Abstract
Prefoldin (PFDN) is a co-chaperone protein that is primarily known for its classic cytoplasmic functions in the folding of actin and tubulin monomers during cytoskeletal assembly. Here, we report a marked increase in prefoldin subunit 1 (PFDN1) levels during the transforming growth factor (TGF)-β1-mediated epithelial-mesenchymal transition (EMT) and in human lung tumor tissues. Interestingly, the nuclear localization of PFDN1 was also detected. These observations suggest that PFDN1 may be essential for important novel functions. Overexpression of PFDN1 induced EMT and cell invasion. In sharp contrast, knockdown of PFDN1 generated the opposite effects. Overexpression of PFDN1 was also found to induce lung tumor growth and metastasis. Further experiments showed that PFDN1 overexpression inhibits the expression of cyclin A. PFDN1 suppressed cyclin A expression by directly interacting with the cyclin A promoter at the transcriptional start site. Strikingly, cyclin A overexpression abolished the above PFDN1-mediated effects on the behavior of lung cancer cells, whereas cyclin A knockdown alone induced EMT and increased cell migration and invasion ability. This study reveals that the TGF-β1/PFDN1/cyclin A axis is essential for EMT induction and metastasis of lung cancer cells.
Introduction
Lung cancer has become one of the most prevalent and lethal cancers worldwide, and metastasis is the main cause of its high mortality rate in patients.1 Detachment from the primary loci and the invasion of cancer cells into the surrounding tissues can be initiated by the loss of cell–cell adhesion and the gain of motility and invasive properties.2 During the past decade, mounting data have shown that epithelial-mesenchymal transition (EMT) is a critical factor contributing to the invasion and distal metastasis of many epithelial-originated cancers. EMT has been characterized to be a fundamental biological event that has important roles in embryonic development, wound healing, chronic fibrosis and cancer metastasis.3 EMT causes the reorganization of the cytoskeleton and endows epithelial cells with a mesenchymal phenotype, which is important for mediating changes in cell identity and behavior. Various factors have been implicated in the control of EMT. Transforming growth factor (TGF)-β1 is one of the strongest inducers of EMT and receives abundant attention owing to its potent pleiotropic effects implicated in a variety of patho-physiological processes, including cancer progression.4 An increasing number of molecules have been identified to be involved in TGF-β1 signaling and associated cellular and biological events. Insulin receptor substrate-1,5 forkhead box transcription factor A2,6 and hepatocyte nuclear factor 67 were shown to be potent EMT suppressors. They are essential for maintaining the epithelial phenotype and are therefore important in the inhibition of EMT and its associated cellular events. Decreased expression levels of these molecules have been linked with TGF-β1-induced EMT, growth and metastasis of lung cancers. In contrast, prostate transmembrane protein, androgen induced-1 is important for the plasticity of epithelial cells and its significant increase is required for TGF-β1-induced EMT in lung cancer cells.8 These findings suggest that a TGF-β1 signaling network is orchestrated to regulate the equilibrium between the epithelial and mesenchymal properties of various cells, which impacts cell fates and behavior decisions. Further investigation into the relevant aspects of TGF-β1 signaling is important for deepening our understanding of EMT and could provide more precise mechanism-based clinical treatment of some cancers.
Chaperone proteins have been shown to be involved in cancer development and progression.9, 10 However, little is known about their roles in TGF-β signaling and the induction of EMT. It has been shown recently that chaperone proteins are involved in the induction of EMT and the metastasis of prostate cancer cells.11, 12 Until recently, the roles of co-chaperones in TGF-β-induced EMT and other relevant patho-physiological processes were poorly understood. Prefoldin (PFDN) is a co-chaperone protein that captures unfolded polypeptides and transfers them to the chaperonin containing tailless complex polypeptide-1.13 PFDN exists in the cytosol as a complex containing six subunits. The involvement of PFDN subunits in cancer progression has been reported in several publications. PFDN subunits can interact with HDAC1 with high affinity in HepG2 hepatocarcinoma cells.14 PFDN4, a subunit of the PFDN complex, is decreased in colorectal cancer and is involved in the inhibition of cell growth and invasiveness.15 Prefoldin subunit 1 (PFDN1) is important in cytoskeletal rearrangement, as the phenotypes caused by PFDN1 depletion were all consistent with that of abnormal cytoskeletal functions.16, 17 Considering these findings and the close relationship between cytoskeletal rearrangement and EMT, we hypothesized that PFDN1 is an essential factor involved in the regulation of EMT and its accompanying biological events.
