Abstract
Tumour suppressor p53 or proto-oncogene MYC is frequently altered in squamous carcinomas, but this is insufficient to drive carcinogenesis. We have shown that overactivation of MYC or loss of p53 via DNA damage triggers an anti-oncogenic differentiation-mitosis checkpoint in human epidermal keratinocytes, resulting in impaired cell division and squamous differentiation. Forkhead box M1 (FOXM1) is a transcription factor recently proposed to govern the expression of a set of mitotic genes. Deregulation of FOXM1 occurs in a wide variety of epithelial malignancies. We have ectopically expressed FOXM1 in keratinocytes of the skin after overexpression of MYC or inactivation of endogenous p53. Ectopic FOXM1 rescues the proliferative capacity of MYC- or p53-mutant cells in spite of higher genetic damage and a larger cell size typical of differentiation. As a consequence, differentiation induced by loss of p53 or MYC is converted into increased proliferation and keratinocytes displaying genomic instability are maintained within the proliferative compartment. The results demonstrate that keratinocyte oncogene-induced differentiation is caused by mitosis control and provide new insight into the mechanisms driving malignant progression in squamous cancer.
Introduction
Although squamous cell carcinomas (SCCs) in different locations such as skin, head and neck or oesophagus are heterogeneous in clinic and prognosis, they share a similar histology with cell morphology reminiscent of the differentiated layers of the epidermis. For this reason they are also referred to as epidermoid carcinomas. In addition, they share similar risk factors that cause genetic damage, including ultraviolet light, human papillomavirus, tobacco and alcohol. Therefore, they might share common or overlapping molecular mechanisms. SCCs are often aggressive and have poor prognosis. Finding common pathways to SCCs would provide a new basis for their diagnosis and treatment.
Human epidermis is a paradigm of self-renewal stratified squamous epithelium highly exposed to mutagenic hazard and frequently affected by cancer. The tumour suppressor protein p53, also known as the guardian of the genome, is mutated in most human skin SCCs (80%),1, 2 although its alteration is not sufficient for the development of epithelial skin cancer.3, 4 Within the same lines, it is well established that proto-oncogene MYC in keratinocytes promotes differentiation instead of proliferation.5, 6, 7, 8 Similarly, overactivation of a variety of cell growth promoters including the DNA replication protein Cyclin E is not tumourigenic when overexpressed in epidermal cells9, 10, 11, 12 (reviewed in Gandarillas13). The cell cycle regulation explaining this resistance of keratinocytes to transformation upon cell cycle deregulation remains intriguing but is critical to understand squamous carcinogenesis. Recently, we have reported that loss of p53 causes squamous differentiation in epidermal human keratinocytes.14 This might explain why inactivation of p53 does not drive skin carcinogenesis by itself and, notably, why sun-exposed healthy skin often contains patches of cells with the mutated protein that cause no clinical impact.15, 16, 17 This finding points at a self-protective response of the epidermis against oncogenic transformation. We have shown that epidermal keratinocytes respond to a differentiation-mitosis checkpoint (DMC) that triggers squamous differentiation in the event of cell cycle deregulation.13, 18 The DMC functions as an oncogene-induced differentiation response (OID).13 Upon hyperactivation of the cell cycle, keratinocytes block cell division and trigger terminal differentiation, although they fail to maintain G2/M arrest (mitotic slippage) and continue DNA replication (endoreplication), become polyploid and significantly increase their size. Differentiating keratinocytes migrate towards the surface of the epidermis and are finally eliminated from the skin by shedding. We have proposed that because of the DMC, precancerous alterations need additional modifications in the mitosis control for epidermal carcinogenesis to occur.14 We now have challenged this model by overexpressing forkhead box M1 (FOXM1) in human keratinocytes after overactivation of conditional MYC or inactivation of endogenous p53.
The FOXM1 transcription factor is a mammalian regulator of cell cycle progression and frequently upregulated in human cancer.19 Although FOXM1 can induce cell cycle progression into the DNA replication S phase (G1/S), it plays a major role in the G2/M transition by the transactivation of regulators of mitosis and cytokinesis such as Cyclin B, Aurora B, Polo-like kinase and CENP.20 FOXM1 is frequently deregulated in SCCs of head and neck and the skin.21, 22
The results herein show that FOXM1, in combination with precancerous cell growth deregulation, allows human keratinocytes to proliferate in spite of accumulating DNA damage and therefore promoting genomic instability. This may explain why mutated p53 and deregulated FOXM1 are both frequently selected in cancer.
Results
FOXM1 rescues the proliferative block caused by inactivation of p53
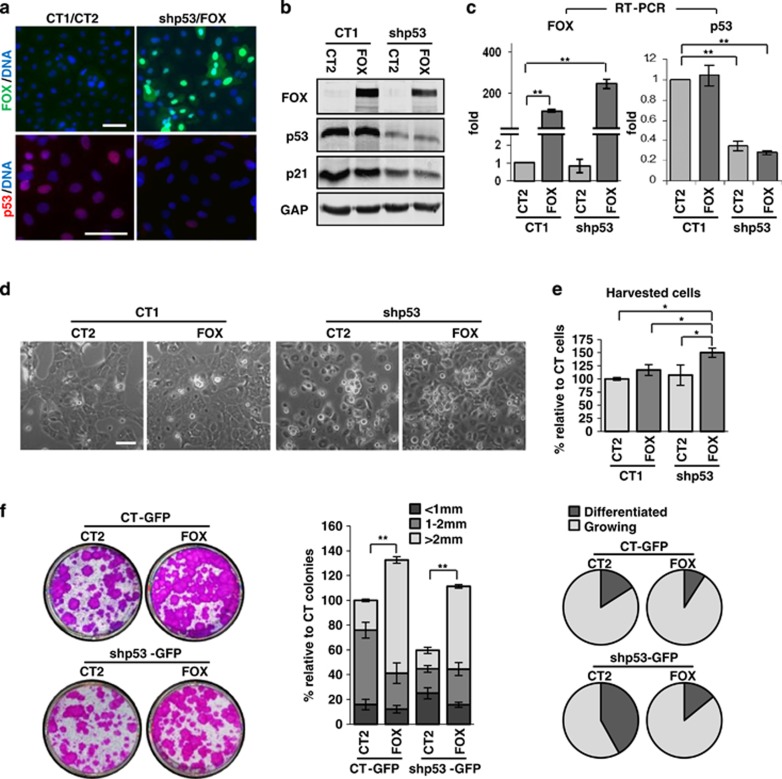
We aimed to investigate whether FOXM1 affects the loss of proliferation potential observed in primary human keratinocytes when the expression of p53 is inhibited. To this end, we silenced p53 by means of a specific lentiviral construct carrying a short hairpin RNA (shRNA; shp53)14, 23 in human keratinocytes (Kshp53) and then overexpressed FOXM1 (Kshp53/FOXM1) by a lentiviral vector. Figures 1a–c and Supplementary Figure 1a show the downregulation of p53 or the overexpression of FOXM1 as determined by immunofluorescence, western blot or real-time PCR. shp53 was delivered into 90–95% of cells (Supplementary Figure 1a).14 Downregulation of p53 not only reduced the amount of protein but also its ability to induce its target gene p21Cip (Figure 1b). As described previously,14 although cell number did not decrease, by 5 days Kshp53 cells halted proliferation as compared with control cells carrying the empty vector (Figure 1d). However, Kshp53/FOXM1 cells continued to proliferate even further than control cells or cells overexpressing FOXM1 only. This observation was confirmed by the striking increase in the cell count of the Kshp53/FOXM1 cultures (Figure 1e). The effect on cell proliferation was confirmed by the inhibition of p53 with a second shRNA (shp53-2, data not shown).
