Abstract
In the event of an indoor release of an environmentally persistent microbial pathogen such as Bacillus anthracis, the potential for human exposure will be considered when remedial decisions are made. Microbial site characterization and clearance sampling data collected in the field might be used to estimate exposure. However, there are many challenges associated with estimating environmental concentrations of B. anthracis or other spore-forming organisms after such an event before being able to estimate exposure. These challenges include: (1) collecting environmental field samples that are adequate for the intended purpose, (2) conducting laboratory analyses and selecting the reporting format needed for the laboratory data, and (3) analyzing and interpreting the data using appropriate statistical techniques. This paper summarizes some key challenges faced in collecting, analyzing, and interpreting microbial field data from a contaminated site. Although the paper was written with considerations for B. anthracis contamination, it may also be applicable to other bacterial agents. It explores the implications and limitations of using field data for determining environmental concentrations both before and after decontamination. Several findings were of interest. First, to date, the only validated surface/sampling device combinations are swabs and sponge-sticks on stainless steel surfaces, thus limiting availability of quantitative analytical results which could be used for statistical analysis. Second, agreement needs to be reached with the analytical laboratory on the definition of the countable range and on reporting of data below the limit of quantitation. Finally, the distribution of the microbial field data and statistical methods needed for a particular data set could vary depending on these data that were collected, and guidance is needed on appropriate statistical software for handling microbial data. Further, research is needed to develop better methods to estimate human exposure from pathogens using environmental data collected from a field setting.
Keywords: analytical methods, environmental monitoring, inhalation exposure
INTRODUCTION
Bacillus anthracis, a Gram-positive bacteria and the causative agent for anthrax, occurs naturally in many soil environments.1 B. anthracis in spore form can persist for many years in several environmental matrices including water and soil2, 3, 4, 5, 6, 7 and can resist heat treatment8 as well as degradation via sunlight.9 Following an intentional wide-area release of B. anthracis or another pathogen, the U.S. Environmental Protection Agency (EPA) would have a role in remediation of contaminated indoor and outdoor areas. The overall objective in such a response would be to protect human health and the environment, and to implement necessary remediation. Site remediation efforts might include both site-characterization sampling to determine the extent of contamination, and clearance sampling to determine efficacy of the cleanup effort.10, 11, 12 Following the 2001 attacks, the National Response Team conducted an assessment of the lessons learned from response at the contaminated sites.13 At the time of the report, findings included limited knowledge of environmental fate and transport of B. anthracis and the need for assessment of technologies that provide more timely detection of spores and at lower levels than currently available.13 Currently, the EPA and Centers for Disease Control and Prevention (CDC) have recommended that clearance goals be set to “no detection of viable spores”.14, 15 However, for anthrax cleanup efforts, the remediation goal continues to be limited by the detection limits of the technologies used to analyze the field samples.16
Decisions regarding remediation of a contaminated site are often risk-driven, but setting specific risk-based cleanup goals for microbial contamination events using data collected from the field have been stymied by unresolved issues. First, there is a lack of consensus on how to address uncertainties and variability in the field data collected at a contaminated site to estimate the environmental concentration. Second, the uncertainties and variability in the environmental concentration data complicate the use of these data for exposure estimation. Currently, many of the available exposure assessment methods are focused on chemical exposures or are not specific to indoor microbial releases;11, 12, 17, 18, 19, 20, 21, 22 it is unknown if these available methodologies are applicable to microbial contamination incidents.
There are numerous factors that can affect the uncertainty (absent or incomplete information) and variability (range of observed differences in a parameter that are a result of chance)18 of the field data collected following a contamination event, and assessing the usability of data for evaluating environmental concentrations can be difficult. For example, the source, type, and magnitude and distribution of contamination may be unknown, and the reliability of the field data may be affected by an inability to control for these factors. This uncertainty and variability creates challenges when using these field data to evaluate environmental concentrations. Specific challenges could include:
Collecting field samples that are appropriate for the intended use.
Conducting laboratory analysis and selecting a reporting format needed for the laboratory data.
Analyzing and interpreting the data set using appropriate statistical techniques.
Using the field data for assessing exposure introduces additional challenges. The lack of guidelines on microbial field data usability make it difficult to use these data to assess exposure. Lacking guidelines for accurately using microbial field data to estimate exposure, chemical guidelines might become potential alternatives. However, considerations for microbial risk assessment differ from chemical assessments in several ways. Pathogens are genetically diverse and often different strains of the same species have varied virulence for human hosts.18 In addition, pathogens have the ability to grow and die-off, whereas while chemicals might degrade or be transformed, they do not multiply.18, 23 Considerations of host immunity and susceptibility such as genetic and acquired differences might not be handled the same way for chemical assessments compared with microbial assessments, which use a more dynamic model.18 Routes of exposure, health endpoints, and the length of time for a health outcome to appear will differ between chemical and microbial exposures.18 Following infection with a microbial agent, transmission might occur between individuals; however, chemicals that have entered the tissue of an individual are typically not transmitted to other individuals (although some exceptions exist).18 Pathogens can also be endemic in the environment, present themselves on a seasonal basis, and might respond differently to environmental treatment options than chemicals.18, 23 Because pathogens might also interact with other species or debris in the environment, detection of pathogens in environmental matrices is complicated. The analytical methods used need to be capable of determining viability and be sensitive enough to detect the pathogen of concern.18
Until these challenges surrounding the uncertainty and variability of the field data can be addressed, estimation of environmental concentrations for the purposes of exposure assessment is limited. This paper summarizes challenges faced in collecting and interpreting microbial field data from a contaminated site and explores the implications of related data limitations when using field data for estimating potential environmental concentrations. Although the paper was written with considerations for B. anthracis contamination, it may also be applicable to other bacterial agents, especially spore formers. Further guidance is needed to inform decision makers how data collected in the field can be used to estimate exposure following a biological release.
CHALLENGES INTERPRETING FIELD DATA
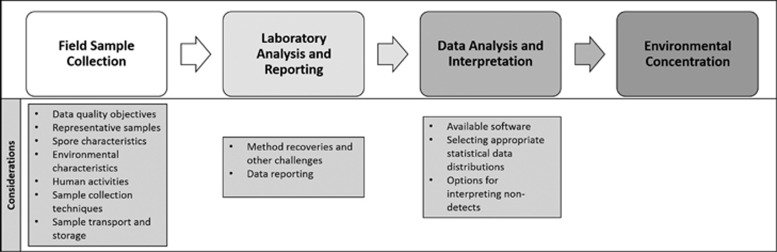
To address the three overarching challenges that were identified above, a discussion of each is presented below. These challenges are broken out by field sample collection (Table 1), laboratory analysis and reporting, and data analysis and interpretation, which might occur during a contamination event (Figure 1).
Table 1. Factors affecting spore recovery and their potential effects.
