible for maintaining the structural integrity of the skin by joining cells together and to the extracellular matrix (ECM) [8, 9]. These studies not only showed the positive effects of BoNTA on HDFs for remodeling skin but also implied the importance of HDFs. In 2016, Zhu et al. proved that topical BoNTA application could enhance the rejuvenation effect of fractional CO2 laser, further indicating that BoNTA can refine skin texture via improving the activity of HDFs [10]. But until now, the molecular mechanisms through which BoNTA could affect HDFs are still not completely understood.
Long noncoding RNAs (lncRNAs) are a group of noncoding RNA transcripts longer than 200 nucleotides which cannot encode proteins [11]. In comparison with protein-coding genes, lncRNAs have limited coding potential and show little evolutionary conservation in sequence. Furthermore, some researchers have detected that lncRNAs expression is more tissue specific and at apparently lower levels [12]. LncRNAs, which were previously thought to be transcriptional “noise,” are now proved to have some functions by regulating gene expression at the epigenetic, transcriptional, and posttranscriptional levels and participating in some biologic functions, such as genomic imprinting, chromosome modification, intranuclear transport, transcriptional activation, and interference [13]. Therefore, the understanding of cellular processes in physiological conditions will not be complete without analyzing the contributions made by lncRNAs. Until now, no information is available regarding the effect of BoNTA on expression profiling of lncRNAs in HDFs.
In this study, we investigated on lncRNA expression signature together with messenger RNA (mRNA) expression profile in BoNTA treated HDFs and confirmed the changing of some differentially expressed lncRNAs and mRNA using qRT-PCR. In conjunction, we also conducted functional analysis using Gene Ontology (GO) analysis and pathway analysis, in which genes are mapped to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
2. Materials and Methods
2.1. Cell Separation and Culture
Normal human skin samples were obtained from the prepuce of young healthy individuals in accordance with the ethics committee approval process of The First Affiliated Hospital of Nanjing Medical University (Nanjing, China). The acquirement of HDFs can be divided into two procedures. Initially dispase enzyme was used to separate the dermis and epidermis, and then collagenase enzyme was used to extract the HDFs. HDFs were grown in Dulbecco's modified Eagle medium (DMEM) with 1% penicillin-streptomycin and 10% fetal bovine serum in an environment of 5% CO2 at 37°C. The cells used in our study were from passages 8–11.
2.2. Group Divisions and Botulinum Toxin Type A (BoNTA) Treatment
In order to study differentially expressed lncRNAs and mRNAs, we separated the cells into two groups, control group and BoNTA group: (1) control group: HDFs were grown in DMEM with 1% penicillin-streptomycin and 10% fetal bovine serum for 5 days and then serum-starved for 4 days, without receiving BoNTA treatment; (2) BoNTA group (48 h): HDFs were grown in DMEM with 1% penicillin-streptomycin and 10% fetal bovine serum for 5 days, serum-starved for 2 days, and then were grown in serum-free DMEM with BoNTA at a dose of 5 U/106 cells for 2 days.
In order to determine whether the changes of RNAs expression in BoNTA treated HDFs were time or dosage dependent, the cells were divided into 4 groups: (1) BoNTA group (24 h): HDFs were grown in DMEM with 1% penicillin-streptomycin and 10% fetal bovine serum for 5 days, serum-starved for 2 days, and then were grown in serum-free DMEM with BoNTA at a dose of 5 U/106 cells for 24 h; (2) BoNTA group (72 h): HDFs were grown in DMEM with 1% penicillin-streptomycin and 10% fetal bovine serum for 5 days, serum-starved for 2 days, and then were grown in serum-free DMEM with BoNTA at a dose of 5 U/106 cells for 72 days; (3) BoNTA group (48 h 2.5 U): HDFs were grown in DMEM with 1% penicillin-streptomycin and 10% fetal bovine serum for 5 days, serum-starved for 48 h, and then were grown in serum-free DMEM with BoNTA at a dose of 2.5 U/106 cells for 48 h; (4) BoNTA group (48 h 7.5 U): HDFs were grown in DMEM with 1% penicillin-streptomycin and 10% fetal bovine serum for 5 days, serum-starved for 2 days, and then were grown in serum-free DMEM with BoNTA at a dose of 7.5 U/106 cells for 48 h.
All groups were rinsed with Phosphate Buffer Solution (PBS) and the medium was changed every day, except during BoNTA treatment. Each method of detection consists of these six groups of cell culture, containing about 1 × 107 cells in each group, and there were at least 3 samples in each group. The BoNTA used in this study was manufactured by Lanzhou Institute of Biological Products Co., Ltd., Lanzhou, China.
2.3. Isolation of RNA and Preparation of Array Hybridization
The RNA extraction was conducted using the TRIZOL reagent and then was dissolved in RNase-free water. The purified labeled genomic DNA was used for quantification and the RNA quantity was decided spectrophotometrically as A260/A280 ratio (1.9–2.1 were obtained). The collected RNAs were stored at −70°C for microarray analysis and qRT-PCR.
2.4. Microarray Analysis of lncRNAs and mRNAs Expression
The Agilent Human lncRNA (8∗60 K) arrays were designed in this experiment for measuring the expression profiles of lncRNAs and mRNAs. The lncRNA sequences were acquired from the following databases: NCBI-RefSeq, NONCODE v4, Ensembl, broad lincRNA, and frnadb v3.4. We conducted the sample labeling, microarray hybridization, and washing according to the manufacturer's proposals. Briefly speaking, we initially transcribed the total RNA to double stranded cDNA, synthesized them into cRNA, and labeled them with Cyanine-3-CTP and then hybridized the labeled cRNAs onto the microarray. We applied the Agilent Scanner G2505C (Agilent Technologies) to scan the arrays after washing.
2.5. Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
In order to confirm the results obtained from microarray, we applied qRT-PCR to remeasure the abundance of differentially expressed lncRNAs and mRNAs selected from microarray analysis. RNAs in the medium were determined based on the protocol of KeyGen Biotech Co., Ltd., Nanjing, Jiangsu, China. In brief, we applied TRIzol to extract the total RNAs from HDFs and then synthesized the cDNAs from the separated RNA using SuperScript III Reverse Transcriptase (KeyGen, China). QRT-PCR was performed on ABI Prism 7700 Sequence Detector (Applied Biosystems). The reactions were performed at 9°C for 10 min, then at 40 cycles at 95°C for 15 s, and followed by at 60°C for 30 s. The 2 (−ΔΔCt) methods were used to evaluate relative quantification of lncRNAs and mRNAs expression. Primer sequences of lncRNAs and mRNAs for qRT-PCR are listed in Table 1. The results were expressed as mean ± SD (standard deviation) of each independent experiment.
Table 1.