Figure 1.
Ectopic expression of FOXM1 rescues the proliferative capacity of human epidermal keratinocytes lacking p53. (a–c) Detection of FOXM1 and p53 in primary keratinocytes infected with shRNA for p53 (shp53-1) or the corresponding empty control vector (plK01, CT1) and FOXM1 (FOX) or the corresponding empty control vector (plVX, CT2) by (a) immunofluorescence (also in Supplementary Figure 1a, DAPI for DNA in blue), (b) western blot and (c) real-time (RT)–PCR (fold differences with respect to control CT1/CT2). Detection of the p53 target p21Cip (p21) is also shown by western blot (b). GAPDH (GAP) was used as loading control. (d) Phase contrast images of keratinocytes 5 days after construct delivery, as indicated. (e) Bar histogram represents the differences in the number of cells collected relative to control (CT1/CT2=100%) 5 days after infections with the indicated constructs. Similar results were obtained when a different shRNA for p53 (shp53-2) was used. (f) Clonogenic capacity of cells plated 5 days after infections with CT-GFP or shp53-GFP and CT2 or FOX (2500 cells per well). Representative wells of triplicate samples are shown (left). Bar histogram represents the differences in large colonies (>2 mm, light grey), medium size colonies (1–2 mm, intermediate grey) or small and differentiated colonies (<1 mm, dark grey) relative to control cells (CT-GFP/CT2=100%). Circle histograms show the proportion of actively growing (light grey) or differentiated (dark grey) colonies. Scale bars: 50 μm; *P<0.05 and **P<0.01. Data are mean±s.e.m. of triplicate samples of representative experiments (n=3). Results are representative of two different strains of keratinocytes from different individuals (N=2).
To investigate whether FOXM1 affected the clonogenic capacity of Kshp53, we performed clonogenicity assays 5 days after introducing shp53-GFP and/or FOXM1 (Figure 1f). These assays allow to detect the effect on cell fate by estimating both the capacity to form colonies and the degree of cell expansion of the stem (large colonies, >2 mm) or the committed progenitor amplifying cells (small colonies, <1 mm).24 Overexpression of FOXM1 in normal keratinocytes increased their clonogenic potential (Figure 1f), consistently with previous observations in cells from oral epithelium.25 However, in the Kshp53 cultures the number of total and proliferative colonies was reduced compared with controls, whereas the number of abortive differentiated colonies was increased. In contrast, Kshp53/FOXM1 cells recovered and improved the potential of control cells. The size and the number of large colonies generated by putative stem cells were greatly increased in the Kshp53/FOXM1 cultures. Interestingly, the size of small colonies, formed by transit amplifying progenitors, was also increased. Thus, ectopic FOXM1 rescues the proliferative and clonogenic capacity of human epidermal keratinocytes that is lost by inactivation of p53. Although Kshp53/FOXM1 cells proliferated more than FOXM1 cells 5 days after infection (Figure 1e), their clonogenicity potential was weaker as the loss of p53 had already driven cell differentiation (Figure 1f).
FOXM1 attenuates the squamous differentiation response caused by loss of p53
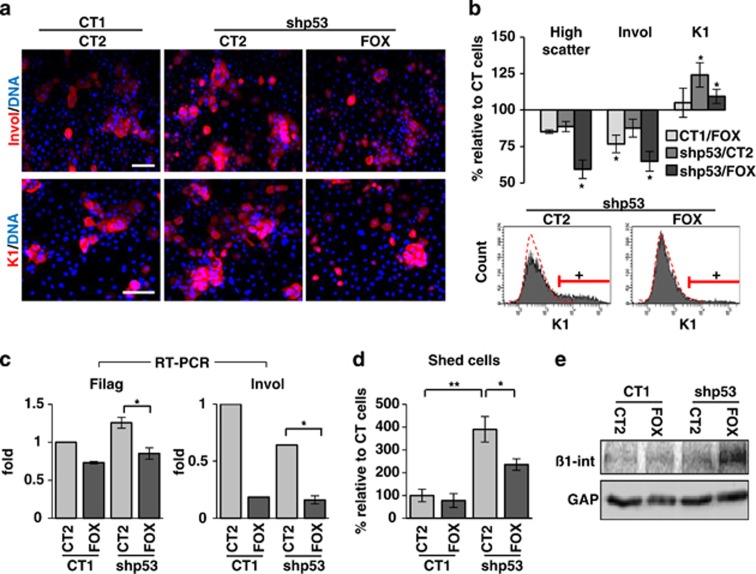
As the ectopic expression of FOXM1 improves the proliferative capacity of Kshp53, we aimed to investigate whether FOXM1 affects their differentiation response. At 3 days after gene delivery the proportion of Kshp53/FOXM1 cells expressing the epidermal differentiation markers involucrin, keratin K1 and filaggrin significantly decreased (Figures 2a–c and Supplementary Figures 1b and c). The inhibition of differentiation by FOXM1 was also observed when the shp53-2 was used (data not shown). Keratinocytes enlarge and become more complex as they differentiate and this can be monitored by flow cytometry by means of increased light scattering.24, 26 Accordingly with the decrease in the expression of differentiation markers, we found that the percent of cells with high scatter parameters typical of differentiation was also decreased in Kshp53/FOXM1 with respect to Kshp53 (Figure 2b). It is important to mention that differentiation of human epidermal keratinocytes increases at cell confluence,27 when keratinocytes are pushed to stratify and detach. Kshp53/FOXM1 cells reached confluence faster and stratified more than controls and certainly than Kshp53 cells (Figure 1d and Supplementary Videos 1 and 2). In addition, at confluence and in spite of a higher cell density, FOXM1 inhibited the initiation of differentiation as the proportion of cell shedding into the medium was attenuated (Figure 2d). Increased stratification and decreased shedding in Kshp53/FOXM1 populations might be related to increased expression of β1 integrin (Figure 2e). The β1 integrins are adhesion molecules that maintain keratinocytes attached to the basement membrane and thus their proliferative potential.28 They are lost at differentiation. Therefore, although FOXM1 did not abolish stratification and differentiation of Kshp53 cells, it significantly rescued the proliferative potential lost by the absence of p53.
Figure 2.
Ectopic expression of FOXM1 inhibits the differentiation response to loss of p53. (a) Immunodetection of the epidermal differentiation markers in red, involucrin (Invol) or keratin K1 (K1) in keratinocytes 3 days after infection with CT1 or shp53-1 and CT2 or FOX (as in Figure 1). DAPI for DNA in blue. (b) Bar histogram shows cells with high size and complexity (light scatter; differentiation), or positive for Invol or K1 relative to control cells (CT1/CT2=100%) as measured by flow cytomety. Bottom histograms show representative plots for the expression of K1 (+, positive cells according to negative isotype antibody control, red broken line). More details in Supplementary Figures 1b and c. (c) Real-time (RT)–PCR for expression of the differentiation markers Filaggrin (Filag) and Invol 2 days after infections as indicated (fold differences relative to control CT1/CT2). (d) Shedding cells collected from the culture supernatant due to differentiation 5 days after infections relative to control (CT1/CT2=100%). (e) Detection by western blot of β1 integrin 5 days after infections as indicated. GAPDH (GAP) was used as loading control. Scale bar: 50 μm; *P<0.05 and **P<0.01. Data are mean±s.e.m. of triplicate samples of representative experiments (n=3). Results are representative of two different strains from different individuals (N=2).
