Abstract
Previous studies showed that members of the Burkholderiales were important in the succession of aerobic, molybdenum-dependent CO oxidizing-bacteria on volcanic soils. During these studies, four isolates were obtained from Kilauea Volcano (Hawai‘i, USA); one strain was isolated from Pico de Orizaba (Mexico) during a separate study. Based on 16S rRNA gene sequence similarities, the Pico de Orizaba isolate and the isolates from Kilauea Volcano were provisionally assigned to the genera Burkholderia and Paraburkholderia, respectively. Each of the isolates possessed a form I coxL gene that encoded the catalytic subunit of carbon monoxide dehydrogenase (CODH); none of the most closely related type strains possessed coxL or oxidized CO. Genome sequences for Paraburkholderia type strains facilitated an analysis of 16S rRNA gene sequence similarities and average nucleotide identities (ANI). ANI did not exceed 95% (the recommended cutoff for species differentiation) for any of the pairwise comparisons among 27 reference strains related to the new isolates. However, since the highest 16S rRNA gene sequence similarity among this set of reference strains was 98.93%, DNA-DNA hybridizations (DDH) were performed for two isolates whose 16S rRNA gene sequence similarities with their nearest phylogenetic neighbors were 98.96 and 99.11%. In both cases DDH values were <16%. Based on multiple variables, four of the isolates represent novel species within the Paraburkholderia: Paraburkholderia hiiakae sp. nov. (type strain I2T = DSM 28029T = LMG 27952T); Paraburkholderia paradisi sp. nov. (type strain WAT = DSM 28027T = LMG 27949T); Paraburkholderia peleae sp. nov. (type strain PP52-1T = DSM 28028T = LMG 27950T); and Paraburkholderia metrosideri sp. nov. (type strain DNBP6-1T = DSM 28030T = LMG 28140T). The remaining isolate represents the first CO-oxidizing member of the Burkholderia cepacia complex: Burkholderia alpina sp. nov. (type strain PO-04-17-38T = DSM 28031T = LMG 28138T).
Keywords: Burkholderia, carbon monoxide, Paraburkholderia, volcanic soils
Introduction
Soils have long been recognized as important biological sinks for carbon monoxide (CO), a critical reactant in the troposphere, yet the microbiology of soil CO oxidizers remains largely unstudied (e.g., Bartholomew and Alexander, 1982; Bender and Conrad, 1994; Conrad, 1996; King, 1999). Early work by Conrad (1988) established rates of atmospheric CO uptake by different soil types along with some of the controls of uptake, but those studies did not identify populations that were active in situ or lead to the isolation and characterization of novel CO oxidizers. More recently, molecular ecological approaches have revealed an unexpectedly large diversity of soil aerobic CO oxidizers, including members of the phylum Actinobacteria and class Ktedonobacteria (phylum Chloroflexi), as well as diverse α-, β-, and δ-Proteobacteria and Euryuarchaeota (e.g., King, 2003a; Dunfield and King, 2004; King and Weber, 2007; King et al., 2008; Weber and King, 2010a,b; Quiza et al., 2014; King and King, 2014a,b; McDuff et al., 2016).
Recent work with north temperate deciduous forest soils has been particularly interesting, since it has revealed that a group of δ-Proteobacteria related to the myxobacterium, Haliangeum ochraceum, plays significant roles in high-affinity atmospheric CO uptake (Quiza et al., 2014). Haliangeum ochraceum is intriguing itself, since it harbors the smallest known cox gene operon comprised only of the three structural genes for carbon monoxide dehydrogenase (CODH) and one accessory gene. Whether this or aspects of its CODH structure are related to its capacity of atmospheric CO uptake is unknown. Likewise, whether this group accounts for atmospheric CO uptake in other soils is unknown.
Previous work with a rapidly developing forest colonizing volcanic cinders identified members of the β-Proteobacteria, and Burkholderiales in particular, as important contributors to the CO-oxidizing community, a community that as a whole was involved with rapid atmospheric CO consumption (King, 2003b; Weber and King, 2010b, 2012). During these studies several novel CO oxidizers were isolated and their ability to consume atmospheric CO was established. Based on a battery of molecular, biochemical, and physiological analyses, we describe here these and a related isolate as new species within the genera Burkholderia and Paraburkholderia (Yabuuchi et al., 1992; Sawana et al., 2014). We propose the new isolates as potential models for understanding atmospheric CO oxidation by a widely distributed group of terrestrial β-Proteobacteria.
Materials and methods
Isolation
Paraburkholderia isolates DNBP6-1T, I2T, PP52-1T, and WAT were isolated by Weber and King (2012) from enrichments initiated with forest soil from a 1959 tephra deposit (Pu'u Puai) located on Kilauea Volcano (19° 24′ 22.5″ N × 155° 15′ 18.2″ W); this site has been described previously (King, 2003b; Gomez-Alvarez et al., 2007; King and Weber, 2008). Burkholderia isolate PO-04-17-38T was isolated by F.A. Rainey (University of Alaska, Anchorage) and colleagues from enrichments initiated with soil obtained above the tree line at an altitude of about 4357 m on Pico de Orizaba (Mexico) a dormant stratovolcano (see Callegan et al., 2008 for additional details on the site).
Three strains (WAT, I2T, and PP52-1T) were enriched in basal salts media (King, 2003a) with various carbon sources (WAT, xylose; I2T pyruvate; PP52-1T; mannose). All enrichments were assayed for CO oxidation after adding CO (100 ppm final concentration) to the headspaces of sealed 160-ml serum bottles, and monitoring headspace concentrations at intervals using gas chromatography (King, 1999). Cultures that oxidized CO were used to inoculate solidified versions of the basal salts media used for the original enrichments. Individual colonies were selected from plates, and used to inoculate small volumes of liquid media that were monitored for CO oxidation using gas chromatography as before. Cultures that oxidized CO were purified further by plating, colony selection, and transfer to liquid media; additional CO uptake assays were conducted as necessary. Strain DNBP6-1T was isolated similarly, except that nutrient broth (0.8 g L−1) containing penicillin (500 μg ml−1) was used for enrichment. Strain PO-04-17-38T was isolated using a medium comprised of 10% R2A (DSMZ medium 830) at 15°C without regard to its capacity for CO oxidation. It was subsequently identified as a CO oxidizer after screening as above. CO uptake capacity for the isolates was assayed following King and King (2014a) with stationary phase liquid cultures (10 ml in 160-ml serum bottles) amended with approximately 200-ppm headspace CO concentrations; cultures were incubated at 30°C with shaking at 200 rpm; cell protein concentrations were determined at the end of the uptake assays using a kit based on bicinchoninic acid (Pierce Protein Research Products; Thermo Scientific).
Molecular phylogenetic characterization
Phylogenetic characterizations were initiated with genomic DNA obtained from 2-ml cell suspensions that were harvested by centrifugation (10,000 × g, 1 min). DNA in cell pellets was extracted using a MoBio Ultraclean Microbial DNA Extraction Kit (MoBio Laboratories, Carlsbad, CA) following the manufacturer's recommendations with the exception of an added freeze (−80°C)-thaw (65°C) step (3 cycles) prior to bead-beating.