Primer sequences for long noncoding RNAs and protein-coding RNAs.
| Gene name | Primer sequences |
|---|---|
| RP11-517C16.2-001 | Forward primer: TTTGTCAACGGGCTCTACCC |
| Reverse primer: TTTGTCAACGGGCTCTACCC | |
|
| |
| FR271872 | Forward primer: CACCTCCTTCCCTGGACTAGA |
| Reverse primer: CACCTCCTTCCCTGGACTAGA | |
|
| |
| LOC283352 | Forward primer: AAGGGTTTATGTGCTCGGAGG |
| Reverse primer: CTGGCTGAGGAGTCTCACTT | |
|
| |
| RP11-401E9.3 | Forward primer: CCAGTCATGCCCATCCAGAA |
| Reverse primer: CCATGCAGCAACTAGCAAAGG | |
|
| |
| FGFR3P | Forward primer: ATGGAAGGCTGCTTCATGCT |
| Reverse primer: GTTTCAAGACCTCAGCGGGA | |
|
| |
| XXbac-BPG16N22.5 | Forward primer: AGACTCAAGGGGACCAGACC |
| Reverse primer: CTGCAGGCAGGTGTATCTCA | |
|
| |
| COL19A1 | Forward primer: TCGAGTACGAAGAAACGCCAA |
| Reverse primer: TGCCACTGACGATCAAACAAA | |
|
| |
| NOS2 | Forward primer: ACATCGACCCGTCCACAGTAT |
| Reverse primer: CAGAGGGGTAGGCTTGTCTC | |
|
| |
| C13orf15 | Forward primer: CTGAATTCTCCAACAGACT |
| Reverse primer: ATGGGAAAGCTTACTGCT | |
|
| |
| E2F1 | Forward primer: AGCTGGACCACCTGATGAAT |
| Reverse primer: GAGGGGCTTTGATCACCATA | |
|
| |
| SPINT1 | Forward primer: AGACTACTGCCTCGCATCCAA |
| Reverse primer: CAAGCAGCCTCCATAAACGAA | |
|
| |
| FOS | Forward primer: TGACAGATACACTCCAAGCGG |
| Reverse primer: GGGAAGCCAAGGTCATCG | |
|
| |
| FCN2 | Forward primer: GTAAAACGACGGCCAGTTATGGCCCTGCTTCTTCCTC |
| Reverse primer: TTCCAGAGTGTGTTCTCCCAC | |
|
| |
| PLAC8 | Forward primer: CCTCTACACTGCCTCAGCATC |
| Reverse primer: GTAAAACGACGGCCAGTTTCTACACAATAAGGGAGGAATGG | |
|
| |
| BIRC5 | Forward primer: AGGACCACCGCATCTCTACAT |
| Reverse primer: AAGTCTGGCTCGTTCTCAGTG | |
2.6. Data Analysis
We applied the Feature Extraction software (version 10.7.1.1, Agilent Technologies) to analyze array images to get raw data and used GeneSpring (version 13.1, Agilent Technologies) to complete the fundamental analysis with the raw data. First of all, the raw data was normalized with the quantile algorithm. The probes which have signed with P (p value ≤0.05 is recommended) were selected for further data analysis. Deferentially expressed lncRNAs and protein-coding RNAs were then identified through fold change. The critical value set for up- and downregulated genes was a fold change ≥ 2.0. Afterwards, GO analysis and KEGG analysis were applied to depict alterations in the gene expression. SPSS 17.0 software (SPSS Inc., Chicago, IL, United States) was used to analyze the statistical data. The differences in RNA expression between the two groups and more than three groups were analyzed using the Student's t-test and one-way ANOVA, separately. p < 0.05 was considered to indicate a statistically significant difference.
3. Results
3.1. Different Expression Profiles of lncRNAs and mRNAs in Control Group and BoNTA Treated Group
Data analysis showed 2124 differentially expressed lncRNAs and 638 differentially expressed mRNAs of the cells in BoNTA (48 h 5 U) treated group (fold change > 2) compared with control group. Hierarchical clustering heat-map showed the expression ratios of mRNAs (Figure 1(a)) and lncRNAs (Figure 1(b)) between the two groups. Of the 2124 differentially expressed lncRNAs, 1122 were upregulated and 1002 were downregulated. Of the 638 differentially expressed mRNAs, 303 were upregulated and 335 were downregulated. The distinctly expressed lncRNAs are listed in Table 2 (fold change > 5) and the distinctly expressed mRNAs are summarized in Table 3 (fold change > 3).
Figure 1.
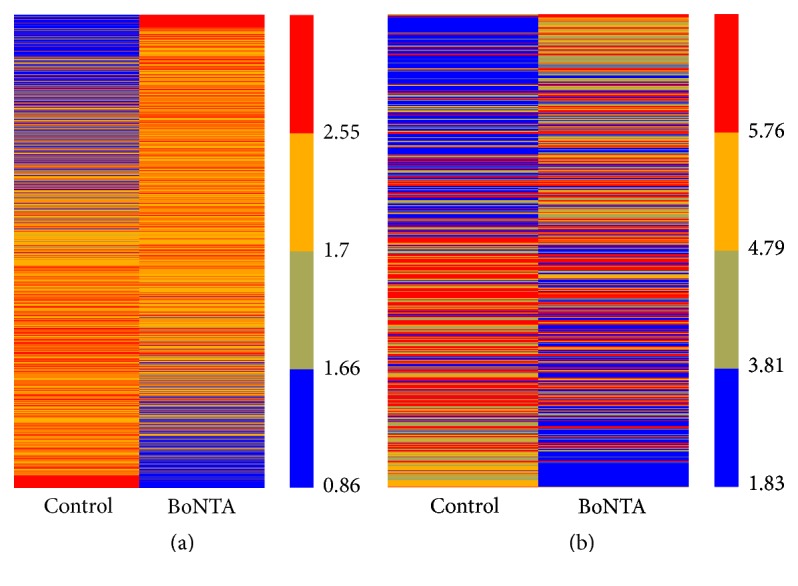
Heat maps presentation of the expression profiling of mRNAs (a) and lncRNAs (b) in different groups of HDFs. Red and blue represent high and low relative expressions, respectively. Control indicates the normal HDFs and BoNTA indicates the HDFs treated by BoNTA (5 U/106 cells 48 h).
Table 2.
