Abstract
Human fibroblast growth factor-1 (FGF-1) has broad therapeutic potential in regenerative medicine but has undesirable biophysical properties of low thermostability and 3 buried cysteine (Cys) residues (at positions 16, 83, and 117) that interact to promote irreversible protein unfolding under oxidizing conditions. Mutational substitution of such Cys residues eliminates reactive buried thiols but cannot be accomplished simultaneously at all 3 positions without also introducing further substantial instability. The mutational introduction of a novel Cys residue (Ala66Cys) that forms a stabilizing disulfide bond (i.e., cystine) with one of the extant Cys residues (Cys83) effectively eliminates one Cys while increasing overall stability. This increase in stability offsets the associated instability of remaining Cys substitution mutations and permits production of a Cys-free form of FGF-1 (Cys16Ser/Ala66Cys/Cys117Ala) with only minor overall instability. The addition of a further stabilizing mutation (Pro134Ala) creates a Cys-free FGF-1 mutant with essentially wild-type biophysical properties. The elimination of buried free thiols in FGF-1 can substantially increase the protein half-life in cell culture. Here, we show that the effective cell survival/mitogenic functional activity of a fully Cys-free form is also substantially increased and is equivalent to wild-type FGF-1 formulated in the presence of heparin sulfate as a stabilizing agent. The results identify this Cys-free FGF-1 mutant as an advantageous “second generation” form of FGF-1 for therapeutic application.
Keywords: FGF-1, disulfide, protein stability, empirical phase diagram, X-ray crystallography, cysteine-free mutant, protein engineering, cystine
Introduction
Cysteine (Cys) residues in proteins can have a pronounced effect on folding equilibrium. Oxidation of 2 Cys residues in close proximity, and with appropriate stereochemistry, in the native state can form a covalent bond (a cystine). Cystines prevent complete unfolding due to residual structure in the denatured state, thus substantially reducing the entropy of the denatured state (while having minimal consequences on the native state).1,2 In such cases, the folding rate constant can be significantly increased, whereas the unfolding rate constant may be largely unperturbed (thereby shifting the folding equilibrium in favor of the native state).
In contrast, buried reduced Cys residues can have an opposite effect on the folding equilibrium. Cys residues are subject to chemical modification (e.g., oxidation) that can substantially alter their physical properties of size and charge. In the native state, a buried residue is largely protected from chemical modification by agents in bulk solvent; however, in the denatured state such buried residues are freely exposed and can readily participate in chemical modification. In the case of Cys residues, the alteration of size and charge associated with oxidative modification can preclude their accommodation within the buried environment of the native state. For example, exposed Cys residues can react to form a mixed disulfide with a thiol compound present in solution. Effective refolding would require that the added bulk of the thiol adduct be successfully accommodated within the core of the protein. Because most protein cores are efficiently packed,3 it is unlikely that such accommodation is possible without significant structural and stability perturbation.4,5 Cys residues can also oxidize to form various acidic forms (e.g., sulfenic, sulfinic, and cysteic acid); thus, refolding would require accommodation of a novel acidic charge within the core of the protein. However, the accommodation of an unpaired charge within the hydrophobic core region is associated with substantial destabilization—in the range of 20–24 kJ/mol.6 Chemical modification of a buried Cys residue is therefore likely to result in an irreversible unfolding pathway. Such irreversibility shifts the folding equilibrium (by Le Chatelier’s principle), continuously driving it toward the unfolded state, and substantially reducing functional half-life.7–9
Buried Cys residues are commonly found in proteins,10,11 and their oxidation has long been known to lead to protein inactivation.7,12–14 Fibroblast growth factor-1 (FGF-1) is a small protein (with a 140 amino acid “mature” form), with therapeutic potential in regenerative medicine,15–18 and contains 3 buried Cys residues (at positions 16, 83, and 117).19 A prior study of Cys positions in wild-type (WT) FGF-1 demonstrated that the functional half-life of FGF-1 in cell culture media can be increased as much as 40-fold by mutational substitution of Cys residues.9 A detailed structural and thermodynamic study also showed that FGF-1 is structurally optimized to accept Cys residues at 2 of these buried positions (Cys16, Cys83)20,21; thus, their mutational substitution is associated with significant thermodynamic destabilization. Unfortunately, WT FGF-1 is characterized by very low overall thermostability22,23; thus, it is unable to accommodate any significantly destabilizing mutation and remain folded. Consequently, it is not feasible to generate a Cys-free form of FGF-1 by simple point mutation at each Cys position and retain a favorable folding equilibrium.21,24
Sequence and structure analysis of the FGF family indicates that one of the Cys residues (Cys83) in FGF-1 can be considered a vestigial half-cystine—such that an Ala66→Cys mutation (Ala66Cys; for facilitation of reading the single letter amino acid code will be used when describing combination mutations) can create a novel cystine with Cys83.25 A query of the UNIPROT database indicates that this novel cystine is not present in any FGF-1 of any species. Although the Ala66Cys mutation introduces an additional Cys residue, the resulting Cys66-Cys83 cystine effectively eliminates one of the 3 native buried Cys residues in WT FGF-1 while simultaneously stabilizing the protein. Based on thermodynamic data for Ala, Val, Ser, and Thr point mutations at the remaining 2 buried Cys positions (Cys16 and Cys117)21 it appeared feasible that point mutations that eliminate Cys residues at positions Cys16 and Cys117, although destabilizing overall, could potentially be combined with the stabilizing Ala66Cys disulfide mutation to generate a Cys-free form of FGF-1 with minimal overall thermodynamic perturbation. Such a mutation should entirely eliminate irreversible unfolding due to modification of buried Cys residues.
In the present report, we describe thermodynamic, biophysical, structural, and functional characterization of a Cys-free mutant form of FGF-1. We show the combination mutant C16S/A66C/C117A achieves an overall thermostability essentially indistinguishable from WT FGF-1, while effectively eliminating all Cys residues. This mutant does, however, manifest acidic (pH 5.0) sensitivity to unfolding that is not apparent in WT FGF-1. An additional point mutation, Pro134Ala, has only a modest increase in thermostability but less acidic pH sensitivity. Assays of cell survival and mitogenicity, using both a BaF3 cell system expressing FGF receptor-1c (FGFR-1c) and 3T3 fibroblasts, demonstrate up to a 3-order of magnitude increase in functional potency for various Cys-free mutants compared to WT FGF-1.
