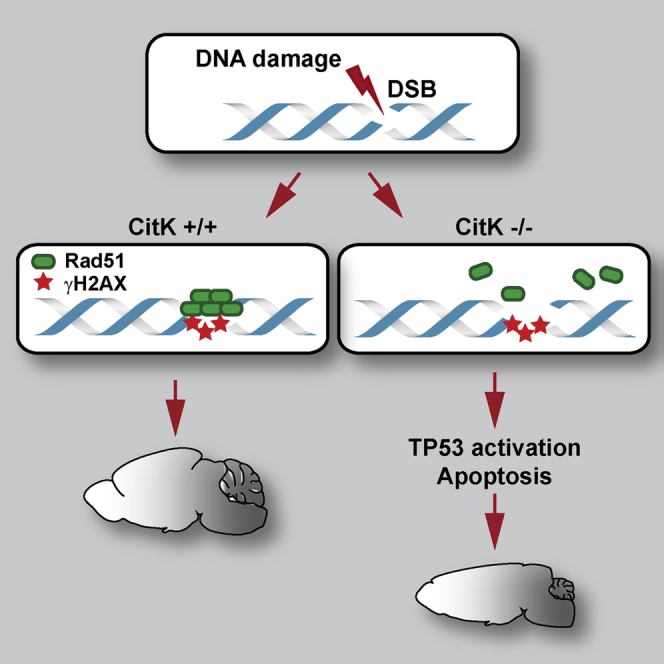
Summary
Mutations in citron (CIT), leading to loss or inactivation of the citron kinase protein (CITK), cause primary microcephaly in humans and rodents, associated with cytokinesis failure and apoptosis in neural progenitors. We show that CITK loss induces DNA damage accumulation and chromosomal instability in both mammals and Drosophila. CITK-deficient cells display “spontaneous” DNA damage, increased sensitivity to ionizing radiation, and defective recovery from radiation-induced DNA lesions. In CITK-deficient cells, DNA double-strand breaks increase independently of cytokinesis failure. Recruitment of RAD51 to DNA damage foci is compromised by CITK loss, and CITK physically interacts with RAD51, suggesting an involvement of CITK in homologous recombination. Consistent with this scenario, in doubly CitK and Trp53 mutant mice, neural progenitor cell death is dramatically reduced; moreover, clinical and neuroanatomical phenotypes are remarkably improved. Our results underscore a crucial role of CIT in the maintenance of genomic integrity during brain development.
Keywords: citron kinase, microcephaly, genome instability, cytokinesis, DNA damage, double strand breaks, RAD51, apoptosis, TP53
Graphical Abstract
Highlights
-
•
CITK is required for chromosome integrity independently of its role in cytokinesis
-
•
CITK binds RAD51 and is required for its normal recruitment to DSBs
-
•
Citron kinase loss leads to TP53-dependent apoptosis of cortical neural progenitors
-
•
TP53 inactivation rescues CITK-dependent microcephaly, ataxia, and epilepsy
Mutations leading to inactivation of the citron kinase protein (CITK) cause primary microcephaly in humans and rodents. Bianchi et al. find a conserved function for CITK in ensuring genomic stability and show that the neurological phenotype caused by CITK loss can be largely reverted by concomitant inactivation of the TP53 gene.
Introduction
Congenital microcephaly (CM) is a heterogeneous group of disorders characterized by reduced head circumference at birth to at least three SDs below the mean (Barbelanne and Tsang, 2014, Cox et al., 2006, Mahmood et al., 2011, Passemard et al., 2013, Woods and Parker, 2013). CM can be the result of non-genetic conditions, such as viral infections or toxic exposure, or can be generated by rare genetic disorders, usually characterized by autosomal recessive inheritance (Passemard et al., 2013). For example, a severe CM has been observed in individuals bearing mutations in genes involved in DNA repair, centrosome function, microtubule organization, and spindle orientation (Faheem et al., 2015, Morris-Rosendahl and Kaindl, 2015). In primary hereditary microcephaly (MCPH), brain volume reduction is the main clinical phenotype, associated with conserved brain architecture and mild to moderate intellectual disability (Kaindl et al., 2009, Passemard et al., 2013).
Recent studies have identified mutations of the CIT gene, leading to loss or inactivation of the citron rho-interacting serine/threonine kinase (henceforth designated as CITK) in patients affected by severe microlissencephaly (Basit et al., 2016, Harding et al., 2016, Li et al., 2016, Shaheen et al., 2016), with strong neuron depletion and a high frequency of binucleated cells (Harding et al., 2016). Consistent with these findings, previous reports had demonstrated that CITK loss in mice and rats (CitK −/−) causes a dramatic form of microcephaly, disproportionate with respect to body size, and an association with ataxia and epilepsy leading to perinatal death (Di Cunto et al., 2000, Sarkisian et al., 2002). In these animal models, neuronal loss is primarily due to massive apoptosis, occurring throughout brain development (Ackman et al., 2007, Di Cunto et al., 2000, Sarkisian et al., 2002, Sgrò et al., 2016).
CITK is a conserved protein involved in midbody maturation and cytokinetic abscission, from insects to mammals (D’Avino et al., 2004, Naim et al., 2004). In CitK −/− mice, cytokinesis failure occurs with high penetrance in neuronal progenitors (Di Cunto et al., 2000, Gai et al., 2011), producing binucleated and polyploid cells. It has therefore been postulated that apoptosis from CITK loss is a consequence of impaired cytokinesis (Di Cunto et al., 2000). However, studies performed in many different models indicate that cytokinesis failure leads more frequently to cell cycle arrest than apoptosis (Ganem et al., 2014, Panopoulos et al., 2014). Thus, it is possible that cytokinesis failure and apoptosis are two independent consequences of CITK loss. In this study, we investigated the basis of apoptosis induction in CitK −/− mice.
Results
CITK Is Required for DNA Integrity in Developing Mouse Brain
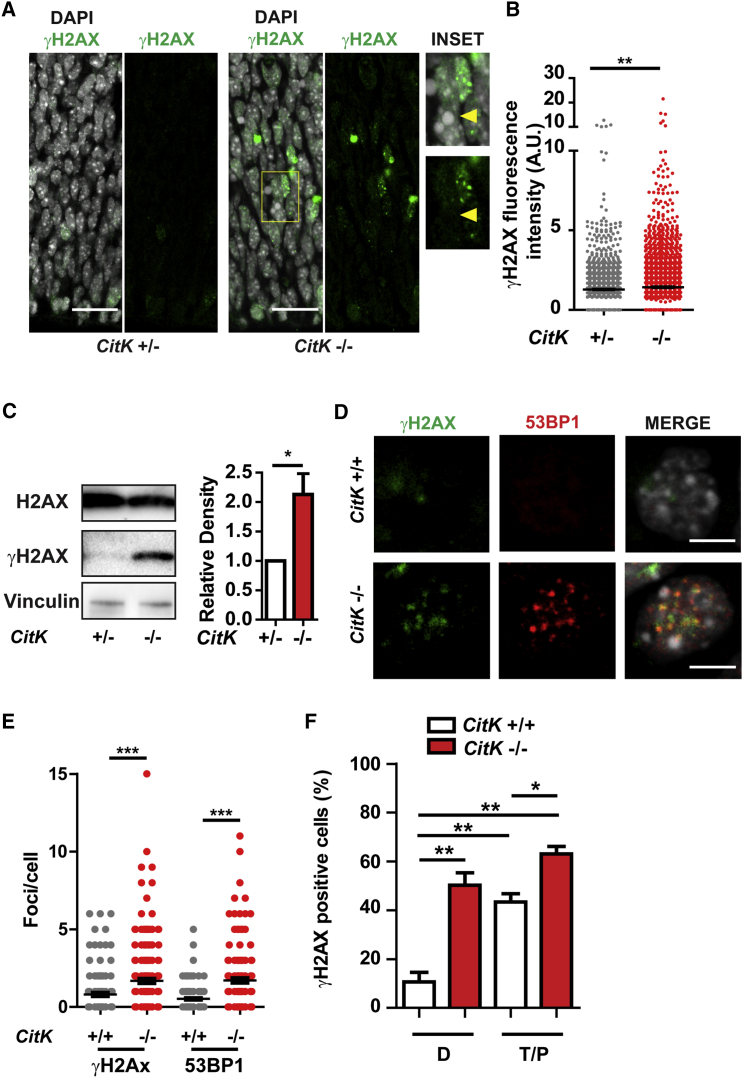
Because, in microcephaly models, apoptosis has frequently been associated with DNA damage (Passemard et al., 2013) and because DNA damage seems to occur at increased frequency in polyploid cells (Zhang et al., 2013), we asked whether developing brains of CitK −/− mice show more DNA lesions than littermate controls. To detect these lesions, we used quantitative immunofluorescence to visualize nuclear foci of phosphorylated histone H2AX (γH2AX), an established marker of DNA damage (Turinetto and Giachino, 2015). This analysis revealed that the developing neocortex of CitK −/− mice exhibits a significant increase in cells with high content of γH2AX foci (Figures 1A and 1B). At embryonic day 14.5 (E14.5), frequent γH2AX-positive cells are found in ventricular and subventricular regions (Figure 1A), which contain most cortical mitotic cells, but also in the intermediate region and cortical plate (not shown), which contain post-mitotic and differentiating cells. Notably, most cortical cells with bright nuclear γH2AX do not show apoptotic morphology, whereas the majority of apoptotic nuclei are γH2AX negative (Figure 1A), suggesting that the increase of γH2AX is not a secondary effect of apoptosis induction. Western blot analysis showed that CitK −/− mice have significantly increased levels of γH2AX in the developing cerebellum (Figure 1C), which is the tissue most severely affected by CITK loss (Di Cunto et al., 2000). In addition, developing cortices of CitK −/− mice display a significant increase of nuclei with 53BP1 foci (Figure S1A), suggesting that the accumulation of γH2AX may derive from increased DNA double-strand breaks (DSBs) (Schultz et al., 2000).