Differentially expressed long noncoding RNAs in HDFs (fold change > 5).
| Probe name | Gene symbol | Expression | Chromosome | Strand | Start | End | Fold change |
|---|---|---|---|---|---|---|---|
| CUST_18266 | RP11-262H14.3-005 | Up | Chr9 | − | 66513657 | 66553531 | 18.14 |
| CUST_28663 | linc-TCTE3-3 | Up | Chr6 | − | 170470884 | 170475667 | 9.33 |
| CUST_73981 | linc-GRAMD3-2 | Up | Chr5 | + | 124372380 | 124486527 | 8.12 |
| CUST_25057 | Z83001.1-003 | Up | Chr11 | − | 31710730 | 31789373 | 7.07 |
| CUST_54220 | RP11-517C16.2-001 | Up | Chr16 | − | 84492865 | 84500967 | 6.13 |
| CUST_77914_PI429545380 | RP11-444D13.1-001 | Up | chr1 | − | 183723553 | 183724072 | 5.70 |
| CUST_42824_PI429545402 | RPS2P41 | Up | chr12 | − | 112317141 | 112318053 | 5.63 |
| CUST_24581_PI429545380 | FR271872 | Up | chr1 | + | 40364066 | 40364332 | 5.55 |
| CUST_27762_PI429545402 | KLRD1 | Up | chr12 | + | 10378664 | 10467608 | 5.45 |
| CUST_42463_PI429545376 | linc-SCGB1D4-2 | Up | chr11 | − | 62178649 | 62179162 | 5.35 |
| CUST_83283_PI429545395 | RSPH10B | Up | chr7 | − | 5995734 | 6002990 | 5.34 |
| CUST_63264_PI429545410 | LOC283352 | Up | chr12 | + | 129594234 | 129597843 | 5.34 |
| CUST_70508_PI429545399 | RP11-401E9.3 | Up | chr10 | − | 7875580 | 7875920 | 5.05 |
| CUST_70526 | MTHFD1 | Down | Chr14 | + | 64924714 | 64926721 | −25.69 |
| CUST_86832 | AC010136.2-001 | Down | Chr2 | + | 218843430 | 218857338 | −8.43 |
| CUST_75452 | PNKP | Down | Chr19 | − | 50369397 | 50370818 | −8.04 |
| CUST_89880 | STPG1 | Down | Chr1 | − | 24717747 | 24742643 | −7.77 |
| CUST_45703 | RP11-643A5.2-002 | Down | Chr15 | − | 54239821 | 54267147 | −6.66 |
| CUST_15063 | FGFR3P | Down | chr6_cox_hap2 | + | 2857852 | 2858455 | −6.52 |
| CUST_29613 | linc-HOXA11 | Down | Chr7 | − | 27226865 | 27232305 | −6.25 |
| CUST_33771 | FR081392 | Down | Chr6 | + | 161943497 | 161943737 | −6.21 |
| CUST_75093 | CTA-292E10.6-001 | Down | Chr22 | + | 29196671 | 29244547 | −6.04 |
| CUST_90031_PI429545395 | RP11-73B2.7 | Down | chr7 | − | 63398282 | 63398887 | −5.63 |
| CUST_81910_PI429545380 | AC007131.2-003 | Down | chr2 | − | 59465851 | 59476702 | −5.5 |
| CUST_92526_PI429545402 | ZFAND6 | Down | chr15 | + | 80364932 | 80413144 | −5.2 |
| CUST_62442_PI429545395 | XXbac-BPG16N22.5 | Down | chr6 | + | 31483755 | 31483988 | −5.14 |
| CUST_19370_PI429545380 | FR316649 | Down | chr3 | + | 188550879 | 188551155 | −5.07 |
Table 3.
Differentially expressed protein-coding RNAs in HDFs (fold change > 3).
| Probe name | Gene symbol | Expression | Chromosome | Strand | Start | End | Fold change |
|---|---|---|---|---|---|---|---|
| A_33_P3367396 | FAM177B | Up | Chr1 | + | 222923479 | 222923538 | 30.05 |
| A_33_P3400699 | SLC26A5 | Up | Chr9 | − | 10 2993259 | 102993200 | 5.16 |
| A_23_P502464 | NOS2 | Up | Chr17 | − | 26083921 | 26083862 | 4.21 |
| A_33_P3423270 | TMEM40 | Up | Chr3 | − | 12775584 | 12775525 | 4.08 |
| A_24_P183128 | PLAC8 | Up | Chr4 | − | 84015842 | 84012077 | 4.03 |
| A_23_P135226 | OR1N2 | Up | Chr9 | + | 125316281 | 125316340 | 3.91 |
| A_23_P153616 | MADCAM1 | Up | Chr19 | + | 505192 | 505251 | 3.89 |
| A_23_P49060 | SPINT1 | Up | Chr15 | + | 41149316 | 41149375 | 3.58 |
| A_24_P10137 | C13orf15 | Up | Chr13 | + | 42042937 | 42044635 | 3.42 |
| A_24_P630490 | DFNB59 | Up | Chr2 | + | 179325165 | 179325759 | 3.38 |
| A_23_P106194 | FOS | Up | Chr14 | + | 75748214 | 75748273 | 3.05 |
| A_23_P313588 | TMPRSS6 | Up | Chr22 | − | 37480125 | 37480066 | 3.04 |
| A_23_P134085 | CNKSR3 | Down | Chr6 | − | 154726608 | 154726549 | −14.99 |
| A_24_P323598 | ESCO2 | Down | Chr8 | + | 27662101 | 27662160 | −5.44 |
| A_23_P216756 | FCN2 | Down | Chr9 | + | 137779164 | 137779223 | −4.27 |