PCR amplification of 16S rRNA genes was performed with primers 27f and 1492r (Lane, 1991). PCR products were visualized using gel electrophoresis (1% agarose) and GelRed stain (Biotium, Inc., Hayward CA). Products of the correct size were purified using a MoBio Ultraclean PCR Cleanup Kit (MoBio Laboratories, Carlsbad, CA). PCR products were sequenced bidirectionally on an ABI model 3130XL at the Louisiana State University Genomics Facility (Baton Rouge, LA). Sequences were assembled and edited using Sequencher v. 4.8 (Gene Codes Corporation, Ann Arbor, MI). Sequences were deposited in Genbank with the following accession numbers: DNBP6-1T, JF763856; I2T, JF763857.1; PO-04-17-38T, JF763852; PP52-1T, JF763849; WAT, JF763851.
The SINA alignment tool (Pruesse et al., 2007) was used to align isolate 16S rRNA gene sequences with sequences derived from their closest phylogenetic neighbors and related taxa [determined from the EZtaxon application (Kim et al., 2012)]. Alignments were adjusted manually as necessary using MEGA7 (Tamura et al., 2013). Maximum likelihood analyses were also performed using MEGA7 with a general time reversible model and 100 and 1,000 bootstrap replicates, respectively.
EZTaxon (Kim et al., 2012) was used to obtain 16S rRNA gene sequences similarities for a set of type strains representing the phylogenetic neighborhood of the CO-oxidizing isolates. Genomes for strains with 16S rRNA gene sequence similarities > 98.0% were then used to generate parallel pairwise comparisons of average nucleotide identity (ANI). ANI was calculated using the comparative genome toolkit from the Integrated Microbial Genomes/Microbiome Samples website (https://img.jgi.doe.gov/cgi-bin/m/main.cgi).
RecA genes were amplified using primers Bur3 (forward) and Bur4 (reverse) to further clarify the phylogenetic positions of isolates DNBP6-1T, I2T, PP52-1T, and WAT; the PCR protocol followed the methods of Payne et al. (2005). Purified amplicons of the correct size (385 bp) were sequenced bi-directionally as above. MUSCLE was used in the MEGA7 platform (Tamura et al., 2013) to align partial recA gene sequences for the isolates' close phylogenetic neighbors. Phylogenetic analyses were also performed using MEGA7. Sequences were deposited in Genbank with the following accession numbers: DNBP6-1T, KY305132; I2T, KY305131; PP52-1T, KY305133; WAT, KY3051130.
Morphological and physiological characterization
Routine microscopy and staining methods were used for basic isolate characterization (Gerhardt et al., 1994). pH ranges suitable for growth were determined by cultivating the isolates in R2A media with pH adjusted between values of 5.5 and 9.5. A phosphate buffer (0.1 M) was used to prepare media with pH values from 5.5 to 6.5; a CO2/sodium bicarbonate/sodium carbonate buffer was used to prepare media with pH values from 7.5 to 9.5. Temperature optima were assessed similarly using cultures grown from 5 to 50°C (R2A medium at pH 6.5).
Sole carbon source metabolism patterns were assessed with Biolog GN2 plates (Biolog, Inc.; Hayward CA, USA) following the manufacturer's recommendations. Sole carbon source assimilation, enzymatic reactions (including oxidase and catalase), nitrate reduction, and other biochemical traits were also assayed with API 20NE strips following the manufacturer's recommendations (bioMérieux SA; Marcy l'Etoile, France). In addition, the ability of isolates to grow with selected sole carbon and energy sources was assessed in liquid culture with the basal salts medium above containing 25 mM of individual carbon sources.
Phospholipid fatty acid characterization
Phospholipid fatty acid analyses for the isolates were carried out by the DSMZ Identification Service using standard extraction and analytical methods (Miller, 1982; Kuykendall et al., 1988). After methylation and gas chromatographic quantitation, individual fatty acids were identified using the standard protocol of the Sherlock Microbial Identification System (MIDI Inc.).
DNA G+C content and DNA-DNA hybridization
DNA base composition (mol% G+C) for all strains was also determined by the Identification Service of the DSMZ (Braunschweig, Germany) using the method of Mesbah et al. (1989). The Identification Service of DSMZ performed DNA-DNA hybridizations for two isolates, Paraburkholderia sp. DNBP6-1T and Paraburkholderia sp. PP52- 1T, with their closest phylogenetic neighbors (P. bryophila LMG 23644T and P. mimosarum DSM 21841T, respectively) using the protocols of Cashion et al. (1977), De Ley et al. (1970) and Huss et al. (1983).
Results and discussion
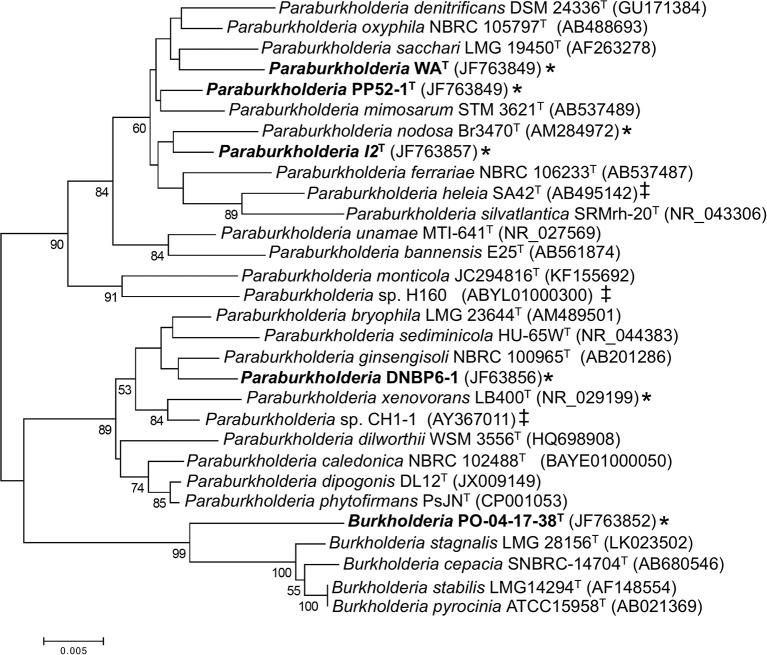
Phylogenetic analyses of 16S rRNA gene sequences (Figure 1) showed that the Pico de Orizaba isolate (PO-04-17-38T) clustered with the genus Burkholderia. Results from analyses conducted with EZtaxon (Kim et al., 2012) further showed that PO-04-17-38T was most closely related to B. stabilis LMG 14294T with a 16S rRNA gene sequence similarity of 97.49%. This level of similarity is considered consistent with species novelty and is less than similarity values (i.e., > 98.7–99.0%) for which DNA-DNA hybridization assays have been proposed for establishing species distinctions (Stackebrandt and Ebers, 2006).
Figure 1.
Maximum likelihood analysis of partial 16S rRNA gene sequences from CO-oxidizing Paraburkholderia and Burkholderia isolates, and related taxa. Bootstrap support ≥ 50% is indicated below the branches. A discrete Gamma distribution was used to model evolutionary rate differences among sites. All gapped positions and positions with missing or ambiguous data were removed, leaving 1263 positions in the final dataset. Asterisks (*) indicate confirmed CO oxidizers; ‡ indicates isolates with form I CO dehydrogenase genes identified in their genome sequences.