Figure 1.
CITK Prevents DNA Damage Accumulation in the Developing Nervous System
(A) Cerebral cortex sections of E14.5 mouse embryos stained anti-γH2AX antibodies and DAPI. The scale bars represent 25 μm.
(B) Quantification of γH2AX fluorescence intensity per cell in sections obtained as in (A).
(C) Western blots of cell lysates from P4 cerebella of CitK −/− and control mice (+/+ and +/−) probed with anti-H2AX and anti-γH2AX antibodies. Quantification of the γH2AX/H2AX ratio is shown.
(D) NPCs from E12.5 embryo cortices stained for γH2AX (green), 53BP1 (red), and DNA (DAPI, blue). The scale bars represent 10 μm.
(E) Quantification of γH2AX foci or 53BP1 foci per cell in NPCs.
(F) Cells stained as in (D) were classified as diploid (D) or tetraploid/polyploid (T/P) by quantification of the DAPI signal (see Experimental Procedures) and as γH2AX positive if they displayed more than five foci/cell.
Two tails unpaired Student’s t test was used for the statistical analysis of these experiments (n = 3–6 per group). Graphs show mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
To elucidate the relationship between DSB accumulation and cytokinesis failure in CitK −/− brains, we examined cultures of cortical neural progenitor cells (NPCs) obtained from E12.5 embryos. Compared to the developing tissue, these cells allow better analysis of γH2AX and 53BP1 foci, as well as definition of nuclear morphology and DNA content. CitK −/− NPCs show a significant increase of γH2AX and 53BP1 foci compared to CitK +/+ controls (Figures 1D and 1E). Moreover, most 53BP1 foci are also positive for γH2AX staining (Figure 1D), consistent with an increase in DSBs frequency in CitK-depleted cells. We next asked whether DNA damage accumulation in CitK −/− NPCs correlates with cytokinesis failure. As previously observed in vivo (Di Cunto et al., 2000), NPC cultures from CitK −/− embryos show a high frequency of binucleated cells and of cells with large nuclei and an increased DNA content, which are likely to result from one or multiple rounds of cytokinesis failures (Figures S1B and S1C). In control cells, the fraction of nuclei with more than five γH2AX foci is significantly higher in tetraploid/polyploid cells compared to diploid cells (Figure 1F). Moreover, tetraploid/polyploid cells of CitK −/− cultures show an even higher prevalence of γH2AX positivity compared to tetraploid/polyploid controls (Figure 1F). Interestingly enough, diploid CitK −/− cells also show a significant increase in the frequency of γH2AX-positive nuclei compared to diploid control cells (Figure 1F). To further elucidate this finding, we analyzed the prevalence of γH2AX-positive nuclei in post-mitotic neurons and in proliferating progenitors, identified by immunostaining with the markers TuJ (TUBB3) and Nestin, respectively (Figures S1D–S1G). This analysis revealed that 58% of CitK −/− diploid precursors and 34% of CitK −/− diploid neurons show more than five foci per nucleus, whereas in the diploid control cells, these frequencies are 17% and 8%, respectively (Figures S1D–S1G). Thus, in developing CitK −/− NPCs, a significant accumulation of DNA lesions occurs in both proliferating and differentiating diploid cells. Because mouse diploid cells are most likely generated by normal cytokinesis, these results suggest that a large fraction of the DSBs burden that characterizes CitK −/− neural progenitors is unrelated to cytokinesis failure.
The Role of CITK in Maintenance of Genome Integrity Is Phylogenetically Conserved and Unrelated to Cytokinesis Control
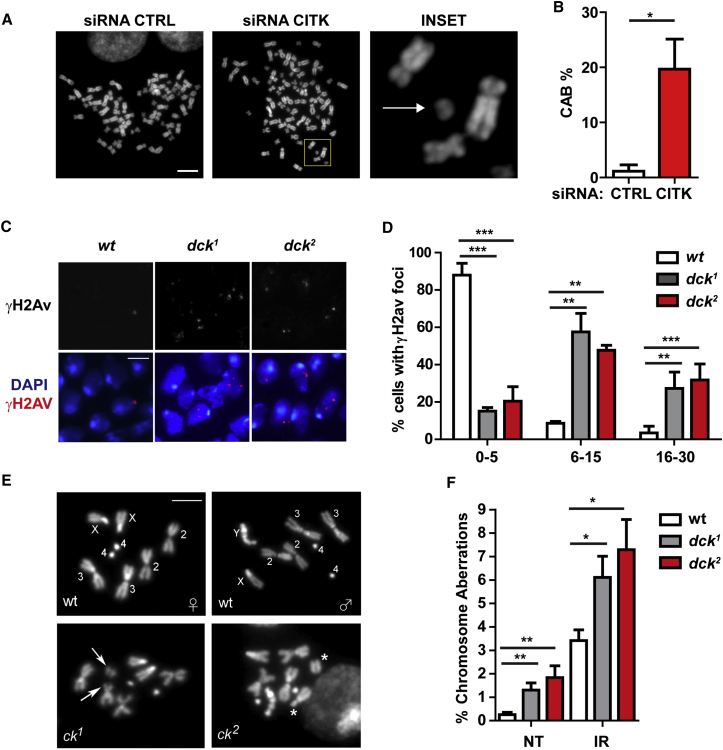
To assess whether CITK loss leads to increased DNA damage also in human cells, we used RNAi to induce CITK depletion in the TP53-proficient medulloblastoma cell line ONS-76 (Moriuchi et al., 1997). We chose these cells because they closely resemble cerebellar granule progenitors, which are severely affected by CITK loss also in humans (Harding et al., 2016). Similar to mouse NPCs, CITK-depleted ONS-76 cells (Figure S2A) display significant increase in γH2AX foci (Figure S2B), as compared to cells expressing control small interfering RNA (siRNA). To evaluate whether the DNA damage detected by γH2AX accumulation leads to chromosome instability, we analyzed metaphase chromosome of CITK-depleted and control ONS-76 cells 48 hr after siRNA transfection. Metaphases of cells transfected with control siRNA show a hyper-triploid chromosome number (range between 74 and 80 chromosomes), with low frequency of chromosome aberrations (CABs) (Figures 2A and 2B). In contrast, metaphases of CITK-depleted ONS-76 cells bearing a chromosome number similar to control cells (which therefore did not fail cytokinesis in the previous mitosis) show a significant increase of CABs (Figures 2A and 2B). This result further indicates that CITK loss favors the accumulation of DSBs independently of cytokinesis failure.
Figure 2.
CITK Has a Phylogenetically Conserved Role in Maintenance of Genome Integrity
(A) DAPI-stained metaphase spreads from control and CITK-depleted ONS-76 cells. An acentric isochromatid fragment is indicated in the inset (arrow).
(B) Frequencies of CABs in mock-treated or CITK-depleted ONS-76 metaphases.
(C) Drosophila brain squashes from wild-type (wt) dck1 and dck2 mutant larvae stained with anti-γH2Av antibodies and DAPI.
(D) Frequencies of negative (0–5 γH2Av foci/cell), moderately positive (6–15 foci/cell), or strongly positive (>15 foci/cell) cells in the samples shown in (C).
(E) Diploid DAPI-stained metaphase spreads from control (wt) and mutant (dck1 and dck2) Drosophila larval brains showing broken chromosomes (arrows and stars).
(F) Frequencies of chromosome aberrations (CABs) in untreated (NT) or irradiated (IR) (4Gy of X-rays) diploid larval brain cells from wild-type or dck mutant larvae.