| A_24_P225616 | RRM2 | Down | Chr2 | + | 10270487 | 10270546 | −4.19 |
| A_23_P15844 | BRIP1 | Down | Chr17 | − | 59760967 | 59760908 | −4.04 |
| A_23_P126212 | CLSPN | Down | Chr1 | − | 36204176 | 36204117 | −3.85 |
| A_33_P3807062 | HJURP | Down | Chr2 | − | 234746088 | 234746029 | −3.84 |
| A_23_P100127 | CASC5 | Down | Chr15 | + | 40917525 | 40917584 | −3.81 |
| A_33_P3257678 | HIST2H3A | Down | Chr1 | + | 149824626 | 149824685 | −3.79 |
| A_33_P3326210 | ESCO2 | Down | Chr8 | + | 27660830 | 27660889 | −3.72 |
| A_23_P8452 | LFNG | Down | Chr7 | + | 2567971 | 2568030 | −3.70 |
| A_23_P51085 | SPC25 | Down | Chr2 | − | 169728015 | 169727956 | −3.61 |
| A_23_P130182 | AURKB | Down | Chr17 | − | 8110917 | 8110655 | −3.59 |
| A_23_P254733 | MLF1IP | Down | Chr4 | − | 185616396 | 185616337 | −3.49 |
| A_23_P133956 | KIFC1 | Down | Chr6 | + | 33374428 | 33374621 | −3.48 |
| A_24_P214231 | STIL | Down | Chr1 | − | 47716850 | 47716791 | −3.45 |
| A_23_P131330 | LRRTM1 | Down | Chr2 | − | 80529291 | 80529232 | −3.45 |
| A_32_P96719 | HIST1H2AL | Down | Chr6 | − | 27833502 | 27833561 | −3.27 |
| A_23_P363174 | GTSE1 | Down | Chr22 | + | 46725391 | 46725450 | −3.23 |
| A_23_P118246 | GINS2 | Down | Chr16 | − | 85711713 | 85711654 | −3.20 |
| A_24_P322354 | SKA1 | Down | Chr18 | + | 47919899 | 47919958 | −3.14 |
| A_23_P118815 | BIRC5 | Down | Chr17 | + | 76220720 | 76220779 | −3.08 |
| A_24_P413884 | CENPA | Down | Chr2 | + | 27016914 | 27016973 | −3.06 |
| A_24_P314571 | SPC24 | Down | Chr19 | − | 11257053 | 11256994 | −3.05 |
| A_23_P99292 | RAD51AP1 | Down | Chr12 | + | 4668182 | 4668241 | −3.01 |
| A_23_P80032 | E2F1 | Down | Chr20 | − | 32264048 | 32263989 | −3.01 |
3.2. QRT-PCR Analysis of lncRNAs and mRNAs Expression
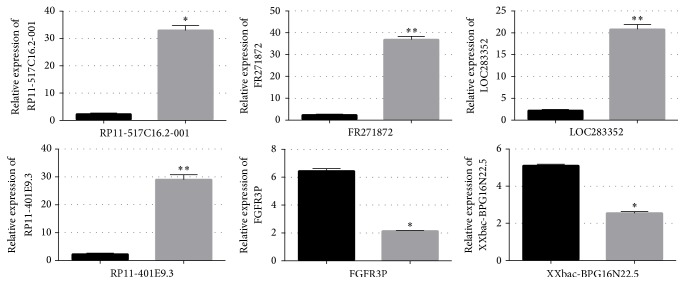
QRT-PCR analysis indicated the transcription of the selected lncRNAs: RP11-517C16.2-001, FR271872, LOC283352, RP11-401E9.3, FGFR3P, XXbac-BPG16N22.5, and mRNAs: collagen 19a1 (COL19A1), nitric oxide synthase 2 (NOS2), chromosome 13 open reading frame 15 (C13orf15), FBJ murine osteosarcoma viral oncogene homolog (FOS), ficolin (collagen/fibrinogen domain containing lectin) 2 (hucolin) (FCN2), serine peptidase inhibitor, Kunitz type 1 (SPINT1), placenta-specific 8 (PLAC8), E2F transcription factor 1 (E2F1), and baculoviral IAP repeat containing 5 (BIRC5), perfectly correlated to microarray results. The qRT-PCR analysis revealed that the expression level of RP11-517C16.2-001, FR271872, LOC283352, and RP11-401E9.3 in BoNTA (5 U 48 h) treated groups was upregulated to 33.479-, 39.519-, 19.713-, 26.362-, and 29.293-fold separately (Figure 2), and the expression level of FGFR3P and XXbac-BPG16N22.5 was downregulated to 2.139- and 2.554-fold, respectively (Figure 2).
Figure 2.
The differential expression level of long noncoding RNAs (lncRNAs) between control and BoNTA (5 U/106 cells 48 h) treated groups was validated by qRT-PCR. Six lncRNAs (RP11-517C16.2-001, FR271872, LOC283352, RP11-401E9.3, FGFR3P, and XXbac-BPG16N22.5) were studied using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control. The heights of the columns in the chart represent the fold changes. Data are the mean ± SEM (n = 6). p < 0.05 was considered to indicate a statistically significant difference compared with control HDFs. ∗p < 0.05 and ∗∗p < 0.01.
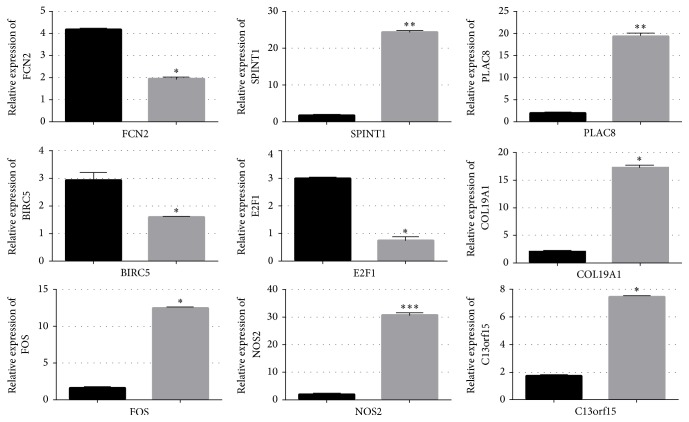
For the mRNAs, in comparison with the control group, the expression level of COL19A1, NOS2, C13orf15, FOS, SPINT1, and PLAC8 was upregulated to 10.331-, 31.374-, 7.534-, 12.573-, 24.758-, and 19.885-fold in BoNTA (5 U 24 h) treated groups, respectively (Figure 3), and the expression level of FCN2, BIRC5, and E2F1 was downregulated to 1.890-, 0.923-, 0.709-fold separately (Figure 3).
Figure 3.