FOXM1 allows Kshp53 to proliferate in spite of genome instability
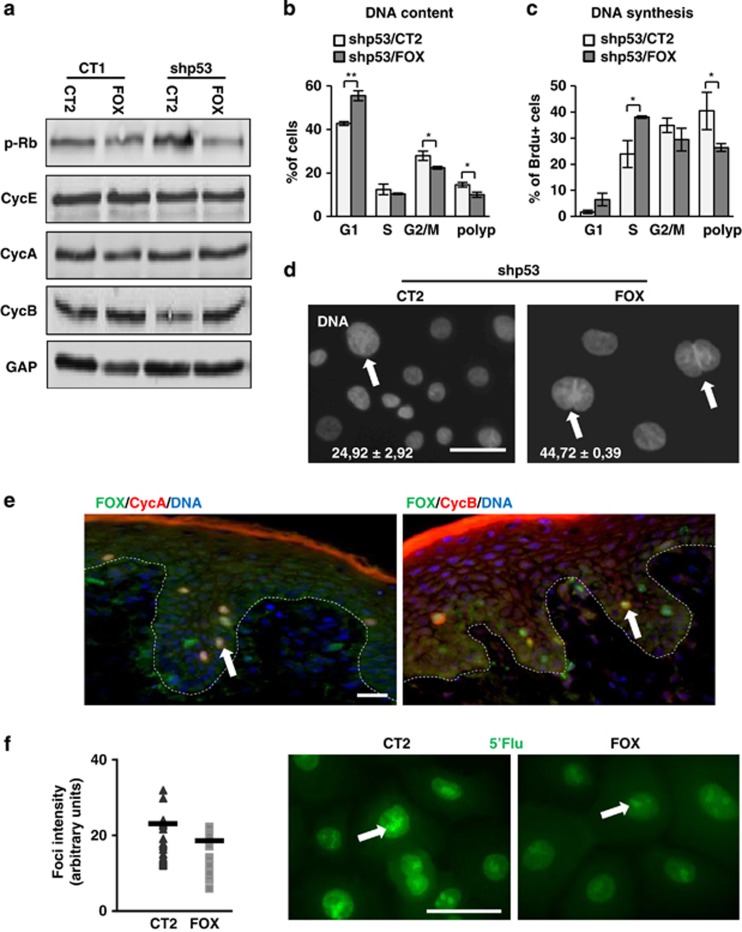
Loss of p53 induces early hyperactivation of the cell cycle that results in a G2/M block and endoreplication in human epidermal keratinocytes, subsequently increasing the population of differentiated polyploid cells.14 The analysis of cell cycle regulators suggested that Kshp53/FOXM1 cells did not accumulate as much in G2/M. Although the expression of the S/G2/M marker phospho-retinoblastoma (p-Rb) was increased in Kshp53 cells compared with controls, it was decreased upon FOXM1 overexpression (Figure 3a). Rb is hyperphosphorylated at entry in S phase and hypophosphorylated after cell division.29 FOXM1 induces Cyclin B among other mitosis/cytokinesis regulators20 to drive cell division. The expression of Cyclin B was reduced in Kshp53 cells and increased in Kshp53/FOXM1 (Figure 3a).14 However, the expression of Cyclin E and Cyclin A remained unchanged, suggesting that the main action of FOXM1 in keratinocytes takes place in mitosis. We also observed changes in the cell cycle 3 days after infection. Kshp53/FOXM1 cells showed a decrease in the percent of cells accumulated in G2/M and in the polyploid region (Figure 3b and Supplementary Figures 2a and b). Accordingly, the proportion of cells returning to G1 increased. DNA synthesis was also measured in these cells by 5-bromodeoxyuridine (BrdU) incorporation (Figure 3c and Supplementary Figure 2c). Similar to the changes observed in the cell cycle, the percent of Kshp53/FOXM1 cells bypassing mitosis into polyploidy was decreased and the percent of BrdU cells in G1/S phase increased as compared with Kshp53 cells. However, cultures carrying only the FOXM1 plasmid displayed an increase in the percent of BrdU-positive cells in G2/M (Supplementary Figure 2c). This suggests that FOXM1 and loss of p53 together accelerated the transition from G2/M to G1. Altogether, the results are consistent with a role of FOXM1 in potentiating keratinocyte cell division downstream of p53. Interestingly, although FOXM1 caused a decrease of polyploidy in Kshp53, yet the number of binucleated cells within the reduced polyploid population was increased (Figure 3d). This suggests that FOXM1 pushes nuclei to divide even when the cell cannot divide anymore, consistent with the role of FOXM1 in mitotic spindle assembly, chromosome alignment and nuclear division.20, 30 The expression of FOXM1 in normal human skin is unknown and we aimed to identify it. As shown in Figure 3e, FOXM1 strikingly accumulated in cells expressing mitotic Cyclins A and B, suggesting that the main function of FOXM1 in keratinocytes in the epidermis is related to mitosis.
Figure 3.
Ectopic expression of FOXM1 suppresses the mitotic block caused by loss of p53 in human epidermal keratinocytes. (a–d) Analyses 3 days after infections with CT1 or shp53-1 and CT2 or FOX (as in Figure 1). (a) Expression of cell cycle regulators p-Rb, Cyclin E (CycE), Cyclin A (CycA) and Cyclin B (CycB) by western blotting. GAPDH (GAP) was used as loading control. (b) Percent of cells in the G1 (2N), S, G2/M (4N) or polyploid (>4N) phases of the cell cycle as determined by flow cytometry. More details in Supplementary Figures 2a and b. (c) Percent of BrdU-positive cells in the G1, S (2N), G2/M (4N) and polyploid (>4N) phases as in (b). More details in Supplementary Figure 2c. (d) Binucleated keratinocytes (white arrows) bearing shp53 and CT2 or FOX as indicated, stained with DAPI for DNA. White numbers are the percent of polyploid cells (nuclei >15 μm) that were binucleated. (e) Immunodetection of endogenous FOX (green), CycA (red, left panel) and CycB (red, right panel) in normal human epidermis by immunofluorescence. DAPI for DNA in blue. Red line is nonspecific staining of the superficial cornified layer. Arrows for cells coexpressing FOX and CycA or CycB. Broken line for the basement membrane. (f) Detection of nascent transcription by 5′-fluorouridine 5′-Flu; green) incorporation 2 days after infections as indicated (nucleolar foci are indicated on representative photographs by white arrows). Dot plot on the left side represents quantitation of the fluorescence intensity of 5′-Flu incorporating single foci. Black bars represent the mean. Scale bars: 50 μm; *P<0.05 and **P<0.01. Data are mean±s.e.m. of triplicate samples of representative experiment (n=3).