Materials and Methods
Mutant Design
Based on our previous studies,25 the introduction of an Ala66Cys mutation yields a novel cystine with Cys83 and also provides ~10kJ/mol gain in overall protein stability. However, no equivalent cystine-forming mutations were identified adjacent to positions Cys16 and Cys117. A systematic mutational analysis of substitutions of Cys16 in WT FGF-1 indicated that the least destabilizing amino acid substitution is Ser, albeit with a loss of ~10 kJ/mol in overall stability.21,24 In contrast, position Cys117 is largely neutral to mutation, although Cys117Ala appears the best choice and provides ~2 kJ/mol increase in stability.21,26 With simple additivity of mutational effects, the combination of Cys16Ser, Ala66Cys, and Cys117Ala offers the possibility of a completely Cys-free form of FGF-1 with minimal perturbation of WT FGF-1 stability. The C16S/A66C/C117A combination mutation was constructed to test this design strategy. Further stability gains were investigated by combining this Cys-free mutant design with additional stabilizing mutations Pro134Ala and Pro134Val.27 Mutant pI values were calculated from the ExPASy Compute pI/Mw tool.28 Major histocompatibility complex 1 (MHC-1) binding predictions were made using the IEDB analysis resource Consensus tool.29 The average predicted affinity for a given non-apeptide centered at the site of mutation was calculated for all MHC alleles. The predicted aggregation potential for WT FGF-1 and the C16S/A66C/C117A/P134A mutant were calculated from the X-ray structures (WT FGF-1 PDB accession 1JQZ) using the AggreScan 3D server.30
Protein Expression and Purification
Recombinant protein expression used a pET21a(+) expression vector (EMD Millipore, Billerica, MA) with a codon-optimized synthetic gene encoding the 140 amino acid “mature” form of human WT FGF-1 and with an N-terminal 6× His tag. The QuickChange™ site-directed mutagenesis protocol (Agilent Technologies, Santa Clara, CA) was used to introduce all FGF mutations and were confirmed by DNA sequencing (Biomolecular Analysis Synthesis and Sequencing Laboratory, Florida State University). Recombinant WT FGF-1 protein was expressed from pET21a(+)/BL21(DE3) Escherichia coli as previously described.31 Recombinant disulfide mutants were expressed from SHuffle T7 Express E coli (New England BioLabs, Ipswich, MA) and Luria broth media. The E coli culture was incubated at 30°C until OD600 = 0.6, at which point the temperature was shifted to 20°C and 1 mM isopropyl-β-D-thio-galactoside was added to induce protein expression with overnight incubation. The expressed protein was purified using sequential column chromatography on Ni-nitrilotriacetic acid affinity resin (Qiagen, Valencia, CA) and heparin Sepharose resin (GE Life Sciences, Pittsburgh, PA). Protein purity was evaluated by gel densitometry of Coomassie blue–stained SDS-PAGE. An extinction coefficient of E280nm (0.1 %, 1 cm) = 1.2631 was used for concentration determination of WT FGF-1 and all mutant proteins.
5,5′-Dithiobis-(2-Nitrobenzoic Acid) Assay
A 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) assay was used to determine the molarity of Cys in each purified mutant protein and thereby quantify the extent of cystine formation. Assays were performed by mixing 100 μL of 0.1-mM (1.6 mg/mL) purified protein with 400-μL DTNB reaction solution (0.75-mM DTNB, 0.1-M sodium phosphate, 7.5-M guanidinium hydrochloride [GuHCl], 1-mM EDTA, pH 8.0). The DTNB reaction mixtures were incubated at room temperature for 15 min, and the absorbance at 412 nm was measured to quantify the amount of reduced 2-nitro-5-thiobenzoate (TNB−) product released (using an extinction coefficient of 13,700 M−1). WT FGF-1 was also assayed as a control (i.e., 3 mol Cys per molar protein basis). In each mutant case, the mole fraction of Cys is 2.0 and the % oxidized cystine was calculated as:
| (1) |
Analytical Heparin Sepharose Chromatography
The chromatographic profiles of purified WT FGF-1 and C16S/A66C/C117A/P134A mutant proteins were characterized on an analytical heparin Sepharose chromatography column. One hundred micrograms of each protein in 2.0 mL of crystal buffer was loaded onto a 10-mL heparin Sepharose CL-6B (GE Healthcare BioSciences AB, Uppsala, Sweden) column (1.6 cm × 5.0 cm) equilibrated to crystal buffer. The column was resolved with a linear gradient of 0–2.0 M NaCl (in crystal buffer) over 20 column volumes, with a flow rate of 1.0 mL/min, using an AKTA FPLC chromatography workstation (GE Healthcare Life Sciences, Pittsburgh, PA).
Empirical Phase Diagrams
Sample Preparation
Purified WT and mutant FGF-1 proteins were dialyzed into 20-mM citrate-phosphate buffer (pH 3.0–8.0) with an overall ionic strength of 0.15 M (by adjusting NaCl concentration) using an 8 kDa molecular weight cutoff membrane tubing (Spectrum Industries Inc., Chippewa Falls, WI). The effects of 3-fold mass addition of heparin sulfate on thermal stabilization of WT FGF-1 and mutant C16S/A66C/C117A/P134A at pH 7.0 were also evaluated by empirical phase diagram (EPD) analysis.
Intrinsic Fluorescence Spectroscopy
All fluorescence (FL) experiments were performed using a PTI QM-40 spectrofluorometer (Photon Technology International, Birmingham, NJ) equipped with a 4-cell position temperature-controlled Pelletier (Quantum Northwest, Liberty Lake, WA), a 75 W Xe lamp, and a R1527 photomultiplier tube. The samples were excited at 280 nm and FL emission was monitored from 290 to 380 nm in 1-nm increments and a 1 s integration time at each wavelength. All experiments were performed using 1-cm quartz cuvettes and 0.1 mg/mL protein concentration. Excitation and emission slits were set to 3 and 4 nm, respectively. The spectrum of the buffer was subtracted from all measurements and theratio of the intensityat305 nm to the intensity at 330 nm was plotted as a function of temperature.
Extrinsic Fluorescence Spectroscopy
Accessibility of hydrophobic moieties of FGF-1 as a function of temperature was assessed using 8-anilino-1-naphthalenesulfonate (ANS; Sigma, St. Louis, MO). ANS (suspended in dimethylsulfoxide) was added to FGF-1 at a 15:1 molar ratio and incubated in the dark for at least 5 min at 10°C. The samples were measured using an excitation wavelength of 372 nm and the emission spectrum was monitored from 400 to 600 nm as a function of temperature (10°C–87.5°C). The excitation and emission slits were set at 3 and 4 nm, respectively. Step size and integration time were 1 nm and 0.5 s, respectively. The spectra were collected at 2.5°C intervals with a 2-min equilibration time at each temperature. Instrument and cuvette setup were identical to the intrinsic FL experiments. FL intensity at 480 nm was plotted as a function of temperature and emission of the buffer-containing ANS was subtracted from all measurements.
Static Light Scattering
Light scattering was measured at 280 nm and quantifying the scattered light at the same wavelength. Data were acquired concurrent with the intrinsic FL emission using another photomultiplier tube oriented 180° from the FL photomultiplier. Excitation and emission slits were set at 3 and 0.25 nm, respectively. The light scattering intensity of the buffer was subtracted from all measurements before data analysis.
EPD Construction
An EPD is a colored map representing the protein structural changes as a function of environmental conditions (i.e., pH and temperature). Protein unfolding was monitored by different methods including intrinsic FL, FL dye binding, and static light scattering (SLS). The color changes correspond to spectroscopic signal changes and are plotted as a function of pH and temperature. Three-index EPDs were constructed as described29 using the MiddaughSuite software. Extrinsic FL values were normalized by min/max normalization before EPD construction.