The differential expression levels of message RNAs (mRNAs) between control and BoNTA (5 U/106 cells 48 h) treated groups of microarray results were validated by qRT-PCR. Nine mRNAs (FCN2, SPINT1, PLAC, BIRC5, E2F1, COL19A1, FOS, NOS2, and C13orf15) were studied using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control. The heights of the columns in the chart represent the fold changes. Data are the mean ± SEM (n = 6). p < 0.05 was considered to indicate a statistically significant difference compared with control HDFs. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
3.3. GO and Pathway Analysis of Deferentially Expressed RNAs
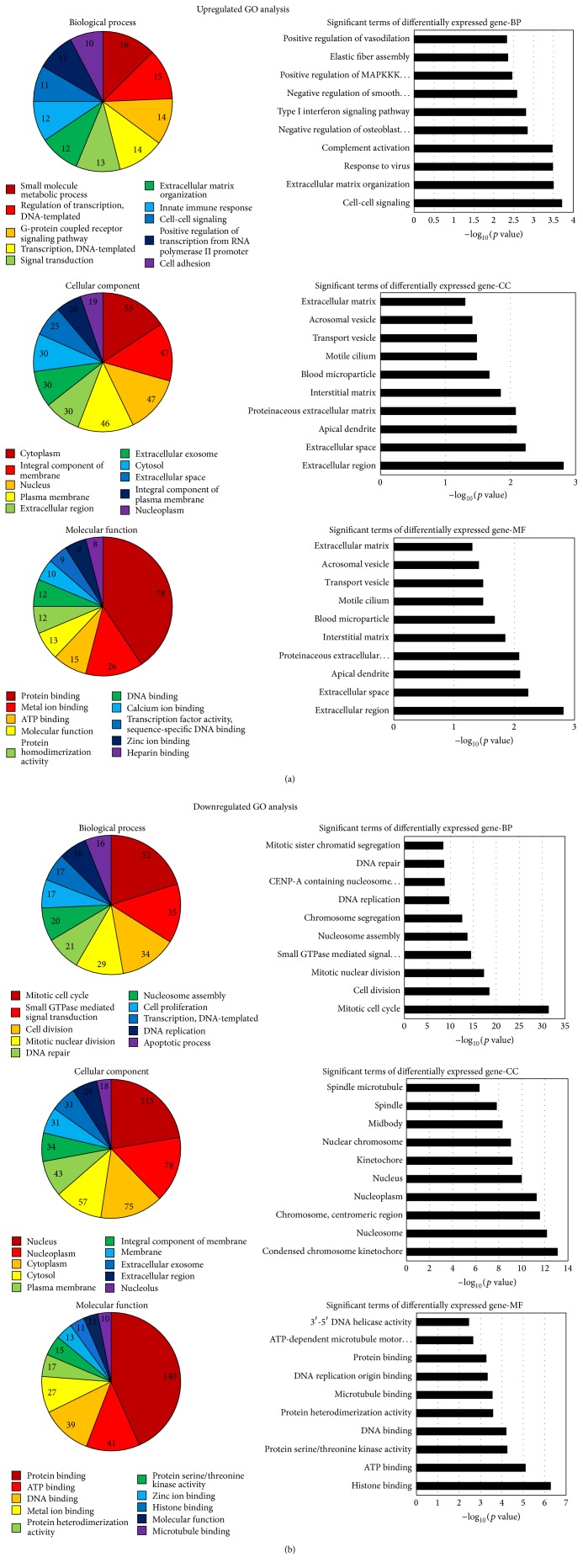
The upregulated genes were involved in 898 biological processes, 171 cellular components, and 236 molecular functions separately. The response to cell-cell signaling (−log10 (p value) = 3.7114) was the most significant term among the biological process category. The extracellular region (−log10 (p value) = 2.8128) was the most represented GO term in the cellular component category. Heparin binding (−log10 (p value) = 3.3714) was the most highly represented term within the molecular component category. The downregulated genes were involved in 880 biological processes, 179 cellular components, and 240 molecular functions, respectively. The most significant term was the response to mitotic cell cycle (−log10 (p value) = 31.3859) among the biological process category. The most represented GO term was the condensed chromosome kinetochore (−log10 (p value) = 13.0822) in the cellular component category. Heparin binding (−log10 (p value) = 6.2714) was the most highly represented term with in the molecular component category. The top ten of the number of deferentially expressed genes and the significance analyzed by GO term are demonstrated in Figure 4.
Figure 4.
Bioinformatic analysis of the differentially expressed genes. The p value denotes the significance of GO terms enrichment in the differentially expressed genes. The lower the p value, the more significant the GO term (p value ≤ 0.05 is recommended). We can choose the target genes for further study based on the combination of the analysis provided by GO and the biologic significance.
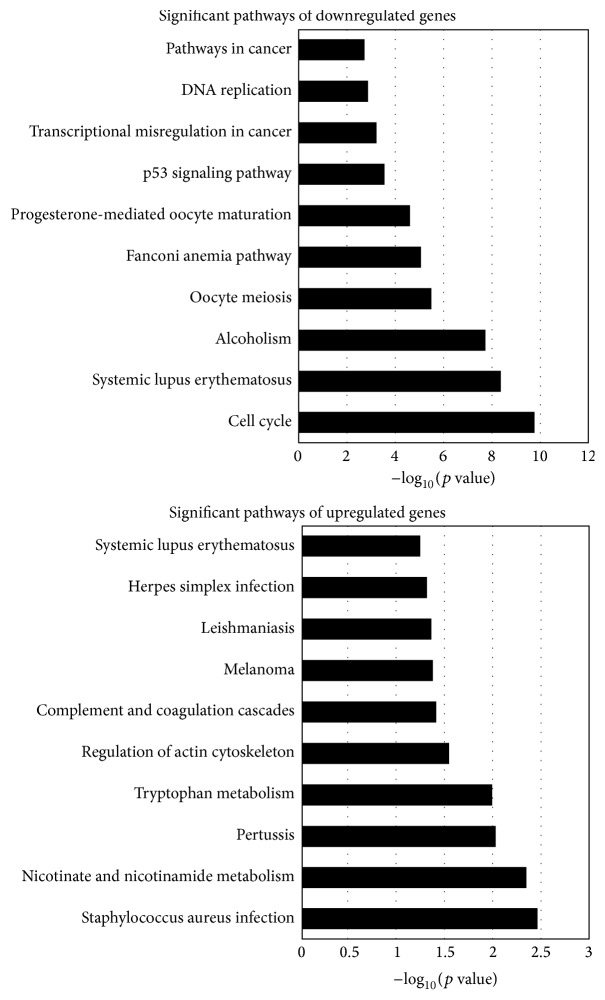
KEGG (Kyoto Encyclopedia of Genes and Genomes) was used to analyze the pathway enrichment. The pathway analysis showed that changed genes participated in the DNA replication, cell cycle, P53 signaling pathway, pathway in cancer, and melanoma. The upregulated genes participated in 157 pathways while downregulated genes participated in 169 pathways. The top ten pathways among them are shown in Figure 5.
Figure 5.
Pathway analysis of the differentially expressed genes. Fisher's exact test was used to select the main pathway, and the significance threshold was defined with p value. The lower the p value, the more significant the pathway (p value ≤0.05 is recommended). We can get some information about the possibility between the differentially expressed genes and the change of cellular pathways.
3.4. The Regulation of BoNTA on the Expression of FGFR3P and COL19A1
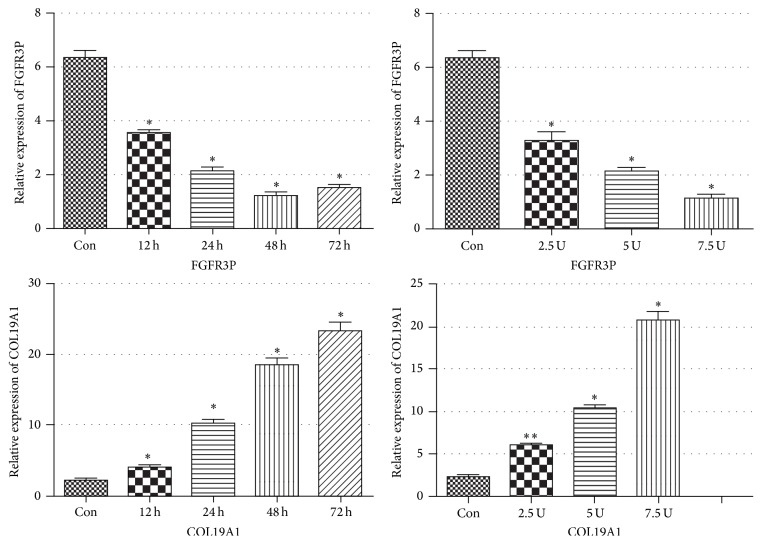
We also performed qRT-PCR to validate the altered lncRNA expression at different time-points and different dosage of BoNTA treatment. We found that the level of FGFR3P reached the lowest at 48 h but indicated an upward trend at 72 h when treated by BoNTA at the dosage of 5 U/106 cells (Figure 6). The level of FGFR3P was gradually increasing with the increasing dosage of BoNTA (at a range from 2.5 U/106 cells to 7.5 U/106 cells). At the same time, the level of COL19A1 in HDFs was gradually increasing with the extension of time treated by BoNTA at the dosage of 5 U/106 cells (Figure 6), and its level showed a trend of increasing when the dosage of BoNTA ranges from 2.5 U/106 cells to 7.5 U/106 cells (Figure 6).