It is worth noting that ectopic expression of FOXM1 reduced the intensity of nascent RNA foci as measured by incorporation of 5′-fluorouridine (Figure 3f). This suggests that FOXM1 function may have a relationship with the inhibition of transcription required for the initiation of mitosis. Therefore, we performed double labelling for Cyclin A, which accumulates mainly in G2/M, and transcription in these cells. The analyses showed that, as expected, in controls transcription was strong in Cyclin A-negative cells and weak in mitotic cells. However, transcription was as weak in interphasic, nonmitotic FOXM1-overexpressing cells (Supplementary Figure 3a). The role of FOXM1 in the inhibition of transcription has not been studied. FOXM1 gene encodes three different protein isoforms obtained by alternative splicing: FOXM1a, FOXM1b and FOXM1c.31 Although we have made used of a DNA construct bearing the FOXM1b isoform, involved in oncogenesis,32, 33 it has been shown that this protein can induce the expression of the other two variants34 (Supplementary Figure 3b). Whereas FOXM1b and FOXM1c are considered transcriptionally active, FOXM1a lacks the transactivation domain and has a negative regulatory function on transcription.31 The expression of the three isoforms might be important for the correct regulation of mitosis. Consistent with a global transcription decrease, we found that keratinocytes overexpressing FOXM1 displayed a lighter optical density (Supplementary Figure 3c). In addition, we observed FOXM1 in the nucleolus, centre of ribosomal RNA transcription (Supplementary Figure 3d). Nucleolar localisation has been previously proposed to have a FOXM1 inhibitory role.35
Kshp53/FOXM1 proliferative keratinocytes increased in size (measured by their forward scatter parameter), in spite of reduced cellular complexity (side scatter parameter; Figure 4a and Supplementary Figure 3e). Cells enlarge in G2 in order to produce two daughters of the same size before cell division, and therefore the mitosis block provokes differentiating keratinocytes to increase in size and this process is stimulated by shp53.14 The higher size of proliferative Kshp53/FOXM1 cells suggests that this protein pushes cell division even though the mitosis block has initiated.
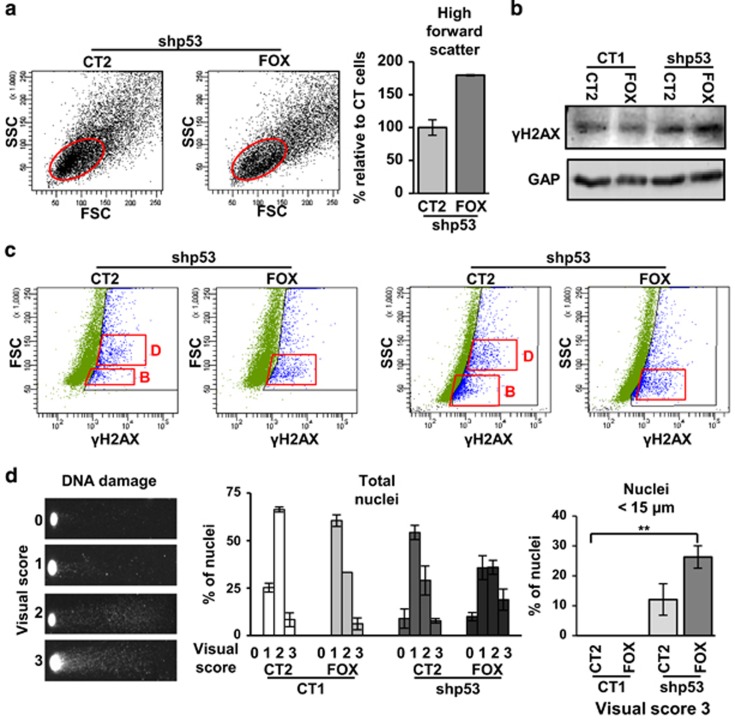
Figure 4.
FOXM1 allows accumulation of DNA damage in proliferative keratinocytes upon p53 inactivation. (a) Dot plots show side and forward scatter parameters for cell complexity and size (side scatter parameter (SSC) and forward scatter parameter (FSC)) by flow cytometry of keratinocytes infected with shp53-1 and CT2 or FOX. Red oval represents the basal proliferative population. Histograms showing the SSC and FSC of proliferative cells (red oval) are shown in Supplementary Figure 3e. Bar histogram indicates basal cells (red oval) with high FSC (according to gate in Supplementary Figure 3e) relative to cells infected with shp53 and CT2 (shp53/CT2=100%). (b) Western blot for γH2AX in keratinocytes 5 days after infections as indicated. GAPDH (GAP) was used as loading control. (c) Dot plots for FSC or SSC (y axis) and γH2AX (x axis) 3 days after infections as indicated. Red squares represent the main γH2AX-positive populations (blue) of differentiating (D) or basal (B) cells. Similar results were obtained when shp53-2 was used. More details in Supplementary Figure 4. (d) DNA damage monitored by comet assays (see Materials and methods). (d, left) Scoring assigned for damage according to the size and intensity of the nuclear tails of keratinocytes (0–3) 5 days after infections. (d, centre) Bar histogram showing the quantitation of DNA damage according to the 4 levels of intensity. (d, right) Bar histogram represents only the percent of small nuclei (<15 μm) with maximum DNA damage intensity level 3. Constructs as indicated. **P<0.01. Data are mean±s.e.m. of triplicate samples of representative experiment (n=3).
The deregulation of the cell cycle in Kshp53 leads to the accumulation of DNA damage because of replication stress. We studied whether FOXM1, by alleviating the mitosis block, allowed keratinocytes to divide in spite of irreparable damage. Expression of the canonical DNA damage marker γH2AX36, 37 was evaluated in Kshp53/FOXM1 cells 5 days after gene delivery and we found a significant increase with respect to control cells (Figure 4b). In Kshp53 cells the increase in γH2AX was detected both in basal and differentiating keratinocytes according to light scatter parameters (Figure 4c and Supplementary Figure 4). However, in Kshp53/FOXM1 cells the accumulation of the γH2AX signal was detected mainly in the basal low scatter population (Figure 4c and Supplementary Figure 4). This result was confirmed by using shp53-2 (data not shown). To assess for actual DNA breaks typical of replication stress37 we performed comet assays. As shown in Figure 4d, inactivation of p53 caused a striking increase of DNA breaks that augmented even further in combination with FOXM1. Interestingly, Kshp53/FOXM1 cells augmented significantly the DNA breaks in smaller nuclei (<15 μm), typical of proliferative cells. These results suggest that ectopic expression of FOXM1 allowed damaged Kshp53 cells to keep dividing and amplifying.