The scale bars in (A) and (C) represent 5 μm. Two tails unpaired Student’s t test was used for the statistical analysis of these experiments (n = 3–4 per group). Graphs show mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
As long as CITK serves an evolutionarily conserved function in the control of final cytokinesis events (D’Avino et al., 2004, Gai et al., 2011, Naim et al., 2004), we asked whether its protective role from DNA damage is also conserved. We thus investigated Drosophila larval brains for the presence of DNA damage foci by immunostaining for the histone variant γH2Av (the Drosophila ortholog of mammalian γH2AX). We compared wild-type larval brains with brains from larvae homozygous for either dck1 or dck2, two mutant alleles in the Drosophila citron kinase gene (also called sticky; see FlyBase; D’Avino et al., 2004, Naim et al., 2004). Because both mutations cause cytokinesis failures, in our comparative analyses, we only considered mutant nuclei that were likely to be diploid. In dck1 and dck2 mutant brains, diploid nuclei with six or more γH2Av foci are significantly more frequent (84% and 80%, respectively) compared to wild-type (12%; Figures 2C and 2D). To confirm and extend these results, we examined diploid metaphases from both wild-type and dck mutant brains for the presence of CABs. Consistent with our previous results (Marzio et al., 2014, Mengoli et al., 2014), wild-type brains show a very low CABs frequency (∼0.003/metaphase). In contrast, brains from larvae homozygous for either dck mutant allele displayed a 4-fold increase in the CABs frequency compared to controls (Figures 2E and 2F). Remarkably, the observed increases in the CABs frequencies are comparable with those seen for DNA repair foci. Because both γH2Av repair foci and CABs are thought to be the generated by DSBs (Durante et al., 2013, Polo and Jackson, 2011), it is likely that wild-type dck protects cells from this type of DNA lesion.
The finding that increases in both foci and CABs are found in diploid cells rules out the possibility that they are the consequence of defects in cytokinesis. Consistent with this conclusion, diploid cells from larval brains of other Drosophila mutants strongly defective in cytokinesis did not show any increase in CABs compared to wild-type controls. For example, in mutants in spaghetti squash (sqh) (encoding a regulatory light chain of the nonmuscle type 2 myosin; Karess et al., 1991) or twinstar (tsr) (that encodes a Drosophila cofilin/ADF homolog; Gunsalus et al., 1995), the frequencies of CABs were not significantly different from controls (Figure 2F; 0.003 and 0.002 CABs/cell for sqh and tsr, respectively; 500 metaphases from at least eight brains examined in each mutant).
To obtain further insight into the mechanisms underlying generation of DNA damage in dck mutants, we asked whether they are sensitive to X-rays, which directly induce DNA breaks. Following irradiation (IR) (2.5 Gy), diploid cells of dck mutants show a 2-fold increase in the CABs frequency compared to controls (Figure 2F). Collectively, these results suggest that dck is involved in repair of DBSs and indicate that this role in maintenance of genome integrity is independent of its well-known function in cytokinesis completion (Bassi et al., 2011, D’Avino et al., 2004, Gai et al., 2011, Naim et al., 2004).
CITK Is Critical for Recovery from DNA Damage and for RAD51 Recruitment at DSBs
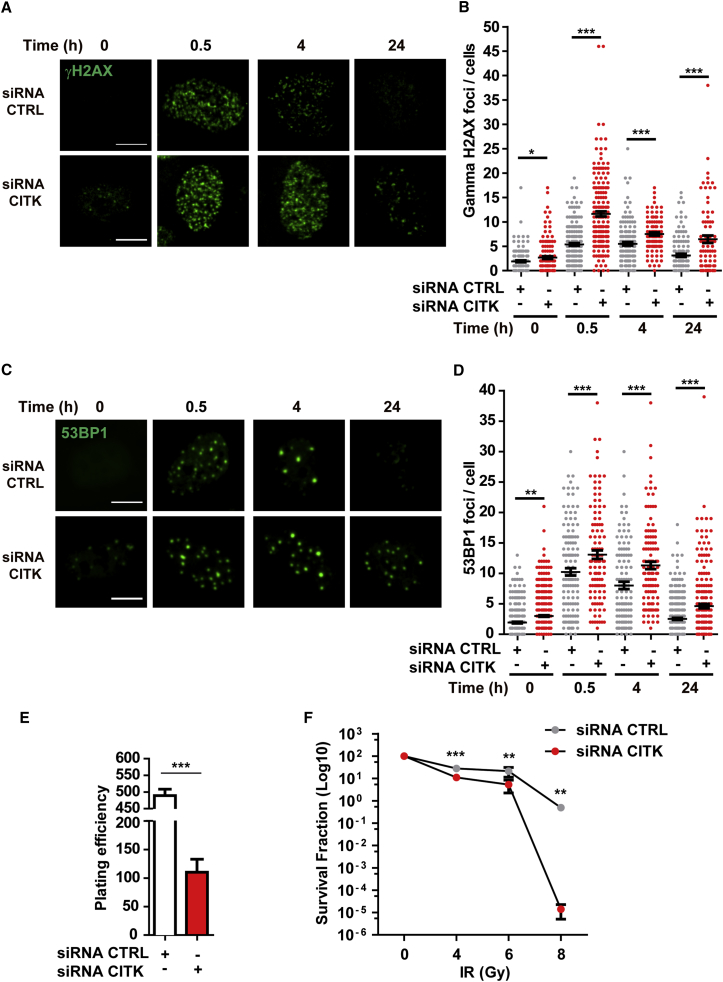
To further analyze the role of CITK in DNA damage response, we used HeLa cells, which are known to be sensitive to CITK depletion (Gai et al., 2011, McKenzie and D’Avino, 2016). We found that RNAi-mediated CITK deficiency increases the number of γH2AX and 53BP1 foci in mononucleated HeLa cells, although to a lesser extent than in NPCs (Figures 3A–3D). DSBs accumulation is already detectable 48 hr after siRNA transfection, a time at which CITK is depleted (Gai et al., 2011), but the cell cycle profile is not yet perturbed by the occurrence of cytokinesis failure (Figure S2D) and accumulation of cells in mitosis (Figure S2E).
Figure 3.
CITK-Depleted HeLa Cells Display Reduced DNA Damage Recovery and Enhanced Radiosensitivity
(A) Representative nuclei of HeLa cells transfected with mock siRNA (CTRL) or with CITK-specific siRNAs; cells were irradiated with 4 Gy and stained with anti-γH2AX antibodies at the indicated post-irradiation times.
(B) Quantification of γH2AX fluorescence intensity per cell in (A).
(C) Representative nuclei of HeLa cells treated as in (A) and stained with anti-53BP1 antibodies at the indicated post-irradiation times.
(D) Quantification of γH2AX fluorescence intensity per cell in (C).
(E) Plating efficiency of control and CITK-depleted HeLa.
(F) Clonogenic survival assay of control and CITK-depleted HeLa cells irradiated with the indicated IR dosage.
Two tails unpaired Student’s t test was used for the statistical analysis of these experiments (n = 3–7 per group). Graphs show mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
To investigate the role of CITK in the recovery from DNA damage induced by ionizing radiations (IRs), control and CITK-depleted HeLa cells were irradiated (4 Gy) and analyzed over 24 hr for the formation and dissolution of γH2AX and 53BP1 foci. After 30 min post-IR, CITK-depleted cells showed a significantly higher frequency of foci compared to mock-treated cells (Figures 3A–3D). Moreover, DNA-damage foci resolved with a slower kinetic than in controls (Figures 3A–3D). The increased sensitivity of CITK-depleted HeLa cells to IR seems not to be affected by cell cycle stage, as increased frequency of foci is observed in both cyclin-B1-negative cells (which are mostly in G1/S phase) and cyclin-B1-positive cells (which are mostly in G2; Figures S2F and S2G). CITK-depleted cells showed reduced plating efficiency in a clonogenic survival assay (Figure 3E), as well as a reduced growth potential when treated with different doses of X-rays (Figure 3F). Altogether, these results are consistent with a role of CITK in DSBs repair.