Figure 6.
The regulation of BoNTA on the expression of FGFR3P and COL19A1 in HDFs. The expressions of FGFR3P and COL19A1 in HDFs different dosage of BoNTA were measured using qRT-PCR. The heights of the columns in the chart represent the fold changes. Data are the mean ± SEM (n = 6). p < 0.05 was considered to indicate a statistically significant difference compared with control HDFs. ∗p < 0.05 and ∗∗p < 0.01.
4. Discussion
Recently, lncRNAs have come into the extent of probing into human biological functions and diseases [13–17]. Until now, only a few lncRNAs had been reported about HDFs, and studies on the expression of lncRNAs in BoNTA treated HDFs were still lacking. In this study, we identified some differentially expressed lncRNAs and mRNAs between the BoNTA treated group and control group by analyzing gene expression profiles. The number of changed lncRNAs was greater than that of mRNAs. Although currently the precise roles of the changed lncRNAs in BoNTA treated HDFs are not clear, lncRNAs have been regarded as vital regulators of gene expression and have various biofunctions. There are a large number of evidences demonstrating that lncRNAs can regulate gene expression by forming RNA-protein, RNA-RNA, DNA-RNA, and protein-DNA interactions [18]. Although no direct relationship was found between the altered lncRNA and mRNA expressions, we are convinced that the clear changes of lncRNAs (RP11-517C16.2-001, FR271872, LOC283352, RP11-401E9.3, FGFR3P, and XXbac-BPG16N22.5) in HDFs are in response to BoNTA and are related to the changes of protein-coding RNAs. We suspected that the decrease of NOS2 indicated the regulation of cell proliferation process [19, 20]. The increase of FOS induced by BoNTA showed the regulation of proliferation, which is also involved in the cellular senescence process of HDFs [21–24]. BIRC5 has been reported to participate in modulation of diverse cellular processes such as proliferation, adhesion, apoptosis, migration and invasion during growth, development, repair, maintenance, and regression of a wide variety of mesenchymal tissues [25–28]. The downregulated BIRC5 induced by BoNTA indicated the decrease of apoptosis in HDFs. PLAC8 has been proved to regulate cell cycle and participate in the regulation of apoptosis and cell division [29–31].
GO database was applied to analyze the function of the differentially expressed genes. The results were divided into three sections: biological process (BP), cellular component (CC), and molecular function (MF). Besides the top ten of the number of differentially expressed genes and the significance analyzed by GO term demonstrated in Figure 4, the analysis showed that the differentially expressed genes were also involved in a variety of other biological functions, such as negative regulation of autophagy (GO: 0010507), positive regulation of collagen biosynthetic process (GO: 0032967), regulation of G1/S transition of mitotic cell cycle (GO: 2000045), and positive regulation of fibroblast proliferation (GO: 0048147). The analysis indicated that the lncRNAs can affect the function of HDFs by regulating the expression profiles of genes related to HDFs. KEGG analysis can provide some suggestive information about the potential relativity of the changed gene expression with the alterations of the pathways. The result of our study indicates several significant pathways related to a variety of functions, such as cell proliferation, cell cycle, apoptosis, and DNA replication. It had been reported that BoNTA can regulate the process of cell cycle and DNA replication by other researchers. For example, G. Karsenty et al. reported in their study that BoNTA obviously reduced LNCaP cell proliferation and increased apoptosis in a dose-dependent manner [32]. Park et al. found that BoNTA upregulates the expression of cell cycle related genes such as RhoA, Rac1, and Cdc42 in a dose-dependent manner [33]. Our prediction results accord with the functional analysis of BoNTA obtained from other investigations.
QRT-PCR was applied to verify the result of microarray analysis. We learned from other studies that BoNTA can regulate cell proliferation and collagen synthesis; the mechanisms may play important roles in skin rejuvenation effects of BoNTA [32–34]. The RNAs chosen for qRT-PCR confirmation meet at least one of two following criteria: 1: the RNAs being at least 2-fold differently expressed in BoNTA treated groups in comparison with the control group according to the data analysis; 2: the RNAs that had been proved in other studies to have a relationship with biological functions such as cell proliferation and collagen synthesis. The qRT-PCR confirmation results of six lncRNAs (RP11-517C16.2-001, FR271872, LOC283352, RP11-401E9.3, FGFR3P, and XXbac-BPG16N22.5) and mRNAs (NOS2, C13orf15, FOS, FCN2, SPINT1, PLAC8, BIRC5, and COL19A1) were consistent with the microarray data which confirmed the reliability of our microarray analysis.
Next, we applied qRT-PCR to further investigate the expression changes of COL19A1 and FGFR3P after BoNTA treatment at different dosages and culture times. COL19A1, as one member of the fibril-associated collagens with interrupted triple helices (FACIT) group, is thought to act as a cross-bridge between extracellular matrix molecules (ECM) and is involved in the formation of the well-known striated fibrils [35]. Bioinformatic analysis reveals that FGFR3P which is located on chromosome 6 is defined as the pseudogene of Fibroblast Growth Factor Receptor 3 (FGFR3), noncoding RNA. FGFR3 is one member of FGFR family and had been reported to participate in the regulation of cell proliferation, apoptosis, migration, and angiogenesis in many cells including HDFs [36–39]. Although pseudogenes have been considered as the remnants of functional genes which have no coding ability over a long period of time, evergrowing number of studies have proved that some pseudogenes have diverse functions, including serving as miRNA decoys, functioning as antisense transcripts, encoding short peptides, and producing siRNAs or proteins [40–43]. Our results showed that downregulating effect of BoNTA on the expression of FGFR3P was time-related. Meanwhile, the dosage of BoNTA at a range from 2.5 U/106 cells to 7.5 U/106 cells increased the expression of FGFR3P and COL19A1 in HDFs as well. We speculated significant change of expression of these RNAs in a time-dependent manner after BoNTA treatment which may be of some clinical significance. Zhu et al. conducted one clinical experiment and found that topical application of BoNTA could enhance the rejuvenation effect of fractional CO2 laser; they also found that best results of skin detection were in the last observation point at 3 months after treatment [34]. Zhu et al. also performed another clinical study and found that intradermal BoNTA injection showed its best rejuvenation results in the last observation point at 12 weeks after treatment as well [44]. The results of the previous clinical investigations showed that BoNTA had a persistent promoting effect of collagen synthesis with the extending of time. Taking these into consideration, we speculate that BoNTA have the antiaging effect not only by relaxing in muscle fiber, but also by promoting activity of HDFs directly. The underlying mechanisms still need further studies.