Isothermal Equilibrium Denaturation
Purified proteins were buffer exchanged into 50-mM sodium phosphate, 100-mM sodium chloride, 10-mM ammonium sulfate, pH 7.5 (“crystallization buffer”) using an 8 kDa molecular weight cutoff membrane tubing (Spectrum Industries Inc., Chippewa Falls, WI). WT FGF-1 contains a single buried Trp residue (Trp107) whose FL is internally quenched in the native state; Trp107 quenching is relatively diminished on denaturation, thereby providing a spectroscopic probe of unfolding.19,23 WT FGF-1 and mutant proteins were equilibrated overnight in crystallization buffer with and without 2-mM dithiothreitol at 298 K in 0.1-M increments of GuHCl (final protein concentration 5 μM) to evaluate the effects of cystine formation on stability. FL intensity was quantified using a Varian Eclipse FL spectrophotometer (Varian Medical Technologies, Palo Alto, CA). Triplicate scans were collected and averaged, and buffer background was collected and subtracted from the protein scans. FL scans were integrated to quantify the total FL as a function of denaturant concentration. Data were analyzed using a 6 parameter 2-state model32:
| (2) |
where [D] is the denaturant concentration, F0N and F0D are the 0 M denaturant intercepts for the native and denatured state baselines, respectively, and SN and SD are the slopes of the native and denatured state baselines, respectively. ΔG0 and m describe the linear function (y-intercept and slope, respectively) of the unfolding free energy versus denaturant concentration. The effect of mutation on the stability of the protein (ΔΔG) is calculated taking the difference between the GuHCl concentration at the midpoint of denaturation (i.e., Cm) for WT and mutant protein and multiplying by the average m-value, as described by Pace and Scholtz33:
| (3) |
where a negative value indicates the mutation is stabilizing in comparison to the WT protein.
Crystallization, X-Ray Diffraction Data Collection and Processing, and Structural Refinement
Mutant proteins were concentrated to 6 mg/mL in crystallization buffer for crystallization trails. Diffraction quality crystals were obtained at 4°C and 25°C incubation from solutions containing 0.7 M–1.0 M sodium citrate, 0.1-M imidazole, pH 8.5, using the hanging drop vapor diffusion method. Crystals were mounted by dipping in the mother liquor mixed with 25% glycerol as cryoprotectant and frozen immediately in liquid nitrogen. Mutant C16S/A66C/C117A/P134A diffraction data were collected at the Southeast Regional Collaborative Access Team 22-ID beam line at the Advanced Photon Source at Argonne National Laboratory using a Mar300 detector (MarUSA, Evanston, IL). Diffraction data were indexed, integrated, and scaled using the HKL2000 software package.34 Other mutants grew as single plate crystals, but diffraction data indicated that all crystals were multi-nucleated, and no single crystals suitable for diffraction were obtained.
The C16S/A66C/C117A/P134A mutant structure was solved by molecular replacement using the PHENIX software package35 and WT FGF-1 coordinates (PDB code 1JQZ) as a search model. The Coot molecular graphics software package36 was used for model building and visualization. Structure refinement was performed using PHENIX and with 5% of reflection data set aside as the test data for free R calculation.37 Refined atomic coordinates and reflection data have been deposited in the RCSB data bank (accession 4YOL).
BaF3/FGFR-1c Cell Culture and Cell Survival/Mitogenic Stimulation
BaF3 murine lymphoid cells were transfected to express the single FGFR-1c isoform so that specific FGFR-1c activation can be quantified.38 Cells were maintained in Roswell Park Memorial Institute 1640 media (Sigma Chemical, St. Louis, MO) supplemented with 10% newborn bovine serum (Sigma Chemical), 50-μM β-mercaptoethanol, 0.5 ng/mL murine recombinant interleukin-3 (PeproTech Inc, Rocky Hill, NJ), 2-mM L-glutamine, penicillin-streptomycin (“BaF3 culture medium”), and G418 antibiotic (600 μg/mL). FGFR-1c expressing BaF3 cells were washed twice in BaF3 “assay media” (“culture media” lacking both murine recombinant interleukin-3 and β-mercaptoethanol) and plated at a density of 30,000 cells per well in a 96-well assay plate in assay media containing heparin sulfate (1 μg/mL) and concentrations of recombinant WT FGF-1 and Cys-free mutants ranging from 0.02 to 5 nM (3.18 × 102–7.95 × 105 pg/mL). The cells were incubated for 36 h, and DNA synthetic activity was determined by adding 1 μCi of 3H-thymidine in 50 μL of BaF3 assay medium to each well. Cells were harvested after 4 h by filtration through glass fiber paper. Incorporated 3H-thymidine was counted on a MicroBeta plate scintillation counter (PerkinElmer, Waltham, MA).
3T3 Fibroblast Mitogenic Stimulation
The mitogenic activity of WT FGF-1 and C16S/A66C/C117A/P134A mutant protein toward NIH 3T3 fibroblasts was performed following a previously described procedure.27 In contrast to BaF3 cells, NIH 3T3 fibroblast cells express heparan sulfate (HS) proteoglycan, and no heparin sulfate was added in this assay. Briefly, NIH 3T3 fibroblasts were plated in Dulbecco’s Modified Eagle’s Medium high glucose 1× (Gibco, Carlsbad, CA) supplemented with 10% (vol/vol) newborn calf serum (Sigma), 100 units/mL of penicillin, 100 μg/mL of streptomycin, 10 μg/mL of gentamicin, and 4 mM L-Glutamine (Gibco) (“serum-rich medium”) in T75 tissue culture flasks (Fisher, Pittsburgh, PA). Cultures were incubated at 37°C with 5% (vol/vol) CO2 supplementation. At ~80% cell confluence, the cells were washed with 5 mL of Tris buffered saline (TBS) (0.14-M NaCl, 5.1-mM KCl, 0.7-mM Na2HPO4, 24.8-mM Trizma base, pH 7.4) and subsequently treated with 5 mL of TrypLE Express™ (Gibco). The trypsinized cells were seeded in T25 tissue culture flasks at a density of 3.0 × 104 cells/cm2 (representing ~20% confluence). Cell quiescence was initiated by serum starvation in high glucose 1× supplemented with 0.5% newborn calf serum, 100 units/mL of penicillin, 100 μg/mL of streptomycin, 10 μg/mL gentamicin, and 4 mM L-Glutamine (“serum starvation media”). Cultures were incubated for 48 h at 37°C. The medium was then aspirated, washed twice with 5-mL TBS, and replaced with fresh serum starvation medium supplemented with WT or mutant FGF-1 protein from 0.006 to 630 nM (1.0 × 102–1.0 × 107 pg/mL, the traditional concentration units for FGF-1 mitogenic assays) and the cultures incubated for an additional 48 h. After this incubation, the medium was aspirated, and the cells were washed twice with 5 mL of TBS. One milliliter of TrypLE Express™ was added to release the cells from the flask surface; subsequently, 2 mL of serum-rich medium was added to dilute and inhibit the trypsin. Triplicate cell samples were counted using a Coulter Counter (Beckman Coulter, Brea, CA). Dose response experiments were performed in quadruplicate, and the cell counts were averaged and normalized to % maximal stimulation.