Cyclin A (also known as cyclinA2) is important in cell cycle regulation and is implicated in cell fate determination. Cyclin A depletion was shown to cause an increase in cytoskeletal rearrangement and cell migration in normal mammary epithelial cells.18 Compared with low invasive or primary tumors, the expression level of cyclin A is markedly lower in a highly invasive colon adenocarcinoma cell line, suggesting that loss of cyclin A is linked to tumor metastasis.19 Cyclin A knockdown was recently reported to induce EMT in transformed mammary epithelial cells.20 However, the mechanism underlying the regulation of cyclin A expression and the involvement of cyclin A in the EMT of other tumor types remains poorly understood.
In this study, we investigated the link between PFDN1 and TGF-β1 signaling, the roles of PFDN1 and its regulatory effect and mechanism of suppressing cyclin A expression during human lung cancer progression. PFDN1 levels can be increased by TGF-β1 stimulation, which promotes migration, invasion, xenograft growth and the metastasis of lung cancer cells. These effects were mediated through the transcriptional inhibition of cyclin A by PFDN1 and the subsequent induction of EMT and associated changes in cell behavior. Our findings underscore the importance of PFDN1 in TGF-β1 signaling and EMT induction linked with lung cancer development and progression.
Results
Increases in PFDN1 levels in tumors and TGF-β1-induced EMT
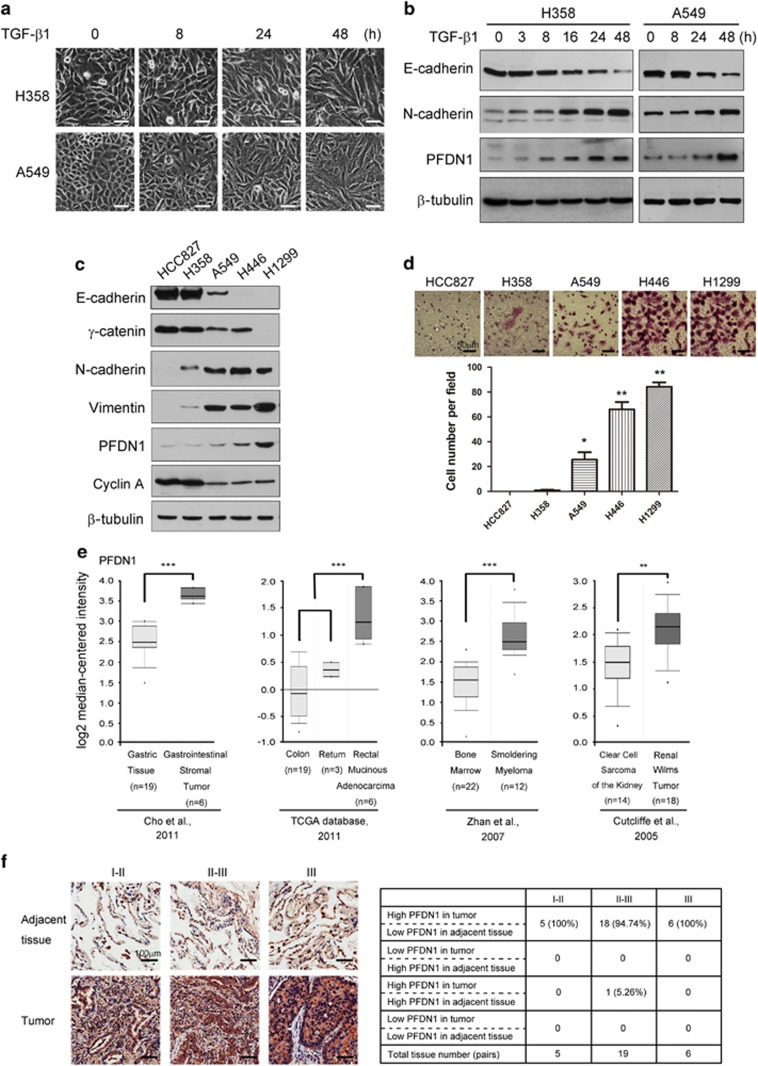
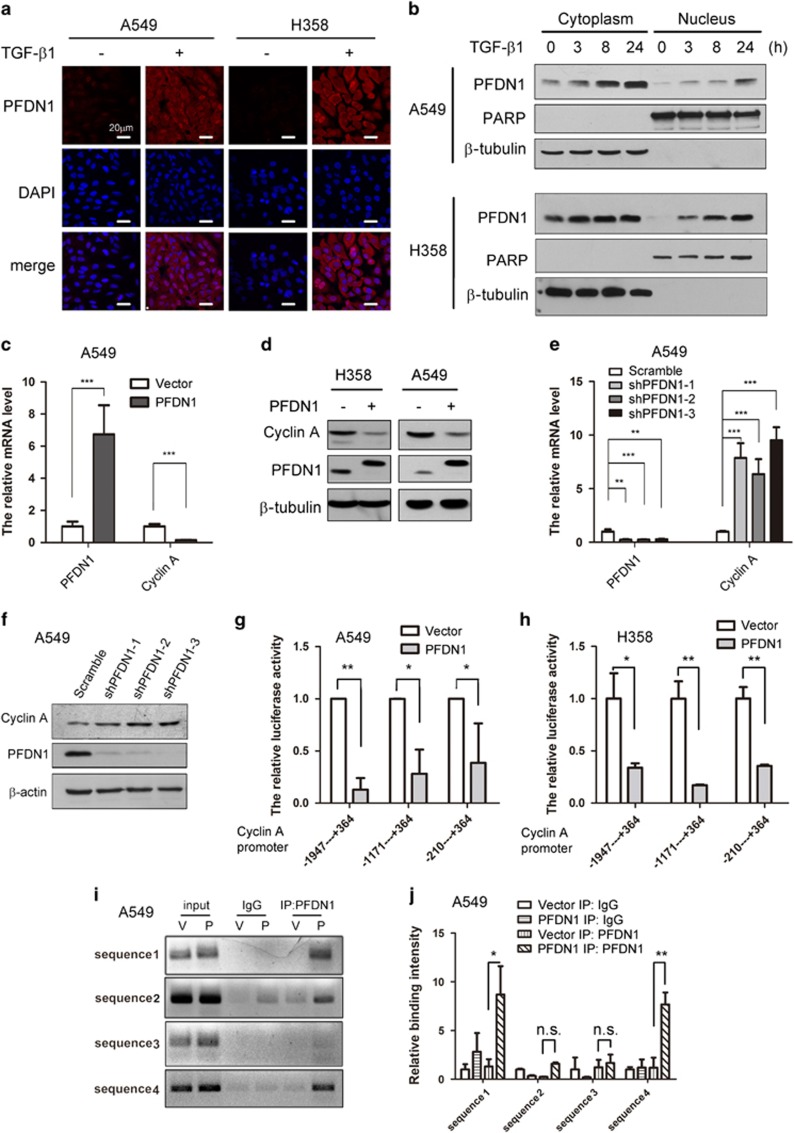
In identifying novel molecules implicated in TGF-β1-mediated EMT in A549 and H358 lung cancer cells (Figure 1a), we observed that PFDN1 levels were markedly increased in both time- and dose-dependent manners (Figure 1b and Supplementary Figure S1a), suggesting that PFDN1 may be involved in EMT and tumor progression. The basal expression levels of PFDN1 in H446 and H1299 are much higher than that in HCC827 human lung cancer cells. PFDN1 levels in A549 and H358 lung cancer cells were moderate. Interestingly, PFDN1 levels are correlated positively and negatively with their degrees of mesenchymal and epithelial phenotypes, respectively (Figure 1c). Consistent with these findings, PFDN1 levels are also correlated with the invasion ability of these cells (Figure 1d). These results suggest that PFDN1 is essential for the mesenchymal phenotype and for the induction of EMT. Analysis of the online Oncomine database revealed that PFDN1 is highly expressed in some tumors (Figure 1e). In addition, PFDN1 is significantly increased in human lung adenocarcinomas ranging from Stage I to Stage III (Figure 1f), suggesting that increased PFDN1 expression is closely associated with the development of lung tumors. No change in the mRNA level of PFDN1 was observed in response to TGF-β1 (Supplementary Figure S1b). Treatment of cells with a proteasome inhibitor (MG132) also had no obvious effect on the TGF-β1-mediated increase in PFDN1 expression levels (Supplementary Figure S1c). However, inhibition of protein synthesis by cycloheximide abolished the increase in PFDN1 induced by TGF-β1 (Supplementary Figure S1d). These data suggest that the TGF-β1-induced increase in PFDN1 occurs at the translational level.
Figure 1.
Increases in PFDN1 levels in TGF-β1-induced EMT and lung cancer samples. (a and b) Increases in PFDN1 levels in TGF-β1-induced EMT in A549 and H358 cells. Cells were treated with TGF-β1 (5 ng/ml) for the indicated time. The changes in cell morphology (a) and the expression levels of E-cadherin, N-cadherin and PFDN1 were determined by immunoblotting (b). (c) The correlations between PFDN1 levels and mesenchymal phenotypes in different lung cancer cell lines were determined by immunoblotting. (d) The invasion ability of different lung cancer cell lines was determined by a Transwell invasion assay. A representative morphology image (upper) and statistical results (lower) are shown. (e) Oncomine box plots of PFDN1 expression levels in multiple human cancers. (f) The correlations between PFDN1 levels and the progressive stages of lung cancers. Left panel, PFDN1 levels of representative lung tumor tissues and adjacent normal lung epithelium were determined by immunohistochemistry. Right panel, statistic data of PFDN1 expression levels.
PFDN1 promotes EMT and associated events in lung cancer cells
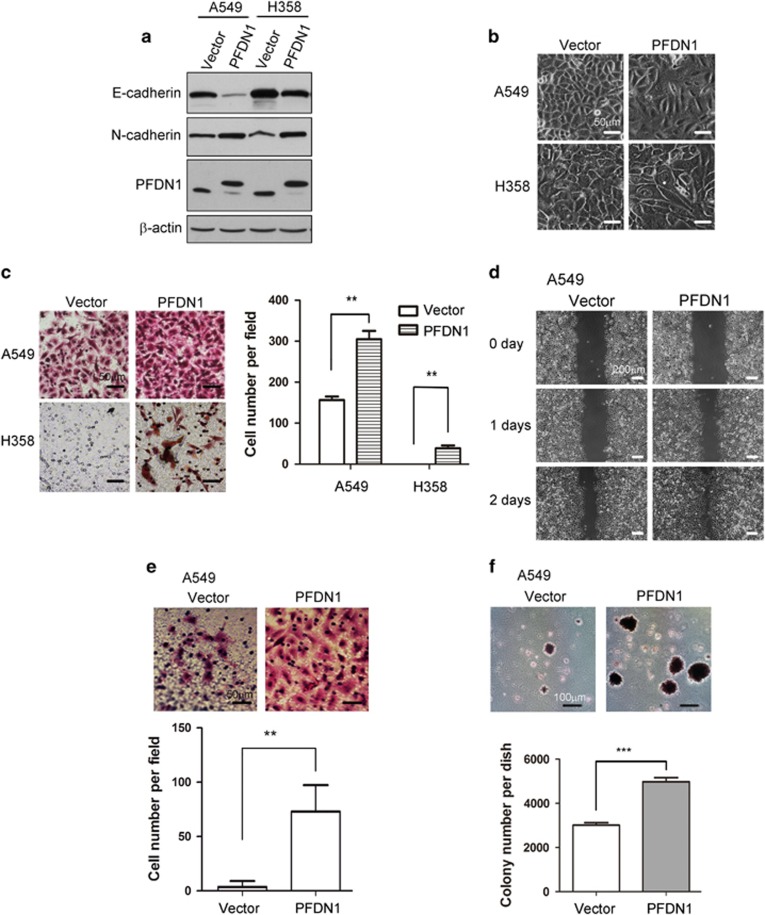
To determine the role of PFDN1 in EMT and EMT-related cell processes, we generated PFDN1-overexpressing lung cancer cell lines. PFDN1 overexpression induced EMT in A549 and H358 cells, as determined by E-cadherin and N-cadherin levels (Figure 2a) and cell morphological changes (Figure 2b). In line with the induction of EMT, increased cell migration (Figures 2c and d), cell invasion ability (Figure 2e) and colony formation of cells in soft agar (Figure 2f) were induced.
Figure 2.
PFDN1 overexpression promotes EMT and increases the motility of lung cancer cells. (a and b) The effect of PFDN1 overexpression on EMT was examined by immunoblotting of E-cadherin and N-cadherin (a) and by changes in cell morphology (b). (c and d) The effect of PFDN1 overexpression on cell migration was examined by a Transwell assay (c) and wound-healing assay (d). (e) The cell invasion ability of PFDN1-overexpressed A549 cells was examined by a matrigel-coated transwell assay. (f) The effect of PFDN1 overexpression on anchorage-independent cell growth was evaluated by a colony-formation assay in soft agar.