| Factors affecting spore recovery/variability factor | Potential effect on recovery efficiency from literature review | Related literature |
|---|---|---|
| DQO | Information on types of microbial data needed to answer the study goal, interpreting the microbial data received from the laboratory, and defining the appropriate statistical distributions that can be used with this type of data are needed to help improve the DQO process. | EPA,24 |
| Representative samples | ||
| Material sampled | Non-porous surfaces generally yield higher spore recovery efficiencies than porous surfaces. Relationships between measures of surface roughness and spore recoveries appear promising, while other surface characteristics may also influence spore recovery such as surface charge and hydrophobicity. | Brown et al.;43 Busher et al.;35 Buttner et al.;31 Buttner et al.;40 Da Silva et al.;36 Edmonds;34 Edmonds et al.;38 Estill et al.;32 Frawley et al.;30 Hodges et al.;41 Hong-Gellar et al.;42 Krauter et al.;39 Herzog et al.;63 National Response Team;27 Probst et al.;37 Valentine;33 |
| Sample area | Devices covering larger areas (wipes and vacuums) likely recover more spores than devices with smaller sampling areas (swabs). Based on spores recovered per area sampled, larger sampling devices will likely have lower detection limits, even the though the smaller sampling devices may recover a higher percentage of the spores available. | Brown et al.;46 Buttner et al.;40 Buttner et al.;44 Edmonds et al.;38 Rose et al.45 |
| Sampling device | For surface sampling, wipes and vacuums are likely to perform better (more positive detections and higher recovery efficiencies) than swabs (especially dry swabs). These trends may differ for spore recoveries on a per-area basis for devices with differing sample areas. However, only swabs and sponge-sticks have been validated (on stainless steel surfaces) for recovering B. anthracis spores. For air sampling, personal breathing zone samplers for air sampling might be more representative of the concentration of spores at the breathing zone. Aggressive air sampling and sampling of HVAC filters has been recommended as methods to help determine indoor extent of contamination. | Calfee et al.;28 EPA;17 EPA;26 Einfield et al.;50 Estill et al.;32 Hodges et al.;41 Hong et al.;25 Rose et al.;45 Sanderson et al.;47 Teshale et al.;48 Valentine,;33 Valiante et al.;49 Van Cuyk et al.29 |
| Sampling device material/characteristics | Most research has focused on swabs with some conflicting findings. Macrofoam swabs are recommended by the CDC and their use has been supported in the literature. The CDC also recommends using cellulose sponge and non-cotton polyester or rayon/polyester blend gauze for sampling smooth, nonporous surfaces for B. anthracis spores. | Budowle et al.;55 Da Silva et al.;51 Da Silva et al.;36 Edmonds et al.;38 Krauter et al.;39 Probst et al.;37 Probst et al.;52 Rose et al.;53 Thomas et al.;54 Valentine;33 |
| Dry vs wet; wetting agent | Wet sampling devices (swabs and wipes) perform better than dry materials. The CDC recommends using neutralizing buffer as the wetting agent, although many researchers have seen benefits with the incorporation of surfactants such as Tween into the wetting agent. | Frawley et al.;30 Rose et al.;53 Sanderson et al.;47 Teshale et al.;48 Valiante et al.49 |
| Spore characteristics, environmental characteristics, and human activities | ||
| Bacillus species/strain | Spore characteristics associated with different species/strains can affect spore recovery efficiency. Limited studies have shown higher recoveries with B. atrophaeus than B. anthracis Sterne and higher recoveries with B. anthracis Sterne than B. anthracis Ames. Caution should be used in extrapolating spore recovery efficiency from one organism to another. | Baron et al.;60 Carrera et al.;56 Chen et al.;58 Da Silva et al.;51 Estill et al.;32 Greenberg et al.;57 Hong-Gellar et al.;42 Probst et al.;37 Probst et al.;52 Thomas et al.;54 Williams et al.59 |
| Spore inoculation method | Spore application methods affect spore recoveries; spores applied via liquid inoculation are more prone to spore clumping. Low concentrations of spores inoculated via dry aerosols can re-disperse. | Edmonds et al.;38 Gao;71 Hodges et al.;64 Lee et al.;72 Probst et al.;52 Rose et al.53 |
| Spore loading | Although not always reported, spore recovery efficiency often increases with higher spore loadings and variability in spore recovery increases at lower spore loadings. | Brown et al.;43 Brown et al.;65 Edmonds et al.;38 Estill et al.;32 Hodges et al.;64 Hodges et al.;41 Krauter et al.;39 Probst et al.;52 Rose et al.;45 Thomas, et al.54 |
| Non-uniform deposition | Spores tend to deposit/accumulate preferentially in certain areas resulting in a patchy spatial distribution that will be reflected in the sampling results. Smaller particles might settle slower than larger particles. | Amidan et al.;66 Baron et al.;60 Brown et al.;43 Hong et al.;67 Hong et al.;25 Sextro et al.68 |
| Surface orientation | Fewer spores are expected to be recovered from vertical and horizontal (face-down) surfaces, which will contribute to the overall (within room) sample variability. Although no testing was reported in the literature, enclosed surfaces, for example, inside of drawers, may also have different spore concentrations than those in more open locations. | Johnson et al.;69 Lewandowski et al.70 |
| Environmental conditions | The influence of temperature and relative humidity was infrequently investigated, although relative humidity apparently affected the recovery of B. atrophaeus spores from stainless steel. Low relative humidity conditions may keep spores from firmly attaching to certain surfaces. | Beecher;73 Busher et al.;35 Einfield et al.;50 Krauter et al.;39 |
| Re-dispersion and transport | Spores may be re-aerosolized via indoor air currents or during sampling activities, possibly affecting spore recovery efficiencies and variability. | Busher et al.;35 Edmonds;34 Estill et al.;32 GAO;71 Sanderson et al.;47 Sextro et al.;68 Sippola et al.;75 Van Cuyk et al.;29 Weis et al.76 |
| Sample collection techniques | Variations in sampling technique may contribute to spore recovery variability. Limited laboratory studies indicate a relatively low (<10%) impact on spore recovery. However, the impacts of sampling technique may be heightened under more difficult field conditions. | Beecher;73 Brown et al.;43 Da Silva et al.;36 Edmonds;34 GAO;71 Hodges et al.;64 Piepel et al.;78 Probst et al.37 |
| Sample transport and storage | Not expected to be a significant source of variability if transport and storage occur at ~5 °C with sample processing within 1 or 2 days of collection. | CDC;79 Hubbard et al.80 |
Abbreviations: CDC, Centers for Disease Control and Prevention; DQO, Data Quality Objective; EPA, U.S. Environmental Protection Agency; HVAC, heating, ventilation, and air conditioning.
Figure 1.
Flowchart of considerations for interpreting environmental field data.
Field Sample Collection Challenges
Determining data quality objectives
Before sample collection, determination of performance or acceptance criteria is needed to design a collection plan that will provide the quantity and quality of data needed to support remediation efforts. EPA's Data Quality Objective (DQO) Process24 is an iterative seven-step planning approach used to prepare for data collection activities in environmental monitoring efforts and research. It provides the criteria for the sampling design, where to collect samples, tolerable decision error rates, and the number of samples to collect. In a real-world response, this process could be used to develop a sampling plan that supports the quantity and quality of data needed for estimation of exposure; however, the document does not provide guidance specific for use of the process for collection of microbial data, such as helping to define the types of microbial data needed to answer the study goal, interpreting the microbial data received from the laboratory, and defining the appropriate statistical distributions that can be used with this type of data.
Representative samples
One key question that might be identified when trying to estimate environmental concentrations is whether these data are representative for the intended purpose and exposure pathways of interest. However, not all data collected for site characterization and clearance sampling will be representative of environmental concentration(s) at the contact point(s) and may not be appropriate for evaluating the environmental concentrations to which people are exposed. For example, following a release, the concentration of spores in the air will decline over time, making it difficult to characterize the room using aerosol concentrations alone.25 Use of a stationary air sampler might not account for exposure at an individual's breathing zone and only offers a snapshot in time. Personal monitors would be needed to measure point-of-contact concentrations.17 Use of a personal breathing zone sampler during sample collection could be more representative of the level of re-aerosolized spores present at that specific breathing zone during remediation efforts. There might also need to be consideration given to the characteristics of the organism being sampled (i.e., viability and spore formation) when sampling methods are selected. However, there is no standard air sampling method to be used in the remediation process and selection of air sampling equipment will be site-specific. Use of aggressive air sampling has been suggested as a potential technique following decontamination to reduce the sampling burden,26, 27 however this technique would not allow estimation of personal exposure. Sampling of heating, ventilation, and air conditioning (HVAC) filters has also been recommended as a method to help determine indoor extent of contamination.25, 27, 28, 29
Another key question that could be asked when using field samples is whether these data collected for one purpose can be used for another purpose. For example, in some cases, samples are collected for an initial site characterization or to determine whether a site is cleared for re-entry. These methods often include surface sampling methods, which are associated with swabs, wipes (including sponges), and high-efficiency particulate air vacuum-filter socks (vacuuming). The device type selected for sampling is often based on the area and surface to be sampled. These methods do not address point-of-contact exposures (concentration at interface of the environment and the exposed person) as are needed for estimation of exposure.17 The type of surface to be sampled,27, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43 the size of the sampling area,38, 40, 44, 45, 46 the type of sampling device selected for sampling,32, 33, 47, 48, 49, 50 the material characteristics of the sampling device,33, 36, 37, 38, 39, 51, 52, 53, 54, 55 and the use of wet vs dry sampling devices30, 47, 48, 49 53 could affect the potential recovery of the spores from the sample, and therefore the potential estimate of the environmental concentration. In addition, only swabs and sponge-sticks on stainless steel surfaces have been validated by CDC as sampling methods for recovering B. anthracis spores.41, 42, 43, 44, 45 Because no device/material/surface combination is capable of achieving 100% recovery, spore loss due to the sampling process might need to be accounted for to prevent bias in the results.
Spore characteristics, environmental characteristics, and human activities
In an incident involving contamination by B. anthracis or other spore-forming organisms, the characteristics of the spores, the method of dissemination, the spore deposition pattern, and the spore load on the surface might not be initially known. However, documenting information on these factors might provide insight on the expected spore recovery from the samples.27 For example, spore adhesion to surfaces and recovery efficiency tend to vary for different Bacillus spp.32, 37, 42, 51, 52, 54, 56, 57, 58, 59, 60 In addition, viability of spores might vary in different matrices and can be impacted by environmental stressors.61 Knowledge of the distribution of the size of the particles that have been released help reduce uncertainty in the sample estimate of estimate of risk.62 Further, data collected in a real-world event could differ from data collected in field research.
