Abstract
Genome-wide association studies suggest that 10q24.32-33 is a risk region for schizophrenia (SCZ). Considering the substantial genetic overlap between SCZ and major affective disorders, we would like to investigate whether the 10q24.32-33 region confers risk of affective disorders. We chose three SCZ genome-wide significant SNPs (rs7914558, rs7085104, and rs11191580) in 10q24.32-33 and collected the statistical data from European and Asian populations to perform systematic meta-analyses, which finally included up to 26,413 cases with affective disorders and 24,849 controls. Meta-analyses showed that all SNPs were nominally associated with major affective disorders. Considering the a priori evidence that these SNPs were associated with the expression of AS3MTd2d3 isoform in the human brain, our data confirms the potential involvement of AS3MTd2d3 in the genetic risk of major affective disorders.
Keywords: 10q24.32–33, AS3MTd2d3, Schizophrenia, Major affective disorders
Introduction
The association between psychotic and affective illnesses has been an area of debate in psychiatry. Within the family members of a proband with schizophrenia (SCZ), there is an increased risk (above the level in the general population) of both bipolar disorder (BPD) and major depressive disorder (MDD) [1,2]. More recently, a population-based cohort study reported that the prevalence of either SCZ or BPD was higher in family members of an index proband regardless of their diagnosis [3], and some genome-wide association studies (GWAS) also suggested shared genetic risk between SCZ and affective disorders (BPD and MDD) [4,5].
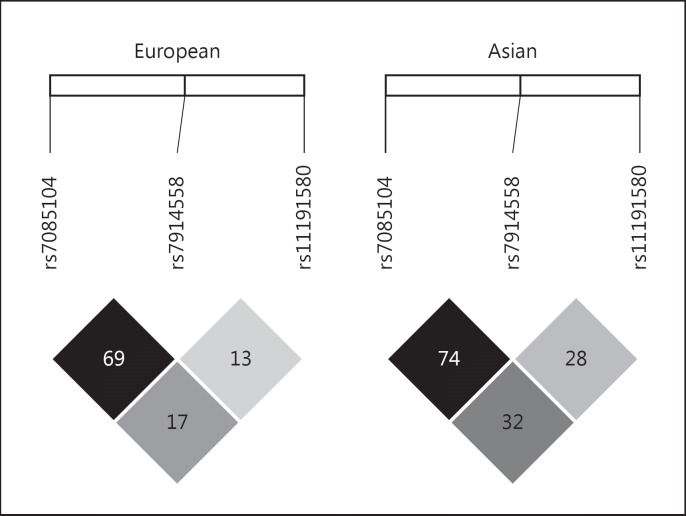
Chromosome 10q24.32-33 is one of the most significant genomic regions for SCZ [6,7,8]. In 2011, rs7914558 and rs11191580 in 10q24.32-33 were significantly associated with SCZ in the GWAS by the Psychiatric Genomics Consortium (PGC1) in populations of European descent [7], and the association became stronger in the later European GWAS meta-analysis (PGC1+SWE) with an increased sample size [6]. In this PGC1+SWE GWAS [6], rs7085104 was the most significant variant among the 10q24.32-33 region. Rs7085104 is in moderate to strong linkage disequilibrium with rs7914558 (r2 = 0.69 in Europeans and r2 = 0.74 in Asians; Fig. 1), although it is in low linkage disequilibrium with rs11191580 (r2 = 0.17 in Europeans and r2 = 0.32 in Asians; Fig. 1). In our previous study, we showed that rs7914558 and rs11191580 were also strongly associated with SCZ in Asian samples [9].
Fig. 1.
Linkage disequilibrium relationship of the included SNPs in Europeans and Asians.
In searching for the molecular mechanisms underlying the genetic risk of SCZ in 10q24.32-33, Li et al. [10] found that the risk allele of rs7085104 was strongly associated with an increased expression of a human-specific AS3MT transcript (AS3MTd2d3), which lacks exon 2 and 3 compared with its full-length form. Rs7914558 and rs11191580 were also associated with AS3MTd2d3 expression, although less significant than rs7085104 (supplementary data in the study by Li et al. [10]).
Considering the genetic overlap between SCZ and major affective disorders, it is compelling to investigate whether the SCZ risk variants in 10q24.32-33 also confer susceptibility to major affective disorders. Therefore, we systematically collected data from the literature to perform meta-analytic associations of the three SNPs in up to 51,262 subjects from diverse ethnic groups, and we found that these SCZ genetic risk variants at 10q24.32-33 were also nominally associated with major affective disorders.