PFDN1 knockdown inhibits EMT and cell motility in lung cancer cells
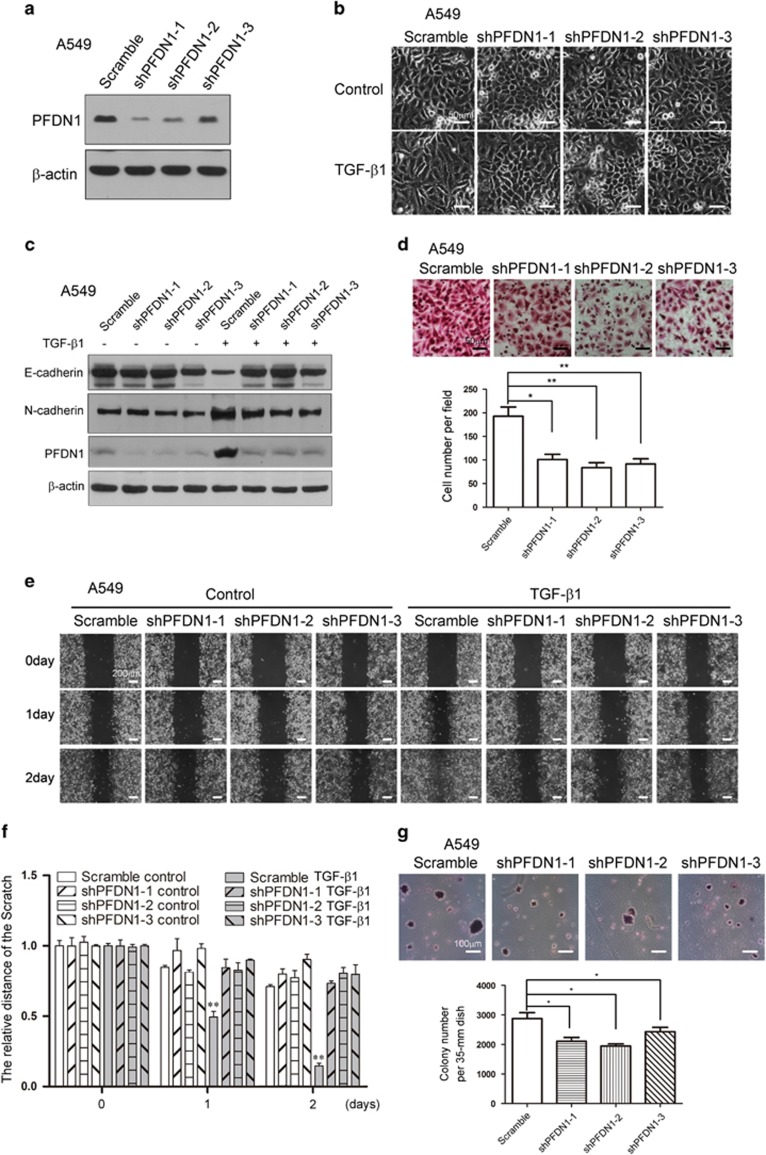
To verify the role of PFDN1, we generated PFDN1-knockdown lung cancer cell lines (Figure 3a). PFDN1 knockdown inhibited TGF-β1-induced EMT, as determined by morphological changes (Figure 3b), E-cadherin and N-cadherin levels (Figure 3c), and cell migration (Figure 3d). PFDN1 knockdown alone also significantly reduced mesenchymal marker protein levels (Supplementary Figure S2a and b). Consistent with these data, PFDN1 knockdown abrogated TGF-β1-induced cell migration (Figures 3e and f) and reduced the anchorage- independent growth of cells (Figure 3g). These results verify the notion that PFDN1 has an important role in EMT and EMT-associated changes in cell behavior.
Figure 3.
PFDN1 knockdown inhibits TGF-β1-mediated EMT and cell motility in lung cancer cells. (a) Stable PFDN1-knockdown A549 cells were generated by lentivirus infection. PFDN1 was examined by immunoblotting. (b and c) PFDN1 knockdown inhibited TGF-β1-induced EMT. PFDN1-knockdown A549 cells were treated with TGF-β1 (2 ng/ml) for 48 h. Cell morphology (b) and levels of N-cadherin and E-cadherin (c) were examined. (d) Cell migration was assessed by a transwell assay. A representative morphology image is shown (upper) and the number of migrated cells per field was determined (lower). (e and f) PFDN1 knockdown inhibited TGF-β1-induced cell migration. Cells were treated with TGF-β1 (2 ng/ml) for the time indicated, the migration of A549 cells was detected by a wound-healing assay (e), and the statistical analysis was assessed to show the migration rate (f). (g) PFDN1 knockdown inhibited anchorage-independent growth in A549 cells, as detected by colony formation in a soft agar assay. A representative image of cell morphology is shown (upper) and number of colonies per 35-mm dish was determined (lower).
PFDN1 promotes the growth and metastasis of lung cancer cells
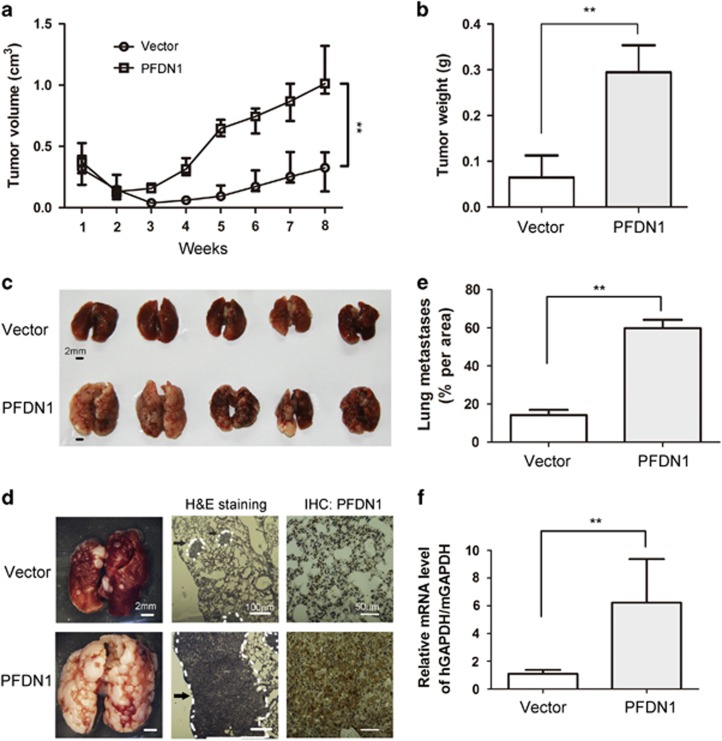
PFDN1-mediated EMT and relevant cell events suggest its important roles in cancer progression. PFDN1 overexpression increased the growth of xenograft lung cancer cells (Figures 4a and b) and the lung metastatic nodule size (Figures 4c and d). The sizes of metastatic areas with larger metastatic nodules were also found to be increased (Figure 4e). The survival of cancer cells by increased resistance to anoikis in the circulating blood is a critical step in the formation of distal metastases, which is indicated by the presence of circulating tumor cells (CTCs) in blood. As shown in Figure 4f, PFDN1 overexpression significantly increased the level of CTCs in blood. These results demonstrate that PFDN1 can function as a lung cancer promoter.
Figure 4.
PFDN1 promotes tumor growth and metastasis. Mice were injected subcutaneously with PFDN1-overexpressed or vector-control A549 cells, and tumor volumes were determined at indicated days. Tumor volume (a) and tumor weight (b) were measured. (c) Mice were tail vein injected with PFDN1-overexpressed A549 cells. The mice were killed 6 weeks after injection and their lungs were harvested. (d) Representative stereoscope images of lung tumors are shown, and metastasis was determined by H&E staining. The expression levels of PFDN1 in lung tumors were detected by immunohistochemistry. Metastasis is indicated by a white dotted line, and arrows indicate exemplary loci of metastasis. (e) The level of metastasis per area was quantified using image software that measures pixel units. (f) Relative CTC levels were determined by real-time PCR. hGAPDH, human GAPDH; mGAPDH, mouse GAPDH.