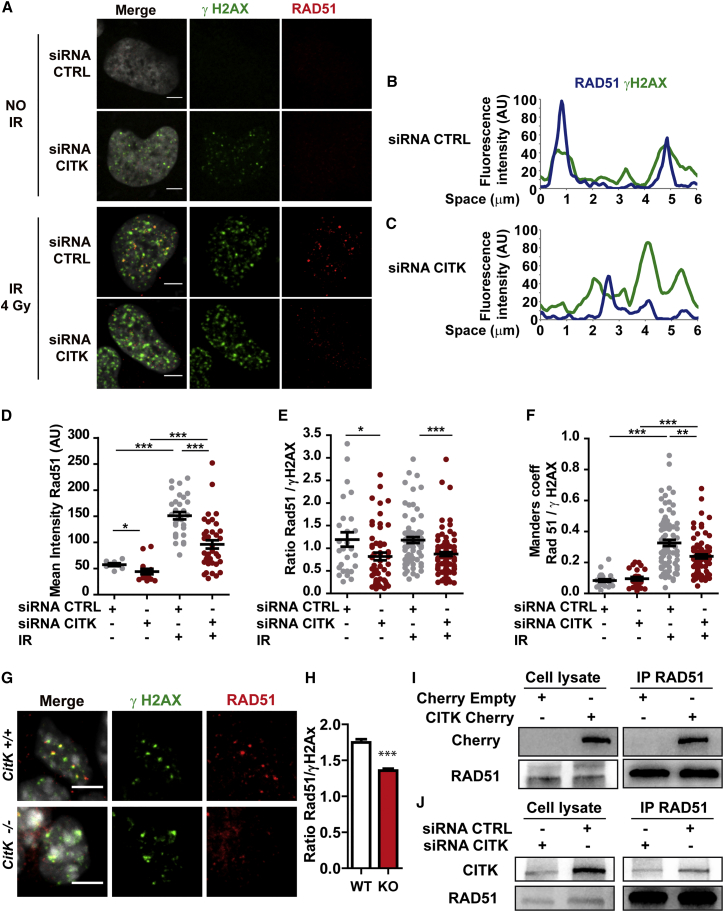
DSBs are mainly repaired by the non-homologous end joining (NHEJ) or homologous recombination (HR) pathways (Ceccaldi et al., 2016). CITK was reported to interact with the KIF4A chromokinesin (Maliga et al., 2013) and with CDKN1B (P27) (Serres et al., 2012), both implicated in HR (See et al., 2010, Wu et al., 2008), suggesting that also CITK could play a role in this process. To investigate the involvement of CITK in proximal DSBs repair events, we evaluated whether 53BP1 (early DSBs marker; Panier and Boulton, 2014), Phospho-RPA (P-RPA), or RAD51 (which both bind sequentially to single-strand DNA during HR; Cejka, 2015) are normally recruited to γH2AX foci in nuclei of IR-treated CITK-depleted and control cells. No differences were observed in relative intensities and co-localization of 53BP1 and γH2AX foci (Figures S3A–S3C) or of P-RPA and γH2AX foci (Figures S3D and S3E), suggesting that the molecular events comprised from early DSBs recognition to end resection are not perturbed by CITK loss. In contrast, in CITK-depleted cells, the intensity of RAD51 signals (Figure 4A and 4D) and the γH2AX/RAD51 fluorescence ratio (Figure 4E) are both substantially reduced compared to controls. Interestingly, we also found that CITK depletion decreases the fraction of RAD51 signals that co-localize with γH2AX foci (Figures 4A–4C), as confirmed by quantitative analysis (Figure 4F).
Figure 4.
CITK-Depleted HeLa Cells Are Defective in RAD51, but Not in 53BP1, Recruitment at γH2AX Foci
(A) HeLa cells stained with anti-γH2AX and anti-RAD51 antibodies before irradiation or 4 hr after irradiation (X-rays, 4 Gy).
(B and C) Examples of co-localization profiles between γH2AX and RAD51 signals in HeLa cells treated as in (A). Fluorescence intensity was plotted for the two channels along a 6-μm-long line, randomly drawn in the nuclei of exemplar cells.
(D–F) Quantitative analysis of the RAD51 signals in cells treated as in (A); each dot corresponds to one nucleus. The dot plot graphs show the fluorescence intensities of the RAD51 signals (D), the RAD51/γH2AX fluorescence ratios (E), and a quantification of co-localization of the γH2AX and RAD51 signals determined by the Manders overlap coefficient (F).
(G) Nuclei from NPCs from E12.5 embryo cortices stained with anti-γH2AX, anti RAD51, and DAPI. The scale bars represent 5 μm.
(H) Quantification of the RAD51/γH2AX fluorescence ratios of (G).
(I) CITK-Cherry or Cherry Empty vectors were overexpressed in 293T cells by transfection, and the lysates were subjected to immunoprecipitation using an anti-RAD51 antibody. Immunoprecipitates (IPs) and whole-cell extracts (cell lysate) were immunoblotted with the indicated antibodies.
(J) Total lysate of ONS-76 medulloblastoma cells, transfected with control or CITK-specific siRNAs, were immunoprecipitated with anti-RAD51 antibodies. Endogenous CITK and RAD51 were then revealed by western blotting (WB). The scale bars represent 5 μm.
Two tails unpaired t test with Welch’s correction was used for the statistical analysis of these experiments (n = 3–4 per group). Graphs show mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
We next asked whether RAD51 recruitment to DSBs is similarly compromised in developing cortex by evaluating the spontaneous DNA-damage foci arising in control and CitK −/− NPCs. Also in this case, we observed a reduced γH2AX/RAD51 fluorescence ratio (Figures 4G and 4H), which is not due to a reduction in the RAD51 intracellular level. Indeed, in developing neural tissues of CitK −/− mice, the RAD51 protein levels are not significantly reduced compared to controls (Figure S3D) as also occurs in CITK-depleted cell cultures (data not shown). To further elucidate the interplay between CITK and RAD51, we explored the possibility that they can form protein complexes. We found that both overexpressed and endogenous CITK can be detected in endogenous RAD51 immunoprecipitations (Figures 4I–4J). The association of the two proteins is not mediated by DNA, as it is not affected by DNase treatment (Figure S4A). However, endogenous RAD51 could be detected in CITK immunoprecipitates only when CITK was overexpressed (Figure S4B). Interestingly, endogenous RAD51 can interact even more efficiently with CITN (an isoform of CIT lacking the amino-terminal kinase domain; Madaule et al., 1995) and with a kinase-dead mutant of CITK (Figure S4C), indicating that the catalytic activity of CITK may modulate the CITK-RAD51 interaction. Altogether, these results suggest that CITK may participate in HR pathway by promoting RAD51 accumulation at end-resected DSB.
CITK Loss Leads to Neural Progenitors’ Apoptosis through TP53 Activation
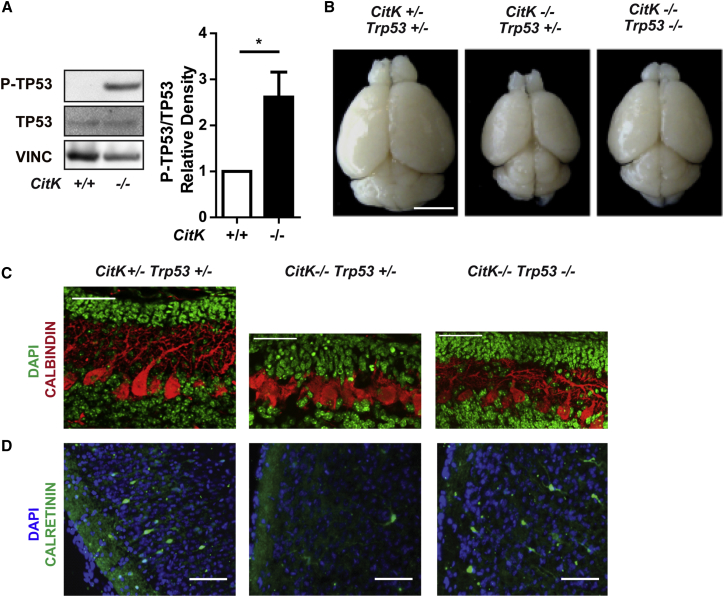
Consistent with the increased levels of DNA damage in CITK-depleted cells, the developing cerebellum of CitK −/− mice displayed an increased activation (phosphorylation) of the ATM kinase compared to non-mutant tissues (Figures S5A and S5B). ATM activation is accompanied by a strong increase of the TP53 phosphorylation at Ser-15 (Figure 5A), a post-translational modification that strongly correlates with TP53 activation (Shieh et al., 1999). These results suggest that the DNA damage arising in developing CitK −/− brains leads to TP53 activation and TP53-dependent apoptosis of neural progenitors and neurons.
Figure 5.
Neuroanatomical Rescue of the CitK Knockout Mice Phenotype by Tpr53 Deficiency
(A) Representative western blots of cell lysates of P4 cerebella from CitK −/− and control mice (+/+) probed with the indicated antibodies. Quantification of the relative density between P-TP53 and TP53 is shown.
(B) Brain morphology in 30-day-old mice of the indicated genotypes.
(C) Frozen sections of P8 cerebella from mice of the indicated genotypes stained with anti-calbindin antibodies and DAPI.
(D) Frozen sections of neocortex from P21 mice of the indicated genotypes stained with anti-Calretinin antibodies and DAPI.
The scale bars correspond to 50 μm in (C) and to 200 μm in (D). Two tails unpaired Student’s t test was used for the statistical analysis of these experiments (n = 3–5 mice per group). Graphs show mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
To test this possibility, we crossed CitK knockout mice into a Trp53-null background (Jacks et al., 1994). The intercross of the two knockout lines produces all possible genotypes with the expected Mendelian frequencies (data not shown). Morphological examination of CitK Trp53 double-knockout (DKO) brains revealed a significant increase in brain size compared with Trp53-proficient CitK −/− animals, although DKO brains are still smaller than littermate controls (Figure 5B). Consistent with this finding, histological examination of DKO mice revealed a significant recovery of cortical (Figure S6A) and cerebellar granular layer thickness (Figure S6B), correlated with good recovery of Purkinje cells’ lining and dendritic arborization (Figure 5C). We also observed a dramatic recovery of calretinin-positive GABA-ergic interneurons throughout the cortex (Figure 5D) and of olfactory granules’ precursors in rostral migratory stream (Figure S6C). Finally, the number of granule cells in the dentate gyrus of the hippocampus is increased, whereas the structure appears to be disorganized (Figure S6D).