Many factors play a role in the efficiency of recovery of spores from surfaces (which have been reported to range from 10 to 50%) and which could include the sampling protocol used, surface type, and type of sample processing used.63 The influence of spore loading on recovery efficiency has been reported to be variable.32, 38, 39, 41, 43, 45, 52, 54, 64, 65 Variability in recovery efficiency has been attributed to non-uniform surface deposition of spores,43 which might occur if settling is not the primary deposition mechanism.60 Spores tend to deposit/accumulate preferentially in certain areas resulting in a patchy spatial distribution.66 In addition, deposition rates will vary with particulate size (single spore estimated to be 1 μm in size compared with >10 μm when clumping of spores occur), with smaller particles settling slower than larger particles.25, 67, 68 Fewer spores are expected to be recovered from vertical and horizontal face-down surfaces compared with horizontal face-up surfaces, contributing to the overall (within room) sample variability.69, 70 Enclosed samples (such as inside drawers) could be contaminated at different levels than more open locations. In addition, spores applied to surfaces via liquid inoculation do not behave the same way as spores applied as dry material; liquid inoculated spores are more prone to clumping and spore agglomeration.38, 52, 53, 64, 71, 72 Recovery efficiency of the sample might also need to consider the number of spores in a clump, which has been estimated by Hong et al.,67 to be 1, 3–14, 6–65, and 13–524 number of spores for 1, 3, 5, and 10 μm clumps, respectively. Adherence or non-adherence of the spores to a surface might also be influenced by temperature and relative humidity.35, 39, 50, 73 Comparison of recovery efficiencies from different studies might be difficult, especially when different methods are used to calculate recovery efficiency.33, 60
Finally, human movement, repeated disturbances, indoor air currents, transport through ventilation systems, movement of indoor equipment, and transfer of contamination to other areas via tracking on footwear impact the spore deposition patterns and reaerosolization potential.29, 32, 35, 68, 71, 74, 75, 76, 77 Reaerosolization of spores from outside the target sampling area might affect sampling results.34, 47 For example, on a tracked floor, loss occurs due to resuspension for the spores, so the concentration on the floor cannot be directly related back to the air concentration.67 There is no guidance on how to quantify these factors mentioned throughout this section which affect the spore recovery efficiency, however, the potential for these factors to influence the sampling results does exist and might be a point for consideration when formulating recommendations or in making decisions.
Sample collection techniques
Variability in sample-collection techniques among different sampling personnel could also affect sample results for both surface samples and samples taken via personal breathing zone monitors. One study suggested that differences arising from sample collection might contribute more to variability than sample processing.78 Although training can improve the consistency among sampling personnel, the variation associated with each individual's technique (i.e., angle, pressure, and changes in sampling direction) must be characterized to quantify the accuracy of the sampling procedure.36, 43, 71 Sampling personnel performance could be hindered by adverse conditions such as excessive heat, high-stress environments, time restrictions, and the use of protective clothing.34 Sampling results could also be affected by areas that are difficult to sample, that are hard to access or that have irregular or uneven surfaces, and by the cross-contamination of spores on surfaces due to reaerosolization of the spores.34, 73 Note that if the sampling method is robust enough, the difference in sampler experience might produce only marginal differences in recovery37 and variability in spore recovery might be attributed more to the surface being sampled or the device being used than to sampler variability.34, 64
Sample transport
Storage temperature and the type of liquid transport media used may play a role in recovery of spores, and might need to be noted when documenting sample results. CDC recommends that surface samples be shipped on ice packs, stored between 2 and 8 °C, and processed within 48 h of collection.79 The spore form of B. anthracis does not require liquid transport media; however, if preservation of the vegetative form is desired, a transport media might be needed.80 Hubbard et al.80 conducted viability testing of B. anthracis Sterne spiked onto swabs stored at −70, 4, 25, and 45 °C using commercial off the shelf storage media (phosphate-buffered saline with 0.1% Triton X-100 (PBST), liquid Amies, Stuart transport media, and dry samples) held for up to 60 days. Nutrient poor media, such as Amies or Stuart media, combined with higher temperatures, promoted sporulation of vegetative cells.80 The study found that recovery was highest when liquid Amies, PBST, and liquid Stuart media were used and samples were stored at 25 °C.80
Laboratory Analysis and Reporting Challenges
Analysis method recoveries and challenges
The literature revealed several reviews which discuss available detection methods for B. anthracis in environmental samples and challenges to using those methods.81, 82, 83, 84 Although the focus of this review is not to discuss all the challenges associated with the different available analytical techniques, common themes included: sample processing as a limiting step to detection;10, 84 being able to distinguish B. anthracis from other closely related strains that may be present in environmental samples, such as B. cereus;81, 82, 83 needing to use a selective agar such as polymixin-lysozyme EDTA-thallous acetate agar to selectively grow B. anthracis and the slow growing times associated with use of conventional analysis methods;84, 85, 86 consideration of organism viability;10 and the need to detect low concentration of the organisms from the samples.81, 83
When using conventional analysis methods, the final colony-forming units (CFU) counts needs to include consideration of the recovery efficiencies of the analysis method used. However, recovery efficiency of the analytical methods commonly used needs to be further investigated.27 Many factors might affect the recovery efficiency of spores from a sample related to the methods used to extract and analyze the sample.31 Spores can remain on the sampling device,36, 37, 64 on the centrifuge tube,34, 36 or on the coupons (i.e., extracted sample material) themselves.36 Other common reasons for differences in spore recovery during extraction and analysis include type and volume of extraction solution used,38, 40, 51 addition of surfactants to wetting agents to improve recovery,33, 36, 41, 45 presence of residual decontamination agent in the sample,35 choice of dissociation method used (vortexing, sonication, and stomaching),31, 51, 53, 70, 87 use of heat shocking to kill background organisms,43, 45 sensitivity of the culture methods,88 and clumping of spores on culture plates.88 Several types of interferences such as the presence of dust, vegetative bacteria, fungal spores, metals, fibers and other materials can affect spore removal from surfaces, extraction of spores from the sampling device, or successful culturing such that detection/identification of the bacteria is affected.31, 35, 41, 43, 50, 89 If recovery efficiencies are not available for the analysis methods used, then research is needed to fill this gap.
Ideally, only one laboratory would be used to analyze all samples in any given contamination incident. However, given the large numbers of samples that could be collected following a contamination incident, using as few different laboratories as possible using the standardized analysis method could help keep results consistent. Use of multiple laboratories might contribute to the overall variability in results, although the within-laboratory variability (between samples) could be greater than the between-laboratory variability.32, 41, 45, 78
Analysis method data reporting
Guidance on how laboratories should report out analytical results is inadequate, but is needed to ensure data are used appropriately and consistently. Determining the format of required analysis data (e.g., quantitative vs semi-quantitative vs qualitative, CFU vs cycle threshold) will influence the type of analysis that is done. When culture analysis is to be used, a better understanding of whether the analysis data can be used quantitatively or qualitatively needs to be established.27 Data from surface/sampling device combinations that have not been validated will limit the quantitative statistical analysis that can be done using the data, as these data might be considered qualitative or semi-quantitative at best. A non-detected culture result does not necessarily mean that the actual sample value is zero, but rather that the viable organism is present below the limit of detection (LOD) of the assay used. Two types of culture methods, spread plating and filter plating, have a LOD of 1 CFU/plate.90 The limit of quantitation (LOQ) or countable range is considered to be between 30 and 300 CFU or 25–250 CFU for spread plates and 20–200 CFU for filter plates,90, 91, 92, 93 although individual methods have identified slightly different ranges for spread plates94, 95, 96 and filter plates.92, 96, 97
Higher variability is expected below the lower end of the LOQ and in part could be due to growth conditions on the plate, influence of competing organisms, dispersion of the cells in the sample, clumping of bacterial cells, interpretation of the results by the analyst, and that one colony does not necessarily mean there is just one cell or spore present.90, 98, 99, 100 At the higher end of the range, colony overlap might prevent accurate counting and therefore could underestimate the count. Those above the range are often labeled as too numerous to count or reported out as >upper limit.90, 97 Another source of uncertainty might include absent or incomplete information; however, methods to handle this type of uncertainty are often not included in the literature or methods concerning microbial data.101
Laboratories analyzing samples sometimes vary on which data are considered and reported, making final interpretation of the data difficult. In addition, reporting of results falling below the LOQ or LOD could vary. In some cases, only plates within the countable range are used to estimate the CFU in the undiluted sample.102 Other analysis methods require data be reported as less than the countable range multiplied by the inverse of the dilution factor93 or to report the data as below the LOQ91 or LOD. Other recommendations have been to: report non-detects as <values/volume filtered for membrane filters;97 to count the actual CFU/ml or 100 ml for spread plates;97 or to report out an estimated count, even if data are below the countable range.90 In some cases, filter plate data have been substituted in place of spread plate data when the data fell below the LOQ to (1) extend the dynamic range of microbial analysis relative to the spread plate analysis and to lower the LOD by analyzing a higher fraction of the undiluted extract and (2) decrease variability in the results when using spread plate data below the defined LOQ. However, there is no guidance on how to handle cases where filter plate data are missing and the spread plate data fall below the LOQ. The number of significant figures used in calculations must also be considered.90 Agreement needs to be reached with the analytical laboratory on the definition of the countable range, as well as how data below the LOQ will be reported (e.g., report as non-detect, report CFU as is). Standardization of the method used to report the data by the analytical laboratories could help provide more repeatable and reliable results. In addition, to ensure data are reported in a consistent manner, establishment of data usability and reporting guidelines are needed.