PFDN1 functions as a transcriptional suppressor of cyclin A
Although the nuclear location and function of PFDN1 in yeast has been reported,21 there are no publications reporting the existence of nuclear PFDN1 in mammalian cells. Interestingly, PFDN1 can be detected in both the cytoplasm and the nucleus. TGF-β1 treatment significantly increased the level of nuclear PFDN1, as determined by immunofluorescent staining (Figure 5a) and immunoblotting (Figure 5b). The nuclear location of PFDN1 suggests that PFDN1 may have novel functions in addition to its cytoplasmic role in the assembly of cytoskeleton. PFDN1 overexpression has an inhibitory effect on the proliferation rate of lung cancer cells (Supplementary Figure S3). PFDN1 overexpression significantly reduced both the mRNA (Figure 5c) and the protein levels (Figure 5d) of cyclin A, whereas PFDN1 knockdown upregulated the mRNA (Figure 5e) and protein levels of cyclin A (Figure 5f). The upregulation of cyclin A by PFDN1 knockdown was also observed in H358 and H1299 cells (Supplementary Figure S4a and b). In addition, TGF-β1 treatment also reduced cyclin A expression at both the mRNA and protein level (Supplementary Figure S5a and b). These results suggest that cyclin A is a downstream target of PFDN1. PFDN1 and PFDN4 have a similar molecular structure,22, 23 and PFDN4 is a transcription factor that regulates the cell cycle.24 Moreover, the detection of nuclear PFDN1 suggests its function as a transcriptional regulator. PFDN1 overexpression inhibited the promoter activity of cyclin A in both A549 (Figure 5g) and H358 cells (Figure 5h). The sequence from −210 bp to 364 bp in the promoter region of cyclin A was essential for PFDN1-mediated suppression of its promoter activity. We performed a Chromatin Immunoprecipitation assay revealing that PFDN1 interacts directly with the promoter of cyclin A around the transcriptional start site (Figures 5i and j). These results indicate that PFDN1 can function as a transcriptional suppressor of cyclin A.
Figure 5.
Nuclear localization of PFDN1 and its suppression of cyclin A expression. (a and b) TGF-β1 increased PFDN1 levels. A549 and H358 cells were treated with TGF-β1 (5 ng/ml) for 24 h. Cytosolic and nuclear PFDN1 were examined by immunofluorescent staining (a) and by immunoblotting of cytosolic and nuclear extracts after the indicated time of treatment (b). Nuclei are stained in blue and PFDN1 is stained in red. Poly ADP-ribose polymerase (PARP) and β-tubulin were used as nuclear and cytosolic marker proteins, respectively. (c) PFDN1 overexpression inhibited the transcriptional expression of cyclin A, as detected by real-time PCR. (d) PFDN1 overexpression reduced cyclin A protein levels, as determined by immunoblotting. (e) PFDN1 knockdown increased cyclin A mRNA levels, as detected by real-time PCR. (f) PFDN1 knockdown increased cyclin A expression levels, as determined by immunoblotting. (g and h) The effect of different sequence regions in the cyclin A promoter on PFDN1-mediated suppression of transcriptional activation of cyclin A. Cyclin A promoters of different lengths were transfected into PFDN1-overexpressed or vector-control A549 cells (g) and H358 cells (h) and luciferase activity was assessed. (i) The interaction of PFDN1 with the cyclin A promoter was detected by a ChIP assay. The PCR products of the cyclin A promoter were analyzed by 1.2% agarose-gel electrophoresis. V, vector-control; P, PFDN1 overexpression. (j) The binding intensity between PFDN1 and cyclin A promoters was determined by a ChIP assay and statistical analysis was performed.
Cyclin A suppresses the PFDN1-induced EMT and cell migration
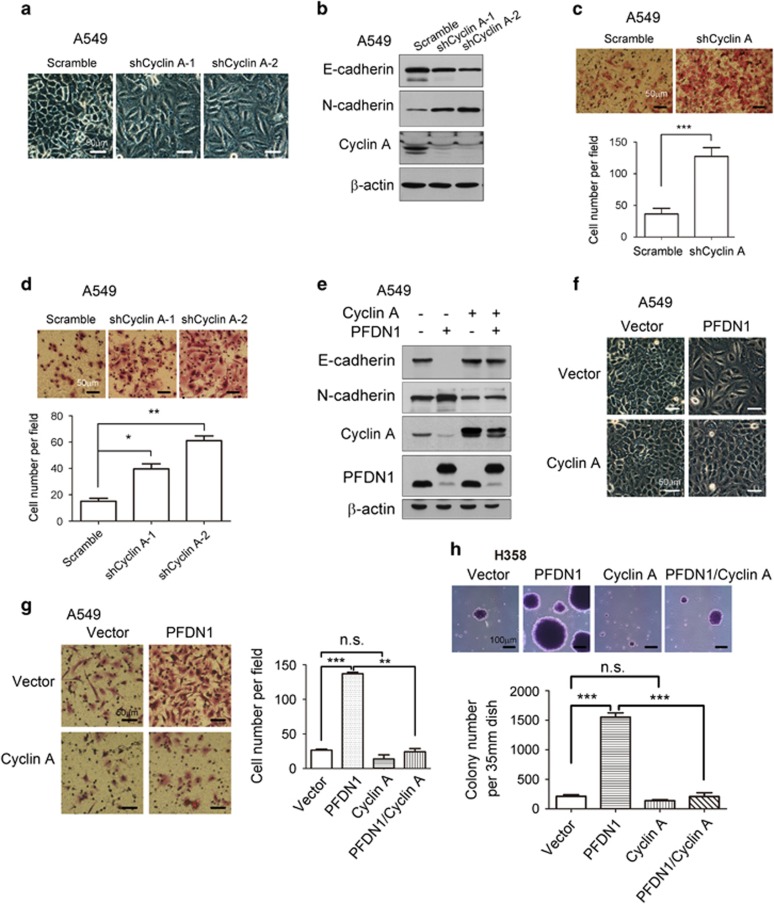
The transcriptional suppression of cyclin A by PFDN1 suggests that cyclin A is important for maintaining the epithelial phenotype of cells. Cyclin A knockdown alone induced EMT as determined by changes in cell morphology (Figure 6a), EMT markers (Figure 6b and Supplementary Figure S6a), cell migration and invasion abilities (Figures 6c and d). Cyclin A knockdown has an inhibitory effect on cell proliferation (Supplementary Figure S6b). Overexpression of cyclin A abrogated the PFDN1-induced EMT (Figure 6e and Supplementary Figure S6c) and cell morphological changes (Figure 6f). Cyclin A overexpression also abrogated PFDN1-induced cell migration (Figure 6g and Supplementary Figure S6d). Meanwhile, cyclin A overexpression suppressed PFDN1-induced anchorage-independent cell growth (Figure 6h). The above results indicate that cyclin A is a downstream target of PFDN1, which acts as a potent suppressor of EMT.
Figure 6.
The role of cyclin A and its effect on PFDN1-induced cellular events. (a) Cyclin A knockdown induced a morphological change characteristic of EMT. (b) Cyclin A knockdown induced changes in the levels of E-cadherin and N-cadherin, as examined by immunoblotting. (c) Cyclin A knockdown increased cell migration, as examined by a Transwell assay. A representative image is shown, and statistical analysis was performed. (d) The effect of cyclin A knockdown on cell invasion was assessed by a matrigel-coated transwell assay. A representative image is shown. A549 cells stably overexpressing both cyclin A and PFDN1 were generated by lentivirus infection. The inhibition of PFDN1-mediated EMT by cyclin A overexpression is shown by the expression levels of E-cadherin and N-cadherin (e) and changes in cell morphology (f). (g) The effect of cyclin A overexpression on PFDN1-mediated cell migration was determined by a transwell assay. A representative image (upper) and statistical analysis are shown (lower). (h) The effect of cyclin A on PFDN1-induced anchorage-independent growth was examined by a colony-formation assay. A representative image (upper) and statistical analysis (lower) are shown.