Phylogenetic analyses also showed that the Kilauea Volcano isolates clustered with the genus Paraburkholderia. Pairwise comparisons revealed that each of the isolates shared 16S rRNA gene sequence similarities no greater than 98.1% with other isolates, a difference consistent with species demarcation (Stackebrandt and Ebers, 2006). However, 16S rRNA gene sequence similarities for the isolates and their closest type species phylogenetic neighbors were somewhat higher: 98.53% for Paraburkholderia sp. I2T vs. P. oxyphila NBRC 105797T; 98.80% for Paraburkholderia sp. WAT vs. P. oxyphila NBRC 105797T; 98.96% for Paraburkholderia sp. PP52-1T vs. P. mimosarum DSM 21841T; and 99.11% for Paraburkholderia sp. DNBP6-1T vs. P. bryophila LMG 223644T.
To determine whether these similarities were consistent with the delineation of novel Paraburkholderia species, 16S rRNA gene sequence similarities were generated using EZTaxon (Kim et al., 2012) for pairs of type species in the phylogenetic neighborhood of the isolates. The maximum sequence similarity in this set of comparisons was 98.93%. For all pairs with similarities ≥ 98.0–98.93%, genome sequences were used to generate a set of average nucleotide identities (ANI; Supplementary Table 1). ANI did not exceed 95% (a recommended lower cutoff for species differentiation; Richter and Rosselló-Móra, 2009; Kim et al., 2014; Yarza et al., 2014) in any comparison (Supplementary Table 1, Supplementary Figure 1). Moreover, a more extensive analysis of ANI and 16S similarities involving multiple genera from multiple phyla yielded comparable resuts (Kim et al., 2014). This observation supports designation of isolates I2T and WAT as novel species based on similarities with their closest phylogenetic neighbor, P. oxyphila NBRC 105797T (98.53 and 98.80%, respectively).
However, similarities for isolates PP52-1T and DNBP6-1T and their closest neighbors exceed 98.3% (see above). DNA-DNA hybridizations (DDH) performed for these two isolates and their nearest neighbors yielded values of 15.8% (Paraburkholderia sp. PP52-1T vs. P. mimosarum DSM 21841T) and 12.4% (Paraburkholderia sp. DNBP6-1T vs. P. bryophila LMG 223644T); these values are consistent with species level demarcation (e.g., Stackebrandt and Ebers, 2006).
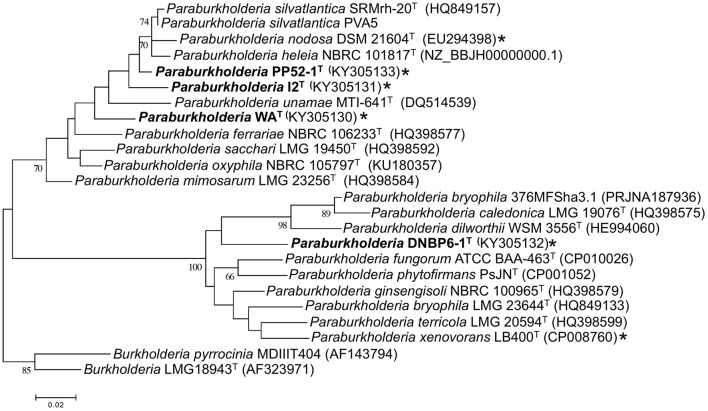
Phylogenetic analyses of partial recA gene sequences were also consistent with species level differentiation (Figure 2). Primers Bur3 and Bur4 have been previously shown to discriminate successfully among numerous Burkholderia (Paraburkholderia) species (Payne et al., 2005; Hall et al., 2015). Although the topologies of phylogenetic trees in this study and that of Payne et al. (2005) did not have strong bootstrap support, the overall topologies were consistent with the topologies of phylogenetic trees based on 16S rRNA gene sequences, and the isolates in this study were clearly distinct from related taxa (Figure 2).
Figure 2.
Maximum likelihood analysis of partial recA gene sequences from CO-oxidizing Paraburkholderia and Burkholderia isolates, and related taxa. Bootstrap support is shown below the branches for values ≥ 50%. All gapped positions and missing data were eliminated resulting in 331 positions for the analysis. *indicates CO oxidier.
BLAST analyses of Burkholderia and Paraburkholderia genomes and previously published culture-based assays of CO oxidation potential (Weber and King, 2012) showed that none of the closest type strain phylogenetic neighbors of the isolates in this study harbored form I carbon monoxide dehydrogenase (cox) genes or oxidized CO. However, genome analyses revealed form I cox genes in P. heleia SA42T, P. nodosa LMG 23741T, Paraburkholderia sp. CH1-1, and Paraburkholderia sp. H160. Previous studies have confirmed CO oxidation by P. nodosa LMG 23741T, P. xenovorans LB400T, and Paraburkholderia sp. LUP (King, 2003a; Weber and King, 2012). All of these isolates are phylogenetically distinct from those in this study (Figure 1).
CO uptake capacity varied substantially among the isolates (nmol CO mg protein−1 h−1; mean ± standard error): PO-04-17-38T, 20.4 ± 0.5 WAT, 27.7 ± 5.0; I2T, 88.8 ± 37.5; PP52-1T, 155.2 ± 24.2; DNBP6-1T, 226.5 ± 85.2. However, these values fall within previously reported ranges for other CO-oxidizing isolates (Weber and King, 2007; King and King, 2014a,b). Although rates for PO-04-17-38T and WAT were relatively low compared to the other isolates, the physiological and ecological significance of these differences are unknown at present.
All isolates were Gram-negative, non-spore forming, non-motile, CO-oxidizing rods [CO oxidation capacity was reported previously by Weber and King (2012)]. All were catalase positive, and all but PP52-1T were oxidase positive (Table 1). Colonies formed on solid pyruvate-yeast extract media [PYE, (Weber and King, 2012)] were circular with entire margins, with white coloration for I2T, DNBP6-1T, and PP52-1T; colonies for WAT and PO-04-17-38T were off-white.
Table 1.