Data Analysis and Interpretation
Available statistical analysis software for microbial data
Guidance is needed appropriate statistical software packages for the intended use with microbial data. Several software products which have been used for microbial data that were mentioned in the literature are briefly described below. This list should not be considered exhaustive.
EPA's ProUCL Version 4.1 software was recently used to investigate several options for interpreting culture-based/microbial count data.103 ProUCL is a statistical software option that provides several state-of-the-art parametric and nonparametric methods such as Kaplan–Meier and ROS, for calculating the 95% upper confidence levels (UCL) on the mean from data sets containing both uncensored and censored data.104 ProUCL software helps identify an appropriate method for the given set of data by applying goodness-of-fit tests relative to a specified distributional model (e.g., normal, lognormal, and gamma) and making recommendations on a method based on the outcome of these tests and other properties of the data (e.g., SD, skewness, sample size, number of less-than values).104 However, ProUCL was not designed for use with microbial data and is not well designed for the calculation of 95% UCL on the mean for parameters measured using count-based analytical methods because (1) counts of zero prohibit ProUCL software from evaluating gamma and lognormal data distributions; (2) count-based data are inherently discrete, but ProUCL software assumes a continuous data distribution; and (3) ProUCL software assumes only inter-sample variation, while count-based data can also consist of random Poisson counting variation.105 Brattin et al.105 developed a statistical analysis approach, similar to ProUCL software, that includes model fitting procedures and 95% UCL on the mean calculations that include the effect of random Poisson counting error. However, with this approach a count of zero would be evaluated with results of zero, and therefore, options for dealing with censored data would likely be limited.
The Visual Sample Plan is a tool that allows for design of a statistical nonparametric and parameteric sampling plans and subsequent statistical analysis of the sample data.106 The tool provides many benefits such as ability to graph hot spot detection and access to statistical and mathematical algorithms. However, the user's guide did not provide specific considerations for use of the tool with microbial data. The Building Restoration Operations Optimization Model (BROOM) tool can be used for collecting, managing, and analyzing data and has been used for collection of data in several microbial related projects. However, one of the studies did not utilize the tool specifically for statistical analysis.26 In addition,it was unclear from the reviewed literature if the other study that used the BROOM tool used it for statistical analysis or not.107
Other statistical software packages mentioned with use of microbial data in the literature identified during this review were MATLAB (Mathworks, Inc., Natick, Massachusetts, USA),108 SYSTAT (Systat Software Inc., San Jose, California, USA),108 SAS statistical software (SAS Institute, Inc),109 SPSS (SPSS Inc., Chicago, Illinois, USA),110 and R (R Foundation for Statistical Computing, Vienna, Austria).111
Selecting appropriate statistical data distributions
Selection of an appropriate statistical distribution for a count-based data set can be problematic. Traditionally, microbial count data have been converted to concentrations by dividing sample volumes for statistical analysis.112 However, microbiological count-based data could vary by several orders of magnitude or might not be uniform in suspension and therefore might not fit the normal distribution used by many traditional parametric statistical tests.112, 113 Use of a lognormal transformation has been suggested for microbiological data.113 Use of discrete rather than continuous distributions could help address the issue. However, the necessity of considering discrete microbial distributions vs continuous distributions might depend on the level of contamination.112 For example, when the contamination level is high, a normal approximation to the Poisson (discrete) distribution might hold, and at low levels of contamination, a heterogeneous distribution of the microbial agent in the matrix becomes more important because the number of pathogens that individuals are exposed to could be very few or zero.18 It has been suggested that when clumping of spores occur, the clumps of spores will have a mixture of different Poisson distributions related to the sizes of the clumps in the sample, assuming the concentration is spread uniformly over the area to be sampled.67 However, when non-uniform concentrations come into play, a negative binomial distribution is often used.67, 114 Non-parametric statistical tests are often used if the data does not fit a normal distribution.113 In addition, because of the differences in the sample area for multiple sample types (swabs, wipes, etc), there may be large differences in the data values when generating summary statistics.103 One possible solution to this issue that has been suggested includes limiting the analysis to a single sampling method that can address the project objectives.103 Finally, the pathogen concentration might increase in number due to growth in the sample, compared with chemical concentrations that might degrade or be transformed but do not multiply.18 The growth of the pathogen in the sample might also cause clustered distributions or clumping of the pathogens in the sample,18 and therefore make recovery difficult to determine. A method to assess both the potential for microbial growth in environmental samples and account for changes in recovery due to microbial growth is needed.
Options for interpreting non-detects
Guidance is needed on how to interpret analytical data falling below the LOQ and therefore reported as not detected (also known as censoring) or below the quantitation limit. Many of the uncertainties associated with interpreting microbial non-detect data might be similar to uncertainties encountered when dealing with chemical concentration data. When encountering non-detect data, a variety of treatment options could be used (Table 2), but the best option depends on goals for use of the data and the required accuracy.17 Traditional options for handling censored data include discarding non-detect data and using simple substitution methods (treating non-detects as zero, half the quantitation limit, at the quantitation limit, or the quantitation limit/√2).17, 113, 115 However, treatment of censored data (data reported as not detected) and data below the LOQ and use of various substitution approaches can result in descriptive statistics that do not adequately represent the true underlying data distribution.116 For example, eliminating non-detects from the data set might not allow for a true interpretation of the data, while substitution method might underestimate or overestimate the mean results.113 Various options for treating censored data have been explored and have continued to evolve in recent years beyond traditional (and problematic) substitution approaches.108, 116, 117, 118, 119 Use of distributional analyses such as log-Probit analysis have also been used in which the non-detects are treated as unknowns while the detects are plotted on a scale.17 More accurate methods for computing statistics are now available in statistical software and include: modern maximum likelihood estimation which uses detected observations and less-than values to generate summary statistics; regression on order statistics (ROS) where non-detects are filled in without assigning the same value; and Kaplan–Meier, which is a nonparametric approach to estimate the cumulative distribution function of data with multiple LODs.116, 120 Imputation methods such as ROS might provide better descriptions of censored data than substitution approaches for non-detects or maximum likelihood estimation for data sets with <50 detected values.108, 120
Table 2. Options for interpreting non-detect data.a.
| Technique | Percent of observations below detection limits | General description | Advantages | Disadvantages | Source |
|---|---|---|---|---|---|
| Discard non-detect entries | NA | Entries with a non-detect value are eliminated. | This approach is simple. | Analysis of results that have been reported as not detected is not possible. The data set may be distorted. | Levine113 |
| Substitution of a value in place of the non-detect value | <15% | Substitute non-detects with zero; half the LOQ or LOD; at the LOQ or LOD; or at the LOQ/√2. Substitution with ½ the LOD has been used frequently in the past for chemical assessments. | Substitution is simple. Treating non-detects as zero reduces overestimation while treating non-detects as the LOD avoids underestimation. | Use of this method could cause the data set to become skewed. Underestimation (with treating non-detects as zero) and overestimation (with treating non-detects at the ½ the DL and at the DL) is possible. | EPA;17 EPA;115 Levine113 |
| Atchison's method | <15% | The mean and variance are adjusted to assume non-detects are zero. Assumption is that microbial data is log normally distributed. | Assumes data below the LOD were actually present, but could not be recorded. | May result in overestimation. | EPA;115 Levine113 |
| Cohen's method | <20% | Uses a maximum likelihood estimation approach to fit a lognormal distribution to the data. Assumes the data follow a normal distribution. | Accounts for data below the LOD. | As the number of observations falling below the LOD increases, the statistical power decreases, and the true significance level increases. observations >20 are required for consistent results. Do not use if >50% of observations are non-detect. The LOD must be the same for all entries. | EPA;115 Levine;113 Helsel116 |
| Kaplan–Meier | <50% | Non-parametric method. Estimates a cumulative distribution function for data that has multiple LODs to compute descriptive statistics. | Does not require a distribution to be specified. Can account for multiple censoring limits. | Used primarily for data with “greater thans”. | Helsel;116 Helsel120 |
| ROS | 50–80% | Imputation method (censored or missing observations are given a value, but not all non-detects are given the same value) which uses probability plot of detects to fill in the non-detect values. | Can be used for data with multiple LODs. Performs better on small sample sizes than MLE and substitution methods or for data that do not fit a distribution. | None given in the cited sources. | Helsel;116 Helsel;120 Wong108 |
| Modern MLE | 50–80% | Uses less-than values (censored values) and detected observations to provide adjusted estimates of the mean and SD that were likely to have produced both detected and non-detected data. Assumes data follow a normal or lognormal distribution. | Accounts for data below the detection level. | Must have an n >50 to use this method. | EPA;115 Helsel;116 Helsel120 |
| Test of proportions | >50% | Non-parametric method. Requires at least 10% of the data be quantified. | Can be used for categorical data (presence/absence). | May not be applicable for composite samples. | EPA;115 Levine113 |
| Log-probit analysis | NA?? | Distributional method. Assumed data has a lognormal probability distribution. Detected values are plotted and percentages of non-detects are accounted for. | More accurate and less biased than substitution. | Requires data to have enough detected observations to define the distribution function with confidence. | EPA17 |
CONSIDERATIONS FOR FIELD STUDIES AND REAL-LIFE CONTAMINATION EVENTS
Several considerations for interpreting field data during future microbial contamination events were identified based on the challenges discussed above. These include considerations for sample collection, data management, and data interpretation. The following sections briefly discuss these considerations in relation to planning sample collection and analysis efforts for a future real-world event.