The interplay between TGF-β1 signaling and PFDN1-cyclin A
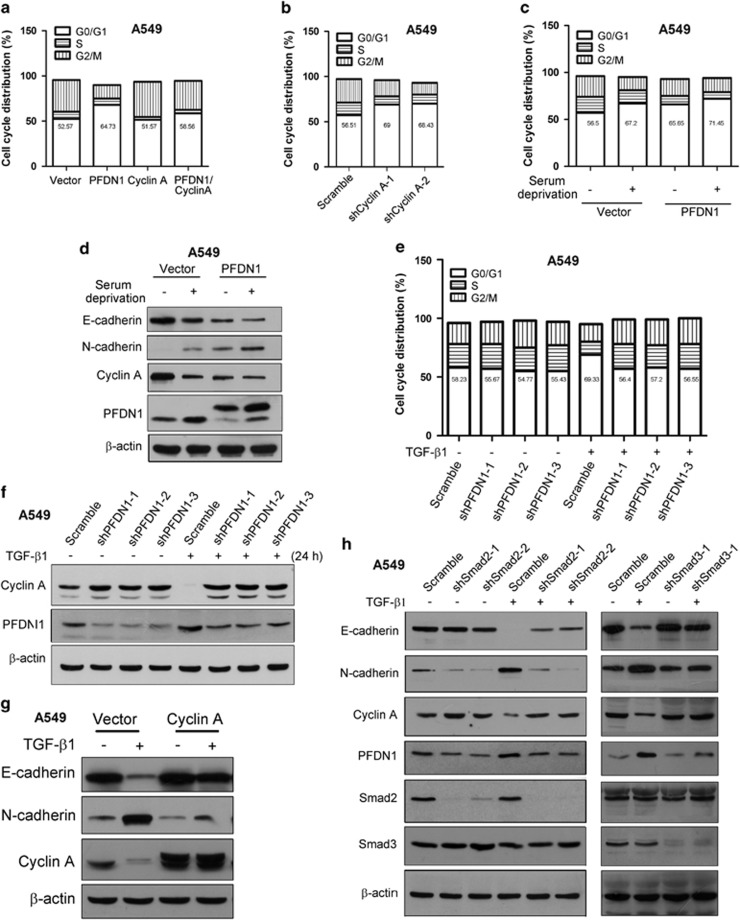
The upregulation of PFDN1 by TGF-β and the effect of PFDN1 in regulating cyclin A and EMT suggest that cell cycle regulation occurs by an interplay between TGF-β1 signaling and PFDN1/cyclin A. Further experiments showed that PFDN1 overexpression alone increased the percentage of cells in G0/G1 phase, whereas cyclin A overexpression blocked this effect (Figure 7a and Supplementary Figure S7a). In contrast, cyclin A knockdown increased the percentage of cells in G0/G1 phase (Figure 7b and Supplementary Figure S7b). PFDN1 overexpression further enhanced the percentage of cells in G0/G1 phase increased by serum starvation (Figure 7c). Consistently, an additive effect of serum starvation and PFDN1 overexpression on the downregulation of cyclin A and EMT induction was observed (Figure 7d). These results suggest that PFDN1 regulates cell cycle distribution and EMT through the inhibition of cyclin A. We also evaluated the effect of PFDN1 knockdown on the TGF-β1-induced increase in G0/G1 phase. PFDN1 knockdown abolished TGF-β1-induced cell cycle changes (Figure 7e) and blocked TGF-β-induced downregulation of cyclin A (Figure 7f). Cyclin A overexpression blocked TGF-β1-induced EMT (Figure 7g), indicating that PFDN1-cyclin A-mediated cell cycle regulation is a critical event controlled by TGF-β1. Further experiments showed that knockdown of either smad2 or smad3 inhibits TGF-β1-induced EMT and expression level changes of PFDN1 and cyclin A (Figure 7h), confirming the involvement of TGF-β1 signaling effectors. As a downstream molecule of TGF-β/Smad signaling, EMT induced by PFDN1 overexpression cannot be blocked by smad2 knockdown (Supplementary Figure S8).
Figure 7.
The interplay between TGF-β1 signaling and PFDN1/cyclin A. The percentage of cells in different phases of the cell cycle under the indicated experimental conditions was determined by flow-cytometric analysis (FACS) of DNA content (a–c and e). (a) The effect of PFDN1 and/or cyclin A overexpression on cell cycle distribution. (b) The effect of cyclin A knockdown on cell cycle distribution. (c) The effects of PFDN1 on cell cycle distribution in serum-starved cell cultures. PFDN1-overexpressed and vector-control A549 cells were serum-starved for 48 h. (d) The effect of PFDN1 on EMT induction and the expression of cyclin A in the absence of serum. The expression levels of cyclin A, PFDN1, E-cadherin and N-cadherin were examined by immunoblotting. (e) The effect of PFDN1 knockdown on the TGF-β1-induced increase in the percentage of cells in G0/G1 phase. Cells were treated with TGF-β1 (2 ng/ml) for 48 h, the percentage of cells in different cell cycles was determined by FACS assay. (f) The effect of PFDN1 knockdown on TGF-β1-induced downregulation of cyclin A levels. Cells were treated with TGF-β1 (2 ng/ml) for 24 h, and the levels of cyclin A and PFDN1 were detected by immunoblotting. (g) The effect of cyclin A overexpression on TGF-β1-induced EMT. The expression of E-cadherin, N-cadherin and cyclin A was determined by immunoblotting. (h) The effects of smad2 or smad3 knockdown on TGF-β1-induced EMT and changes in expression levels of PFDN1 and cyclin A. The levels of E-cadherin, N-cadherin, PFDN1, cyclin A, smad2 and smad3 were examined by immunoblotting.
Cyclin A suppresses PFDN1-induced tumor growth and metastasis
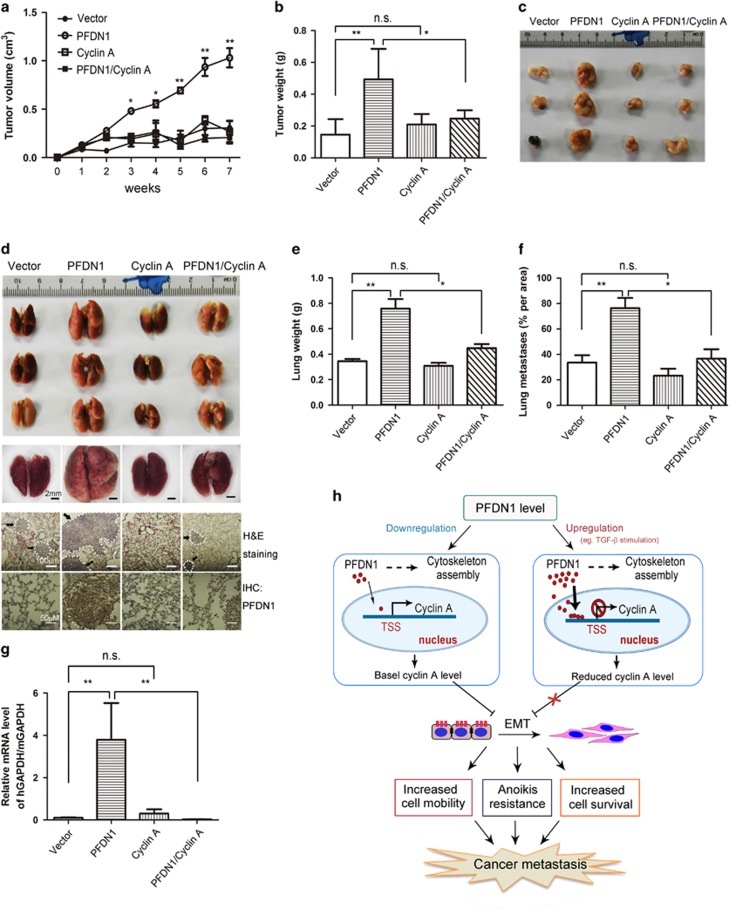
The implication of cyclin A in PFDN1-mediated EMT suggests a role of cyclin A in controlling lung tumor progression. Cyclin A overexpression abolished the PFDN1-mediated growth of xenograft tumors, as indicated by tumor volume (Figure 8a), tumor weight (Figure 8b) and gross tumor morphology (Figure 8c). In a model of metastatic lung cancer wherein animals were injected with tumor cells in the tail vein, cyclin A overexpression suppressed the growth of metastatic tumors induced by PFDN1 overexpression, as indicated by representative lung images and hematoxylin and eosin staining (Figure 8d), lung weight (Figure 8e) and relative metastasis area (Figure 8f). The PFDN1-mediated increase in CTCs was also abrogated by cyclin A overexpression (Figure 8g). To summarize our findings, a schematic illustration is provided (Figure 8h). The expression level of PFDN1 is significantly increased in lung tumors and in TGF-β1-induced EMT. PFDN1 can also be found in the cell nucleus, where it plays a role in transcriptional regulation. Nuclear PFDN1 binds and interacts with the cyclin A promoter at its transcriptional start site, which suppresses the expression of cyclin A and leads to EMT and increased invasion and metastatic growth of lung cancer cells.
Figure 8.