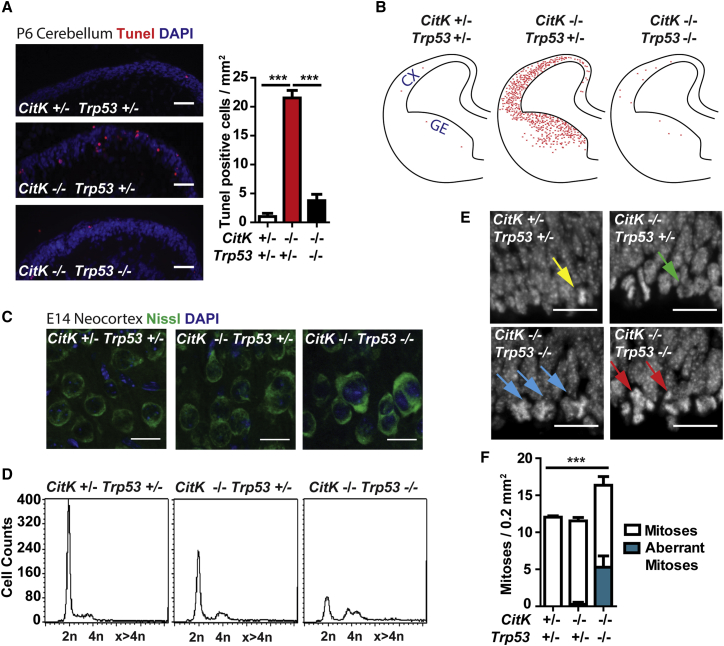
To investigate whether the anatomical rescue obtained by TP53 loss in CitK −/− animals is due to reduced apoptosis, TUNEL assay was performed on developing brains. The frequency of apoptotic cells was dramatically reduced in both cerebellum (Figure 6A) and telencephalon (Figure 6B) of DKO mice compared to CitK −/− Trp53 +/− mice.
Figure 6.
Apoptosis of CitK-Null Neural Progenitors Is TP53 Dependent
(A) Frozen sections of cerebella from P6 mice of the indicated genotypes stained with TUNEL assay (apoptotic cells, red) and DAPI. Quantification of apoptotic cells in the indicated genotypes is shown. The scale bars represent 50 μm.
(B) Distribution of TUNEL-positive cells in coronal sections of E14.5 embryonic brains; CX, neocortex; GE, ganglionic eminence.
(C) Examples of cortical plate cells in developing neocortices of mice of the indicated genotypes, stained with Nissl and DAPI. The scale bars represent 25 μm.
(D) Flow-cytometric analysis of DNA content in postnatal cortices (P14) from mice of the indicated genotypes.
(E) DAPI-stained ventricular regions of E14.5 cortices from embryos of the indicated genotypes. Note the presence of tripolar metaphases (blue arrows) and anaphases (red arrows); bipolar metaphases and anaphases are indicated by yellow and green arrows, respectively. The scale bars represent 10 μm.
(F) Quantification of normal and aberrant mitoses in the samples shown in (E).
Two tails unpaired Student’s t test was used for the statistical analysis of these experiments (n = 3 mice per group). Graphs show mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Conversely, histological examination (Figure 6C) and flow cytometry (Figure 6D) revealed that DKO brains exhibit frequencies of binucleated/multinucleated cells even higher than those seen in CitK −/− Trp53 +/− mice. Similarly, the frequency of aberrant mitoses, such as tripolar metaphases and anaphases, was strongly increased in DKO brains compared to both CitK +/− Trp53 +/− control brains and CitK −/− Trp53 +/− brains (Figures 6E and 6F). Importantly, developing cerebellum and neocortex of DKO mice showed levels of DNA damage (γH2AX levels) and ATM activation comparable to those detected in TP53-proficient CitK −/− samples (Figures S7 and S5, respectively), providing further support to the conclusion that DNA damage is a primary outcome of CITK deficiency and not a consequence of apoptosis.
CITK Loss Activates TP53-Dependent and TP53-Independent Transcriptional Responses
To further characterize the molecular events produced by CITK loss, we used RNA sequencing (RNA-seq) to compare the gene expression profiles of CitK −/− Trp53 +/−, CitK +/− Trp53 −/−, and DKO mice with the profile of their CitK +/− Trp53 +/− littermates, which were used as controls. As for the biochemical analyses, we analyzed gene expression in the developing cerebellum. Samples were collected at postnatal day 4 (P4), when the ratio between proliferating neuronal precursors and post-mitotic neurons in CitK mutant and control mice is still comparable (Di Cunto et al., 2000).
As expected, a comparison of cerebella from the four genotypes revealed that the CitK knockout is associated with decreased expression of neurogenesis-related genes, consistent with a strong loss of post-mitotic neurons (Sgrò et al., 2016). Indeed, the keyword “neurogenesis” was the most statistically enriched (p = 1.9 E−11) in the annotation of the 354 genes downregulated in CitK −/− Trp53 +/− mice compared with their CitK +/− Trp53 +/− littermates (Table S1); conversely, the keyword “p53 signaling pathway” was significantly enriched in the upregulated genes (p = 7.4 E–4; Table S1). A comparison of DKO samples with CitK +/− Trp53 +/− controls or with CitK +/− Trp53 −/− samples provided information on the genes whose expression is modulated by CITK loss in a TP53-independent fashion (Table S1). An analysis of the gene lists generated by these comparisons confirmed that the altered expression of neurogenesis genes is not a direct consequence of CITK loss but is instead a likely consequence of TP53-dependent apoptosis. Indeed, only 26 genes were downregulated in DKO samples compared with either CitK +/− Trp53 +/− or CitK +/− Trp53 −/− samples (Table S1), and these genes did not include a significant number of the neurogenesis functions downregulated in CitK −/− Trp53 +/− brains (Table S1). As expected, genes of the TP53-effector pathway are not significantly upregulated in DKO brains (Table S1). In particular, we did not observe upregulation of TP53-related and DNA-damage-related pro-apoptotic genes, such as Pmaip1/Noxa (Oda et al., 2000, Schuler et al., 2003), Ccnd1 (Kranenburg et al., 1996), Ccng1 (Okamoto and Prives, 1999), and Trp53cor1/LincRNA-p21 (Hall et al., 2015, Huarte et al., 2010). However, Cdkn1a and Trp73, two genes related to DNA damage and to TP53 function, were overexpressed in both DKO and CitK −/− Trp53 +/− samples, indicating that their induction by CITK deficiency is largely TP53-independent. Cdkn1a induction was confirmed at the protein level (Figure S6C). Interestingly, the other genes upregulated independently of TP53 in CITK-deficient samples include Wif1, S100B, MyoD1, and Cend1 (Table S1), which are implicated in cell cycle exit. Thus, in parallel with a TP53-dependent apoptotic program, loss of CITK activates a TP53-independent response that may promote cell cycle exit.
Trp53 Inactivation Restores Clinical and Behavioral Defects of CitK-Null Mice
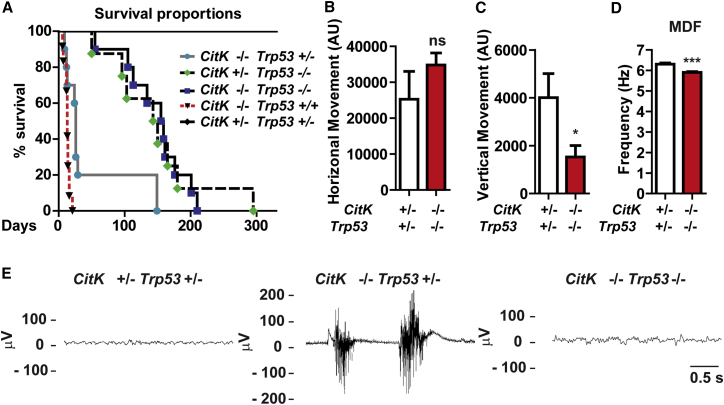
Trp53-proficient CitK −/− mice die of lethal epileptic seizures within the first 2 weeks after birth, whereas DKO mice display strongly improved phenotype, with a lifespan almost indistinguishable from that of their CitK +/− Trp53 −/− littermates (p = 0.97; Figure 7A). Interestingly, even the deletion of a single Trp53 allele is sufficient to extend the survival of CitK −/− mice (p = 0.0028; Figure 7A). The ataxic phenotype of CitK −/− Trp53 +/− mice is largely corrected in their DKO littermates (Movies S1, S2, and S3), which display normal frequency of horizontal movements (Figure 7B) and only a reduction in frequency of vertical movements (Figure 7C). Moreover, electroencephalographic (EEG) recordings are consistent with a strong functional rescue. In particular, in DKO mice, the bursts of high-amplitude poly-spikes that characterize the seizures observed in CitK −/− Trp53 +/− mice were completely absent (Figure 7D). Accordingly, we never observed behavioral alterations compatible with seizures. A reduction of the mean dominant frequency (MDF) of theta region was the only difference that we could detect between the EEG pattern of DKO and CitK +/− Trp53 +/− mice (Figure 7E).