Methods
Search Strategy
The search strategy was designed to identify all sources of published data in the literature. Electronic searches without language or date restriction were carried out in PubMed (1966-present), Web of Science (1899-present) and Embase (1974-present).
Search terms containing “10q24.32-33,” “rs7085104,” “rs7914558,” or “rs11191580,” and the terms “bipolar,” “depression,” and “GWAS” were used in all databases to identify relevant studies, along with a representative search strategy used for the PubMed query. Once these publications had been collected, their bibliographies were then searched for additional references. All articles identified through the searching process were evaluated based on the title and abstract. Clearly irrelevant studies were excluded from further consideration. The remaining articles received a full-text review. The last search was undertaken on August 21, 2016.
Eligibility Criteria
The studies eligible for the meta-analysis satisfy the following criteria: (1) case-control or family-based studies were included; (2) eligible studies should have case status defined with having a diagnosis of BPD or MDD according to the Diagnostic and Statistical Manual of Mental Disorders, fourth edition (DSM-IV) or related measurements; (3) studies included should have no overlap with each other.
Data Extraction
The data from each study were extracted using a standardized data extraction form. Descriptive information extracted contained (1) basic information with the name of the first author and year of publication, (2) methodology including study design, sample size, and definition of case status, and (3) sample characteristics such as collection area and ethnicity.
Statistical Analysis
We used odds ratio (OR) and standard error from each individual study to estimate heterogeneity between samples and to calculate the pooled OR and 95% confidence interval (CI) in the combined samples. To combine the results from individual samples, we first used Cochran's (Q) χ2 test to calculate the heterogeneity between studies. This test is a weighted sum of the squares of deviations of individual OR estimates from the overall estimate. If pQ < 0.10, the heterogeneity was considered statistically significant. In the presence of heterogeneity among individual studies, we used random effects models to combine the samples and to calculate the OR and the corresponding 95% CIs; for homogeneous studies, a fixed-effect model was used. The meta-analyses were performed using the classical inverse variance weighted methods in RevMan, version 5.3.5 (http://tech.cochrane.org/revman/download). The pooled ORs and the 95% CIs were graphically presented using a forest plot. Each study and its weight were represented by a square with the corresponding size in the plot.
Results
Literature Search and Eligible Studies
A flow chart illustrating the literature search and selection process is presented in Figure 2. The initial literature search identified 45 references. We removed 29 references with obviously irrelevant titles and reviewed the abstracts of the remaining 16 studies. This process left 14 potentially eligible studies. The full texts of the 14 publications were screened to examine their eligibilities based on the inclusion criteria. Among them, 9 studies were excluded because of the overlapped samples and lack of analyses of the SNPs of interest (rs7914558, rs7085104, and rs11191580). Therefore, a total of 5 studies with independent samples were considered eligible for the present meta-analysis [4,5,11,12,13]. Characteristics of the 5 included samples are listed in Table 1. Overall, the meta-analysis provided 11,883 BPD cases, 14,530 MDD cases, and 24,849 controls, and this sample size yielded >90% power to detect a significant association (p < 0.05) for any of the three SNPs assuming a commonly observed OR (1.10) in psychiatric genetic studies.
Fig. 2.
Literature search flow chart.
Table 1.