The effect of cyclin A on PFDN1-induced tumor growth and metastasis. (a–c) PFDN1- and cyclin A-overexpressed A549 cells were injected subcutaneously into nude mice. (a) Tumor volume was examined at the indicated time. (b) Seven weeks later, the mice were killed and the tumors were harvested and weighed. Data represent means±s.d. of three independent experiments. (c) A representative set of tumor images from three independent experiments is shown. (d–g) PFDN1- and/or cyclin A-overexpressed A549 cells were inoculated into nude mice by tail vein injection. Six weeks later, the mice were killed and the lungs were harvested. A representative set of lung images from three independent experiments is shown (d, upper). A magnified representative lung is shown under stereoscope (d, middle). The lung tissues were stained by H&E staining and PFDN1 was detected by immunohistochemistry (d, lower). Arrows indicate exemplary loci of metastasis. The effect of PFDN1 and /or cyclin A overexpression on lung weight of mice was examined (e). The metastasis per area was quantified using image software that measures pixel units (f). (g) Cyclin A overexpression abolished the PFDN1-mediated increase in CTCs. The relative level of CTCs was examined by real-time PCR. (h) A schematic illustration of PFDN1-mediated effects. PFDN1 is located in both the cytosol and nuclei of cells. A significant increase in the level of PFDN1 in lung cancers was observed. TGF-β1 can stimulate a marked increase in the expression levels of PFDN1 in lung cancer cells. Nuclear PFDN1 interacts with the promoter of cyclin A and suppresses its transcriptional expression, causing a decrease in cyclin A levels and relieving its suppression of EMT and the invasive migration and growth of lung cancer cells. TSS, transcriptional start site.
Discussion
PFDN1, a small molecule of 14 kDa, is an important subunit of the PFDN complex. Eukaryotic PFDN possesses two α subunits (PFDN3 and PFDN5) and four β subunits (PFDN1, PFDN2, PFDN4 and PFDN6).25 In addition to their roles in the PFDN complex, individual PFDN subunits have been shown to possess other functions. For instance, PFDN3, PFDN4 and PFDN5 have been identified as transcriptional regulators.24, 26, 27 PFDN3 and PFDN5 subunits have been shown to form complexes with proteins other than chaperones.28, 29 The involvement of different PFDN subunits in cancer-related biological events has been reported in the past several years. These subunits include the PFDN2, PFDN3, PFDN4, PFDN5 and PFDN6.15, 30, 31, 32, 33 As PFDN1 is significantly increased during TGF-β1-induced EMT in lung cancer cells, we hypothesized that PFDN1 may also be linked with lung cancer development and progression, and this was supported by our results. Our studies revealed that PFDN1 is an essential contributing factor in the transcriptional repression of cyclin A expression and the induction of the mesenchymal phenotype. Although it has been shown that PFDN subunits can have individual roles outside of the PFDN complex, it is not clear if the involvement of PFDN1 in tumor development and progression are complex-dependent.
Tumor growth is an outcome influenced by multiple factors, such as cell proliferation, cell survival, the tumor microenvironment and angiogenesis.34, 35, 36 Growth arrest is generally regarded as a precondition for cell differentiation and transdifferentiation.37 It has become increasingly clear that the induction of EMT generally presupposes cell growth arrest or decelerated proliferation, which is particularly prominent in TGF-β1-induced EMT in non-cancer cells. For instance, TGF-β1-induced EMT is highly correlated with growth arrest in Mv1Lu and AML12 cells.38, 39 The finding that PFDN1 induces EMT and inhibits cell proliferation also suggests a link between cell growth arrest and the induction of EMT. EMT also confers cells with anoikis resistance, which is important for anchorage-independent growth in soft agar and for the survival of CTCs in the blood. This is likely a major factor contributing to the increased metastatic growth of lung cancer cells induced by PFDN1 overexpression. The fact that PFDN1 promotes anchorage-independent proliferation, increased CTC levels, growth and the metastatic formation of lung tumors suggests that PFDN1 is a pro-survival factor for lung cancer cells.
The finding of PFDN1 in both the cytosol and nucleus and our data indicating that PFDN1 levels can be significantly increased by TGF-β1 during EMT induction imply that PFDN1 may have independent and different roles in the cytosol and nucleus. The cytoplasmic and nuclear functions of PFDN1 may also be coupled in a way as to favor cytoskeleton dynamics and cell homeostasis, which has been suggested by other published findings.40 It has been reported that dynamic reorganization of actin is linked with the morphology, migration and invasion of cancer cells.41, 42 Tubulin has been shown to be essential in cell–cell adhesion formation and EMT.43, 44 In addition, the nuclear function of PFDN in eukaryotes has been reported. Nuclear PFDN4 was found to act as a transcription factor.24 PFDN5 was shown to be involved in the transcriptional repression of c-MYC.27 The function of nuclear PFDN1 in yeast has been reported,45 but the function of nuclear PFDN1 in mammalian cells was previously unknown. This study revealed a role for nuclear PFDN1 as a transcriptional suppressor.
The findings that PFDN1 increases the viability of lung cancer cells and induces growth arrest at G0/G1 phase imply a relationship between the role of PFDN1 and cell cycle control, which is also consistent with the cell cycle-dependent induction of EMT by TGF-β1.39 It has been reported that the expression of cyclin A during G1 phase is very low and is induced upon entry into S phase.46, 47, 48 In addition, downregulation of cyclin A has been reportedly linked with cell cycle arrest at G0/G1, the resistance to anoikis and the control of cell motility and invasiveness.20, 49, 50 Thus, we hypothesized that PFDN1 acts through cyclin A to control EMT and the invasive and metastatic growth of lung cancer cells, which was verified by further investigations. The present study showed that PFDN1 downregulates cyclin A through transcriptional suppression of cyclin A expression. As a cell cycle regulator, cyclin A functions as a critical element in both TGF-β1- and PFDN1-mediated EMT and EMT-related lung cancer progression. As cyclin A can block PFDN1-induced growth, metastasis and tumorsphere formation of lung cancer cells, cyclin A may function as a potent suppressor of lung cancer progression. Cyclin A levels are significantly higher in human primary colorectal tumors than their corresponding metastases,19 which also suggest that cyclin A is a tumor suppressor. Because TGF-β1 induces sustained and significant increases in PFDN1 levels and TGF-β1-induced EMT is dependent on PFDN1, these results also suggest that PFDN1 can function in both TGF-β-dependent and -independent manners. Our data also suggest a novel molecular link between TGF-β signaling and lung cancer development and progression.
The findings that PFDN1 is highly expressed in several types of tumor tissues (Figure 1e) and breast cancer cells (Supplementary Figure S9) suggest that PFDN1 promotes lung cancer progression. The fact that PFDN1 is significantly increased in all the stages of lung cancer cells (Figure 1f) and that PFDN1 promotes tumor growth and lung cancer metastasis (Figure 4) suggest that PFDN1 is involved in the early stage of lung cancer development. PFDN1 overexpression induces the formation of larger tumorspheres of lung cancer cells (Supplementary Figure S10a and b), indicating that PFDN1 overexpression enhances the self-renewal property and tumorigenicity of lung cancer cells. Thus, PFDN1 may be used as prognostic marker for lung cancer.
In summary, our data show that PFDN1 is significantly increased in lung cancers. Increased expression of PFDN1 is also associated with TGF-β1-induced EMT and EMT-related changes in cell behavior. PFDN1 functions as a tumor promoter that stimulates the invasive growth and distal metastasis of lung cancer cells via EMT induction. PFDN1 was also detected in the nuclei of cancer cells. Nuclear PFDN1 binds with and suppresses the promoter activity of cyclin A, causing transcriptional downregulation of cyclin A expression. The suppression of cyclin A expression reduced the proliferation rate of lung cancer cells, promoted their EMT, and increased cell migration and resistance to anoikis, causing invasive growth and distal metastasis of lung cancer cells. Thus, PFDN1 may be an important candidate target molecule for potential consideration in clinic diagnosis and treatment of lung cancers.
Materials and methods
Cell culture
A549, H358, HCC827, H446 and H1299 human lung cancer cells and HEK293T cells were purchased from the American Type Culture Collection (Manassas, VA, USA). A549 and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (Thermo Scientific Hyclone, Rockford, IL, USA) containing 10% fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin (Life Technologies Gibco, Grand Island, NY, USA). H358, HCC827, H446 and H1299 were cultured in RPMI medium 1640 (Thermo Scientific Hyclone) containing the same supplements. All cells were incubated in a humidified atmosphere with 5% CO2 at 37°C.
Reagents
Human recombinant TGF-β1 was purchased from Chemicon (Millipore, Billerica, MA, USA). MG132 (M7449) and cycloheximide (46401) were purchased from Sigma (St Louis, MO, USA). Matrigel matrix (356231) was purchased from BD Biocoat (Franklin Lakes, NJ, USA). Lentivirus system plasmids pCDH-CMV-MCSEF1-Puro, Δ8.9, pVSVG, psPAX2, pMD2.G and pLKO.1-TRC were purchased from Addgene (Cambridge, MA, USA). Antibody information is listed in supplementary Table S1.
RNA isolation and real-time PCR
PCR assays were conducted as previously described.7 All values were normalized against the glyceraldehyde-3-phosphate dehydrogenase. The primers used for real-time PCR are listed in supplementary Table S2.