Figure 7.
TP53 Inactivation Rescues the Clinical Phenotype of CitK Knockout Mice
(A) Survival curves of mice of the indicated genotypes.
(B and C) Spontaneous horizontal (B) and vertical (C) movements in 2-month-old mice of the indicated genotypes.
(D) Mean dominant frequency (MDF) of theta band, measured on EEG recording in 4-week-old mice of the indicated genotypes.
(E) EEG recordings in 12-day-old mice of the indicated genotypes.
Two tails unpaired Student’s t test was used for the statistical analysis of these experiments reported in (B) and (C) (n = 3–10 mice per group). Graphs show mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ns, not significant.
In conclusion, the recovery of the neurological phenotype in DKO mice indicates that the neural progenitors with abnormal DNA content and elevated DNA damage, which would normally be eliminated through TP53-dependent apoptosis, are capable of differentiating and integrating into relatively normal neural circuits if TP53 is concomitantly lost.
Discussion
This study provides insight into the role of CITK in cortical development and microcephaly. So far, the phenotype produced by CITK loss has been ascribed to massive apoptosis, directly or indirectly generated by cytokinesis failure (Di Cunto et al., 2000, Cunto et al., 2002, Sarkisian et al., 2002). This view was essentially based on the finding that CitK −/− mice and rats display cytokinesis failure and apoptosis in developing neural tissues (Di Cunto et al., 2000, Cunto et al., 2002, Sarkisian et al., 2002), a phenotype recently confirmed in human microcephaly patients with CIT/CITK mutations (Basit et al., 2016, Harding et al., 2016, Li et al., 2016, Shaheen et al., 2016). However, the mechanistic link between cytokinesis failure and apoptosis remained controversial. Data obtained from non-tumorigenic cell lines and from primary cells (Ganem et al., 2014, Kuffer et al., 2013, Panopoulos et al., 2014) indicate that, rather than apoptosis, the main outcome of cytokinesis failure is G1 arrest, produced by TP53 activation (Kuffer et al., 2013), cell senescence (Panopoulos et al., 2014), or activation of the Hippo pathway (Ganem et al., 2014). On the other hand, it is reasonable to hypothesize that cells failing to arrest after abortive cytokinesis would give rise to tetraploid or polyploid cells, which can lead to DNA damage (Ganem and Pellman, 2012, Hayashi and Karlseder, 2013, Zhang et al., 2013) and, consequently, to apoptosis (Hall et al., 2015).
We showed that diploid cells from neuronal progenitor cultures from CitK −/− mice exhibit a significant increase in DNA repair foci compared to diploid cells of CitK +/+ control mice. Also CITK-depleted cells from human medulloblastoma displayed a substantial increase in the frequencies of CABs, with respect to mock-treated cells. Because the NPC and human medulloblastoma that we analyzed are unlikely to be the products of defective cytokinesis, these results strongly suggest that, in both cell types, DNA damage arises independently of cytokinesis. Moreover, we demonstrated that Drosophila diploid cells from dck mutant brains exhibit an increase in both DNA repair foci and CABs compared to diploid cells of wild-type brains. Collectively, these results indicate that, in a variety of systems, depletion of CITK leads to DNA damage independently of cytokinesis.
It has been shown that DNA damage can arise in cells that remain arrested in a prometaphase-metaphase stage for long periods, suggesting that a bock in metaphase can cause DNA lesions (Hayashi and Karlseder, 2013, Orth et al., 2012, Pilaz et al., 2016). This possibility, however, is excluded for Drosophila, as our previous work showed that diploid cells of dck1 and dck2 mutant brains proceed normally though mitosis; mutant cells show mitotic indices and relative frequencies of prometaphases, metaphases, and anaphases that are fully comparable to those of wild-type controls (Naim et al., 2004). This possibility is also excluded for CITK-depleted HeLa cells, as they show DNA damage when the cell cycle is not significantly perturbed (Figure S3). Altogether, these results indicate that the evolutionarily conserved role of CITK in the maintenance of DNA integrity is largely independent of its role in cytokinesis.
Abundant evidence indicates that γH2AX repair foci and CABs are generated by DSBs (Durante et al., 2013, Polo and Jackson, 2011), suggesting that citron kinase protects cells from this type of DNA lesion. How does loss of CITK lead to these lesions? In theory, CITK loss could increase DSBs formation, decrease DSBs repair, or both. The relatively slow recovery of CITK-depleted cells from IR (Figure 3) suggests a deficiency in DSB repair. The reported interaction of CITK with KIF4A (Wu et al., 2008) and p27 (Serres et al., 2012) has further suggested that CITK may play a role in HR, as both proteins were previously implicated in this pathway. Consistent with this possibility, CITK-deficient cells displayed a specific reduction in RAD51 recruitment at γH2AX foci, whereas they showed normal recruitment of 53BP1 and P-RPA at these foci. Moreover, CITK and endogenous RAD51 co-purify in immunoprecipitation experiments, either when CITK is overexpressed or when it is present at normal background levels, supporting the idea of a functional interaction between these proteins. However, despite this interaction, we were not able to detect CITK accumulation at γH2AX foci (data not shown), suggesting that CITK is not directly required for RAD51 recruitment at foci. Our results also suggest that the CITK-RAD51 association can be modulated by CITK kinase activity (Figure S4). A crucial role of CITK catalytic activity would be consistent with the high fraction of kinase-inactivating mutations found in microcephaly patients with mutations in the CIT/CITK gene (Basit et al., 2016, Harding et al., 2016, Li et al., 2016, Shaheen et al., 2016). Therefore, it will be very important to determine whether RAD51 is a direct CITK substrate.
Functional impairment of HR due to defective RAD51 recruitment to DNA lesions provides a possible explanation for DSBs formation in CITK-depleted cells. However, it must be considered that DNA lesions and IR sensitivity caused by CITK loss occur not only in cell cycle phases characterized by RAD51 expression but also in G1 and post-mitotic cells, in which RAD51 expression is very low. Therefore, future studies should clarify whether the NHEJ pathway is also compromised by CITK loss.
Our results indicate that pathogenesis of the CIT/CITK-dependent microcephaly syndrome is more complicated than previously thought. We have clearly shown that TP53 activation is a fundamental culprit of microcephaly. Indeed, similar to other mouse models bearing mutations in DNA repair genes (Frank et al., 2000, Frappart et al., 2005, Gao et al., 2000, Pao et al., 2014), apoptosis and most aberrant phenotypic traits of CitK −/− brains are reverted by TP53 inactivation. In particular, the rescue of lethality correlates with the disappearance of epileptic seizures (Figure 7E) and could be explained by the recovery of GABA-ergic interneurons (Figure 5), which play a crucial role in control of brain excitability (Powell, 2013) and are severely affected by CITK loss (Figure S5; Muzzi et al., 2009). However, despite apoptosis elimination and remarkable neurological recovery, brain volume of CITK-null mice is only partially restored by Trp53 inactivation. This finding suggests that, in CitK −/− mice, TP53-independent reduction of cell proliferation may contribute to microcephaly. Consistent with this possibility, we detected in DKO developing cerebella a TP53-independent growth-inhibitory signature, characterized by increased p21 and p73 expression. This growth-suppressive program could be activated by cytokinesis failure, as shown in other cell types (Ganem et al., 2014, Panopoulos et al., 2014). However, gene expression analysis in DKO cerebella did not provide specific indications about this issue. In particular, we could not detect a significant activation of the Hippo pathway (Ganem et al., 2014). If growth suppression does not result from cytokinesis failure, it might be triggered by TP53-independent events activated by DNA damage, as those described in mammalian cells treated with mutagens (Reinhardt et al., 2007). Thus, the mechanism underlying the growth-suppressive pathway in CIT/CITK-dependent microcephaly remains an open issue.
In conclusion, our study reveals a conserved function for CIT in ensuring genomic stability and supports the notion that blocking TP53 signaling during development can attenuate the phenotypic consequences of at least a subset of mutations leading to severe microcephaly.
Experimental Procedures
Experimental Animal Work
Experiments involving samples from mutant and control mice have been performed conforming to the Italian laws on animal experimentation and under the supervision of the veterinary service of our animal facility. The corresponding experimental protocols have been approved by the Italian Ministry of Health, Department of Public Veterinary Health with approval number 343/2015-PR, released on 08/05/2015.
Statistical Analysis
Statistical analyses were performed using Microsoft Office Excel or Graphpad (GraphPad Software). Unpaired Student’s t test and Welch’s unequal variances t test was used for p values determination. Values represented the mean and the SE of at least three independent experiments.
Additional materials and methods are described in the Supplemental Experimental Procedures.