Overexpression of FOXM1 drives MYC-induced differentiation into proliferation
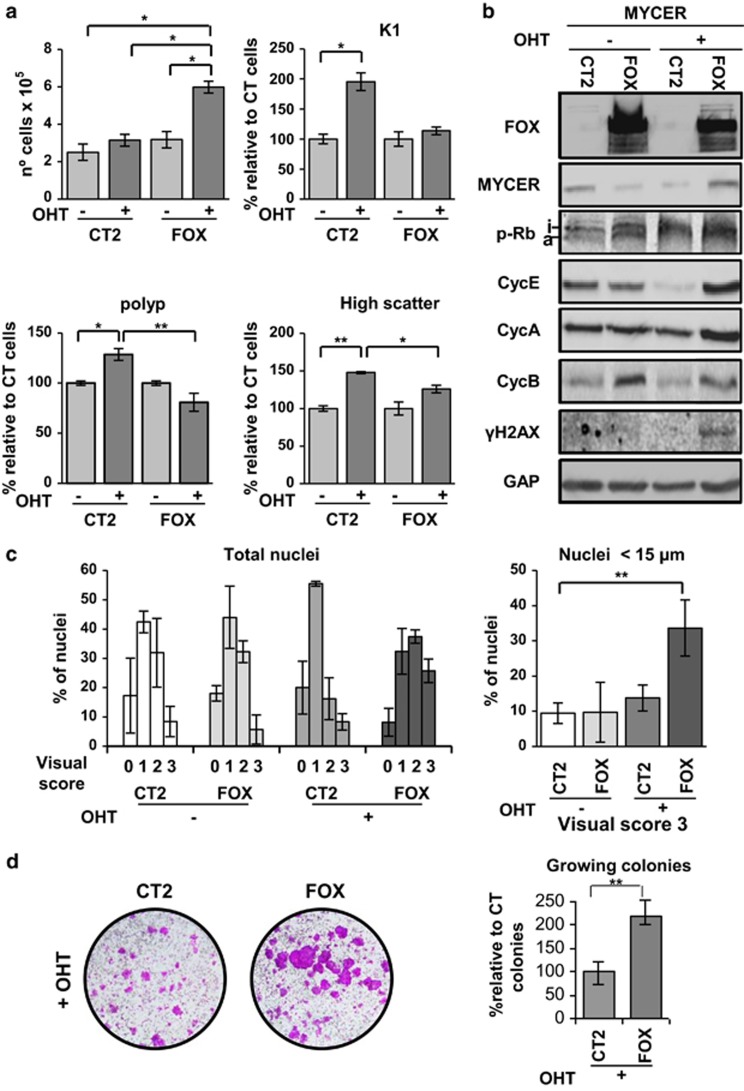
In primary keratinocytes, the activation of MYC drives the cell cycle in 1–2 days and epidermal differentiation in 5–6 days.5, 12, 38 We infected human epidermal keratinocytes with a retroviral construct carrying a conditional form of human MYC (MYCER) whose protein product is regulated by 4-hydroxytamoxifen (OHT),39 and then infected them with the lentiviral construct bearing FOXM1. Cells expressing MYCER (KMYCER) differentiated when treated with OHT for 5 days (Figure 5a). The number of KMYCER cells collected after 5 days of OHT treatment was doubled when FOXM1 was ectopically expressed (Figure 5a and Supplementary Figure 5a). FOXM1 also significantly suppressed MYC-induced differentiation as monitored by the reduced expression of the squamous terminal marker keratin K1 (Figure 5a) and the decrease in light scattering (Figure 5a and Supplementary Figure 5b). We also observed that FOXM1 influenced the cell cycle parameters of KMYCER cells. Induction of MYCER with OHT drove keratinocytes into the polyploid compartment, whereas ectopic expression of FOXM1 significantly decreased the polyploid population (Figure 5a and Supplementary Figure 5c). The expression of cell cycle regulators Cyclin E, Cyclin A and Cyclin B was reduced in KMYCER treated with OHT 3 days, whereas phospho-Rb was inactivated (Figure 5b), consistent with mitotic block slippage.12 However, ectopic expression of FOXM1 in KMYCER cells recovered the expression of Cyclin E and the mitotic Cyclins A and B (Figure 5b). In addition, MYC deregulation causes replication stress and DNA damage14 and we observed a stronger accumulation of the DNA damage marker γH2AX when FOXM1 was overexpressed (Figure 5b and Supplementary Figure 5d). Consistently and as observed for shp53, in KMYCER small nuclei typical of proliferative cells contained strikingly more damage in comet assays when FOXM1 was overexpressed (Figure 5c and Supplementary Figure 5e).
Figure 5.
FOXM1 drives MYC-induced differentiation into increased proliferation. (a) MYCER keratinocyte cultures plated at high density (see Supplementary Materials and methods) and infected with CT2 or FOX for 5 days in the presence (+) or absence (−) of 4- hydroxytamoxifen (OHT) as indicated. Bar histograms represent: number of cells harvested (top left), percent of cells expressing keratin K1 (K1; top right), differences in the proportion of polyploid cells relative to control (bottom left; histograms in Supplementary Figure 5c) and differences in the proportion of cells with high scatter parameters relative to control (bottom right; dot plots in Supplementary Figure 5b). Controls are cells infected with CT2, nontreated with OHT; CT2/-OHT=100%). (b) Western blotting for FOX, MYCER, p-Rb (a, activated; i, inactivated), Cyclin E (CycE), Cyclin A (CycA), Cyclin B (CycB) and γHA2X in MYCER keratinocytes in the presence or absence of exogenous FOX and in the presence or absence of OHT as indicated. GAPDH (GAP) was used as loading control. (c) DNA damage monitored by comet assays after 5 days in the presence or absence of OHT as indicated. Left bar histogram shows the quantitation of DNA damage according to the 4 levels of DNA damage intensity described in Figure 4d. Right bar histogram shows the percent of small nuclei (<15 μm) with maximum DNA damage intensity level 3. Representative pictures are shown in Supplementary Figure 5e. (d) Clonogenicity assays of keratinocytes plated at low density (see Supplementary Materials and methods), sequentially infected with MYCER and CT2 or FOX and cultured in the presence of OHT for 10 days. Bar histograms represent large growing colonies when FOX is ectopically expressed relative to MYCER+OHT cells infected with control vector (CT2/+OHT=100%). Clonogenicity in the absence of OHT is shown in Supplementary Figure 5f. *P<0.05 and **P<0.01. Data are mean±s.e.m. of triplicate (n=3) or duplicate (n=2) samples of representative experiments. Results are representative of two different strains from different individuals (N=2).
FOXM1 also recovered the colony-forming potential of keratinocytes that is lost by MYC activation (Figure 5d).5 Overexpression of FOXM1 in KMYCER cells treated with OHT not only rescued the deleterious effect of MYC on colony growth, but also boosted this capacity beyond controls (Figure 5d and Supplementary Figure 5f).
Altogether, the results strongly indicate that FOXM1 switches the activation of the cell cycle by MYC from differentiation to proliferation in spite of increased replication stress and DNA damage.
Discussion
Induction of squamous differentiation in the epidermis by proto-oncogene MYC and other oncogenic molecules is a long standing paradox5, 6, 7, 8, 13 in spite of the important implications to squamous cancer. Clearly, MYC does not promote sustained cell division in keratinocytes. The finding that replication stress in keratinocytes triggers the mitosis checkpoints and in turn squamous differentiation (DMC) suggests that mitosis control might be the limiting factor in epidermal carcinogenesis.14 Our hypothesis was therefore that reinforcement of the mitosis machinery might inhibit differentiation and imbalance the keratinocyte decision making towards proliferation. Our present results upon ectopic expression of FOXM1 in keratinocytes strongly support this model.
FOXM1 controls a set of mitotic genes and has been proposed to be a mitosis master gene.30 FOXM1 promotes the transcription of genes linked to execution of mitosis and proper chromosome segregation.20, 30 Here we have shown that FOXM1 drives proliferation of keratinocytes, although cell cycle entry was barely affected. In contrast, the cell cycle shortened because of a shorter G2/M phase. In addition, we found FOXM1 to accumulate in mitotic keratinocytes in human epidermis.