Phenotypic characteristics of CO-oxidizing Paraburkholderia and Burkholderia isolates and phylogenetic neighbors.
| Trait | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Motility | − | − | − | − | − | − | − | nr | − | − | − |
| Oxidase reaction | − | + | + | + | + | − | + | + | + | + | + |
| Nitrate reduction | + | + | − | + | + | − | − | v | − | + | + |
| G+C content (mol%) | 63.7 | 63.1 | 60.3 | 64.9 | 64.9 | 64 | 62 | 64.8 | 68−69 | 66.8 | 67 |
| Temp optimum (°C) | 30 | 30 | 30 | 30 | 25 | 30 | nr | nr | 20−24 | nr | nr |
| Range | 15− <40 | 15− <40 | 15−40 | 15−45 | 5− <45 | 15−40 | nr | nr | 6−32 | nr | nr |
| pH optimum | 6.5 | 5 | 7.5 | 6.5 | 6.5 | nr | nr | nr | 4.7−5.2 | nr | nr |
| Range | 5.7− <8.5 | 5.7−8.0 | 5.7−8.5 | 5.7−8.5 | 5.7−8.0 | 3.5−8.5 | nr | nr | 3.2−6.6 | nr | nr |
| Enzyme reactions: | |||||||||||
| Arginine dihydrolase | − | − | ± | − | − | − | − | − | − | − | − |
| Gelatinase | − | − | − | + | − | − | − | − | − | + | + |
| Esculin hydrolysis | − | − | − | + | − | + | − | − | − | − | − |
| β-galactosidase | − | − | + | − | + | − | + | − | − | − | + |
| Assimilation: | |||||||||||
| Glucose | + | + | − | + | − | + | + | + | + | + | + |
| Arabinose | + | + | − | − | − | − | + | + | − | + | + |
| Mannose | + | + | − | + | − | + | + | v | + | + | + |
| Mannitol | + | + | + | + | − | + | + | + | + | + | + |
| N-acetylglucosamine | + | + | + | + | − | + | + | + | − | + | + |
| Malate | − | − | − | + | − | − | + | + | + | + | + |
| Gluconate | − | + | − | + | − | + | + | v | − | + | + |
| Caprate | + | + | − | + | − | + | + | − | + | + | + |
| Adipate | + | − | − | ± | − | + | v | − | + | + | + |
| Maltose | ± | + | − | + | − | + | − | − | + | − | − |
| Citrate | − | − | − | ± | − | + | + | − | + | + | + |
| Phenylacetate | + | − | − | + | − | + | − | v | + | + | + |
1 = PP52-1T, 2 = I2T, 3 = DNBP6-1T, 4 = WAT, 5 = PO-04-17-38T, 6 = P. oxyphila OX-01T (Otsuka et al., 2011); 7 = P. bryophila LMG 23644T (Vandamme et al., 2007a); 8 = P. mimosarum LMG 23256T (Chen et al., 2006); 9 = B. stabilis LMG 14294T (Henry et al., 2001); 10 = B. ambifaria LMG19812T (Coenye et al., 2001); 11 = B. diffusa LMG24065T (Vanlaere et al., 2008a). All isolates were catalase positive; nr = not reported; v = reported as variable among strains, type strain response not indicated; ± = variable for multiple trials.
Three isolates (PO-04-17-38T, PP52-1T, and WAT) grew with pH optima at 6.5, while the optimum for the remaining two was 7.5; all grew at pH 5.7 and showed an upper limit of pH 8–pH 8.5 (Table 1). Isolate PO-04-17-38T grew with a notably lower temperature optimum (25°C) and minimum (5°C) than for other isolates in this study, but its response to temperature was similar to that of its closest phylogenetic neighbor, Burkholderia stabilis LMG 14294T (Table 1). The remaining isolates grew optimally at 30°C with a minimum at 15°C and an upper limit of 40°–45°C; similar values were observed for their phylogenetic neighbors (Table 1).
Positive reactions on Biolog GN-2 plates varied from a low of 20 substrates for PO-04-17-38T (mostly sugars and a few simple organic acids) to 72 of 95 substrates for DNBP6-1T (Supplementary Table 2). With the exception of PO-04-17-38T, the isolates used a variety of sugars, sugar derivatives, organic acids, and amino acids that reflected the broad substrate utilization patterns reported for Paraburkholderia (e.g., Chen et al., 2007; Vandamme et al., 2007b; Compant et al., 2008; Vanlaere et al., 2008b; Aizawa et al., 2011; Otsuka et al., 2011). Nonetheless, substrate use differed for each of the isolates, and for three of the isolates when compared with their phylogenetic neighbors. Previously published BIOLOG GN-2 substrate utilization reactions facilitated comparisons between strains PP52-1T and P. mimosarum DSM 21841T, I2T and P. nodosa LMG 23741T, and DNBP6-1T and P. bryophila LMG 23644T (Supplementary Table 2). At least 10% of the 95 substrate reactions differed in each of these paired comparisons. It must be noted however that strain variability and variability among assays could reduce or increase these differences.
Results from API 20NE strips also revealed differences in substrate assimilation among the CO-oxidizing isolates and some of their close phylogenetic neighbors. Notably, isolate PO-04-17-38T did not assimilate any of the substrates in the panel, while its nearest phylogenetic neighbor, B. stabilis LMG 14294T, assimilated all but mannitol, N-acetylglucosamine and gluconate (Table 1). The lack of substrate assimilation by PO-04-17-38T contrasts with its ability to oxidize substrates in Biolog GN2 plates (Supplementary Table 2), and to grow in liquid culture with arabinose, glucose, mannitol, and mannose (Supplementary Table 3). The lack of substrate assimilation by PO-04-17-38T in the API 20NE panel was repeatable, however, which suggests that assay conditions for the API tests do not reliably reflect the capacity of PO-04-17-38T to use substrates.
Strain I2T differed from its closest phylogenetic neighbor, P. oxyphila OX-01T, in its ability to assimilate arabinose, but not adipate, citrate or phenylacetate (Table 1). Strain PP52-1T differed from its closest phylogenetic neighbor, P. mimosarum DSM 21841T, in its ability to assimilate mannose, caprate, adipate, and phenylacetate, but not malate (Table 1). Strain DNBP6-1T differed from its closest phylogenetic neighbor, P. bryophila LMG 23644T, in its inability to assimilate glucose, arabinose, mannose, malate, gluconate, caprate, or citrate (Table 1). Strain WAT differed from its closest phylogenetic neighbor, P. oxyphila OX-01T, in its inability to assimilate malate (Table 1).
The ability of the isolates to grow on various substrates in liquid culture (Supplementary Table 3) largely paralleled observations from Biolog GN2 plates (Supplementary Table 2). Strain PO-041783T grew with the fewest substrates (11 of 39, mostly sugars and a few organic acids) and was inhibited by several, while the Paraburkholderia isolates grew with 19–22 of 39 substrates. None of the strains were able to grow with glycine, phthalate, or solvents and alcohols, but two strains (I2T and WAT) were able to use dimethylamine and trimethylamine.
Clear distinctions were observed for the fatty acid compositions of each of the CO-oxidizing strains relative to compositions reported for each of their closest phylogenetic neighbors (Table 2). Strain PP52-1T contained greater amounts of C16:0, C17:0cyclo, and C16:1−2OH, and lesser amounts of C18:1ω7c fatty acids than P. mimosarum DSM21841T; PP52-1T also lacked C16:0−2OH. Strain I2T contained modest levels of C16:1−2OH, which was absent from P. oxyphila OX-01T, and considerably less C18:1ω7c. Strain WAT also contained C18:1−2OH and modest levels of C16:1−2OH, which were lacking in P. oxyphila OX-01T lipids. Strain DNBP6-1T contained notably higher amounts of C17:0cyclo and C19:0cycloω8c, and lower amounts of C18:1ω7c and isoC15:0−2OH/C16:1ω7c (sum feature 3) than P. bryophila LMG 23644T; strain DNBP6-1T also lacked C16:0−2OH. Strain PO-141738T contained sum feature 3, C18:1ω7c and C16:1−2OH, which were absent in B. stabilis LMG 14294T, but it lacked C16:0−2OH, C16:1ω7c, C18:1, and C18:1−2OH.