Sample Collection Considerations
Before an event, planning for the response and remediation efforts for a real-world scenario could entail establishing, which sampling device and surface combinations will provide the best data for the intended purpose. This could be accomplished through research to develop sampling device/surface combinations for different scenarios. For example, research into modeling dispersion of spores into a homogenously mixed room has recommended that 10–25 samples be taken in each room to be sampled from untracked floors, HVAC filters, and walls while targeting specific particulate sizes (3, 5, and 10 μM).25 Sego et al.121 have proposed a sampling model using Bayesian techniques which combines judgement samples taken in high risk areas and random sampling in low-risk areas and provide an estimate of the number of samples needed to provide confidence in decisions regarding clearing a room. Hong et al.62 have also used Bayesian techniques to integrate fate, transport, and risk assessment models. From the results of their study, they have recommended that environmental sampling can be limited to determination of concentration of spores on the floor if the particle size distribution is known. However, if the particle size distribution is not known, then multiple sample locations and an account of air circulation and particle density measurements would also be needed.62 Stuart and Wilkening61 have suggested the need to better understand degradation of agent viability and subsequent modeling of viability in different environments both before an event and after an event. In addition, establishment of a DQO process24 for microbial agents is needed. This process could include a clear definition of the samples needed to support environmental concentration estimates (and subsequent exposure assessment) and steps to meet established performance criteria. Additional validation work is needed to determine collection and recovery efficiency for the common indoor surface/sampling devices,27, 122 including identification of the appropriate type of air samplers that could be used for estimating human exposure. Where possible, keeping sample-collection personnel teams the same throughout the entirety of the response might improve consistency during collection. For example, this might include ensuring that sampling personnel have adequate training, understand the sampling procedures, and can reproduce the sampling method during the sampling phases.
Noting the challenges encountered during sampling for a real-world event might help to account for unusual results. For example, a recent study set out to determine risk-informed environmental concentration standards for mitigating prior exposures (retrospective risk) and future exposures (prospective risk) to B. anthracis in an indoor room.67 The study provided a recommended approach for translating results from environmental sampling for B. anthracis into risk from exposure. However, owing to the many uncertainties in the assumptions used for the calculated values and the variance associated within the model, additional research is needed on reliable sampling techniques to depict a more accurate representation of a real-world scenario.67
Data Management Considerations
Before a real-world event, establishment of methods for management and quality assurance of all data collected is crucial to ensuring reliability and reproducibility of the results. The data management process might include standardized collection and organization of not only field data including descriptive metadata, but also reporting of the laboratory results. Some of the elements of metadata might include the sample number, the sampling device used, the sample location and orientation, the sample size, the type of surface sampled, time the sample was taken, surface properties, and characteristics.36 In addition, metadata such as reporting names of sample personnel who collected the sample and any issues occurring during collection of that sample (such as debris on the surface or irregular surface types) might aid in interpreting irregular results. Handheld devices that are durable, lightweight, and easy to use could be used to enter data as it is collected when possible. Tools such as Scribe,123 Environmental Quality Information System,124 and BROOM26 are examples of tools that have been used to manage environmental data.
Determination of the format of the data (CFU vs cycle threshold, qualitative vs quantitative data etc) reported by the laboratories could be determined before a sampling event. Establishment of guidelines on how to interpret microbial data before an event is also needed to determine how the data can be used. In addition, a way to manage and integrate the results collected in field with the analytical data reported by the laboratories is needed. Ideally, a common database could receive data directly uploaded from the handheld field device. Fields for the analytical results could be pre-built into this database so that laboratory personnel could directly enter the results for each corresponding sample. An easily accessible database could serve as the main source for storage of data related to the event. Ideally, the database would also allow options for printing and sorting of data, and integration of the data with Geographic Information System technology for purposes of mapping the results.
Data Interpretation Considerations
Interpretation of results might require a multi-disciplinary team including coordination of efforts by both sampling teams and laboratory personnel.27 Before an event, guidance is needed on how to handle data below the detection limit. Herzog et al.63 has recommended that both instrument (evaluation using pure culture) and environmental (evaluation using spiked environmental matrices; LOD) are needed to determine exposure using quantitative microbial risk assessment. In addition, the appropriate statistical distribution for count-based data will vary for each set of data, and will need to be determined during analysis of the data. Recently, several options for interpreting culture-based/microbial count data using spread plate counts below the established LOQs were evaluated by the EPA using air and surface sample data collected from a field study.103 Methods that were evaluated included: substituting zero, substituting half the LOQ (15 CFU), treating detects below the LOQ as detected, censoring data at the LOQ (30 CFU), and censoring data at the LOD. Major conclusions of the study were that censoring approaches might be more useful than substitution approaches and that the distribution of the data and statistical methods needed for a particular data set could vary depending on the data that was collected. The study aimed to use ProUCL software to examine how applying different distributions to the same data set affect the final result. Unfortunately, ProUCL software does not include an option for evaluating the Poisson distribution, as recommended by Brattin et al.105 Statistical software that includes the distributions amendable to microbial count-based data, including the Poisson distribution, are needed.
CONCLUSIONS
This paper summarized challenges faced in collecting and interpreting microbial field data from a contaminated site and explores the implications of related data limitations when using field data for estimating potential environmental concentrations. These challenges included (1) collecting environmental field samples that are adequate for the intended purpose; (2) conducting the appropriate type of laboratory analysis and selecting a reporting format for the laboratory data; and (3) analyzing and interpreting the data based on appropriate statistical techniques. Several key findings were noted during this effort. First, the only surface/sampling device combinations that have been validated (by CDC) are swabs and sponge-sticks on stainless steel surfaces. Thus, statistical analyses might be limited due to lack of available quantitative method results. Validation of additional surface/sampling device combinations is needed. Agreement also needs to be reached with the analytical laboratory on the definition of the countable range, as well as how data below the LOQ will be reported. Also, guidance is needed on appropriate statistical software packages for the intended use with microbial count data. In addition, the distribution of the microbial field data and statistical methods needed for a particular data set could vary depending on the data that were collected. Without proper guidance and tools, decision makers might have to use the best available methods for estimating environmental concentrations (and subsequent exposure) rather than the most applicable ones.19 Additional guidance is needed on how exposure estimated from environmental data can be integrated into decisions regarding site characterization and clearance efforts.
Acknowledgments
The U.S. EPA, through its Office of Research and Development, funded and managed this effort under the Battelle/Chemical, Biological, Radiological, and Nuclear Defense Information and Analysis Center Contract No. SP0700-00-D-3180, Delivery Order 0679, Technical Area Task 886. Input was also received from the CDC. The contents do not necessarily reflect the views of the U.S. government. Reference herein to any specific commercial product, service by trade name, trademark, manufacturer, and so on. does not constitute or imply its endorsement, recommendation, or favoring by the U.S. government. The following individuals are recognized for their contributions: Hiba Ernst (EPA), Deborah McKean (EPA), Wendy O'Brien (EPA), Angela Weber (CDC), Stephen Morse (CDC), Laura Rose (CDC), Sean Shadomy (CDC), and Marti Sinclair (Alion Science and Technology).
Footnotes
The authors declare no conflict of interest.