Characteristics of included samples in this meta-analysis
| Data source |
|||||
|---|---|---|---|---|---|
| Ruderfer et al. [5], 2014 | Saito et al. [11], 2014 | Grigoroiu-Serbanescu et al. [13], 2015 | PGC Cross-Disorder Group [4], 2013 | CONVERGE Consortium [12], 2015 | |
| Phenotype | BPD | BPD | BPD | MDD | MDD |
| Diagnostic criteria | DSM-IV, DSM-IIR, RDC | DSM-IV | DSM-IV | DSM-IV | DSM-IV |
| Cases, n | 10,410 | 1,012 | 461 | 9,227 | 5,303 |
| Controls, n | 10,700 | 993 | 436 | 7,383 | 5,337 |
| Genotyping method | Multiple | Sequenom iPLEX | Illumina | Multiple | Illumina Hi-Seq |
| rs7914558 | |||||
| p value | 0.0198 | 0.387 | 0.300 | 0.00245 | 0.255 |
| OR | 1.051 | 1.060 | 1.100 | 1.074 | 1.036 |
| 95% CI | 1.008–1.096 | 0.931–1.206 | 0.918–1.317 | 1.026–1.125 | 0.981–1.095 |
| rs7085104 | |||||
| p value | 0.00571 | N/A | N/A | 0.00198 | 0.0861 |
| OR | 1.063 | N/A | N/A | 1.078 | 1.054 |
| 95% CI | 1.018–1.110 | N/A | N/A | 1.028–1.130 | 0.998–1.113 |
| rs11191580 | |||||
| p value | 0.0120 | 0.391 | 0.240 | 0.0220 | 0.0231 |
| OR | 1.098 | 1.064 | 1.244 | 1.098 | 1.075 |
| 95% CI | 1.021–1.181 | 0.918–1.232 | 0.913–1.695 | 1.013–1.190 | 1.012–1.143 |
BPD, bipolar disorder; MDD, major depressive disorder; DSM-IV, Diagnostic and Statistical Manual of Mental Disorders, fourth edition; DSM-IIR, Diagnostic and Statistical Manual of Mental Disorders, second edition, revised; RDC, Research Diagnostic Criteria; OR, odds ratio; CI, confidence interval; N/A, not available.
Meta-Analysis of rs7914558 with Major Affective Disorders
A total of 3 BPD and 2 MDD studies were included to perform meta-analysis of rs7914558 [4,5,11,12,13]. Due to the fact that no significant heterogeneity between samples was observed (p = 0.875, I2 = 0), a classical fixed effect model was adopted to combine the individual samples. For BPD, our results showed that rs7914558 was nominally associated with the disease in a total of 11,883 patients and 12,129 normal controls (p = 0.0081, OR = 1.054). For MDD, we found that rs7914558 was also nominally associated with the illness in a total of 14,530 patients and 12,720 normal controls (p = 0.0017, OR = 1.074). We then merged BPD and MDD cases as a single phenotype, namely “major affective disorders.” In this analysis, the association for rs7914558 was strengthened in a total of 26,413 cases and 24,849 controls (p = 4.15 × 10-5, OR = 1.056). The forest plot of the meta-analysis between rs7914558 and major affective disorders is presented in Figure 3a.
Fig. 3.
Forest plot of meta-analysis for the included SNPs with major affective disorders: rs7914558 (a), rs7085104 (b), and rs11191580 (c). The results for BPD samples are marked in dark gray, and results for MDD samples are marked in light gray.
Meta-Analysis of rs7085104 with Major Affective Disorders
Rs7085104 was analyzed in three studies [4,5,12]. We adopted a classical fixed effect model to combine the individual samples given no significant heterogeneity was observed (p = 0.819, I2 = 0). An overall analysis showed that rs7085104 was strongly associated with major affective disorders in a total of 24,940 patients and 23,420 normal controls (p = 6.39 × 10-6, OR = 1.066), which was in the same direction of allelic effects with a priori SCZ analysis [6]. The forest plot of the meta-analysis between rs7085104 and major affective disorders is presented in Figure 3b.
Meta-Analysis of rs11191580 with Major Affective Disorders
A total of three BPD and two MDD studies were included to perform meta-analysis of rs11191580 [4,5,11,12,13]. A classical fixed effect model was adopted to combine the individual samples since no significant heterogeneity was observed (p = 0.900, I2 = 0). For BPD, rs11191580 showed nominal association in a total of 11,883 patients and 12,129 controls (p = 0.0043, OR = 1.097). For MDD, this SNP was also associated with the illness in a total of 14,530 patients and 12,720 normal controls (p = 0.0012, OR = 1.084). When we merged BPD and MDD samples, the association was further strengthened in a total of 26,413 cases and 24,849 controls (p = 1.62 × 10-5, OR = 1.089). The forest plot of the meta-analysis between rs11191580 and major affective disorders is presented in Figure 3c.
Discussion
Chromosome 10q24.32-33 is one of the most significant loci for SCZ in world populations [6,7,8,9,14,15]. In searching for the molecular mechanisms underlying the risk of psychiatric disorders conferred by 10q24.32-33 region, Li et al. [10] previously found that the risk SNPs (e.g., rs7085104) were significantly associated with the expression of AS3MTd2d3, a truncated isoform of AS3MT. Intriguingly, the expression of AS3MTd2d3 was also significantly increased in patients with MDD than healthy controls [10]. AS3MTd2d3 expression is also higher in patients with BPD [10], but such increase did not achieve statistical significance, which is likely due to limited sample size.