Table 2.
Fatty acid composition for novel Paraburkholderia and Burkholderia isolates and related strains.
| Fatty acid | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| C14:0 | 4.8 | 3.6 | 5.2 | 4.00 | 3.7 | 1.0 | 4.7 | 1.2 | 4.4 | 3.4 |
| C14:0−3OH | 4.5 | 5.2 | 4.7 | 4.80 | 4.7 | 4.7 | 5.3 | 6.3 | 5.4 | 7.2 |
| C16:0 | 29.1 | 19.1 | 23.5 | 18.3 | 20.9 | 22.6 | 18.3 | 24.2 | 25.6 | 15.0 |
| C16:0−2OH | – | 1.6 | – | – | – | 1.5 | – | – | 4.5 | – |
| C16:0−3OH | 3.1 | 4.2 | 3.6 | 4.00 | 3.7 | 3.9 | 3.7 | 4.3 | 5.7 | 5.9 |
| C17:0 | 0.1 | – | – | – | 0.1 | – | 0.1 | – | – | – |
| C17:0cyclo | 14.0 | 3.3 | 21.9 | 18.40 | 27.7 | 11.0 | 16.2 | 13.7 | 22.3 | 5.1 |
| C18:0 | 0.2 | – | 0.2 | – | 0.3 | – | 0.2 | 0.6 | – | 1.6 |
| C19:0iso | 0.2 | – | 0.4 | – | 0.5 | – | 0.1 | 0.6 | – | – |
| C19:0cycloω8c | 2.0 | 1.8 | 10.8 | 2.70 | 22.2 | 2.7 | 6.9 | 9.8 | 15.3 | 1.7 |
| C16:1ω5c | – | – | 0.6 | – | 0.6 | – | 0.6 | – | – | – |
| C16:1ω7c | – | – | – | – | – | – | – | – | 3.3 | 22.5 |
| C16:1−2OH | 5.4 | 0.9 | 5.3 | – | 3.0 | 2.1 | 4.6 | 5.2 | – | 1.5 |
| anteisoC17:1ω9c | 0.1 | – | 0.1 | – | – | – | 0.1 | – | – | – |
| C18:1 | – | – | – | – | – | – | – | – | 10.6 | – |
| C18:1ω5c | 0.1 | – | 0.2 | – | 0.1 | – | 0.2 | – | – | – |
| C18:1ω7c | 21.8 | 45.6 | 17.8 | 34.60 | 6.9 | 22.6 | 31.8 | 17.7 | – | 32.1 |
| C18:1−2OH | 0.8 | 1.0 | – | – | 0.6 | – | 1.1 | – | 2.4 | 2.4 |
| C18:1−11methylω7c | – | – | 0.2 | – | 0.6 | – | 0.1 | 0.5 | – | – |
| C20:2ω6.9c | – | – | 0.2 | – | 0.4 | – | – | 0.7 | – | – |
| C16:1ω7c/isoC15:0−2OH | 13.1 | 12.6 | 3.7 | 6.40 | 2.7 | 22.2 | 5.8 | 9.8 | – | – |
| Total | 99.3 | 98.9 | 99.6 | 93.10 | 98.7 | 94.0 | 99.8 | 94.3 | 99.5 | 97.9 |
Strain designations as follows: 1, PP52-1T; 2, P. mimosarum LMG 23256T (Chen et al., 2006); 3, I2T; 4, P. oxyphila OX-01T (Otsuka et al., 2011); 5, DNBP6-1T; 6, P. bryophila LMG 23644T (Lu et al., 2012); 7, WAT; 8, PO-04-17-38T; 9, B. stabilis LMG 14294T(Henry et al., 2001); 10, B. ambifaria LMG19812T (Coenye et al., 2001).
G+C contents varied between 60.3 and 64.9 mol% for the various CO-oxidizing isolates. These values were consistent with results reported for other Paraburkholderia and Burkholderia (Table 1).
The isolates obtained in this study share multiple characteristics with members of the genera Burkholderia and Paraburkholderia into which they were placed on the basis of 16S rRNA gene analyses (Figure 1). Although several CO-oxidizing members of Paraburkholderia have been identified previously (King, 2003a), PO-04-17-38T represents the first CO-oxidizing member of the Burkholderia, and in particular the Burkholderia cepacia complex, a group that harbors a number of important pathogens (Peeters et al., 2016). This observation is notable, since genomic sequencing of a large number of Burkholderia has yet to reveal any putative CO oxidizers, while at least 5 putative CO oxidizers have been identified among the Paraburkholderia. In addition, a phylogenetic analysis has shown that the PO-04-17-38T coxL gene clusters most closely with coxL from isolate DBNP6-1T, which suggests that a horizontal gene transfer event from Paraburkholderia to PO-04-17-38T might account for its apparently unusual capacity to oxidize CO.
The collective phenotypic, physiological, phylogenetic and biochemical results indicate that the CO-oxidizing Burkholderia and Paraburkholderia strains isolated during this study represent novel species, for which the following designations are proposed: Burkholderia alpina sp. nov. (the type strain is PO-04-17-38T = DSM 28031T = LMG 28138T); Paraburkholderia hiiakae sp. nov. (the type strain is I2T = DSM 28029T = LMG 27952T); Paraburkholderia paradisi sp. nov. (the type strain is WAT = DSM 28027T = LMG 27949T); Paraburkholderia metrosideri sp. nov. (the type strain is DNBP6-1T = DSM 28030T = LMG 28140T); Paraburkholderia peleae sp. nov. (the type strain is PP52-1T = DSM 28028T = LMG 27950T).
Description of Burkholderia alpina sp. nov.
Burkholderia alpina (al.pi'na. L. fem. adj. alpina, pertaining to the Alps and generally from or inhabiting mountainous regions, especially above the tree line, alpina referring to an isolate from an alpine altitude).
Cells are Gram-negative, non-sporing, non-motile rods, catalase, and oxidase positive. Colonies are circular, entire, off-white. The following carbon sources supported growth at 25 mM in a basal salts medium: alanine, arabinose, galactose, glucose, glutamate, β-hydroxybutyrate, lactate, mannitol, mannose, pyruvate, ribose, and tartrate. The following carbon sources did not support growth: acetate, aspartate, benzoate, betaine, citrate, dimethylamine, formate, gluconate, glucuronate, glycine, α-keto-glutarate, isopropanol, lactose, malonate, maltose, methanol, methylamine, phenylalanine, phthalate, propionate, succinate, trimethylamine, and valine. Weak growth was observed with fructose, glycerol, malate, and proline. Temperature optimum 25°C with growth at 5°C and at 35°C but not 45oC. pH optimum 6.5 with growth at pH 5.7 but not pH 8.5. Carbon monoxide is oxidized aerobically. The major cellular fatty acids (≥1% of the total) include: C14:0, C14:0−3OH, C16:0, C17:0cyclo, C16:1−2OH, C16:0−3OH, C18:1−ω7c, C19:0−cycloω8c, and iso-C15:0−2OH/C16:1−ω7c. The DNA G+C content of the type strain is 64.9 mol%. The type strain, PO-04-17-38T (= DSM 28031T = LMG 28138T), was isolated from volcanic soils from Pico de Orizaba (Mexico). The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain PO-04-17-38T is JF763852.1.