Overexpression of FOXM1 in freshly isolated keratinocytes rescued the proliferative loss caused by inactivation of p53 or by hyperactivation of MYC. Keratinocytes overcame the DMC and continued to proliferate. Concomitantly, FOXM1 provoked a decrease in general transcription. Transcription is inactive during mitosis but very active during keratinocyte differentiation involving massive protein production. The lower level of nascent RNAs upon ectopic FOXM1 might be because of a transcription inhibitory function of a FOXM1 isoform31 contributing to coordinate the end of the S phase with mitosis. Possibly as a consequence of decreased global transcription, keratinocytes overexpressing FOXM1 displayed a clearer, less granulated and complex cytoplasm, reminiscent of ‘clear cell carcinomas' of poor prognosis, some of which have been associated with FOXM1 overexpression.40
Our results altogether support a key role of FOXM1 in driving epidermal cell division, consistent with observations in keratinocytes of oral epithelium.25 To note, we have recently shown that FOXM1 mediates the function of the epidermal growth factor receptor in keratinocyte proliferation.41 In our present study, although FOXM1 did not abolish differentiation, it did increase the proportion of binucleated cells within the remaining polyploid population, indicating that it pushed nuclear division even when cytokinesis was irreversibly blocked by terminal differentiation.
The results have implications in cancer beyond the role of FOXM1 in keratinocytes. The inhibition of squamous differentiation by this key mitotic regulator and the resulting imbalance in cell decision making supports a model where mitosis control (DMC) is critical in squamous homeostasis. We have shown that gain of MYC or loss of p53 provokes mitosis slippage and differentiation via DNA damage.14 The results herein show that FOXM1 overexpressing proliferative keratinocytes accumulate DNA damage. Altogether, the results suggest that ectopic FOXM1 by driving entry in mitosis shortens the repair G2 phase and prevents cells with irreparable damage to undergo terminal differentiation (Figure 6). This supports a role for the DMC as a protective mechanism against irreparable genetic damage5, 12, 13, 14 by which precancerous cells are eliminated by stratification and shedding.42 We have proposed that additional alterations in the DMC might render cells harbouring mutations in cell growth control (p53, MYC) capable to divide (Figure 6). As a consequence, precancerous cells would be selected for, leading to malignant transformation.14 This may explain why deregulation of p53 or MYC is frequently found in skin carcinomas, even though they are not considered initiators of cancer and they trigger the mitosis checkpoints.
Figure 6.
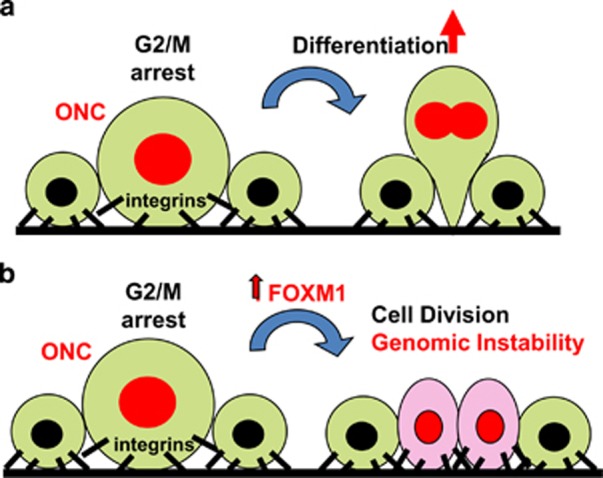
Model for the action of ectopic FOXM1 in p53- or MYC-mutant keratinocytes. (a) Cells upon potentially oncogenic alterations causing cell cycle deregulation and replication stress (ONC) and accumulation of irreparable DNA damage (red nuclei) block in mitosis unable to divide.14 Prolonged G2/M block allows cell size increase and triggers terminal epidermal differentiation, resulting in downregulation of integrins,52 irreversible suppression of cytokinesis and stratification.12, 26 (b) Ectopic expression of FOXM1 pushes damaged keratinocytes to progress in mitosis and achieve cytokinesis in spite of an initial mitosis pause, giving rise to two slightly larger daughter cells that do not downregulate integrins. This leads to expansion of cells bearing genetic damage and to genomic instability.
In summary, FOXM1 drove the cell cycle deregulation caused by MYC or loss of p53 from differentiation into increased proliferation. In part, this process might be mediated by cell adhesion. In steady-state epidermis keratinocyte proliferation within the basal layer is tightly maintained by cell adhesion β4 or β1 integrin complexes.28 We found that ectopic FOXM1 in keratinocytes recovered the expression of β1 integrins upon the loss of p53. Integrin recovery might be a direct effect of FOXM1 or an indirect effect because of sustained proliferation. However, it is interesting that FOXM1 knockout mice suffered from severe abnormalities in lung development because of the downregulation of β1 integrins.43 Keratinocytes arrested in mitosis because of irreparable DNA damage might stratify by lateral pressure from more adherent proliferative neighbour cells (Figure 6).14 FOXM1 cells with a basal-like morphology and high integrin expression displayed a higher size and a higher level of DNA damage typical of the onset of differentiation. These results suggest that FOXM1 might drive cells that were committed to initiate differentiation into cell division. FOXM1 might raise the threshold of genetic damage allowed for keratinocytes to divide even when the mitotic checkpoints delay mitosis (Figure 6). As a result, FOXM1 would extend the number of cell divisions in a high genomic instability context. Interestingly, most squamous carcinomas of the skin and head and neck still conserve a differentiated component and we have evidence that this paradox might respond to alterations in mitosis control.44
In conclusion, in the context of overexpression of MYC or inactivation of p53, persistent expression of FOXM1 may result in the creation of a pool of genomically instable cells with the capacity to divide, even if they are committed to leave the basal layer of the epidermis. This may fix precancerous mutations and give rise to cell clones with malignant potential. These results support a model where the DMC is key in coordinating keratinocyte proliferation with differentiation. This novel checkpoint might exist in developmental tissues, particularly in those where mitotic slippage leads to polyploidy.13 A variety of mammalian tissues have been so far found to undergo endoreplication.45 Very recently, the greatly expanding mammary epithelium has been shown to become binucleated at lactancy.46 The challenge now is dissecting the DMC whose alterations might give rise to genome instability and cancer.
Materials and methods
Cell culture, plasmids and viral infections
Ethical permission for this study was requested, approved and obtained from the Ethical Committee for Clinical Research of Cantabria Council, Spain. In all cases, human tissue material discarded after surgery was obtained with written consent presented by clinicians to the patients, and it was treated anonymously.
Primary keratinocytes were isolated from neonatal human foreskin and cultured in the presence of a mouse fibroblast feeder layer (inactivated by mitomycin C) in Rheinwald FAD medium as described (10% serum and 1.2 mM Ca+2).5, 47 Low passages (1–4) of keratinocytes were utilised.