References
- Van Ert MN, Easterday WR, Huynh LY, Okinaka RT, Hugh-Jones ME, Ravel J et al. Global genetic population structure of Bacillus anthracis. PLoS One 2007; 2: e461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minett FC. Sporulation and viability of B. anthracis in relation to environmental temperature and humidity. J Comp Pathol 1950; 60: 161–176. [DOI] [PubMed] [Google Scholar]
- Manchee RJ, Broster MG, Melling J, Henstridge RM, Stagg AJ. Bacillus anthracis on Gruinard Island. Nature 1981; 294: 254–255. [DOI] [PubMed] [Google Scholar]
- Turnbull PC (ed). Anthrax in Humans and Animals. World Health Organization: Geneva. 2008. [PubMed] [Google Scholar]
- Dragon DC, Rennie RP. The ecology of anthrax spores: tough but not invincible. Can Vet J 1995; 36: 295–301. [PMC free article] [PubMed] [Google Scholar]
- EPAPersistence of Categories A and B Select Agents in Environmental Matrices. U.S. Environmental Protection Agency: Cincinnati, OH, USA, EPA/600/R-14/074, 2014. [Google Scholar]
- EPALiterature Review on Mechanism That Affect Persistence of Bacillus Anthracis in Soils. U.S. Environmental Protection Agency: Cincinnati, OH, USA, EPA 600/R-14/216, 2014. [Google Scholar]
- Patel KD, Bhanshali FC, Chaudhary AV, Ingle SS. A new enrichment method for isolation of Bacillus thuringiensis from diverse sample types. Appl Biochem Biotechnol 2013; 170: 58–66. [DOI] [PubMed] [Google Scholar]
- EPAInvestigation Report: Investigation of Simulated Sunlight in The Inactivation of B. Anthracis and B. Subtilis On Outdoor Materials. U.S. Environmental Protection Agency: Washington, D.C., USA, EPA/600/R-10/048, 2010. [Google Scholar]
- EPASelected Analytical Methods for Enviromental Remediation and Recovery. U.S. Environmental Protection Agency: Cincinnati, OH, USA, EPA/600/R-12-555, 2012. [Google Scholar]
- EPARisk Assessment Guidance for Superfund Volume I Human Health Evaluation Manual (Part A) Interim Final. U.S. Environmental Protection Agency, Office of Emergency and Remedial Response: Washington, D.C., USA, EPA/540/1-89/002, 1989. [Google Scholar]
- EPAFramework for Ecological Risk Assessment. U.S. Environmental Protection Agency, Risk Assessment Forum: Washington, D.C., USA, EPA/630/R-92/001, 1992. [Google Scholar]
- NRTObservations and Lessons Learned From Anthrax Responses. U.S. National Response Team. 2004. [Google Scholar]
- EPACDCInterim Clearance Strategy for Environments Contaminated with Bacillus anthracis. U.S. Environmental Protection Agency and Centers for Disease Control and Prevention. 2012. [Google Scholar]
- NRCReopening Public Facilities After A Biological Attack: A Decision Making Framework. Committee on Standards and Policies for Decontaminating Public Facilities Affected by Exposure to Harmful Biological Agents: How Clean is Safe? National Research Council. National Academies Press: Washington, D.C., USA. 2005. [Google Scholar]
- Price PN, Sohn MD, Lacommare KS, McWilliams JA. Framework for evaluating anthrax risk in buildings. Environ Sci Technol 2009; 43: 1783–1787. [DOI] [PubMed] [Google Scholar]
- EPAGuidelines For Exposure Assessment. U.S. Environmental Protection Agency, Risk Assessment Forum: Washington, D.C., USA, EPA/600/Z-92/001, 1992. [Google Scholar]
- EPAMicrobial Risk Assessment Guideline: Pathogenic Microorganisms with Focus on Food and Water., U.S. Environmental Protection Agency and U.S. Department of Agriculture/Food Safety and Inspection Service. Interagency Microbial Risk Assessment Guideline Workgroup, EPA/100/J-12/001, 2012.
- NRCExposure Science in the 21st Century: A Vision and a Strategy. National Research Council. National Academy Press: Washington, D.C., USA. 2012. [PubMed] [Google Scholar]
- NRCScience and Decisions, Advancing Risk Assessment. National Research Council. National Academy Press: Washington, D.C., USA. 2009. [Google Scholar]
- NRCRisk Assessment in the Federal Government: Managing the Process. National Research Council. National Academy Press: Washington, D.C., USA. 1983. [PubMed] [Google Scholar]
- ILSIRevised Framework For Microbial Risk Assessment, An Ilsi Risk Science Institute Workshop Report. International Life Sciences Institute. ILSI Press: Washington, D.C., USA. 2000. [Google Scholar]
- Thompson KM, Armstrong RE, Thompson DF. Bayes, Bugs, and Bioterrorists: Lessons Learned From The Anthrax Attacks, National Defense University Center for Technology and National Security, 2005.
- EPAGuidance on Systematic Planning Using the Data Quality Objectives Process: EPA QA/G-4. U.S. Environmental Protection Agency: Washington, D.C., USA, EPA/240/B-06/001, 2006. [Google Scholar]
- Hong T, Gurian PL. Characterizing bioaerosol risk from environmental sampling. Environ Sci Technol 2012; 46: 6714–6722. [DOI] [PubMed] [Google Scholar]
- EPABio-Response Operational Testing and Evaluation (BOTE) Project Phase 1: Decontamination Assessment. U.S. Environmental Protection Agency: Washington, D.C., USA, EPA/600/R-13/168, 2013. [Google Scholar]
- NRTTechnical Assistance For Anthrax Response: Interim Final Draft, National Response Team, 2005.
- Calfee MW, Rose LJ, Tufts J, Morse S, Clayton M, Touati A et al. Evaluation of sampling methods for Bacillus spore-contaminated HVAC filters. J Microbiol Methods 2014; 96: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cuyk S, Deshpande A, Hollander A, Franco DO, Teclemariam NP, Layshock JA et al. Transport of Bacillus thuringiensis var. kurstaki from an outdoor release into buildings: pathways of infiltration and a rapid method to identify contaminated buildings. Biosecur Bioterror 2012; 10: 215–227. [DOI] [PubMed] [Google Scholar]
- Frawley DA, Samaan MN, Bull RL, Robertson JM, Mateczun AJ, Turnbull PC. Recovery efficiencies of anthrax spores and ricin from nonporous or nonabsorbent and porous or absorbent surfaces by a variety of sampling methods. J Forensic Sci 2008; 53: 1102–1107. [DOI] [PubMed] [Google Scholar]
- Buttner MP, Cruz-Perez P, Stetzenbach LD. Enhanced detection of surface-associated bacteria in indoor environments by quantitative PCR. Appl Environ Microbiol 2001; 67: 2564–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estill CF, Baron PA, Beard JK, Hein MJ, Larsen LD, Rose L et al. Recovery efficiency and limit of detection of aerosolized Bacillus anthracis Sterne from environmental surface samples. Appl Environ Microbiol 2009; 75: 4297–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine NB, Butcher MG, Su YF, Jarman KH, Matzke M, Webb-Robertson BJ et al. Evaluation of sampling tools for environmental sampling of bacterial endospores from porous and nonporous surfaces. J Appl Microbiol 2008; 105: 1107–1113. [DOI] [PubMed] [Google Scholar]
- Edmonds JM. Efficient methods for large-area surface sampling of sites contaminated with pathogenic microorganisms and other hazardous agents: current state, needs, and perspectives. Appl Microbiol Biotechnol 2009; 84: 811–816. [DOI] [PubMed] [Google Scholar]
- Busher A, Nobel-Wang J, Rose L. Surface sampling Emanuel P, Roos JW, Niyogi K Sampling for Biological Agents in the Environment. ASM Press: Washington, DC, USA. 2008, pp 95–131. [Google Scholar]
- Da Silva SM, Urbas AA, Filliben JJ, Morrow JB. Recovery balance: a method for estimating losses in a Bacillus anthracis spore sampling protocol. J Appl Microbiol 2013; 114: 807–818. [DOI] [PubMed] [Google Scholar]
- Probst A, Facius R, Wirth R, Moissl-Eichinger C. Validation of a nylon-flocked-swab protocol for efficient recovery of bacterial spores from smooth and rough surfaces. Appl Environ Microbiol 2010; 76: 5148–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds JM, Collett PJ, Valdes ER, Skowronski EW, Pellar GJ, Emanuel PA. Surface sampling of spores in dry-deposition aerosols. Appl Environ Microbiol 2009; 75: 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauter PA, Piepel GF, Boucher R, Tezak M, Amidan BG, Einfeld W. False-negative rate and recovery efficiency performance of a validated sponge wipe sampling method. Appl Environ Microbiol 2012; 78: 846–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner MP, Cruz P, Stetzenbach LD, Klima-Comba AK, Stevens VL, Emanuel PA. Evaluation of the Biological Sampling Kit (BiSKit) for large-area surface sampling. Appl Environ Microbiol 2004; 70: 7040–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges LR, Rose LJ, O'Connell H, Arduino MJ. National validation study of a swab protocol for the recovery of Bacillus anthracis spores from surfaces. J Microbiol Methods 2010; 81: 141–146. [DOI] [PubMed] [Google Scholar]
- Hong-Geller E, Valdez YE, Shou Y, Yoshida TM, Marrone BL, Dunbar JM. Evaluation of Bacillus anthracis and Yersinia pestis sample collection from nonporous surfaces by quantitative real-time PCR. Lett Appl Microbiol 2010; 50: 431–437. [DOI] [PubMed] [Google Scholar]