Results
Mutant Protein Design, Expression, Purification, and Oxidization
Predicted pI values for all mutants were within 0.05 pH units of the WT FGF-1 protein (Supplementary Table 1), indicating that the mutations had minimal effect on overall charge. The analysis of predicted MHC-1 affinities for all nonapeptide epitopes within FGF-1 is shown in Supplementary Figure 1. All epitopes involving sites of mutation are predicted to have average MHC-1 affinities of >2 × 104 nM (i.e., medium-to-low affinity). The various point mutations used in construction of Cys-free FGF-1 were characterized for their effect on MHC-1 binding affinity and were predicted to have a minimal effect in each case (Supplementary Fig. 1). Mutation sites are located at solvent inaccessible positions, and the predicted aggregation potential in response to mutation, in each case, indicated minimal effect (Supplementary Fig. 2).
The SHuffle T7 Express E coli is a genetically engineered strain with an oxidative intracellular environment that promotes cystine formation in expressed proteins. The DTNB assay showed that the C16S/A66C/C117A and C16S/A66C/C117A/P134A mutant proteins each had 2 mol equivalents of Cys—indicating essentially 100% cystine formation. Mutant C16S/A66C/C117A/P134V had ~60% cystine formation. SDS-PAGE analysis of mutants C16S/A66C/C117A and C16S/A66C/C117A/P134A under nonreducing conditions showed a single protein band corresponding to a uniformly oxidized state. However, SDS-PAGE for mutant C16S/A66C/C117A/P134V under nonreducing conditions exhibited a doublet indicating a mixture of oxidized and reduced forms (with the slower-migrating band being the reduced form). Subsequently, mutant C16S/A66C/C117A/P134V was air oxidized at room temperature for 2 weeks, resulting in >90% disulfide formation by DTNB assay.
Analytical Heparin Sepharose Chromatography
The elution peak of WT FGF-1 from analytical heparin Sepharose chromatography occurred at 1.26 M NaCl; whereas, the elution peak of the C16S/A66C/C117A/P134A mutant occurred at 1.18 M NaCl (Supplementary Fig. 3).
Crystallization and X-Ray Structure Determination
Crystals for each Cys-free mutant were obtained; however, single crystals suitable for diffraction studies were obtained only for the C16S/A66C/C117A/P134A mutant. A 1.97 Å resolution data set, with excellent signal-to-noise and data completion, was collected (Table 1). This mutant crystallized in C-centered orthorhombic space group C2221 with 2 molecules in the asymmetric unit—as observed for WT FGF-1 (PDB code: 1JQZ). A straightforward molecular replacement solution was found using the 1JQZ WT FGF-1 structure as the search model. The 2FO−FC difference electron density map at each site of mutation permitted unambiguous assignment of the specific mutant rotamer, and formation of the Cys66-Cys83 cystine was confirmed. The mutant X-ray structure refined to excellent crystallographic residual and stereochemistry (Table 1).
Table 1.
X-ray Diffraction Data Collection, Processing, and Structure Refinement of Mutant C16S/A66C/C117A/P134A
| Data Collection and Processing | |
|---|---|
| Space group | C2221 |
| Cell constants | |
| a (Å) | 75.6 |
| b (Å) | 97.8 |
| c (Å) | 107.9 |
| α, β, γ (°) | 90 |
| Max resolution (Å) | 1.97 |
| Mosaicity range (°) | 0.69 |
| Redundancy | 9.7 |
| Mol/ASU | 2 |
| Matthews coeff. (Å3/Da) | 2.98 |
| Total reflections | 272,211 |
| Unique reflections | 28,191 |
| I/σ (overall) | 39.1 |
| I/σ (highest shell) | 9.0 |
| Completion overall (%) | 97.7 |
| Completion highest shell (%) | 99.2 |
| R-merge overall (%) | 9.4 |
| R-merge highest shell (%) | 40.5 |
|
| |
| Refinement | |
|
| |
| Nonhydrogen protein atoms | 2374 |
| Solvent molecules/ion | 334 |
| Rcryst (%) | 0.160 |
| Rfree (%) | 0.199 |
| RMS deviation bond length (Å) | 0.007 |
| RMS deviation bond angle (°) | 1.06 |
| Ramachandran favored (%) | 95.9 |
| Ramachandran outliers (%) | 0.8 |
| PDB accession | 4YOL |
RMS, root mean square.
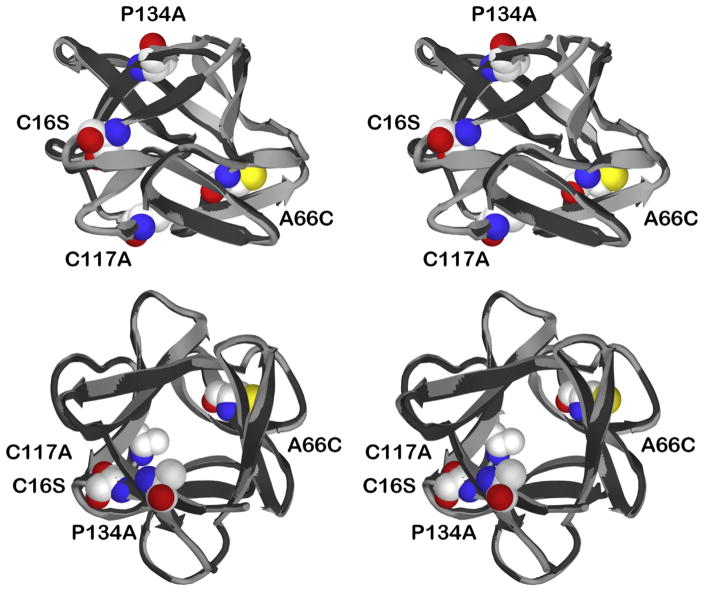
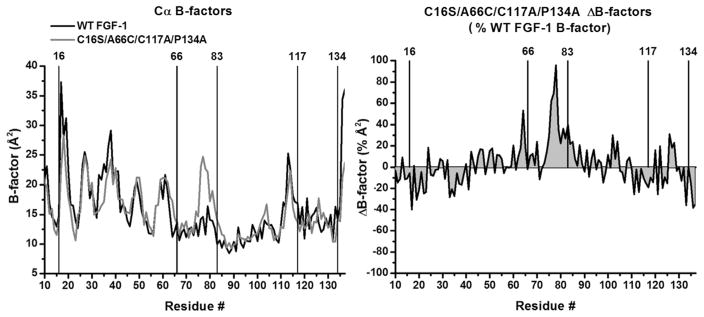
A comparison of the refined 1.97 Å resolution X-ray structure of the C16S/A66C/C117A/P134A mutant with WT FGF-1 (PDB accession 1JQZ) shows that FGF-1 is largely unperturbed in response to the mutations (Fig. 1). An overlay of the mutant main chain atom coordinates with WT FGF-1, over residue positions 11–137 (i.e., the fundamental β-trefoil structure, omitting the N-terminal His tag and flexible N-terminal residues 1–10), yields a root mean square deviation of 0.28 Å (Table 2). An examination of main chain atom perturbation within a 6 Å radius from the site of mutation indicates that the structural environment surrounding each mutation is similarly unperturbed. The greatest structural perturbation is not associated with a mutation site, per se, rather it is associated with the oxidized half-cystine that is created at position Cys83. Specifically, residue positions 76–80 (adjacent to position Cys83) exhibit a root mean square deviation of 0.51 Å (i.e., the largest positional deviation of any region of the mutant structure). Furthermore, a comparison of Cα B-factors for mutant and WT FGF-1 indicates a conservation of values over the entire structure with the exception of a general increase in Cα B-factors over the region 75–80 (Fig. 2).