For gene delivery in primary keratinocytes the following constructs driven by constitutive promoters were used. (1) Retroviral: pBabe empty vector (CT-pb) and pBabe-MYCER;5, 14, 39 MYC fusion protein with the ligand binding domain of a mutant oestrogen receptor that responds to 4-OH-hydroxytamoxifen (OHT; Research Biochemicals International, Natick, MA, USA). (2) Lentiviral: empty plKO1 (CT1; Sigma-Aldrich, Inc., St Louis, MO, USA), empty pLVTHM-GFP (CT-GFP) and three different constructs coding for shRNAs against p53 with different target sequences: pLVUH-shp53-GFP (shp53-GFP), pLKO1-p53-shRNA-427 (shp53-1) and pLKO1-p53-shRNA-941 (shp53-2), all from Addgene (Cambridge, MA, USA);48 empty pLVX (CT2) and pLVX-FOXM1 (FOX).49
Lentiviral production was performed by transient transfection of 293T cells as previously described14 (see Supplementary Materials and methods). For infections with CT-pb, MYCER, CT2 and FOX, keratinocytes were cultured in FAD medium. After cell selection (CT-pb and MYCER) or confluency (CT2 and FOX), cells were transferred to low-calcium concentration medium (<0.1 mM; Keratinocyte Media 2, Promocell, Heidelberg, Germany), following the manufacturer's instructions, and lentiviral infections with CT2 and FOX or CT1 and shp53 plasmids respectively were performed. MYCER was activated by addition of 100 nM OHT (Sigma-Aldrich, Inc.) to the culture medium 24 h after the infection with CT2 or FOX for the periods of time indicated. When necessary, MYCER cells were cultured in high calcium medium after infection with CT2 and FOX by adding CaCl2 (1.2 mM) in order to allow stratification.
Clonogenicity assays were made as described previously24 (see also Supplementary Materials and methods). Quantitation of cell shedding was measured by counting cells detached into the culture medium. Data were obtained from duplicate samples and normalised to controls.
Antibodies
Primary and secondary antibodies utilised in this study are listed in Supplementary Materials and methods.
Flow cytometry
Keratinocytes were harvested, fixed and stained for DNA synthesis, involucrin and keratin K1 as previously described.14 All antibody stainings were controlled by the use of similar concentration of isotype-negative immunoglobulins (mouse or rabbit serum). After staining, cells were firmly resuspended and filtered through a 70 μM mesh to minimise the presence of aggregates and then analysed on a Becton Dickinson FACSCanto (Franklin Lakes, NJ, USA). A total of 10 000 events were gated and acquired.
Immunodetection
For immunofluorescence, keratinocytes were grown on glass coverslips, fixed and stained as previously described.12 For determination of protein expression, cells were washed with phosphate-buffered saline, lysed and subjected to SDS–PAGE electrophoresis and western blotting as previously described.12
Real-time PCR
Total RNA was isolated and reverse-transcribed using NucleoSpin RNA (Macherey-Nagel, Düren, Germany) and the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) according to the manufacturer's instructions. The cDNAs (2 μl) were amplified by real-time PCR (Bio-Rad iQ SYBR Green supermix) and normalised to β-actin mRNA levels. Primers utilised in this study are listed in Supplementary Materials and methods.
Run-on transcription assay and comet assay
For immunodetection of nascent RNA, primary keratinocytes were cultured for 20' with 5'-fluorouridine (Sigma-Aldrich, Inc.), as described in Rosa-Garrido et al.50 (see Supplementary Materials and Methods).
Alkaline comet assays were performed as described previously.51 A visual score related to the level of DNA damage observed was assigned to each tail. Scoring was performed by blind counting.
Confocal microscopy
For Supplementary Video 1, keratinocytes were grown on glass coverslips, fixed and stained as previously described.14 Z-stack 3D digital images were reconstructed after frame collection by confocal microscopy (Nikon A1R, Melville, NY, USA; 20 × numerical aperture (NA) 0.75) and processed by NIS Elements software (AR, 3.2 64 bits; Nikon) as indicated in the legend of Supplementary Video 1.
Time-lapse videos
For Supplementary Video 2, keratinocytes 5 days after infection were treated with NucBlue Live ReadyProbes Reagent (Life Technologies, Carlsbad, CA, USA), filmed by time-lapse imaging for 12 h and photographed every 7 min.
Statistical analyses
Results were obtained with two different keratinocyte strains from different individuals (N=2). Data are average of triplicate samples of representative experiments (n=3). Exclusion of samples was carried out based on the appearance of the negative control sample. Standard deviation and variance were calculated and served as estimates of variation within each group of data. For statistical comparison of groups with similar variance, a homoscedastic t-test was performed. For statistical comparison of groups with diverging variance, a heteroscedastic t-test was applied. A P-value of <0.05 was considered statistically significant.
Acknowledgments
This work was funded by Instituto de Salud Carlos III ISCIII FIS/FEDER Grants PI11/02070 and PI14/00900 (to AG) and National Institutes of Health, NIH Awards R03 AR049420 (to SWS) and R01 AR052889 (to JTE).
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Shea CR, McNutt NS, Volkenandt M, Lugo J, Prioleau PG, Albino AP. Overexpression of p53 protein in basal cell carcinomas of human skin. Am J Pathol 1992; 141: 25–29. [PMC free article] [PubMed] [Google Scholar]
- Brash DE. Roles of the transcription factor p53 in keratinocyte carcinomas. Br J Dermatol 2006; 154(Suppl 1): 8–10. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery Jr CA, Butel JS et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992; 356: 215–221. [DOI] [PubMed] [Google Scholar]
- Kemp CJ, Donehower LA, Bradley A, Balmain A. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell 1993; 74: 813–822. [DOI] [PubMed] [Google Scholar]
- Gandarillas A, Watt FM. c-Myc promotes differentiation of human epidermal stem cells. Genes Dev 1997; 11: 2869–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waikel RL, Wang XJ, Roop DR. Targeted expression of c-Myc in the epidermis alters normal proliferation, differentiation and UV-B induced apoptosis. Oncogene 1999; 18: 4870–4878. [DOI] [PubMed] [Google Scholar]
- Watt FM, Frye M, Benitah SA. MYC in mammalian epidermis: how can an oncogene stimulate differentiation? Nat Rev Cancer 2008; 8: 234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt A, Frye M, Herold S, Benitah SA, Braun K, Samans B et al. Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J Cell Biol 2006; 172: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias E, Miliani de Marval PL, Senderowicz A, Cullen J, Rodriguez-Puebla ML. Expression of CDK4 or CDK2 in mouse oral cavity is retained in adult pituitary with distinct effects on tumorigenesis. Cancer Res 2008; 68: 162–171. [DOI] [PubMed] [Google Scholar]
- Pierce AM, Fisher SM, Conti CJ, Johnson DG. Deregulated expression of E2F1 induces hyperplasia and cooperates with ras in skin tumor development. Oncogene 1998; 16: 1267–1276. [DOI] [PubMed] [Google Scholar]
- Ruiz S, Santos M, Segrelles C, Leis H, Jorcano JL, Berns A et al. Unique and overlapping functions of pRb and p107 in the control of proliferation and differentiation in epidermis. Development 2004; 131: 2737–2748. [DOI] [PubMed] [Google Scholar]
- Freije A, Ceballos L, Coisy M, Barnes L, Rosa M, De Diego E et al. Cyclin E drives human keratinocyte growth into differentiation. Oncogene 2012; 31: 5180–5192. [DOI] [PubMed] [Google Scholar]