Author Contributions
Conceptualization, F.T.B., M. Gatti, S.B., A.M., and F.D.C.; Investigation, F.T.B., C.T., Y.L., F.V., C.M., N.E.-A., L.P., M. Gai, G.E.B., A.M.A.C., F.S., G.P., A.C., F.N., and L.T.; Resources, S.G. and S.O.; Formal Analysis, F.T.B., U.A., and F.D.C.; Writing – Original Draft, F.T.B. and F.D.C.; Writing – Review & Editing, F.T.B. and F.D.C.; Supervision, F.D.C.; Funding, M. Gatti, S.B., and F.D.C.
Acknowledgments
We thank Dr. Maurizio Giustetto (Department of Neuroscience, Turin) and Dr. Roberto Piva for providing us reagents, Dr. Alessandro Fioravanti and Dr. Juan Carlos Cutrin for the histology, Dr. Paolo Provero for help on the statistical analysis, and Dr. Luigi Varesio (Gaslini Hospital, Genova). This work was supported by the Telethon Foundation through grant nos. GGP12095 and GGP13081 to F.D.C., by the Associazione Italiana per la Ricerca sul Cancro (AIRC) with grants IG17527 to F.D.C. and IG16020 to M. Gatti, and by a PRIN grant to S.B. C.M. was supported by a Teresa Ariaudo fellowship from Istituto Pasteur-Fondazione Cenci Bolognetti.
Published: February 14, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, one table, three movies, and a supplemental data file and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2017.01.054.
Contributor Information
Federico Tommaso Bianchi, Email: federico.bianchi@unito.it.
Ferdinando Di Cunto, Email: ferdinando.dicunto@unito.it.
Supplemental Information
Column A: genes up-regulated in CitK -/- p53 +/-, as compared to both CitK +/- p53 +/- cerebella and CitK +/- p53 -/- cerebella; Column B: genes down-regulated in CitK -/- p53 +/-, as compared to both CitK +/- p53 +/- cerebella and CitK +/- p53 -/-; Column C: genes upregulated in CitK -/- p53 -/-, as compared to both CitK +/- p53 +/- cerebella and CitK +/- p53 -/-; Column D: genes down-regulated in CitK -/- p53 -/-, as compared to both CitK +/- p53 +/- cerebella and CitK +/- p53 -/-. Significantly overrepresented Gene Ontology kyewords for each list are at the bottom of each list. Enrichment P-values were obtained using DAVID software (https://david.ncifcrf.go) with Benjamini-Hochberg correction. The results of all the pairwise comparisons are reported in the Supplemental Data S1 file.
References
- Ackman J.B., Ramos R.L., Sarkisian M.R., Loturco J.J. Citron kinase is required for postnatal neurogenesis in the hippocampus. Dev. Neurosci. 2007;29:113–123. doi: 10.1159/000096216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbelanne M., Tsang W.Y. Molecular and cellular basis of autosomal recessive primary microcephaly. BioMed Res. Int. 2014;2014:547986. doi: 10.1155/2014/547986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basit S., Al-Harbi K.M., Alhijji S.A., Albalawi A.M., Alharby E., Eldardear A., Samman M.I. CIT, a gene involved in neurogenic cytokinesis, is mutated in human primary microcephaly. Hum. Genet. 2016;135:1199–1207. doi: 10.1007/s00439-016-1724-0. [DOI] [PubMed] [Google Scholar]
- Bassi Z.I., Verbrugghe K.J., Capalbo L., Gregory S., Montembault E., Glover D.M., D’Avino P.P. Sticky/Citron kinase maintains proper RhoA localization at the cleavage site during cytokinesis. J. Cell Biol. 2011;195:595–603. doi: 10.1083/jcb.201105136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R., Rondinelli B., D’Andrea A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P. DNA end resection: nucleases team up with the right partners to initiate homologous recombination. J. Biol. Chem. 2015;290:22931–22938. doi: 10.1074/jbc.R115.675942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Jackson A.P., Bond J., Woods C.G. What primary microcephaly can tell us about brain growth. Trends Mol. Med. 2006;12:358–366. doi: 10.1016/j.molmed.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Cunto F.D., Imarisio S., Camera P., Boitani C., Altruda F., Silengo L. Essential role of citron kinase in cytokinesis of spermatogenic precursors. J. Cell Sci. 2002;115:4819–4826. doi: 10.1242/jcs.00163. [DOI] [PubMed] [Google Scholar]
- D’Avino P.P., Savoian M.S., Glover D.M. Mutations in sticky lead to defective organization of the contractile ring during cytokinesis and are enhanced by Rho and suppressed by Rac. J. Cell Biol. 2004;166:61–71. doi: 10.1083/jcb.200402157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cunto F., Imarisio S., Hirsch E., Broccoli V., Bulfone A., Migheli A., Atzori C., Turco E., Triolo R., Dotto G.P. Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron. 2000;28:115–127. doi: 10.1016/s0896-6273(00)00090-8. [DOI] [PubMed] [Google Scholar]
- Durante M., Bedford J.S., Chen D.J., Conrad S., Cornforth M.N., Natarajan A.T., van Gent D.C., Obe G. From DNA damage to chromosome aberrations: joining the break. Mutat. Res. 2013;756:5–13. doi: 10.1016/j.mrgentox.2013.05.014. [DOI] [PubMed] [Google Scholar]
- Faheem M., Naseer M.I., Rasool M., Chaudhary A.G., Kumosani T.A., Ilyas A.M., Pushparaj P., Ahmed F., Algahtani H.A., Al-Qahtani M.H., Saleh Jamal H. Molecular genetics of human primary microcephaly: an overview. BMC Med. Genomics. 2015;8(Suppl 1):S4. doi: 10.1186/1755-8794-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank K.M., Sharpless N.E., Gao Y., Sekiguchi J.M., Ferguson D.O., Zhu C., Manis J.P., Horner J., DePinho R.A., Alt F.W. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- Frappart P.O., Tong W.M., Demuth I., Radovanovic I., Herceg Z., Aguzzi A., Digweed M., Wang Z.Q. An essential function for NBS1 in the prevention of ataxia and cerebellar defects. Nat. Med. 2005;11:538–544. doi: 10.1038/nm1228. [DOI] [PubMed] [Google Scholar]
- Gai M., Camera P., Dema A., Bianchi F., Berto G., Scarpa E., Germena G., Di Cunto F. Citron kinase controls abscission through RhoA and anillin. Mol. Biol. Cell. 2011;22:3768–3778. doi: 10.1091/mbc.E10-12-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Pellman D. Linking abnormal mitosis to the acquisition of DNA damage. J. Cell Biol. 2012;199:871–881. doi: 10.1083/jcb.201210040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Cornils H., Chiu S.Y., O’Rourke K.P., Arnaud J., Yimlamai D., Théry M., Camargo F.D., Pellman D. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell. 2014;158:833–848. doi: 10.1016/j.cell.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Ferguson D.O., Xie W., Manis J.P., Sekiguchi J., Frank K.M., Chaudhuri J., Horner J., DePinho R.A., Alt F.W. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- Gunsalus K.C., Bonaccorsi S., Williams E., Verni F., Gatti M., Goldberg M.L. Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J. Cell Biol. 1995;131:1243–1259. doi: 10.1083/jcb.131.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J.R., Messenger Z.J., Tam H.W., Phillips S.L., Recio L., Smart R.C. Long noncoding RNA lincRNA-p21 is the major mediator of UVB-induced and p53-dependent apoptosis in keratinocytes. Cell Death Dis. 2015;6:e1700. doi: 10.1038/cddis.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding B.N., Moccia A., Drunat S., Soukarieh O., Tubeuf H., Chitty L.S., Verloes A., Gressens P., El Ghouzzi V., Joriot S. Mutations in citron kinase cause recessive microlissencephaly with multinucleated neurons. Am. J. Hum. Genet. 2016;99:511–520. doi: 10.1016/j.ajhg.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M.T., Karlseder J. DNA damage associated with mitosis and cytokinesis failure. Oncogene. 2013;32:4593–4601. doi: 10.1038/onc.2012.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D., Khalil A.M., Zuk O., Amit I., Rabani M. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Remington L., Williams B.O., Schmitt E.M., Halachmi S., Bronson R.T., Weinberg R.A. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Kaindl A.M., Passemard S., Gressens P. Autosomal recessive primary microcephalies (MCPH) Eur. J. Paediatr. Neurol. 2009;13:458. doi: 10.1016/j.ejpn.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Karess R.E., Chang X.J., Edwards K.A., Kulkarni S., Aguilera I., Kiehart D.P. The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell. 1991;65:1177–1189. doi: 10.1016/0092-8674(91)90013-o. [DOI] [PubMed] [Google Scholar]
- Kranenburg O., van der Eb A.J., Zantema A. Cyclin D1 is an essential mediator of apoptotic neuronal cell death. EMBO J. 1996;15:46–54. [PMC free article] [PubMed] [Google Scholar]
- Kuffer C., Kuznetsova A.Y., Storchová Z. Abnormal mitosis triggers p53-dependent cell cycle arrest in human tetraploid cells. Chromosoma. 2013;122:305–318. doi: 10.1007/s00412-013-0414-0. [DOI] [PubMed] [Google Scholar]