- Brown GS, Betty RG, Brockmann JE, Lucero DA, Souza CA, Walsh KS et al. Evaluation of rayon swab surface sample collection method for Bacillus spores from nonporous surfaces. J Appl Microbiol 2007; 103: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Buttner MP, Cruz P, Stetzenbach LD, Klima-Comba AK, Stevens VL, Cronin TD. Determination of the efficacy of two building decontamination strategies by surface sampling with culture and quantitative PCR analysis. Appl Environ Microbiol 2004; 70: 4740–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose LJ, Hodges L, O'Connell H, Noble-Wang J. National validation study of a cellulose sponge wipe-processing method for use after sampling Bacillus anthracis spores from surfaces. Appl Environ Microbiol 2011; 77: 8355–8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GS, Betty RG, Brockmann JE, Lucero DA, Souza CA, Walsh KS et al. Evaluation of vacuum filter sock surface sample collection method for Bacillus spores from porous and non-porous surfaces. J Environ Monit 2007; 9: 666–671. [DOI] [PubMed] [Google Scholar]
- Sanderson WT, Hein MJ, Taylor L, Curwin BD, Kinnes GM, Seitz TA et al. Surface sampling methods for Bacillus anthracis spore contamination. Emerg Infect Dis 2002; 8: 1145–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshale EH, Painter J, Burr GA, Mead P, Wright SV, Cseh LF et al. Environmental sampling for spores of Bacillus anthracis. Emerg Infect Dis 2002; 8: 1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiante DJ, Schill DP, Bresnitz EA, Burr GA, Mead KR. Responding to a bioterrorist attack: environmental investigation of anthrax in New Jersey. Appl Occup Environ Hyg 2003; 18: 780–785. [DOI] [PubMed] [Google Scholar]
- Einfield W, Boucher RM, Tezak MS, Wilson MC, Brown GS. Evaluation of Surface Sampling Method Performance for Bacillus Spores on Clean And Dirty Outdoor Surfaces. Sandia National Laboratories: Albuquerque, NM and Livermore, CA, New Mexico, SAND 2011–4085, 2011. [Google Scholar]
- Da Silva SM, Filliben JJ, Morrow JB. Parameters affecting spore recovery from wipes used in biological surface sampling. Appl Environ Microbiol 2011; 77: 2374–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A, Facius R, Wirth R, Wolf M, Moissl-Eichinger C. Recovery of Bacillus spore contaminants from rough surfaces: a challenge to space mission cleanliness control. Appl Environ Microbiol 2011; 77: 1628–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose L, Jensen B, Peterson A, Banerjee SN, Srduino MJ. Swab materials and Bacillus anthracis spore recovery from nonporous surfaces. Emerg Infect Dis 2004; 10: 1023–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P, Mujawar MM, Upreti R, Sekhar AC. Improved recovery of Bacillus spores from nonporous surfaces with cotton swabs over foam, nylon, or polyester, and the role of hydrophilicity of cotton in governing the recovery efficiency. Appl Environ Microbiol 2013; 79: 381–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budowle B, Schutzer SE, Burans JP, Beecher DJ, Cebula TA, Chakraborty R et al. Quality sample collection, handling, and preservation for an effective microbial forensics program. Appl Environ Microbiol 2006; 72: 6431–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera M, Zandomeni RO, Sagripanti JL. Wet and dry density of Bacillus anthracis and other Bacillus species. J Appl Microbiol 2008; 105: 68–77. [DOI] [PubMed] [Google Scholar]
- Greenberg DL, Busch JD, Keim P, Wagner DM. Identifying experimental surrogates for Bacillus anthracis spores: a review. Investig Genet 2010; 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Driks A, Tawfiq K, Mallozzi M, Patil S. Bacillus anthracis and Bacillus subtilis spore surface properties and transport. Colloids Surf B Biointerfaces 2010; 76: 512–518. [DOI] [PubMed] [Google Scholar]
- Williams G, Linley E, Nicholas R, Baillie L. The role of the exosporium in the environmental distribution of anthrax. J Appl Microbiol 2013; 114: 396–403. [DOI] [PubMed] [Google Scholar]
- Baron PA, Estill CF, Deye GJ, Hein MJ, Beard JK, Larsen LD et al. Development of an aerosol system for uniformly depositing Bacillus anthracis spore particles on surfaces. Aerosol Sci Technol 2008; 43: 159–172. [Google Scholar]
- Stuart AL, Wilkening DA. Degradation of biological weapons agents in the environment: implications for terrorism response. Environ Sci Technol 2005; 39: 2736–2743. [DOI] [PubMed] [Google Scholar]
- Hong T, Gurian PL. Updating a B. anthracis risk model with field data from a bioterrorism incident. Environ Sci Technol 2015; 49: 6701–6711. [DOI] [PubMed] [Google Scholar]
- Herzog AB, McLennan SD, Pandey AK, Gerba CP, Haas CN, Rose JB et al. Implications of limits of detection of various methods for Bacillus anthracis in computing risks to human health. Appl Environ Microbiol 2009; 75: 6331–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges LR, Rose LJ, Peterson A, Noble-Wang J, Arduino MJ. Evaluation of a macrofoam swab protocol for the recovery of Bacillus anthracis spores from a steel surface. Appl Environ Microbiol 2006; 72: 4429–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GS, Betty RG, Brockmann JE, Lucero DA, Souza CA, Walsh KS et al. Evaluation of a wipe surface sample method for collection of Bacillus spores from nonporous surfaces. Appl Environ Microbiol 2007; 73: 706–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amidan BG, Pulsipher BA, Matzke BD. Statistical Analysis of Second Indoor Bio-release Field Evaluation Study at Idaho National Laboratory. Prepared for the U.S. Department of Energy: Richland, WA, USA. 2009. Report no.: PNNL-18932. [Google Scholar]
- Hong T, Gurian PL, Ward NF. Setting risk-informed environmental standards for Bacillus anthracis spores. Risk Anal 2010; 30: 1602–1622. [DOI] [PubMed] [Google Scholar]
- Sextro RG, Lorenzetti DM, Sohn MD, Thatcher TL. Modeling the spread of anthrax in buildings. In: 9th International Proceedings of Indoor Air. Indoor Air, 2002, Santa Cruz, California, USA. 2002.
- Johnson L, Smith ML, Begin M, Fraser B, Miller JD. Remediating office environments of spore-forming bacteria. J Occup Environ Hyg 2010; 7: 585–592. [DOI] [PubMed] [Google Scholar]
- Lewandowski R, Kozlowska K, Szpakowska M, Stepinska M, Trafny EA. Use of a foam spatula for sampling surfaces after bioaerosol deposition. Appl Environ Microbiol 2010; 76: 688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAOAnthrax DHS Faces Challenges in Validating Methods for Sample Collection and Analysis. U.S. Government Accountability Office: Washington, D.C., USA, GAO-12-488, 2012. [Google Scholar]
- Lee SD, Ryan SP, Snyder EG. Development of an aerosol surface inoculation method for Bacillus spores. Appl Environ Microbiol 2011; 77: 1638–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecher DJ. Forensic application of microbiological culture analysis to identify mail intentionally contaminated with Bacillus anthracis spores. Appl Environ Microbiol 2006; 72: 5304–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPAResuspension and Tracking of Particulate Matter From Carpet Due to Human Activity. U.S. Environmental Protection Agency: Washington, D.C., USA. 2007. [Google Scholar]
- Sippola MR, Sextro RG, Thatcher TL. Measurements and modeling of deposited particle transport by foot traffic indoors. Environ Sci Technol 2014; 48: 3800–3807. [DOI] [PubMed] [Google Scholar]
- Weis CP, Intrepido AJ, Miller AK, Cowin PG, Durno MA, Gebhardt JS et al. Secondary aerosolization of viable Bacillus anthracis spores in a contaminated US Senate Office. J Am Med Assoc 2002; 288: 2853–2858. [DOI] [PubMed] [Google Scholar]
- Guh A, Heyman ML, Barden D, Fontana J, Hadler JL. Lessons learned from the investigation of a cluster of cutaneous anthrax cases in Connecticut. JPHMP 2010; 16: 201–210. [DOI] [PubMed] [Google Scholar]
- Piepel GF, Amidan BG, Hu R. Laboratory studies on surface sampling of Bacillus anthracis contamination: summary, gaps and recommendations. J Appl Microbiol 2012; 113: 1287–1304. [DOI] [PubMed] [Google Scholar]
- CDCSurface Sampling Procedures For Bacillus Anthracis Spores From Smooth, Non-porous Surfaces. Centers for Disease Control and Prevention: Atlanta, GA, USA. 2012. [Google Scholar]
- Hubbard K, Pellar G, Emanuel P. Suitability of commercial transport media for biological pathogens under nonideal conditions. Int J Microbiol 2011; 2011: 463096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Mohan KV, Atreya CD. Detection technologies for Bacillus anthracis: prospects and challenges. J Microbiol Methods 2010; 82: 1–10. [DOI] [PubMed] [Google Scholar]
- Edwards KA, Clancy HA, Baeumner AJ. Bacillus anthracis: toxicology, epidemiology and current rapid-detection methods. Anal Bioanal Chem 2006; 384: 73–84. [DOI] [PubMed] [Google Scholar]
- Irenge LM, Gala JL. Rapid detection methods for Bacillus anthracis in environmental samples: a review. Appl Microbiol Biotechnol 2012; 93: 1411–1422. [DOI] [PubMed] [Google Scholar]
- Silvestri EE, Perkins SD, Feldhake D, Nichols T, Schaefer FW. Recent literature review of soil processing methods for recovery of Bacillus anthracis spores. Ann Microbiol 2014; 65: 1215–1226. [Google Scholar]
- Turnbull PC. Definitive identification of Bacillus anthracis—A review. J Appl Microbiol 1999; 87: 237–240. [DOI] [PubMed] [Google Scholar]
- Titball RW, Turnbull PC, Hutson RA. The monitoring and detection of Bacillus anthracis in the environment. Soc Appl Bacteriol Symp Ser 1991; 20: 9S–18S. [PubMed] [Google Scholar]
- Rastogi VK, Wallace L, Smith LS, Pfarr J. Surface Sampling-based Decontamination Studies And Protocol For Determining Sporicidal Efficacy Of Gaseous Fumigants On Military-relevant Surfaces. Edgewood Chemical Biological Center, U.S. Army Research, Development and Engineering Command, Aberdeen Proving Ground: MD, 2008..