Description of Paraburkholderia hiiakae sp. nov.
Burkholderia hiiakae (hi.i.a'kae. Hawaiian N.L. fem. gen. n. from Hiiakaikapoliopele, a Hawaiian goddess of hula dancers, chants and sorcery, hiiakae, honoring the Hawaiian goddess of hula dancers).
Cells are Gram-negative, non-sporing, non-motile rods, catalase, and oxidase positive. Colonies are circular, entire, white. The following carbon sources supported growth at 25 mM in a basal salts medium: alanine, arabinose, aspartate, benzoate, citrate, formate, gluconate, glucose, α-keto-glutarate, glycerol, glutamate, β-hydroxybutyrate, lactate, malate, mannitol, phenylalanine, proline, propionate, ribose, succinate, and tartrate. The following carbon sources did not support growth: arabinose, aspartate, betaine, glucuronate, glutamate, glycine, isopropanol, lactose, maltose, methanol, methylamine, phthalate, and valine. Weak growth was observed with dimethylamine, fructose, galactose, malonate, mannose, and trimethylamine. Temperature optimum 30°C with growth at 15°C and no growth at 40°C. pH optimum 7.5 with growth at pH 5.7 but not pH 8.5. Carbon monoxide is oxidized aerobically. The major cellular fatty acids (≥1% of the total) include: C14:0, C14:0−3OH, C16:0, C17:0cyclo, C16:1−2OH, C16:0−3OH, C18:1−ω7c, C19:0−cycloω8c, C18:1−2OH, and iso-C15:0−2OH/C16:1−ω7c. The DNA G+C content of the type strain is 63.1 mol%. The type strain, I2T (= DSM 28029T = LMG 27952T), was isolated from volcanic soils from Kilauea Volcano (Hawai‘i, USA). The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain I2T is JF763857.1.
Description of Paraburkholderia metrosideri sp. nov.
Paraburkholderia metrosideri (me.tro.si.de'ri. N.L. fem. gen. n. metrosideri of Metrosideros, the genus Metrosideros polymorpha, the ohia tree).
Cells are Gram-negative, non-sporing, non-motile rods, catalase, and oxidase positive. Colonies are circular, entire, white. The following carbon sources supported growth at 25 mM in a basal salts medium: alanine, arabinose, benzoate, betaine, fructose, galactose, gluconate, glucuronate, glucose, α-keto-glutarate, glycerol, β-hydroxybutyrate, lactate, lactose, malate, mannitol, mannose, proline, propionate, pyruvate, ribose, succinate, and tartrate. The following carbon sources did not support growth: aspartate, citrate, dimethylamine, formate, glycine, isopropanol, methanol, methylamine, phenylalanine, phthalate, trimethylamine, and valine. Weak growth was observed with acetate, glutamate, malonate, and maltose. Temperature optimum 30°C with growth at 15°C and at 40°C but not 45°C. pH optimum 7.5 with growth at pH 5.7 and pH 8.5 but not 9.5. Carbon monoxide is oxidized aerobically. The major cellular fatty acids (≥1% of the total) include: C14:0, C14:0−3OH, C16:0, C17:0cyclo, C16:1−2OH, C16:0−3OH, C18:1−ω7c, C19:0−cycloω8c, and iso-C15:0−2OH/C16:1−ω7c. The DNA G+C content of the type strain is 60.3 mol%. The type strain, DNBP6-1T (= DSM 28030T = LMG 28140T), was isolated from volcanic soils from Kilauea Volcano (Hawai‘i, USA). The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain DNBP6-1T is JF763856.1.
Description of Paraburkholderia paradisi sp. nov.
Paraburkholderia paradisi (L. masc. gen. n. pa.ra.di'si of paradise).
Cells are Gram-negative, non-sporing, non-motile rods, catalase, and oxidase positive. Colonies are circular, entire, off-white. The following carbon sources supported growth at 25 mM in a basal salts medium: alanine, benzoate, citrate, dimethylamine, fructose, galactose, gluconate, glucose, glutamate, α-keto-glutarate, glycerol, glutamate, β-hydroxybutyrate, lactate, mannitol, mannose, proline, pyruvate, succinate, trimethylamine, and valine. The following carbon sources did not support growth: arabinose, aspartate, betaine, glycine, isopropanol, lactose, maltose, methanol, methylamine, phenylalanine, phthalate, and ribose. Weak growth was observed with acetate, formate, glucuronate, malate, malonate, propionate, and tartrate. Temperature optimum 30°C with growth at 15°C and at growth at 45°C but not 50°C. pH optimum 6.5 with growth at pH 5.7 and pH 8.5 but not 9.5. Carbon monoxide is oxidized aerobically. The major cellular fatty acids (≥1% of the total) include: C14:0, C14:0−3OH, C16:0, C17:0cyclo, C16:1−2OH, C16:0−3OH, C18:1−ω7c, C19:0−cycloω8c, C18:1−2OH, and iso-C15:0−2OH/C16:1−ω7c. The DNA G+C content of the type strain is 64.9 mol%. The type strain, WAT (= DSM 28027T = LMG 27949T), was isolated from volcanic soils from Kilauea Volcano (Hawai‘i, USA). The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain WAT is JF763851.1.
Description of Paraburkholderia peleae sp. nov.
Paraburkholderia peleae (pe.le'ae. Hawaiian N.L. gen. n. from Pelehonuamea, Pele of the sacred land, Hawaiian goddess of volcanoes, peleae honoring the goddess Pele).
Cells are Gram-negative, non-sporing, non-motile rods, catalase positive, and oxidase negative. Colonies are circular, entire, white. The following carbon sources supported growth at 25 mM in a basal salts medium: acetate, alanine, arabinose, aspartate, betaine, fructose, galactose, gluconate, glucose, α-keto-glutarate, glycerol, β-hydroxybutyrate, lactate, malate, malonate, mannitol, mannose, proline, propionate, pyruvate, ribose, succinate, and tartrate. The following carbon sources did not support growth: benzoate, citrate, dimethylamine, formate, glucuronate, glutamate, glycine, isopropanol, lactose, maltose, methanol, methylamine, phenylalanine, phthalate, trimethylamine, and valine. Temperature optimum 30°C with growth at 15°C and no growth at 40°C. pH optimum 6.5 with growth at pH 5.7 but not pH 8.5. Carbon monoxide is oxidized aerobically. The major cellular fatty acids (≥1% of the total) include: C14:0, C14:0−3OH, C16:0, C17:0cyclo, C16:1−2OH, C16:0−3OH, C18:1−ω7c, C19:0−cycloω8c, and iso-C15:0−2OH/C16:1−ω7c. The DNA G+C content of the type strain is 63.7 mol%. The type strain, PP52-1T (= DSM 28028T = LMG 27950T), was isolated from volcanic soils from Kilauea Volcano (Hawai‘i, USA). The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain PP52-1T is JF763849.1.