- Li H., Bielas S.L., Zaki M.S., Ismail S., Farfara D., Um K., Rosti R.O., Scott E.C., Tu S., Chi N.C. Biallelic mutations in citron kinase link mitotic cytokinesis to human primary microcephaly. Am. J. Hum. Genet. 2016;99:501–510. doi: 10.1016/j.ajhg.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaule P., Furuyashiki T., Reid T., Ishizaki T., Watanabe G., Morii N., Narumiya S. A novel partner for the GTP-bound forms of rho and rac. FEBS Lett. 1995;377:243–248. doi: 10.1016/0014-5793(95)01351-2. [DOI] [PubMed] [Google Scholar]
- Mahmood S., Ahmad W., Hassan M.J. Autosomal recessive primary microcephaly (MCPH): clinical manifestations, genetic heterogeneity and mutation continuum. Orphanet J. Rare Dis. 2011;6:39. doi: 10.1186/1750-1172-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga Z., Junqueira M., Toyoda Y., Ettinger A., Mora-Bermúdez F., Klemm R.W., Vasilj A., Guhr E., Ibarlucea-Benitez I., Poser I. A genomic toolkit to investigate kinesin and myosin motor function in cells. Nat. Cell Biol. 2013;15:325–334. doi: 10.1038/ncb2689. [DOI] [PubMed] [Google Scholar]
- Marzio A., Merigliano C., Gatti M., Vernì F. Sugar and chromosome stability: clastogenic effects of sugars in vitamin B6-deficient cells. PLoS Genet. 2014;10:e1004199. doi: 10.1371/journal.pgen.1004199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie C., D’Avino P.P. Investigating cytokinesis failure as a strategy in cancer therapy. Oncotarget. 2016;7:87323–87341. doi: 10.18632/oncotarget.13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengoli V., Bucciarelli E., Lattao R., Piergentili R., Gatti M., Bonaccorsi S. The analysis of mutant alleles of different strength reveals multiple functions of topoisomerase 2 in regulation of Drosophila chromosome structure. PLoS Genet. 2014;10:e1004739. doi: 10.1371/journal.pgen.1004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriuchi S., Shimizu K., Miyao Y., Kishima H., Okawa M., Hayakawa T. Decreased N-myc expression in human medulloblastoma cell lines during differentiation. Anticancer Res. 1997;17(1A):301–306. [PubMed] [Google Scholar]
- Morris-Rosendahl D.J., Kaindl A.M. What next-generation sequencing (NGS) technology has enabled us to learn about primary autosomal recessive microcephaly (MCPH) Mol. Cell. Probes. 2015;29:271–281. doi: 10.1016/j.mcp.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Muzzi P., Camera P., Di Cunto F., Vercelli A. Deletion of the citron kinase gene selectively affects the number and distribution of interneurons in barrelfield cortex. J. Comp. Neurol. 2009;513:249–264. doi: 10.1002/cne.21927. [DOI] [PubMed] [Google Scholar]
- Naim V., Imarisio S., Di Cunto F., Gatti M., Bonaccorsi S. Drosophila citron kinase is required for the final steps of cytokinesis. Mol. Biol. Cell. 2004;15:5053–5063. doi: 10.1091/mbc.E04-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda E., Ohki R., Murasawa H., Nemoto J., Shibue T., Yamashita T., Tokino T., Taniguchi T., Tanaka N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Prives C. A role of cyclin G in the process of apoptosis. Oncogene. 1999;18:4606–4615. doi: 10.1038/sj.onc.1202821. [DOI] [PubMed] [Google Scholar]
- Orth J.D., Loewer A., Lahav G., Mitchison T.J. Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol. Biol. Cell. 2012;23:567–576. doi: 10.1091/mbc.E11-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S., Boulton S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- Panopoulos A., Pacios-Bras C., Choi J., Yenjerla M., Sussman M.A., Fotedar R., Margolis R.L. Failure of cell cleavage induces senescence in tetraploid primary cells. Mol. Biol. Cell. 2014;25:3105–3118. doi: 10.1091/mbc.E14-03-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao G.M., Zhu Q., Perez-Garcia C.G., Chou S.J., Suh H., Gage F.H., O’Leary D.D., Verma I.M. Role of BRCA1 in brain development. Proc. Natl. Acad. Sci. USA. 2014;111:E1240–E1248. doi: 10.1073/pnas.1400783111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passemard S., Kaindl A.M., Verloes A. Microcephaly. Handb. Clin. Neurol. 2013;111:129–141. doi: 10.1016/B978-0-444-52891-9.00013-0. [DOI] [PubMed] [Google Scholar]
- Pilaz L.J., McMahon J.J., Miller E.E., Lennox A.L., Suzuki A., Salmon E., Silver D.L. Prolonged mitosis of neural progenitors alters cell fate in the developing brain. Neuron. 2016;89:83–99. doi: 10.1016/j.neuron.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S.E., Jackson S.P. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell E.M. Interneuron development and epilepsy: early genetic defects cause long-term consequences in seizures and susceptibility. Epilepsy Curr. 2013;13:172–176. doi: 10.5698/1535-7597-13.4.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt H.C., Aslanian A.S., Lees J.A., Yaffe M.B. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11:175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisian M.R., Li W., Di Cunto F., D’Mello S.R., LoTurco J.J. Citron-kinase, a protein essential to cytokinesis in neuronal progenitors, is deleted in the flathead mutant rat. J. Neurosci. 2002;22:RC217. doi: 10.1523/JNEUROSCI.22-08-j0001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M., Maurer U., Goldstein J.C., Breitenbücher F., Hoffarth S., Waterhouse N.J., Green D.R. p53 triggers apoptosis in oncogene-expressing fibroblasts by the induction of Noxa and mitochondrial Bax translocation. Cell Death Differ. 2003;10:451–460. doi: 10.1038/sj.cdd.4401180. [DOI] [PubMed] [Google Scholar]
- Schultz L.B., Chehab N.H., Malikzay A., Halazonetis T.D. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See W.L., Miller J.P., Squatrito M., Holland E., Resh M.D., Koff A. Defective DNA double-strand break repair underlies enhanced tumorigenesis and chromosomal instability in p27-deficient mice with growth factor-induced oligodendrogliomas. Oncogene. 2010;29:1720–1731. doi: 10.1038/onc.2009.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serres M.P., Kossatz U., Chi Y., Roberts J.M., Malek N.P., Besson A. p27(Kip1) controls cytokinesis via the regulation of citron kinase activation. J. Clin. Invest. 2012;122:844–858. doi: 10.1172/JCI60376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgrò F., Bianchi F.T., Falcone M., Pallavicini G., Gai M., Chiotto A.M., Berto G.E., Turco E., Chang Y.J., Huttner W.B., Di Cunto F. Tissue-specific control of midbody microtubule stability by Citron kinase through modulation of TUBB3 phosphorylation. Cell Death Differ. 2016;23:801–813. doi: 10.1038/cdd.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R., Hashem A., Abdel-Salam G.M., Al-Fadhli F., Ewida N., Alkuraya F.S. Mutations in CIT, encoding citron rho-interacting serine/threonine kinase, cause severe primary microcephaly in humans. Hum. Genet. 2016;135:1191–1197. doi: 10.1007/s00439-016-1722-2. [DOI] [PubMed] [Google Scholar]
- Shieh S.Y., Taya Y., Prives C. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turinetto V., Giachino C. Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015;43:2489–2498. doi: 10.1093/nar/gkv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods C.G., Parker A. Investigating microcephaly. Arch. Dis. Child. 2013;98:707–713. doi: 10.1136/archdischild-2012-302882. [DOI] [PubMed] [Google Scholar]
- Wu G., Zhou L., Khidr L., Guo X.E., Kim W., Lee Y.M., Krasieva T., Chen P.L. A novel role of the chromokinesin Kif4A in DNA damage response. Cell Cycle. 2008;7:2013–2020. doi: 10.4161/cc.7.13.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.Z., Leibowitz M.L., Pellman D. Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 2013;27:2513–2530. doi: 10.1101/gad.229559.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Column A: genes up-regulated in CitK -/- p53 +/-, as compared to both CitK +/- p53 +/- cerebella and CitK +/- p53 -/- cerebella; Column B: genes down-regulated in CitK -/- p53 +/-, as compared to both CitK +/- p53 +/- cerebella and CitK +/- p53 -/-; Column C: genes upregulated in CitK -/- p53 -/-, as compared to both CitK +/- p53 +/- cerebella and CitK +/- p53 -/-; Column D: genes down-regulated in CitK -/- p53 -/-, as compared to both CitK +/- p53 +/- cerebella and CitK +/- p53 -/-. Significantly overrepresented Gene Ontology kyewords for each list are at the bottom of each list. Enrichment P-values were obtained using DAVID software (https://david.ncifcrf.go) with Benjamini-Hochberg correction. The results of all the pairwise comparisons are reported in the Supplemental Data S1 file.