In conclusion, for the first time, our current study identified the changes of expression profiles of lncRNAs in BoNTA treated HDFs and found that BoNTA dynamically regulated the expression of COL19A1 and FGFR3P in HDFs, indicating the potential role of several lncRNAs in BoNTA treated HDFs. Therefore, further explorations are warranted to discover the mechanisms behind the dynamic changes BoNTA induced on COL19A1 and FGFR3P and the findings obtained in this study should lay foundations for further studies into the potential roles of these altered lncRNAs in BoNTA treated HDFs.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grants 81573072 and 81301384) and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
Competing Interests
The authors report no conflict of interests.
References
- 1.Fagien S. Botox for the treatment of dynamic and hyperkinetic facial lines and furrows: adjunctive use in facial aesthetic surgery. Plastic and Reconstructive Surgery. 1999;103(2):701–713. doi: 10.1097/00006534-199902000-00055. [DOI] [PubMed] [Google Scholar]
- 2.Said S. Z., Meshkinpour A., Carruthers A., Carruthers J. Botulinum toxin A: its expanding role in dermatology and esthetics. American Journal of Clinical Dermatology. 2003;4(9):609–616. doi: 10.2165/00128071-200304090-00003. [DOI] [PubMed] [Google Scholar]
- 3.Carruthers A., Carruthers J. Clinical indications and injection technique for the cosmetic use of botulinum A exotoxin. Dermatologic Surgery. 1998;24(11):1189–1194. doi: 10.1111/j.1524-4725.1998.tb04097.x. [DOI] [PubMed] [Google Scholar]
- 4.Seyler T. M., Smith B. P., Marker D. R., et al. Botulinum neurotoxin as a therapeutic modality in orthopaedic surgery: more than twenty years of experience. The Journal of Bone & Joint Surgery—American Volume. 2008;90(supplement 4):133–145. doi: 10.2106/jbjs.h.00901. [DOI] [PubMed] [Google Scholar]
- 5.Alvarez C. M., Tredwell S. J., Keenan S. P., et al. Treatment of idiopathic clubfoot utilizing botulinum A toxin: a new method and its short-term outcomes. Journal of Pediatric Orthopaedics. 2005;25(2):229–235. doi: 10.1097/01.bpo.0000149861.50400.db. [DOI] [PubMed] [Google Scholar]
- 6.Kapoor R., Shome D., Jain V., Dikshit R. Facial rejuvenation after intradermal botulinum toxin: is it really the botulinum toxin or is it the pricks? Dermatologic Surgery. 2010;36, supplement 4:2098–2105. doi: 10.1111/j.1524-4725.2010.01703.x. [DOI] [PubMed] [Google Scholar]
- 7.Oh S.-H., Lee Y., Seo Y.-J., et al. The potential effect of botulinum toxin type A on human dermal fibroblasts: an in vitro study. Dermatologic Surgery. 2012;38(10):1689–1694. doi: 10.1111/j.1524-4725.2012.02504.x. [DOI] [PubMed] [Google Scholar]
- 8.Chou H.-Y., Lee C., Pan J.-L., et al. Enriched astaxanthin extract from haematococcus pluvialis augments growth factor secretions to increase cell proliferation and induces MMP1 degradation to enhance collagen production in human dermal fibroblasts. International Journal of Molecular Sciences. 2016;17(6, article 955) doi: 10.3390/ijms17060955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi M. S., Yoo M. S., Son D. J., et al. Increase of collagen synthesis by obovatol through stimulation of the TGF-β signaling and inhibition of matrix metalloproteinase in UVB-irradiated human fibroblast. Journal of Dermatological Science. 2007;46(2):127–137. doi: 10.1016/j.jdermsci.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J., Ji X., Li M., et al. The efficacy and safety of fractional CO2 laser combined with topical type a botulinum toxin for facial rejuvenation: a randomized controlled split-face study. BioMed Research International. 2016;2016:7. doi: 10.1155/2016/3853754.3853754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nitsche A., Stadler P. F. Evolutionary clues in lncRNAs. Wiley Interdisciplinary Reviews: RNA. 2017;8(1) doi: 10.1002/wrna.1376.e1376 [DOI] [PubMed] [Google Scholar]
- 12.Gibb E. A., Brown C. J., Lam W. L. The functional role of long non-coding RNA in human carcinomas. Molecular Cancer. 2011;10, article 38 doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilusz J. E., Sunwoo H., Spector D. L. Long noncoding RNAs: functional surprises from the RNA world. Genes and Development. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen Q., Carninci P. Expression specificity of disease-associated lncRNAs: toward personalized medicine. Current topics in microbiology and immunology. 2016;394:237–258. doi: 10.1007/82_2015_464. [DOI] [PubMed] [Google Scholar]
- 15.Montes M., Lund A. H. Emerging roles of lncRNAs in senescence. FEBS Journal. 2016;283(13):2414–2426. doi: 10.1111/febs.13679. [DOI] [PubMed] [Google Scholar]
- 16.Ma Q., Chang H. Y. Single-cell profiling of lncRNAs in the developing human brain. Genome Biology. 2016;17(1, article 68) doi: 10.1186/s13059-016-0933-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lv M., Xu P., Wu Y., et al. LncRNAs as new biomarkers to differentiate triple negative breast cancer from non-triple negative breast cancer. Oncotarget. 2016;7(11):13047–13059. doi: 10.18632/oncotarget.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guttman M., Rinn J. L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okayama H., Saito M., Oue N., et al. NOS2 enhances KRAS-induced lung carcinogenesis, inflammation and microRNA-21 expression. International Journal of Cancer. 2013;132(1):9–18. doi: 10.1002/ijc.27644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haag D., Zipper P., Westrich V., et al. Nos2 inactivation promotes the development of medulloblastoma in Ptch+/− mice by deregulation of Gap43-dependent granule cell precursor migration. PLoS Genetics. 2012;8(3) doi: 10.1371/journal.pgen.1002572.e1002572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C. S., Bae I.-H., Han J., et al. Compound K inhibits MMP-1 expression through suppression of c-Src-dependent ERK activation in TNF-α-stimulated dermal fibroblast. Experimental Dermatology. 2014;23(11):819–824. doi: 10.1111/exd.12536. [DOI] [PubMed] [Google Scholar]
- 22.Lephart E. D. Protective effects of equol and their polyphenolic isomers against dermal aging: microarray/protein evidence with clinical implications and unique delivery into human skin. Pharmaceutical Biology. 2013;51(11):1393–1400. doi: 10.3109/13880209.2013.793720. [DOI] [PubMed] [Google Scholar]