Author contributions
CW collected samples, enriched isolates, characterized isolates, analyzed data, and wrote the manuscript. GK collected samples, enriched isolates, characterized isolates, analyzed data, and wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer DS and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
We acknowledge support from National Science Foundation awards MCB-0348100 and DEB-1146444. We thank S.M. LaBorde, C. Landry, and C. Judd for technical support. We thank C. King for isolation of WAT and assistance with characterization; we thank F.A. Rainey for initial isolation of P. alpina PO-04-17-38T. The authors would like to thank the reviewers for their helpful and thoughtful comments which improved the article.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00207/full#supplementary-material
References
- Aizawa T., Vijarnsorn P., Nakajima M., Sunairi M. (2011). Burkholderia bannensis sp. nov., an acid-neutralizing bacterium isolated from torpedo grass (Panicum repens) growing in highly acidic swamps. Int. J. Syst. Evol. Microbiol. 61, 1645–1650. 10.1099/ijs.0.026278-0 [DOI] [PubMed] [Google Scholar]
- Bartholomew G. W., Alexander M. (1982). Microorganisms responsible for the oxidation of carbon monoxide in soil. Environ. Sci. Technol. 16, 300–301. 10.1021/es00099a013 [DOI] [PubMed] [Google Scholar]
- Bender M., Conrad R. (1994). Microbial oxidation of methane, ammonium and carbon monoxide, and turnover of nitrous oxide and nitric oxide in soils. Biogeochemistry 27, 97–112. 10.1007/BF00002813 [DOI] [Google Scholar]
- Callegan R. P., Nobre M. F., McTernan P. M., Battista J. R., Navarro-González R., McKay C. P., et al. (2008). Description of four novel psychrophilic, ionizing radiation-sensitive Deinococcus species from alpine environments. Int. J. Syst. Evol. Microbiol. 58, 1252–1258. 10.1099/ijs.0.65405-0 [DOI] [PubMed] [Google Scholar]
- Cashion P., Hodler-Franklin M. A., McCully J., Franklin M. (1977). A rapid method for base ratio determination of bacterial DNA. Anal. Biochem. 81, 461–466. 10.1016/0003-2697(77)90720-5 [DOI] [PubMed] [Google Scholar]
- Chen W.-M., de Faria S. M., James E. K., Elliott G. N., Lin K.-Y., Chou J.-H., et al. (2007). Burkholderia nodosa sp. nov., isolated from root nodules of the woody Brazilian legumes Mimosa bimucronata and Mimosa scabrella. Int. J. Syst. Evol. Microbiol. 57, 1055–1059. 10.1099/ijs.0.64873-0 [DOI] [PubMed] [Google Scholar]
- Chen W.-M., James E. K., Coenye T., Chou J.-H., Barrios E., de Faria S. M., et al. (2006). Burkholderia mimosarum sp. nov., isolated from root nodules of Mimosa spp. from Taiwan and South America. Int. J. Syst. Evol. Microbiol. 56, 1847–1851. 10.1099/ijs.0.64325-0 [DOI] [PubMed] [Google Scholar]
- Coenye T., Mahenthiralingam E., Henry D., LiPuma J. J., Laevens S., Gillis M., et al. (2001). Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 51, 1481–1490. 10.1099/00207713-51-4-1481 [DOI] [PubMed] [Google Scholar]
- Compant S., Nowak J., Coenye T., Clement C., Barka E. A. (2008). Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 32, 607–626. 10.1111/j.1574-6976.2008.00113.x [DOI] [PubMed] [Google Scholar]
- Conrad R. (1988). Biogeochemistry and ecophysiology of atmospheric CO and H2. Adv. Microb. Ecol. 10, 231–283. 10.1007/978-1-4684-5409-3_7 [DOI] [Google Scholar]
- Conrad R. (1996). Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60, 609–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ley J., Cattoir H., Reynaerts A. (1970). The quantitative measurement of DNA hybridization from renaturation rates. Eur. J. Biochem. 12, 133–142. 10.1111/j.1432-1033.1970.tb00830.x [DOI] [PubMed] [Google Scholar]
- Dunfield K., King G. M. (2004). Molecular analysis of carbon monoxide-oxidizing bacteria associated with recent Hawaiian volcanic deposits. Appl. Environ. Microbiol. 70, 4242–4248. 10.1128/AEM.70.7.4242-4248.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt P., Murray R. G. E., Wood W. A., Krieg N. R. (1994). Methods for General and Molecular Bacteriology. Washington, DC: American Society for Microbiology. [Google Scholar]
- Gomez-Alvarez V., King G. M., Nüsslein K. (2007). Comparative bacterial diversity in recent Hawaiian volcanic deposits of different ages. FEMS Microbiol. Ecol. 60, 60–73. 10.1111/j.1574-6941.2006.00253.x [DOI] [PubMed] [Google Scholar]
- Hall C. M., Busch J. D., Shippy K., Allender C. J., Kaestli M., Mayo M., et al. (2015). Diverse Burkholderia species isolated from soils in the southern United States with no evidence of B. pseudomallei. PLoS ONE 10:e0143254. 10.1371/journal.pone.0143254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry D. A., Mahenthiralingam E., Vandamme P., Coenye T., Speert D. P. (2001). Phenotypic methods for determining genomovar status of the Burkholderia cepacia complex. J. Clin. Microbiol. 39, 1073–1078. 10.1128/JCM.39.3.1073-1078.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss V. A. R., Festl H., Schleifer K. H. (1983). Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst. Appl. Microbiol. 4, 184–192. 10.1016/S0723-2020(83)80048-4 [DOI] [PubMed] [Google Scholar]
- Kim M., Oh H.-S., Park S.-C., Chun J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. 10.1099/ijs.0.059774-0 [DOI] [PubMed] [Google Scholar]
- Kim O. S., Cho Y. J., Lee K., Yoon S. H., Kim M., Na H., et al. (2012). Introducing EzTaxon: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. 10.1099/ijs.0.038075-0 [DOI] [PubMed] [Google Scholar]
- King C. E., King G. M. (2014a). Description of Thermogemmatispora carboxidovorans sp. nov., a carbon monoxide-oxidizing member of the Ktedonobacteria isolated from a geothermally-heated biofilm, and CO oxidation by members of the Ktedonobacteria. Int. J. Syst. Evol. Microbiol. 64, 1244–1251. 10.1099/ijs.0.059675-0 [DOI] [PubMed] [Google Scholar]
- King C. E., King G. M. (2014b). Thermomicrobium carboxidum sp. nov. and Thermorudis peleae gen. nov., sp. nov., carbon monoxide-oxidizing bacteria isolated from geothermally heated biofilms. Int. J. Syst. Evol. Microbiol. 64, 2586–2592. 10.1099/ijs.0.060327-0 [DOI] [PubMed] [Google Scholar]
- King G. M. (1999). Attributes of atmospheric carbon monoxide oxidation in Maine forest soils. Appl. Environ. Microbiol. 65, 5257–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. M. (2003a). Molecular and culture based analyses of aerobic carbon monoxide oxidizer diversity. Appl. Environ. Microbiol. 69, 7257–7265. 10.1128/AEM.69.12.7257-7265.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. M. (2003b). Contributions of atmospheric CO and hydrogen uptake to microbial dynamics on recent Hawaiian volcanic deposits. Appl. Environ. Microbiol. 69, 4067–4075. 10.1128/AEM.69.7.4067-4075.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. M., Weber C. F. (2007). Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 5, 107–118. 10.1038/nrmicro1595 [DOI] [PubMed] [Google Scholar]