- Gandarillas A. The mysterious human epidermal cell cycle, or an oncogene-induced differentiation checkpoint. Cell Cycle 2012; 11: 4507–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije A, Molinuevo R, Ceballos L, Cagigas M, Alonso-Lecue P, Rodriguez R et al. Inactivation of p53 in human keratinocytes leads to squamous differentiation and shedding via replication stress and mitotic slippage. Cell Rep 2014; 9: 1349–1360. [DOI] [PubMed] [Google Scholar]
- Jonason AS, Kunala S, Price GJ, Restifo RJ, Spinelli HM, Persing JA et al. Frequent clones of p53-mutated keratinocytes in normal human skin. Proc Natl Acad Sci USA 1996; 93: 14025–14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZP, Ahmadian A, Ponten F, Nister M, Berg C, Lundeberg J et al. Benign clonal keratinocyte patches with p53 mutations show no genetic link to synchronous squamous cell precancer or cancer in human skin. Am J Pathol 1997; 150: 1791–1803. [PMC free article] [PubMed] [Google Scholar]
- le Pelletier F, Soufir N, de La Salmoniere P, Janin A, Basset-Seguin N. p53 Patches are not increased in patients with multiple nonmelanoma skin cancers. J Invest Dermatol 2001; 117: 1324–1325. [DOI] [PubMed] [Google Scholar]
- Gandarillas A, Molinuevo R, Freije A, Alonso-Lecue P. The mitosis-differentiation checkpoint, another guardian of the epidermal genome. Mol Cell Oncol 2015; 2: e997127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta 2007; 1775: 92–102. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol 2005; 7: 126–136. [DOI] [PubMed] [Google Scholar]
- Teh MT, Wong ST, Neill GW, Ghali LR, Philpott MP, Quinn AG. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res 2002; 62: 4773–4780. [PubMed] [Google Scholar]
- Teh MT, Hutchison IL, Costea DE, Neppelberg E, Liavaag PG, Purdie K et al. Exploiting FOXM1-orchestrated molecular network for early squamous cell carcinoma diagnosis and prognosis. Int J Cancer 2013; 132: 2095–2106. [DOI] [PubMed] [Google Scholar]
- Rodriguez R, Rubio R, Gutierrez-Aranda I, Melen GJ, Elosua C, Garcia-Castro J et al. FUS-CHOP fusion protein expression coupled to p53 deficiency induces liposarcoma in mouse but not in human adipose-derived mesenchymal stem/stromal cells. Stem Cells 2011; 29: 179–192. [DOI] [PubMed] [Google Scholar]
- Jones PH, Watt FM. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell 1993; 73: 713–724. [DOI] [PubMed] [Google Scholar]
- Gemenetzidis E, Elena-Costea D, Parkinson EK, Waseem A, Wan H, Teh MT. Induction of human epithelial stem/progenitor expansion by FOXM1. Cancer Res 2010; 70: 9515–9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J, Freije A, Ruiz M, Coulon V, Sanz JR, Chiesa J et al. A mitosis block links active cell cycle with human epidermal differentiation and results in endoreplication. PLoS One 2010; 5: e15701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec AS, Delcourt P, Dewailly E, Bidaux G. Optimal differentiation of in vitro keratinocytes requires multifactorial external control. PLoS One 2013; 8: e77507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM. Role of integrins in regulating epidermal adhesion, growth and differentiation. EMBO J 2002; 21: 3919–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deursen JM. Rb loss causes cancer by driving mitosis mad. Cancer Cell 2007; 11: 1–3. [DOI] [PubMed] [Google Scholar]
- Costa RH. FoxM1 dances with mitosis. Nat Cell Biol 2005; 7: 108–110. [DOI] [PubMed] [Google Scholar]
- Kong X, Li L, Li Z, Le X, Huang C, Jia Z et al. Dysregulated expression of FOXM1 isoforms drives progression of pancreatic cancer. Cancer Res 2013; 73: 3987–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer 2007; 7: 847–859. [DOI] [PubMed] [Google Scholar]
- Duarte B, Miselli F, Murillas R, Espinosa-Hevia L, Cigudosa JC, Recchia A et al. Long-term skin regeneration from a gene-targeted human epidermal stem cell clone. Mol Ther 2014; 22: 1878–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halasi M, Gartel AL. A novel mode of FoxM1 regulation: positive auto-regulatory loop. Cell Cycle 2009; 8: 1966–1967. [DOI] [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Major ML, Raychaudhuri P. New and unexpected: forkhead meets ARF. Curr Opin Genet Dev 2005; 15: 42–48. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273: 5858–5868. [DOI] [PubMed] [Google Scholar]
- Lecona E, Fernandez-Capetillo O. Replication stress and cancer: it takes two to tango. Exp Cell Res 2014; 329: 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandarillas A, Davies D, Blanchard JM. Normal and c-Myc-promoted human keratinocyte differentiation both occur via a novel cell cycle involving cellular growth and endoreplication. Oncogene 2000; 19: 3278–3289. [DOI] [PubMed] [Google Scholar]
- Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res 1995; 23: 1686–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue YJ, Xiao RH, Long DZ, Zou XF, Wang XN, Zhang GX et al. Overexpression of FoxM1 is associated with tumor progression in patients with clear cell renal cell carcinoma. J Transl Med 2012; 10: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll SW, Stuart PE, Swindell WR, Tsoi LC, Li B, Gandarillas A et al. The EGF receptor ligand amphiregulin controls cell division via FoxM1. Oncogene 2015; 35: 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandarillas A, Molinuevo R, Freije A, Alonso-Lecue P. The mitosis-differentiation checkpoint, another guardian of the epidermal genome. Mol Cell Oncol 2015; 2: e997127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IM, Ramakrishna S, Gusarova GA, Yoder HM, Costa RH, Kalinichenko VV. The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J Biol Chem 2005; 280: 22278–22286. [DOI] [PubMed] [Google Scholar]
- Alonso-Lecue P, Coulon V, Ceballos L, Lopez-Aventin D, Molinuevo R, García-Valtuille A et al. Bypass of mitotic block in response to cell cycle stress leads to genomic instability and malignant progression of squamous carcinoma cells of the skin (submitted). [DOI] [PMC free article] [PubMed]
- Orr-Weaver TL. When bigger is better: the role of polyploidy in organogenesis. Trends Genet 2015; 31: 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios AC, Fu NY, Jamieson PR, Pal B, Whitehead L, Nicholas KR et al. Essential role for a novel population of binucleated mammary epithelial cells in lactation. Nat Commun 2016; 7: 11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinwald JG. Methods for clonal growth and serial cultivation of normal human epidermal keratinocytes and mesothelial cells. In: Baserga R (ed) Cell Growth and Division. IRL Press: Oxford, 1989, pp 81–94. [Google Scholar]
- Kim JS, Lee C, Bonifant CL, Ressom H, Waldman T. Activation of p53-dependent growth suppression in human cells by mutations in PTEN or PIK3CA. Mol Cell Biol 2007; 27: 662–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toll A, Real FX. Somatic oncogenic mutations, benign skin lesions and cancer progression: where to look next? Cell Cycle 2008; 7: 2674–2681. [DOI] [PubMed] [Google Scholar]
- Rosa-Garrido M, Ceballos L, Alonso-Lecue P, Abraira C, Delgado MD, Gandarillas A. A cell cycle role for the epigenetic factor CTCF-L/BORIS. PLoS One 2012; 7: e39371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie A, Gutierrez O, Fernandez-Luna JL. PAR bZIP-bik is a novel transcriptional pathway that mediates oxidative stress-induced apoptosis in fibroblasts. Cell Death Differ 2009; 16: 838–846. [DOI] [PubMed] [Google Scholar]
- Hotchin NA, Gandarillas A, Watt FM. Regulation of cell surface beta 1 integrin levels during keratinocyte terminal differentiation. J Cell Biol 1995; 128: 1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.