- 23.Hwang Y. P., Choi J. H., Kim H. G., et al. Cultivated ginseng suppresses ultraviolet B-induced collagenase activation via mitogen-activated protein kinases and nuclear factor κB/activator protein-1-dependent signaling in human dermal fibroblasts. Nutrition Research. 2012;32(6):428–438. doi: 10.1016/j.nutres.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Quan T., Qin Z., Xu Y., et al. Ultraviolet irradiation induces CYR61/CCN1, a mediator of collagen homeostasis, through activation of transcription factor AP-1 in human skin fibroblasts. Journal of Investigative Dermatology. 2010;130(6):1697–1706. doi: 10.1038/jid.2010.29. [DOI] [PubMed] [Google Scholar]
- 25.Pu F., Shao Z., Yang S., et al. Association between functional variants in BIRC5/survivin gene 3′ untranslated region and mRNA expression in lymphoblastoid cell lines. Oncology Letters. 2015;10(4):2319–2322. doi: 10.3892/ol.2015.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lepri S. R., Zanelatto L. C., da Silva P. B. G., Sartori D., Ribeiro L. R., Mantovani M. S. The effects of genistein and daidzein on cell proliferation kinetics in HT29 colon cancer cells: the expression of CTNNBIP1 (β-catenin), APC (adenomatous polyposis coli) and BIRC5 (survivin) Human Cell. 2014;27(2):78–84. doi: 10.1007/s13577-012-0051-6. [DOI] [PubMed] [Google Scholar]
- 27.Wang L., Huang J., Jiang M., Sun L. Survivin (BIRC5) cell cycle computational network in human no-tumor hepatitis/cirrhosis and hepatocellular carcinoma transformation. Journal of Cellular Biochemistry. 2011;112(5):1286–1294. doi: 10.1002/jcb.23030. [DOI] [PubMed] [Google Scholar]
- 28.Lamers F., Van Der Ploeg I., Schild L., et al. Knockdown of survivin (BIRC5) causes apoptosis in neuroblastoma via mitotic catastrophe. Endocrine-Related Cancer. 2011;18(6):657–668. doi: 10.1530/ERC-11-0207. [DOI] [PubMed] [Google Scholar]
- 29.Li M., Liu D., Wang L., Wang W., Wang A., Yao Y. Expression of placenta-specific 8 in human oocytes, embryos, and models of in vitro implantation. Fertility and Sterility. 2016;106(3):781.e2–789.e2. doi: 10.1016/j.fertnstert.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 30.Kaistha B. P., Lorenz H., Schmidt H., et al. PLAC8 localizes to the inner plasma membrane of pancreatic cancer cells and regulates cell growth and disease progression through critical cell-cycle regulatory pathways. Cancer Research. 2016;76(1):96–107. doi: 10.1158/0008-5472.CAN-15-0216. [DOI] [PubMed] [Google Scholar]
- 31.Sweeney T. E., Khatri P. Comprehensive validation of the FAIM3:PLAC8 ratio in time-matched public gene expression data. American Journal of Respiratory and Critical Care Medicine. 2015;192(10):1260–1261. doi: 10.1164/rccm.201507-1321le. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karsenty G., Rocha J., Chevalier S., et al. Botulinum toxin type A inhibits the growth of LNCaP human prostate cancer cells in vitro and in vivo. Prostate. 2009;69(11):1143–1150. doi: 10.1002/pros.20958. [DOI] [PubMed] [Google Scholar]
- 33.Park T. H., Park J. H., Chang C. H., Rah D. K. Botulinum toxin a upregulates Rac1, Cdc42, and RhoAgene expression in a dose-dependent manner: in vivo and in vitro study. Journal of Craniofacial Surgery. 2016;27(2):516–520. doi: 10.1097/scs.0000000000002272. [DOI] [PubMed] [Google Scholar]
- 34.Zhu J., Ji X., Li M., et al. The efficacy and safety of fractional CO2 laser combined with topical type A botulinum toxin for facial rejuvenation: a randomized controlled split-face study. BioMed Research International. 2016;2016:7. doi: 10.1155/2016/3853754.3853754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su J., Gorse K., Ramirez F., Fox M. A. Collagen XIX is expressed by interneurons and contributes to the formation of hippocampal synapses. Journal of Comparative Neurology. 2010;518(2):229–253. doi: 10.1002/cne.22228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X., Jing X., Cheng X., et al. FGFR3 promotes angiogenesis-dependent metastasis of hepatocellular carcinoma via facilitating MCP-1-mediated vascular formation. Medical Oncology. 2016;33(5, article no. 46) doi: 10.1007/s12032-016-0761-9. [DOI] [PubMed] [Google Scholar]
- 37.Burren A., Neuditschko M., Signer-Hasler H., et al. Genetic diversity analyses reveal first insights into breed-specific selection signatures within Swiss goat breeds. Animal Genetics. 2016;47(6):727–739. doi: 10.1111/age.12476. [DOI] [PubMed] [Google Scholar]
- 38.Smith L. B., Belanger J. M., Oberbauer A. M. Fibroblast growth factor receptor 3 effects on proliferation and telomerase activity in sheep growth plate chondrocytes. Journal of Animal Science and Biotechnology. 2012;3(1, article no. 39) doi: 10.1186/2049-1891-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keegan K., Johnson D., Williams L. T., Hayman M. J. Characterization of the FGFR‐3 gene and its gene product. Annals of the New York Academy of Sciences. 1991;638(1):400–402. doi: 10.1111/j.1749-6632.1991.tb49052.x. [DOI] [PubMed] [Google Scholar]
- 40.Li W., Yang W., Wang X.-J. Pseudogenes: pseudo or real functional elements? Journal of Genetics and Genomics. 2013;40(4):171–177. doi: 10.1016/j.jgg.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Poliseno L. Pseudogenes: newly discovered players in human cancer. Science Signaling. 2012;5(242, article re5) doi: 10.1126/scisignal.2002858. [DOI] [PubMed] [Google Scholar]
- 42.Gray T. A., Wilson A., Fortin P. J., Nicholls R. D. The putatively functional Mkrn1-p1 pseudogene is neither expressed nor imprinted, nor does it regulate its source gene in trans. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(32):12039–12044. doi: 10.1073/pnas.0602216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coin L., Durbin R. Improved techniques for the identification of pseudogenes. Bioinformatics. 2004;20, supplement 1:i94–i100. doi: 10.1093/bioinformatics/bth942. [DOI] [PubMed] [Google Scholar]
- 44.Zhu J., ji X., Xu Y., et al. The efficacy of intradermal injection of type A botulinum toxin for facial rejuvenation. Dermatologic Therapy. 2017;30(1) doi: 10.1111/dth.12433.e12433 [DOI] [PubMed] [Google Scholar]