Figure 1.
Relaxed stereo ribbon diagram overlay of WT FGF-1 and C16S/A66C/C117A/P134A mutant X-ray structures. The C16S/A66C/C117A/P134A mutant structure (light gray) is overlaid onto the WT FGF-1 structure (black). The images include a “side” view (top panel) and a “top” view (parallel to the pseudo 3-fold axis of cyclic symmetry; bottom panel). Also shown are space-filling representations of the mutant residues Cys16Ser, Ala66Cys, Cys117Ala, and Pro134Ala.
Table 2.
Main Chain Atom Deviation in the C16S/A66C/C117A/P134A X-ray Structure Compared to WT FGF-1
| Position | RMS deviation (Å)a |
|---|---|
| Cys16Ser | 0.34 |
| Ala66Cys | 0.25 |
| Cys83 (ox) | 0.29 |
| Cys117Ala | 0.20 |
| Pro134Ala | 0.25 |
| Residues 76–80 | 0.51 |
| Residues 11–137 | 0.28 |
Root mean square (RMS) deviations are calculated for all main chain atoms within 6 Å of the indicated positions.
Figure 2.
X-ray structure B-factor comparison of WT FGF-1 and C16S/A66C/C117A/P134A mutant. Left panel: an overlay of Cα B-factors for WT FGF-1 (1JQZ, molecule A; black line) and C16S/A66C/C117A/P134A (molecule A; gray line). The location of the mutation sites (as well as the oxidized half-cystine at Cys83) is indicated. Right panel: a ΔB-factor plot scaled by the WT FGF-1 B-factor ((mutant–WT FGF-1)/WT FGF-1). The 2 graphs demonstrate broad conservation of Cα B-factors in response to mutation, with the exception of a demonstrable increase in B-factors for the region 75–80 in the C16S/A66C/C117A/P134A mutant.
Isothermal Equilibrium Denaturation
Thermodynamic data for both reduced and oxidized forms of the different Cys-free mutants were determined by isothermal equilibrium denaturation using GuHCl (Table 3). Included in this table are previously reported thermodynamic data for WT FGF-139 and point mutants Cys16Ser,24 Ala66Cys (under oxidizing condition),25 Cys117Ala,21 and Pro134Val.27 A comparison of the stability data for reduced and oxidized forms of all mutants shows a decrease in the midpoint of denaturation Cm and ΔΔG values on chemical reduction—indicating loss of a stabilizing cystine between Cys66 and Cys83 (consistent with the X-ray structure data of C16S/A66C/C117A/P134A and previously reported X-ray structure of oxidized Ala66Cys25). The slope of the thermodynamic m-values (i.e., the sensitivity of ΔG to denaturant) is lower for all oxidized mutant forms; whereas, the m-value of all reduced forms approximates the m-value of the (fully unfoldable) WT FGF-1 protein. Thus, the decreased m-value of the oxidized forms is due to residual structure in the denatured state (due to cystine formation) that prevents complete unfolding. There is good agreement between the experimental ΔΔG values for the C16S/A66C/C117A and C16S/A66C/C117A/P134V combination mutations and the simple sum of previously reported ΔΔG values for the individual point mutations, indicating that the sites of mutations are largely independent of each other (consistent with the nonlocalized distribution of the mutation sites in the X-ray structure; Fig. 1).
Table 3.
Isothermal Equilibrium Denaturation Data for WT FGF-1 and Mutant Proteins in Crystallization Buffer
| Protein | Cm (M) | m-value (kJ mol−1 M−1) | ΔG (kJ mol−1) | ΔΔGa (kJ mol−1) | ΔΔG (pred.)b (kJ mol−1) |
|---|---|---|---|---|---|
| WT FGF-1c | 1.29 ± 0.01 | 20.3 ± 0.7 | 26.6 ± 0.9 | – | |
| Ala66Cys (ox)d | 1.84 ± 0.01 | 16.6 ± 1.0 | 30.5 ± 1.9 | −10.2 | |
| Cys16Sere | 12.1 | ||||
| Cys117Alaf | 1.32 ± 0.01 | 20.5 ± 0.3 | 27.0 ± 0.4 | −0.6 | |
| Pro134Ala | |||||
| Pro134Valg | −8.8 | ||||
| C16S/A66C/C117A (ox) | 1.24 ± 0.01 | 13.0 ± 0.3 | 16.1 ± 0.3 | 0.8 | 1.3 |
| C16S/A66C/C117A (red) | 1.12 ± 0.01 | 16.6 ± 0.4 | 18.6 ± 0.3 | 3.1 | |
| C16S/A66C/C117A/P134A (ox) | 1.39 ± 0.02 | 12.4 ± 0.2 | 17.2 ± 0.2 | −1.6 | N.D. |
| C16S/A66C/C117A/P134A (red) | 1.26 ± 0.01 | 17.0 ± 0.4 | 21.4 ± 0.4 | 0.6 | |
| C16S/A66C/C117A/P134V (ox) | 1.83 ± 0.01 | 13.9 ± 0.2 | 25.4 ± 0.5 | −9.2 | −7.5 |
| C16S/A66C/C117A/P134V (red) | 1.69 ± 0.01 | 17.2 ± 0.3 | 29.1 ± 0.3 | −7.5 |
The single letter amino acid code is used in naming combination mutants.
ΔΔG = (Cm WT − Cm mut) (mWT + mmut)/2.
Calculated from the sum of individual point mutations.
Dubey et al.39
Lee and Blaber.25
Determined from differential scanning calorimetry in 20 mM N-(2-acetamido) iminodiacetic acid (ADA), 0.1 M NaCl, pH 6.6 (“ADA buffer”)/0.7M GuHCl buffer.24
Xia et al.21
Determined in ADA buffer.27
Empirical Phase Diagram
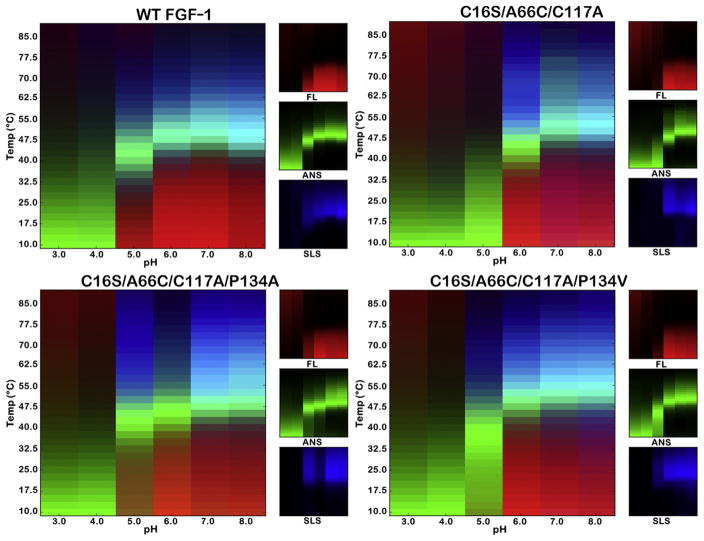
EPDs for WT FGF-1 and mutant proteins were determined over a pH range of 3.0–8.0 and a temperature range of 10°C–85°C, quantifying intrinsic FL, extrinsic FL (ANS binding), and SLS properties (Fig. 3). Taken together, the 3 different probes provide a comprehensive view of the protein structural state, including unfolding, molten globule structure, and aggregation, over a broad regime of temperature and pH.