- King G. M., Weber C. F. (2008). Interactions between bacterial carbon monoxide and hydrogen consumption and plant development on recent volcanic deposits. ISME J. 2, 195–203. 10.1038/ismej.2007.101 [DOI] [PubMed] [Google Scholar]
- King G. M., Weber C. F., Nanba K., Sato Y., Ohta H. (2008). Atmospheric CO and hydrogen uptake and CO oxidizer phylogeny for Miyake-jima, Japan volcanic deposits. Microb. Environ. 23, 299–305. 10.1264/jsme2.ME08528 [DOI] [PubMed] [Google Scholar]
- Kuykendall L. D., Roy M. A., O'Neill J. J., Devine T. E. (1988). Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int. J. Syst. Bacteriol. 38, 358–361. [Google Scholar]
- Lane D. (1991). 16s/23s rRNA Sequencing. West Sussex: John Wiley & Sons. [Google Scholar]
- Lu P., Zheng L.-Q., Sun J.-J., Liu H.-M., Li S.-P., Hong Q., et al. (2012). Burkholderia zhejiangensis sp. nov., a methyl parathion-degrading bacterium isolated from a wastewater-treatment system. Int. J. Syst. Evol. Microbiol. 62, 1337–1341. 10.1099/ijs.0.035428-0 [DOI] [PubMed] [Google Scholar]
- McDuff S., King G. M., Neupane S., Myers M. R. (2016). Isolation and characterization of extremely halophilic CO-oxidizing Euryarchaeota from hypersaline cinders, sediments and soils and description of a novel CO oxidizer, Haloferax namakaokahaiae Mke2.3. FEMS Microbiol. Ecol. 92:fiw028. 10.1093/femsec/fiw028 [DOI] [PubMed] [Google Scholar]
- Mesbah M., Premachandran U., Whitman W. (1989). Precise measurement of the G+C content of deoxyribonucleic acid by high performance liquid chromatography. Int. J. Syst. Bacteriol. 39, 159–167. 10.1099/00207713-39-2-159 [DOI] [Google Scholar]
- Miller L. T. (1982). Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxyl acids. J. Clin. Microbiol. 16, 584–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y., Muramatsu Y., Nakagawa Y., Matsuda M., Nakamura M., Murata H. (2011). Burkholderia oxyphila sp. nov., isolated from acidic forest soil that catabolizes (+)-catechin and its putative aromatic derivatives. Int. J. Syst. Evol. Microbiol. 61, 249–254. 10.1099/ijs.0.017368-0 [DOI] [PubMed] [Google Scholar]
- Payne G. W., Vandamme P., Morgan S. H., LiPuma J. J., Coenye T., Weightman A. J., et al. (2005). Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl. Environ. Microbiol. 71, 3917–3927. 10.1128/AEM.71.7.3917-3927.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters C., Meier-Kolthoff J. P., Verheyde B., De Brandt E., Cooper V. S., Vandamme P. (2016). Phylogenomic study of Burkholderia glathei-like organisms, proposal of 13 novel Burkholderia species, and emended descriptions of Burkholderia sordidicola, Burkholderia zhejiangensis, and Burkholderia grimmiae. Front. Microbiol. 7:877. 10.3389/fmicb.2016.00877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B., Ludwig W., Peplies J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiza L., Lalonde I., Guertin C., Constant P. (2014). Land-use influences the distribution and activity of high affinity CO-oxidizing bacteria associated to the type I-coxL genotype in soil. Front. Microbiol. 5:271. 10.3389/fmicb.2014.00271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M., Rosselló-Móra R. (2009). Shifting the genomic gold standard for species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawana A., Adeolu M., Gupta R. (2014). Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 5:429. 10.3389/fgene.2014.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E., Ebers J. (2006). Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 8, 6–9. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P., Govan J. R. W., LiPuma J. J. (2007b). Diversity and role of Burkholderia spp., in Burkholderia: Molecular Microbiology and Genomics, eds Coenye T., Vandamme P. (Wymondham: Horizon Bioscience; ), 1–28. [Google Scholar]
- Vandamme P., Opelt K., Knöchel N., Berg C., Schönmann De Brandt E., Eberl L., et al. (2007a). Burkholderia bryophila sp. nov., and Burkholderia megapolitana sp. nov,. moss-associated species with antifungal and plant-growth-promoting properties. Int. J. Syst. Evol. Microbiol. 57, 2228–2235. 10.1099/ijs.0.65142-0 [DOI] [PubMed] [Google Scholar]
- Vanlaere E., LiPuma J. J., Baldwin A., Henry D., De Brandt E., Mahenthiralingam E., et al. (2008a). Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int. J. Syst. Evol. Microbiol. 58, 1580–1590. 10.1099/ijs.0.65634-0 [DOI] [PubMed] [Google Scholar]
- Vanlaere E., Roelof van der Meer J., Falsen E., Falcao Salles J., de Brandt E., Vandamme P. (2008b). Burkholderia sartisoli sp. nov., isolated from a polycyclic aromatic hydrocarbon-contaminated soil. Int. J. Syst. Evol. Microbiol. 58, 420–423. 10.1099/ijs.0.65451-0 [DOI] [PubMed] [Google Scholar]
- Weber C. F., King G. M. (2007). Physiological, ecological and phylogenetic characterization of Stappia, a marine CO-oxidizing bacterial genus. Appl. Environ. Microbiol. 73, 1266–1276. 10.1128/AEM.01724-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C. F., King G. M. (2010a). Quantification of Burkholderia coxL in Hawaiian volcanic deposits. Appl. Environ. Microbiol. 76, 2212–2217. 10.1128/AEM.01861-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C. F., King G. M. (2010b). Distribution and diversity of Carbon monoxide-oxidizing bacteria and bulk bacterial communities across a succession gradient on a Hawaiian volcanic deposit. Environ. Microbiol. 12, 1855–1867. 10.1111/j.1462-2920.2010.02190.x [DOI] [PubMed] [Google Scholar]
- Weber C. F., King G. M. (2012). The phylogenetic distribution and ecological role of carbon monoxide oxidation in the genus Burkholderia. FEMS Microbiol. Ecol. 79, 167–175. 10.1111/j.1574-6941.2011.01206.x [DOI] [PubMed] [Google Scholar]
- Yabuuchi E., Kosako Y., Oyaizu H., Yano I., Hotta H., Hashimoto Y., et al. (1992). Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 36, 1251–127. 10.1111/j.1348-0421.1992.tb02129.x [DOI] [PubMed] [Google Scholar]
- Yarza P., Yilmaz P., Pruesse E., Glöckner F. O., Ludwig W., Schleifer K.-H., et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645. 10.1038/nrmicro3330 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.