In sum, the present study confirms that 10q24.32-33 is a susceptibility locus for major affective disorders. These results also suggest the involvement of AS3MTd2d3 in the risk of major affective disorders.
Statement of Ethics
The study was approved by the internal review board of Kunming Institute of Zoology, Chinese Academy of Sciences.
Disclosure Statement
The authors declare no conflict of interest.
Acknowledgement
The authors are deeply grateful for the authors of previous studies who shared the genotype data. This work was supported by CAS Pioneer Hundred Talents Program (to M.L.).
References
- 1.Taylor MA. Are schizophrenia and affective disorder related? A selective literature review. Am J Psychiatry. 1992;149:22–32. doi: 10.1176/ajp.149.1.22. [DOI] [PubMed] [Google Scholar]
- 2.Berrettini WH. Are schizophrenic and bipolar disorders related? A review of family and molecular studies. Biol Psychiatry. 2000;48:531–538. doi: 10.1016/s0006-3223(00)00883-0. [DOI] [PubMed] [Google Scholar]
- 3.Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF, Hultman CM. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruderfer DM, Fanous AH, Ripke S, McQuillin A, Amdur RL; Schizophrenia Working Group of Psychiatric Genomics Consortium, Bipolar Disorder Working Group of Psychiatric Genomics Consortium, Cross-Disorder Working Group of Psychiatric Genomics Consortium, Gejman PV, O'Donovan MC, Andreassen OA, Djurovic S, Hultman CM, Kelsoe JR, Jamain S, Landen M, Leboyer M, Nimgaonkar V, Nurnberger J, Smoller JW, Craddock N, Corvin A, Sullivan PF, Holmans P, Sklar P, Kendler KS. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatry. 2014;19:1017–1024. doi: 10.1038/mp.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schizophrenia Psychiatric Genome-Wide Association Study Consortium Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao X, Luo XJ, Chang H, Liu Z, Li M. Evaluation of European schizophrenia GWAS loci in Asian populations via comprehensive meta-analyses. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-9990-3. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Jaffe AE, Straub RE, Tao R, Shin JH, Wang Y, Chen Q, Li C, Jia Y, Ohi K, Maher BJ, Brandon NJ, Cross A, Chenoweth JG, Hoeppner DJ, Wei H, Hyde TM, McKay R, Kleinman JE, Weinberger DR. A human-specific AS3MT isoform and BORCS7 are molecular risk factors in the 10q24.32 schizophrenia-associated locus. Nat Med. 2016;22:649–656. doi: 10.1038/nm.4096. [DOI] [PubMed] [Google Scholar]
- 11.Saito T, Kondo K, Iwayama Y, Shimasaki A, Aleksic B, Yamada K, Toyota T, Hattori E, Esaki K, Ujike H, Inada T, Kunugi H, Kato T, Yoshikawa T, Ozaki N, Ikeda M, Iwata N. Replication and cross-phenotype study based upon schizophrenia GWASs data in the Japanese population: support for association of MHC region with psychosis. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:421–427. doi: 10.1002/ajmg.b.32246. [DOI] [PubMed] [Google Scholar]
- 12.CONVERGE Consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grigoroiu-Serbanescu M, Diaconu CC, Heilmann-Heimbach S, Neagu AI, Becker T. Association of age-of-onset groups with GWAS significant schizophrenia and bipolar disorder loci in Romanian bipolar I patients. Psychiatry Res. 2015;230:964–967. doi: 10.1016/j.psychres.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Guan F, Wei S, Feng J, Zhang C, Xing B, Zhang H, Gao C, Yang H, Li S. Association study of a new schizophrenia susceptibility locus of 10q24.32-33 in a Han Chinese population. Schizophr Res. 2012;138:63–68. doi: 10.1016/j.schres.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 15.Guan F, Zhang T, Li L, Fu D, Lin H, Chen G, Chen T. Two-stage replication of previous genome-wide association studies of AS3MT-CNNM2-NT5C2 gene cluster region in a large schizophrenia case-control sample from Han Chinese population. Schizophr Res. 2016;176:125–130. doi: 10.1016/j.schres.2016.07.004. [DOI] [PubMed] [Google Scholar]