Figure 3.
pH versus temperature EPDs of WT FGF-1 and Cys-free disulfide mutants. EPDs were determined for WT FGF-1, C16S/A66C/C117A, C16S/A66C/C117A/P134A, and C16S/A66C/C117A/P134V Cys-free mutants. EPDs as a function of pH and temperature were constructed from FL, ANS dye binding, and SLS data in citrate-phosphate buffer for the indicated proteins (see Methods). In this tricolor diagram, the red color indicates the regime of temperature and pH stability of the native state.
Intrinsic FL of the aromatic residues in WT FGF-1 and Cys-free mutants was used as a probe of change in tertiary structure. The ratio between FL intensity at 305 nm and 330 nm (I305/I330) was quantified as a function of temperature. The FL signal for WT FGF-1 at pH 3.0–4.0 exhibits no clear temperature-induced transition, indicating that the protein is unfolded under low pH conditions. For Cys-free mutants, the FL signal suggests that these proteins are also completely unfolded between pH 3.0–4.0. The FL signal of WT FGF-1 between pH 5.0–8.0 indicates a highly cooperative unfolding transition in the region of 40°C–47.5°C, with a pH of maximum stability around 7.0 (although the unfolding transition temperature is a relatively constant value over pH 6.0–8.0). At pH 5.0, the C16S/A66C/C117A/P134A mutation exhibits an FL signal more similar to native WT FGF-1; however, the other mutations exhibit an FL signal characteristic of unfolded protein.
ANS is a fluorophore that can bind to protein in apolar regions in the molten globule state and provides a useful probe highlighting the unfolding transitions during thermally induced unfolding. The binding properties of ANS are similar for WT FGF-1 and the mutant proteins. Over the range of pH 3.0–4.0, ANS binds weakly to unfolded protein at low temperature and subsequently dissociates from the protein as temperature elevates. Greater than pH 5.0–8.0, ANS does not bind to folded protein at low temperature, which results in a low extrinsic FL signal. As temperature increases, ANS binds to partially heat-denatured protein and the FL signal increases. This process is followed by a subsequent decrease in the signal as the ANS dissociates from fully unfolded protein at high temperature.
Heat-induced protein aggregation is monitored by SLS. There is no significant change in light scattering for WT FGF-1 and mutant proteins at pH 3.0 and pH 4.0, indicating that the acid-induced unfolding state does not aggregate with temperature (consistent with visual inspection of all proteins). Greater than pH 5.0–8.0, as temperature elevates above the cooperative unfolding transition, there is a significant increase in light scattering corresponding to aggregate formation for all proteins. This is followed at even higher temperature by a reduction in light scattering as macroscopic protein aggregation takes place.
EPDs for WT FGF-1 and mutant C16S/A66C/C117A/P134A in the presence and absence of 3-fold mass heparin sulfate were determined at pH 7.0 (i.e., the pH in the EPD profile closest to the physiological condition) (Supplementary Fig. 4). The EPD data indicate a similar magnitude of thermal stabilization for both proteins in response to heparin sulfate complexation; however, the SLS data suggest a reduced potential for thermally induced aggregation with the C16S/A66C/C117A/P134A mutant.
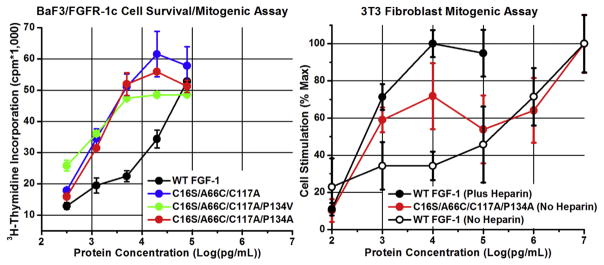
BaF3/FGFR-1c Cell Survival/Mitogenic Stimulation
The cell survival/mitogenic response of BaF3 cells expressing FGFR-1c toward WT FGF-1 and mutant proteins, in the presence of stabilizing heparin sulfate (and excluding reducing agent in the media) is shown in Figure 4. The assay is able to quantify cell survival/mitogenic response of WT FGF-1 and mutant proteins over a concentration range of ~20–5000 pM (3.18 × 102–7.95 × 104 pg/mL). The BaF3 cells have increasing survival/mitogenic response with increasing WT FGF-1 protein concentration, with maximum stimulation occurring at the highest-tested concentration. The Cys-free mutants as a group exhibited ~15-fold greater activity compared to WT FGF-1, and the BaF3 cells approach maximum survival/growth stimulation with mutant protein concentrations of ~320 pM (5.0 × 103 pg/mL).
Figure 4.
Biological activity of WT FGF-1 and Cys-free mutants in BaF3/FGFR1c cell survival/mitogenic and NIH 3T3 fibroblast mitogenic assays. The induced cell survival/proliferation in the BaF3/FGFR-1c cell assay is quantified by radioactive 3H-thimidine incorporation (CPM). The mitogenic activity in the NIH 3T3 cell assay is determined by cell count and normalized to maximum mitogenic activity. The protein concentration is plotted as log (pg/mL) in both assays. The BaF3/FGFR-1c cell system lacks HS proteoglycan, and the assay is performed in the presence of heparin sulfate (see Methods). The NIH 3T3 cell system expresses HS proteoglycan, and the assay for WT FGF-1 was performed in the presence and absence of heparin sulfate (see Methods). The single letter amino acid code is used in this figure. Error bars are standard deviation values.
3T3 Fibroblast Mitogenic Stimulation
The mitogenic activity of WT FGF-1 and mutant C16S/A66C/C117A/P134A is shown in Figure 4. Unlike BaF3 cells, 3T3 fibroblasts express cell surface HS, and therefore, there is no need for exogenous heparin sulfate for formation of the FGF-1/FGFR/HS ternary signal transduction complex. However, exogenous heparin sulfate has a pronounced effect on WT FGF-1 mitogenic activity—increasing the effective mitogenic activity by ~3-orders of magnitude.9 The mitogenic activity of the C16S/A66C/C117A/P134A mutant in the absence of exogenous heparin sulfate approximates that of WT FGF-1 in the presence of added heparin sulfate. Over the concentration range of ~60–250 pM (1 × 103–4 × 103 pg/mL), the C16S/A66C/C117A/P134A mutant has ~100-fold greater mitogenic activity than WT FGF-1 formulated in the absence of heparin sulfate.