Plasmid construction and RNA interference assays
The primers used for amplification of homo PFDN1 and cyclin A are listed in supplementary table S3. Both human PFDN1 and cyclin A were generated by ligating the full-length open-reading frame into the pCDH-Puro-IRES-GFP vector (Addgene). For knockdown of PFDN1 and cyclin A, several independent small hairpin hRNA sequences targeting these proteins were ligated into pLKO.1 vectors (Addgene), with a scrambled small hairpin RNA sequence serving as the control. Small hairpin RNA sequences are listed in Supplementary Table S4.
Stable transfection
High-titer lentivirus was packaged in HEK293T cells using calcium chloride transfection. Viral supernatants were collected twice at 24 h intervals after transfection, filtered and applied to lung cancer cells in the presence of 10 μg/ml polybrene for 16 h. Cells were selected with puromycin for at least 1–2 weeks. Gene overexpression and gene knockdown were confirmed by immunoblotting using specific antibodies.
Preparation of cell lysates and western blotting
Cell lysates were prepared and western blots were performed as previously described.51 β-actin or β-tubulin was used as a loading control.
Preparation of subcellular fractionation
Subcellular fractionation was performed as previously described.51
Immunofluorescent staining
Cells were cultured on glass slides and immunofluorescent staining was performed as previously described.8
Immunohistochemistry staining
Thirty pairs of human lung carcinoma tissues and the relevant adjacent tissues were purchased from Outdo Biotechnology (Shanghai, China). The product ID of the tissue microarray is HLug-Ade 060PG-01. This assay was performed as previously described.8
Cell migration assays
A transwell assay and wound-healing assay were used to determine cell migration ability. The assays were performed as previously described.8
Cell invasive migration assay
The cell invasion assay was performed using Transwell migration chambers (8 μm pore size; Corning Costar, NY, USA) according to the vendor's instructions as previously described.52
Soft agar assay
A soft agar assay was used to determine the anchorage-independent growth of cells, as previously described.7
Animal experiments
Female athymic BALB/c nude mice (4–6 weeks of age) were purchased from Shanghai Laboratory Animal Center (Shanghai, China). All animal experiments were conducted as previously described,52 which are under the ethical guidelines of the Institute of Biochemistry and Cell Biology. Mice were randomly separated into each group, independent samples were collected from groups of at least five mice, and all experiments were performed at least three times. Animals were euthanized and excluded from the analysis when one of the following signs of morbidity was observed: significant tumor ulceration (>1 cm), severe injure that induce secondary infection and dehydration and inability to move, feed and defecate.
Detection of circulating tumor cells
The assay was performed as previously described.52 The relative amount of CTCs was examined by detecting the ratio of human glyceraldehyde-3-phosphate dehydrogenase expression to mouse glyceraldehyde-3-phosphate dehydrogenase expression by real-time quantitative PCR. The primers used for this assay are listed in supplementary Table S2.
Luciferase reporter assay
The human cyclin A promoter (containing 1 GC box and 1 TATAAT box) was cloned into the pGL3 basic firefly luciferase vector (Thermo Scientific) and then transfected into PFDN1-overexpressed cells. The specific primers for promoter sequences are listed in supplementary Table S5. Cell lysates were prepared 24 h after transfection in reporter lysis buffer (Promega, Madison, WI, USA) and luciferase substrate was added. The luciferase activity was recorded on a luminometer (Veritas, Promega).
Chromatin immunoprecipitation assay
The assay was performed with an EZ-Zyme Chromatin Prep Kit (Millipore), according to the manufacturer's protocol.52 Anti-PFDN1 antibody was used to precipitate DNA cross-linked with PFDN1, and normal rabbit IgG was used in parallel as a control. Enriched DNA was then used as a template to assess the binding intensity of PFDN1 to putative binding sites in the cyclin A promoter. Primers used in this assay are listed in supplementary Table S2.
Cell cycle analysis
Cells were trypsinized and gently collected and fixed with 70% ethanol overnight at −20 °C. After washing with phosphate-buffered saline containing 20 mM EDTA, cells were treated with 0.1 mg/ml RNase A for 1 h at 37 °C and were then stained with 0.1 mg/ml propidium iodide. The DNA content of cells was measured using flow cytometry (FACS Calibur, Becton Dickinson, Franklin Lakes, NJ, USA).
Ethical approval
All animal experiments were performed in accordance with a protocol approved by the Shanghai Experimental Animal Center of the Chinese Academy of Sciences (Approval Number: SIBCB-NAF-15-002-S305-021). The lung cancer tissue chip (Hlug-Ade 060PG-01) was purchased from Shanghai Outdo Biotechnology Company. The archived samples from Shanghai National Engineering Research Center for the biological chromatin Immunoprecipitation assay were anonymous, and informed consent was not required.
Statistical analysis
All samples used for statistical analysis were from at least three independent experiments performed in duplicates or up to five parallel controls. For experiments with two groups, statistical significance was determined by Student's t-test. For experiments with more than three groups, statistical analyses were performed with analysis of variance followed by post hoc pairwise comparisons. The data shown are means±s.d. of three independent experiments. The P-value of <0.05 was considered statistically significant. The P-values were designated as *P<0.05, ** P<0.01, *** P<0.005. n.s., non significant.
Acknowledgments
This study was supported by the Natural Science Foundation of China (81472603). We thank Xinwang Yuan, Xiaojie Guo and Guoqing Cai for helpful discussion and technical assistance. We are especially grateful to Dr Lingling Chen for generous support to this work.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016; 66: 7–30. [DOI] [PubMed] [Google Scholar]
- Katsuno Y, Lamouille S, Derynck R. TGF-beta signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol 2013; 25: 76–84. [DOI] [PubMed] [Google Scholar]
- Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol 2011; 27: 347–376. [DOI] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15: 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang DM, Wang CM, Hu Y, Liu AH, Zhang YL et al. Insulin receptor substrate-1 suppresses transforming growth factor-beta1-mediated epithelial-mesenchymal transition. Cancer Res 2009; 69: 7180–7187. [DOI] [PubMed] [Google Scholar]
- Tang YN, Shu GW, Yuan XW, Jing NH, Song JG. FOXA2 functions as a suppressor of tumor metastasis by inhibition of epithelial-to-mesenchymal transition in human lung cancers. Cell Res 2011; 21: 316–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan XW, Wang DM, Hu Y, Tang YN, Shi WW, Guo XJ et al. Hepatocyte nuclear factor 6 suppresses the migration and invasive growth of lung cancer cells through p53 and the inhibition of epithelial-mesenchymal transition. J Biol Chem 2013; 288: 31206–31216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, He K, Wang DM, Yuan XW, Liu Y, Ji HB et al. TMEPAI regulates EMT in lung cancer cells by modulating the ROS and IRS-1 signaling pathways. Carcinogenesis 2013; 34: 1764–1772. [DOI] [PubMed] [Google Scholar]
- Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene 2013; 32: 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Xiong G, Fu H, Evers BM, Zhou BP, Xu R. Chaperone Hsp47 drives malignant growth and invasion by modulating an ECM gene network. Cancer Res 2015; 75: 1580–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota M, Bishop JL, Nip KM, Zardan A, Takeuchi A, Cordonnier T et al. Hsp27 regulates epithelial mesenchymal transition, metastasis, and circulating tumor cells in prostate cancer. Cancer Res 2013; 73: 3109–3119. [DOI] [PubMed] [Google Scholar]
- Shiota M, Zardan A, Takeuchi A, Kumano M, Beraldi E, Naito S et al. Clusterin mediates TGF-beta-induced epithelial-mesenchymal transition and metastasis via Twist1 in prostate cancer cells. Cancer Res 2012; 72: 5261–5272. [DOI] [PubMed] [Google Scholar]
- Kubota H, Hynes G, Carne A, Ashworth A, Willison K. Identification of six Tcp-1-related genes encoding divergent subunits of the TCP-1-containing chaperonin. Curr Biol 1994; 4: 89–99. [DOI] [PubMed] [Google Scholar]