- Almeida JL, Harper B, Cole KD. Bacillus anthracis spore suspensions: determination of stability and comparison of enumeration techniques. J Appl Microbiol 2008; 104: 1442–1448. [DOI] [PubMed] [Google Scholar]
- Perry KA, O'Connell HA, Rose LJ, Noble-Wang JA, Arduino MJ. Storage effects on sample integrity of enviromental surface sampling speciments with Bacillus anthracis spores. Biosafety 2013; S1: 002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton S. Accuracy of plate counts. J Valid Technol 2011; 17: 42–46. [Google Scholar]
- FDA. Aerobic plate count Maturin L, Peeler JT. Bacteriological Analytical Manual Chap 3 Food and Drug Administration, 2001. Available at: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm063346.htm. Accessed 23 September 2015. [Google Scholar]
- USP <55> Biological indicators- Resistance performance test. USP 34, United States Pharmacopeia, 2011, pp 50–52.
- SMC9215 Heterotrophic Plate Count. Standard Methods for the Examination of Water and Wastewater, Standard Methods Committee, 2011..
- USP <1227> Validation of microbial recovery from pharmacopeial articles. USP 34, United States Pharmacopeia, 2011, pp 783–786.
- Breed RS, Dotterrer WD. The number of colonies allowable on satisfactory agar plates. J Bacteriol 1916; 1: 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark HF, Geldreich EE, Jeter HL, Kabler PW. The membrane filter in sanitary bacteriology. Public Health Rep 1951; 66: 951–977. [PMC free article] [PubMed] [Google Scholar]
- ASTMStandard Practice for Determining Microbial Colony Counts from Waters Analyzed by Plating Methods. D5464–93, 2004.
- Sutton S. Counting colonies. Pharm Forum Newsl 2006; 12: 2–11. [Google Scholar]
- Dufour AP, Strickland ER, Cabelli VJ. Membrane filter method for enumerating Escherichia coli. Appl Environ Microbiol 1981; 41: 1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronewold AD, Wolpert RL. Modeling the relationship between most probable number (MPN) and colony-forming unit (CFU) estimates of fecal coliform concentration. Water Res 2008; 42: 3327–3334. [DOI] [PubMed] [Google Scholar]
- Clough HE, Clancy D, O'Neill PD, Robinson SE, French NP. Quantifying uncertainty associated with microbial count data: a Bayesian approach. Biometrics 2005; 61: 610–616. [DOI] [PubMed] [Google Scholar]
- EPAMethod 1603: Escherichia coli (E. coli) in Water by Membrane Filtration Using Modified Membrane-theromtolerant Escherichia coli Agar (Modified mTEC). U.S. Environmental Protection Agency: Washington, D.C., USA. 2009. [Google Scholar]
- EPAEvaluation of Options for Interpreting Environmental Microbiology Field Data Results having Low Spore Counts. U.S. Environmental Protection Agency: Washington, D.C., USA, EPA/600/R-14/331, 2014. [Google Scholar]
- EPAProUCL Version 4.1 User Guide (Draft), Statistical Software for Environmental Applications for Data Sets with and without Nondetect Observations. U.S. Environmental Protection Agency, Office of Research and Development: Washington, D.C., USA, EPA/600/R-07/041, 2010. [Google Scholar]
- Brattin W, Barry T, Foster S. Estimation of the upper confidence limit on the mean of datasets with count-based concentration values. Hum Ecol Risk Assess 2012; 18: 435–455. [Google Scholar]
- Matzke BD, Wilson JE, Newburn LL, Dowson ST, Hathaway JE, Sego LH et al. Visual Sample Plan Version 7.0 User's Guide. Pacific Northwest National Laboratory: Richland, Washington, USA. 2014. [Google Scholar]
- LLNL. An action plan to reopen a contaminated airport. Lawrence Livermore National Laboratories, Sci Technol Rev, 2006. Available at https://str.llnl.gov/str/Dec06/pdfs/12_06.2.pdf. Accessed 23 September 2015.
- Wong M, Kumar L, Jenkins TM, Xagoraraki I, Phanikumar MS, Rose JB. Evaluation of public health risks at recreational beaches in Lake Michigan via detection of enteric viruses and a human-specific bacteriological marker. Water Res 2009; 43: 1137–1149. [DOI] [PubMed] [Google Scholar]
- Pessi AM, Suonketo J, Pentti M, Kurkilahti M, Peltola K, Rantio-Lehtimaki A. Microbial growth inside insulated external walls as an indoor air biocontamination source. Appl Environ Microbiol 2002; 68: 963–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payment P, Locas A. Pathogens in water: value and limits of correlation with microbial indicators. Ground Water 2011; 49: 4–11. [DOI] [PubMed] [Google Scholar]
- Copeland WK. mcaGUI: microbial community analysis R-Graphical User Interface (GUI). Bioinformatics 2012; 28: 2198–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petterson SR, Teunis PF, Ashbolt NJ. Modeling virus inactivation on salad crops using microbial count data. Risk Anal 2001; 21: 1097–1108. [DOI] [PubMed] [Google Scholar]
- Levine AD, Harwood VJ, Fox GA. Collecting, Exploring, and Interpreting Microbiological Data Associated with Reclaimed Water Systems. Water Reuse Foundation: Alexandria, VA, USA. 2009. [Google Scholar]
- Hoffman D. Negative binomial control limits for count data with extra-Poisson variation. Pharm Stat 2003; 2: 127–132. [Google Scholar]
- EPAData Quality Assessment: Statistical Methods for Practitioners, U.S. Environmental Protection Agency, EPA QA/G-9S, 2006.
- Helsel DR. More than obvious: better methods for interpreting nondetect data. Environ Sci Tecnol 2005; 39: 419A–423A. [DOI] [PubMed] [Google Scholar]
- Gleit A. Estimation for small normal data sets with detection limits. Environ Sci Tecnol 1985; 19: 1201–1206. [DOI] [PubMed] [Google Scholar]
- El-Shaarawi AH. Inferences about the mean from censored water quality data. Water Res 1989; 24: 685–690. [Google Scholar]
- Smeets PW, van Dijk JC, Stanfield G, Rietveld LC, Medema GJ. How can the UK statutory Cryptosporidium monitoring be used for Quantitative Risk Assessment of Cryptosporidium in drinking water? J Water Health 2007; 5 (Suppl 1): 107–118. [DOI] [PubMed] [Google Scholar]
- Helsel DR. Statistics for Censored Environmental Data Using Minitab(R) and R. 2nd edn. John Wiley & Sons, Inc.: New Jersey, USA. 2012. [Google Scholar]
- Sego LH, Anderson KK, Matzke BD, Sieber K, Shulman S, Bennett J et alAn environmental sampling model for combining judgement and randomly placed samples. Pacific Northwest National Laboratory for the U.S. Department of Energy, PNNL-16636, 2007..
- GAOAnthrax Detection: Agencies Need To Validate Sampling Activities In Order To Increase Confidence In Negative Results. U.S. Government Accountability Office: Washington, D.C., USA, GAO-05-251, 2005. [Google Scholar]
- EPAScribe, U.S. Environmental Protection Agency, Environmental Response Team, 2015. Available at: http://www.epaosc.org/site/site_profile.aspx?site_id=ScribeGIS. Accessed 22 September 2015.
- EPAEnviromental Data Submission, U.S. Environmental Protection Agency, Region 4. SESDGUID-106-R0, 2010.