Discussion
Although WT FGF-1 has diverse potential in regenerative medicine, its utility as a practical therapeutic is negatively impacted by its intrinsic biophysical properties. Specifically, the combination of low thermodynamic stability (ΔGunfolding = 21.3 kJ/mol in the absence of phosphate or sulfate ion23 and 26.6 kJ/mol in the presence of such ions25) and 3 buried Cys residues (i.e., reactive thiols) results in an oxidation-driven irreversible denaturation pathway that significantly reduces the effective functional half-life.9,14,24,40 Heparin sulfate binds FGF-1 and stabilizes its thermal unfolding by approximately 20°C (~20 kJ/mol).22 The addition of porcine heparin sulfate to FGF-1 has been a widely practiced formulation solution to counteract the low stability of FGF-1. Heparin sulfate, however, has a number of drawbacks including cost, heterogeneity, potential for infectious agents, allergic response, stimulation of bleeding, and thrombocytopenia.41,42 Point mutations are an obvious means by which to eliminate such buried Cys residues; however, the structure of WT FGF-1 is highly optimized to accommodate Cys residues at positions 16 and 8320 such that a combination of optimal point mutations at these 2 positions (i.e., Cys16Ser and Cys83Thr) destabilizes the protein by 13.2 kJ/mol21 (Cys117 substitutions by Ala, Ser, Val, or Thr are essentially neutral). Thus, fully one-half of the stabilizing free energy is lost by point mutation to eliminate buried thiols in WT FGF-1 resulting in a highly destabilized and aggregation-prone protein.
The discovery that Cys83 can participate as a half-cystine on mutation of adjacent residue Ala66 to Cys, with an overall increase in stability of ~10 kJ/mol,25 provides an opportunity to effectively eliminate all buried reactive thiols in FGF-1 while maintaining near-WT thermostability. The simplest manifestation of mutant design to achieve this goal is the C16S/A66C/C117A combination mutation. The current report tests this design hypothesis, and the resulting thermodynamic analysis of this mutation indicates an essentially neutral overall effect on thermodynamic stability (i.e., ΔΔG = 0.8 kJ/mol [Table 3]). Comprehensive analysis of the effect of pH and temperature on protein conformation (EPD analysis) identifies an apparent increase in acid denaturation sensitivity (at pH 5.0) for this mutation. Subsequently, additional stabilizing mutations at position 134 (Pro134Ala and Pro134Val) were added to the C16S/A66C/C117A mutant to evaluate possible recovery of stability at pH 5.0. The C16S/A66C/C117A/P134A combination mutant exhibits improvement of pH 5.0 stability (approaching that of WT FGF-1) and an overall increase in thermostability of 2.4 kJ/mol. Addition of the Pro134Val mutation increased stability by 10.0 kJ/mol, but pH 5.0 sensitivity was surprisingly unaltered. The most likely basis for observed differences in acid stability between the Pro and Ala residues at position 134 is an alteration in solvent structure in the region of position 134 because neither Pro nor Ala alters H-bond or charge in this region. However, Pro could also affect the local β-strand structure compared to Ala and this may also influence local solvent/H-bond effects; further work is needed to understand this effect.
The design of the C16S/A66C/C117A/P134A Cys-free FGF-1 mutant followed a hierarchical process. The best mutant substitution (as regards thermodynamic stability) for each individual Cys residue was identified and combined (including a novel cystine mutation). Additional thermodynamic stability was achieved by combining with stabilizing point mutations located at solvent inaccessible positions. Thus, the combination mutations were engineered so as to limit alteration in surface properties of the protein, thereby minimizing changes in aggregation potential. Alteration in the potential MHC-1 binding affinity of mutant epitopes was also characterized and determined to be minimal. This general design strategy may be readily adapted to other proteins.
FGF-1 is a pan-FGFR activator (able to activate all FGFR splice variants) and the structural dynamics of WT FGF-1 have been implicated in this functional property.43,44 A relationship often exists between thermodynamic stability and the structural dynamics of a protein, such that thermostable proteins require higher temperatures for equivalent functional dynamics compared to their mesophile counterpart.45,46 Thus, a Cys-free mutant form of FGF-1 that retains overall WT stability and dynamics is desirable. In this regard, the C16S/A66C/C117A/P134A mutant is only slightly more stable than WT FGF-1 and possesses a very similar EPD profile. X-ray structure data for this mutant indicates accommodation of mutations with only minor structural perturbation, including limited B-factor changes. These limited structural perturbations are associated with formation of the cystine at position Cys83. In this regard, the efficiency of formation of this cystine from the SHuffle T7 Expression host was only 60% with the most thermostable C16S/A66C/C117A/P134V mutant. To investigate this effect further, an additional stabilizing mutation, Lys12Val, was added to this mutation. This point mutation should provide an additional 9.3 kJ/mol of thermostability.27 Subsequent expression of this stabilizing mutation yielded only 30% efficiency in formation of the cystine bond. Thus, efficient formation of the Cys66-Cys83 cystine requires structural dynamics best achieved by WT equivalent thermostability.
Although none of the mutations in the present study involve positions known to interact with HS, the analytical heparin Sepharose chromatography indicates a modest reduction in HS affinity for the C16S/A66C/C117A/P134A mutant (i.e., a reduction in the elution peak from 1.26 to 1.18 M NaCl; Supplementary Fig. 3). Thus, the aforementioned alteration in structural dynamics, involving an increase in B-factors and minor structural perturbation of positions 75–80, may influence the HS-binding site structure. In this regard, the region 75–80 is essentially antipodal to the HS-binding site of WT FGF-1; thus, such influence is an unexpected distal structural effect. The slight reduction in HS affinity for the C16S/A66C/C117A/P134A mutant does not appear to be significant as regards the BaF3/FGFR-1c and 3T3 fibroblast functional assays. Additionally, EPD analysis of the effects of heparin sulfate on protein stabilization shows an essentially equivalent stability effect when comparing WT FGF-1 and the C16S/A66C/C117A/P134A mutant (Supplementary Fig. 4).
Cys mutation eliminates thiol-mediated irreversibility and extends the functional half-life of WT FGF-1.9 In the present report, we evaluate the practical implications for the effective functional activity in cell culture—using 2 different cell systems and with particular focus on the C16S/A66C/C117A/P134A combination mutant (which largely avoids the pH 5.0 sensitivity of the other mutant forms while maintaining overall WT FGF-1 thermostability and efficient cystine formation). The BaF3/FGFR-1c assay indicates an increase of ~100-fold in functional activity for the C16S/A66C/C117A/P134A mutant; whereas, the 3T3 fibroblast assay indicates an increase of ~15-fold in activity. The discrepancy between these functional assays is likely due to the BaF3/FGFR-1c assay responding to both increased cell survival and mitogenic response; whereas, the 3T3 fibroblast assay is a measure of mitogenic response. WT FGF-1 exhibits substantially different mitogenic activity in the 3T3 fibroblast assay depending on whether heparin sulfate is added. Heparin sulfate stabilizes WT FGF-1 by approximately 20°C, or ~20 kJ/mol9,22,23 and minimizes oxidized thiol-induced unfolding and sensitivity to proteases. The C16S/A66C/C117A/P134A mutant in the absence of added heparin sulfate exhibits a mitogenic activity in the 3T3 fibroblast assay similar to WT FGF-1 in the presence of heparin sulfate. Because the stability of the C16S/A66C/C117A/P134A mutant is increased by only 2 kJ/mol in comparison to WT FGF-1, the increase in mitogenic potency is consistent with the elimination of reactive thiols and an associated increase in functional half-life.