- Farooq M, Hozzein WN, Elsayed EA, Taha NA, Wadaan MA. Identification of histone deacetylase 1 protein complexes in liver cancer cells. Asian Pac J Cancer Prev 2013; 14: 915–921. [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Mimori K, Nishida N, Tokuoka M, Akita H et al. Abnormal expression of PFDN4 in colorectal cancer: a novel marker for prognosis. Ann Surg Oncol 2010; 17: 3030–3036. [DOI] [PubMed] [Google Scholar]
- Cao S, Carlesso G, Osipovich AB, Llanes J, Lin Q, Hoek KL et al. Subunit 1 of the prefoldin chaperone complex is required for lymphocyte development and function. J Immunol 2008; 181: 476–484. [DOI] [PubMed] [Google Scholar]
- Lundin VF, Srayko M, Hyman AA, Leroux MR. Efficient chaperone-mediated tubulin biogenesis is essential for cell division and cell migration in C. elegans. Dev Biol 2008; 313: 320–334. [DOI] [PubMed] [Google Scholar]
- Bendris N, Cheung CT, Leong HS, Lewis JD, Chambers AF, Blanchard JM et al. Cyclin A2, a novel regulator of EMT. Cell Mol Life Sci 2014; 71: 4881–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsic N, Bendris N, Peter M, Begon-Pescia C, Rebouissou C, Gadea G et al. A novel function for Cyclin A2: control of cell invasion via RhoA signaling. J Cell Biol 2012; 196: 147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CT, Bendris N, Paul C, Hamieh A, Anouar Y, Hahne M et al. Cyclin A2 modulates EMT via beta-catenin and phospholipase C pathways. Carcinogenesis 2015; 36: 914–924. [DOI] [PubMed] [Google Scholar]
- Munnia A, Schutz N, Romeike BF, Maldener E, Glass B, Maas R et al. Expression, cellular distribution and protein binding of the glioma amplified sequence (GAS41), a highly conserved putative transcription factor. Oncogene 2001; 20: 4853–4863. [DOI] [PubMed] [Google Scholar]
- Siegert R, Leroux MR, Scheufler C, Hartl FU, Moarefi I. Structure of the molecular chaperone prefoldin: unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell 2000; 103: 621–632. [DOI] [PubMed] [Google Scholar]
- Martin-Benito J, Boskovic J, Gomez-Puertas P, Carrascosa JL, Simons CT, Lewis SA et al. Structure of eukaryotic prefoldin and of its complexes with unfolded actin and the cytosolic chaperonin CCT. EMBO J 2002; 21: 6377–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima M, Kano Y, Nohno T, Namba M. Cloning of cDNA with possible transcription factor activity at the G1-S phase transition in human fibroblast cell lines. Acta Med Okayama 1996; 50: 73–77. [DOI] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL et al. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 1998; 93: 863–873. [DOI] [PubMed] [Google Scholar]
- Kim SY, Kim JC, Kim JK, Kim HJ, Lee HM, Choi MS et al. Hepatitis B virus X protein enhances NFkappaB activity through cooperating with VBP1. BMB Rep 2008; 41: 158–163. [DOI] [PubMed] [Google Scholar]
- Satou A, Taira T, Iguchi-Ariga SM, Ariga H. A novel transrepression pathway of c-Myc. Recruitment of a transcriptional corepressor complex to c-Myc by MM-1, a c-Myc-binding protein. J Biol Chem 2001; 276: 46562–46567. [DOI] [PubMed] [Google Scholar]
- Narita R, Kitaura H, Torii A, Tashiro E, Miyazawa M, Ariga H et al. Rabring7 degrades c-Myc through complex formation with MM-1. PLoS One 2012; 7: e41891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya H, Iseda T, Hino O. Identification of a novel protein (VBP-1) binding to the von Hippel-Lindau (VHL) tumor suppressor gene product. Cancer Res 1996; 56: 2881–2885. [PubMed] [Google Scholar]
- Alldinger I, Dittert D, Peiper M, Fusco A, Chiappetta G, Staub E et al. Gene expression analysis of pancreatic cell lines reveals genes overexpressed in pancreatic cancer. Pancreatology 2005; 5: 370–379. [DOI] [PubMed] [Google Scholar]
- Asperger A, Renner C, Menzel M, Gebhardt R, Meixensberger J, Gaunitz F. Identification of factors involved in the anti-tumor activity of carnosine on glioblastomas using a proteomics approach. Cancer Invest 2011; 29: 272–281. [DOI] [PubMed] [Google Scholar]
- Lopez V, Gonzalez-Peramato P, Suela J, Serrano A, Algaba F, Cigudosa JC et al. Identification of prefoldin amplification (1q23.3-q24.1) in bladder cancer using comparative genomic hybridization (CGH) arrays of urinary DNA. J Transl Med 2013; 11: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrov DA, Barnes CL, Smith LE, Binns S, Brusko TM, Brown AC et al. Characterization of HKE2: an ancient antigen encoded in the major histocompatibility complex. Tissue Antigens 2007; 69: 181–188. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Ma X, Aoki T, Tsuruyama T, Narumiya S. Definition of Prostaglandin E2-EP2 signals in the colon tumor microenvironment that amplify inflammation and tumor growth. Cancer Res 2015; 75: 2822–2832. [DOI] [PubMed] [Google Scholar]
- Yin X, Luistro L, Zhong H, Smith M, Nevins T, Schostack K et al. RG7212 anti-TWEAK mAb inhibits tumor growth through inhibition of tumor cell proliferation and survival signaling and by enhancing the host antitumor immune response. Clin Cancer Res 2013; 19: 5686–5698. [DOI] [PubMed] [Google Scholar]
- Adler JJ, Johnson DE, Heller BL, Bringman LR, Ranahan WP, Conwell MD et al. Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases. Proc Natl Acad Sci USA 2013; 110: 17368–17373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lei WW, Wang XD, Tang YN, Song JG. Glucocorticoid induces mesenchymal-to-epithelial transition and inhibits TGF-β1-induced epithelial-to-mesenchymal transition and cell migration. FEBS Letters 2010; 584: 4646–4654. [DOI] [PubMed] [Google Scholar]
- Yang YN, Pan XC, Lei WW, Wang J, Song JG. Transforming growth factor-beta1 induces epithelial-to-mesenchymal transition and apoptosis via a cell cycle-dependent mechanism. Oncogene 2006; 25: 7235–7244. [DOI] [PubMed] [Google Scholar]
- Millan-Zambrano G, Chavez S. Nuclear functions of prefoldin. Open Biol 2014; 4: 140085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberg A, Kitzing T, Grosse R. Nucleating actin for invasion. Nat Rev Cancer 2011; 11: 177–187. [DOI] [PubMed] [Google Scholar]
- Yilmaz M, EMT Christofori G. the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev 2009; 28: 15–33. [DOI] [PubMed] [Google Scholar]
- Barcellos KS, Bigarella CL, Wagner MV, Vieira KP, Lazarini M, Langford PR et al. ARHGAP21 protein, a new partner of alpha-tubulin involved in cell-cell adhesion formation and essential for epithelial-mesenchymal transition. J Biol Chem 2013; 288: 2179–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple RA, Matrone MA, Cho EH, Balzer EM, Vitolo MI, Yoon JR et al. Epithelial-to-mesenchymal transition promotes tubulin detyrosination and microtentacles that enhance endothelial engagement. Cancer Res 2010; 70: 8127–8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan-Zambrano G, Rodriguez-Gil A, Penate X, de Miguel-Jimenez L, Morillo-Huesca M, Krogan N et al. The prefoldin complex regulates chromatin dynamics during transcription elongation. PLoS Genet 2013; 9: e1003776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henglein B, Chenivesse X, Wang J, Eick D, Brechot C. Structure and cell cycle-regulated transcription of the human cyclin A gene. Proc Natl Acad Sci USA 1994; 91: 5490–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs HW, Keidel E, Lehner CF. A complex degradation signal in Cyclin A required for G1 arrest, and a C-terminal region for mitosis. EMBO J 2001; 20: 2376–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg AR, Zindy F, Le Deist F, Mouly H, Metezeau P, Brechot C et al. Overexpression of human cyclin A advances entry into S phase. Oncogene 1995; 10: 1501–1509. [PubMed] [Google Scholar]
- Kim HR, Lin HM, Biliran H, Raz A. Cell cycle arrest and inhibition of anoikis by galectin-3 in human breast epithelial cells. Cancer Res 1999; 59: 4148–4154. [PubMed] [Google Scholar]
- Wang D, Sun SQ, Yu YH, Wu WZ, Yang SL, Tan JM. Suppression of SCIN inhibits human prostate cancer cell proliferation and induces G0/G1 phase arrest. Int J Oncol 2014; 44: 161–166. [DOI] [PubMed] [Google Scholar]
- Wang DM, Zhou YR, Lei WW, Zhang KH, Shi J, Hu Y et al. Signal transducer and activator of transcription 3 (STAT3) regulates adipocyte differentiation via peroxisome-proliferator-activated receptor gamma (PPARgamma). Biol Cell 2010; 102: 1–12. [DOI] [PubMed] [Google Scholar]
- Tang YN, Ding WQ, Guo XJ, Yuan XW, Wang DM, Song JG. Epigenetic regulation of Smad2 and Smad3 by profilin-2 promotes lung cancer growth and metastasis. Nat Commun 2015; 6: 8230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.