Overall, a Cys-free form of FGF-1 (C16S/A66C/C117A/P134A) was developed with minimal structural and thermodynamic perturbation. This mutant exhibits broad biophysical properties (as quantified by EPDs) that are essentially indistinguishable from WT FGF-1. The elimination of buried Cys residues results in a mitogenic potency in the absence of exogenously added heparin sulfate that approximates that of the WT FGF-1 formulated in the presence of heparin sulfate. The results therefore identify this mutant as an excellent candidate as a “second generation” FGF-1 for therapeutic application.
Supplementary Material
Acknowledgments
This work was supported by a Research Support Agreement from Trefoil Therapeutics, LLC. Xue Xia was supported by the Department of Biomedical Sciences, College of Medicine, Florida State University and NIH grant HL105732 (David M. Ornitz). Use of the instrument resources within the Protein Biology Laboratory of the FSU College of Medicine is acknowledged.
Abbreviations used
- FGF-1
fibroblast growth factor-1
- WT
wild-type
- DTNB
5,5′-dithiobis(2-nitrobenzoic acid)
- GuHCl
guanidine hydrochloride
- ANS
1-anilino-8-naphthalene sulfonate
- SLS
static light scattering
- EPD
empirical phase diagram
- HS
heparan sulfate
- MHC
major histocompatibility complex
Footnotes
Conflicts of interest: Michael Blaber acknowledges equity ownership in Trefoil Therapeutics, LLC.
This article contains supplementary material available from the authors by request or via the Internet at http://dx.doi.org/10.1016/j.xphs.2016.02.010.
References
- 1.Pace CN, Grimsley GR, Thomson JA, Barnett BJ. Conformational stability and activity of ribonuclease T1 with zero, one, and two intact disulfide bonds. J Biol Chem. 1988;263(24):11820–11825. [PubMed] [Google Scholar]
- 2.Hinck AP, Truckses DM, Markley JL. Engineered disulfide bonds in staphylococcal nuclease: effects on the stability and conformation of the folded protein. Biochemistry. 1996;35(32):10328–10338. doi: 10.1021/bi960309o. [DOI] [PubMed] [Google Scholar]
- 3.Richards FM. The interpretation of protein structures: total volume, group volume distributions and packing density. J Mol Biol. 1974;82:1–14. doi: 10.1016/0022-2836(74)90570-1. [DOI] [PubMed] [Google Scholar]
- 4.Karpusas M, Baase WA, Matsumura M, Matthews BW. Hydrophobic packing in T4 lysozyme probed by cavity-filling mutants. Proc Natl Acad Sci U S A. 1989;86:8237–8241. doi: 10.1073/pnas.86.21.8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim WA, Farruggio DC, Sauer RT. Structural and energetic consequences of disruptive mutations in a protein core. Biochemistry. 1992;31:4324–4333. doi: 10.1021/bi00132a025. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen DM, Leila Reynald R, Gittis AG, Lattman EE. X-ray and thermodynamic studies of staphylococcal nuclease variants I92E and I92K: Insights into polarity of the protein interior. J Mol Biol. 2004;341(2):565–574. doi: 10.1016/j.jmb.2004.05.066. [DOI] [PubMed] [Google Scholar]
- 7.Perry LJ, Wetzel R. The role of cysteine oxidation in the thermal inactivation of T4 lysozyme. Protein Eng. 1987;1(2):101–105. doi: 10.1093/protein/1.2.101. [DOI] [PubMed] [Google Scholar]
- 8.Lepock JR, Frey HE, Hallewell RA. Contribution of conformational stability and reversibility of unfolding to the increased thermostability of human and bovine superoxide dismutase mutated at free cysteines. J Biol Chem. 1990;265(35):21612–21618. [PubMed] [Google Scholar]
- 9.Lee J, Blaber M. The interaction between thermodynamic stability and buried free cysteines in regulating the functional half-life of fibroblast growth factor-1. J Mol Biol. 2009;393:113–127. doi: 10.1016/j.jmb.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 10.Nagano N, Ota M, Nishikawa K. Strong hydrophobic nature of cysteine residues in proteins. FEBS Lett. 1999;458(1):69–71. doi: 10.1016/s0014-5793(99)01122-9. [DOI] [PubMed] [Google Scholar]
- 11.Fomenko DE, Marino SM, Gladyshev VN. Functional diversity of cysteine residues in proteins and unique features of catalytic redox-active cysteines in thiol oxidoreductases. Mol Cell. 2008;26(3):228–235. [PMC free article] [PubMed] [Google Scholar]
- 12.Kanarek L, Bradshaw RA, Hill RL. Regeneration of active enzyme from the mixed disulfide of egg white lysozyme and cystine. J Biol Chem. 1965;240(6):PC2755–PC2757. [PubMed] [Google Scholar]
- 13.Buckley WT, Milligan LP. Participation of cysteine and cystine in inactivation of tyrosine aminotransferase in rat liver homogenates. Biochem J. 1978;176(2):449–454. doi: 10.1042/bj1760449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ortega S, Schaeffer M-T, Soderman D, et al. Conversion of cysteine to serine residues alters the activity, stability, and heparin dependence of acidic fibroblast growth factor. J Biol Chem. 1991;266:5842–5846. [PubMed] [Google Scholar]
- 15.Fitzpatrick LR, Jakubowska A, Martin GE, Davis M, Jaye MC, Dionne CA. Acidic fibroblast growth factor accelerates the healing of acetic-acid-induced gastric ulcers in rats. Digestion. 1992;53:17–27. doi: 10.1159/000200967. [DOI] [PubMed] [Google Scholar]
- 16.Mellin TN, Mennie RJ, Cashen DE, et al. Acidic fibroblast growth factor accelerates dermal wound healing. Growth Factors. 1992;7:1–14. doi: 10.3109/08977199209023933. [DOI] [PubMed] [Google Scholar]
- 17.Mellin TN, Cashen DE, Ronan JJ, Murphy BS, DiSalvo J, Thomas KA. Acidic fibroblast growth factor accelerates dermal wound healing in diabetic mice. J Invest Dermatol. 1995;104:850–855. doi: 10.1111/1523-1747.ep12607026. [DOI] [PubMed] [Google Scholar]
- 18.Schumacher B, Pecher P, von Specht BU, Stegmann T. Induction of neoangiogenesis in ischemic myocardium by human growth factors: first clincal results of a new treatment of coronary heart disease. Circulation. 1998;97(7):645–650. doi: 10.1161/01.cir.97.7.645. [DOI] [PubMed] [Google Scholar]
- 19.Blaber M, DiSalvo J, Thomas KA. X-ray crystal structure of human acidic fibroblast growth factor. Biochemistry. 1996;35:2086–2094. doi: 10.1021/bi9521755. [DOI] [PubMed] [Google Scholar]
- 20.Lee J, Blaber SI, Dubey VK, Blaber M. A polypeptide “building block” for the β-trefoil fold identified by “top-down symmetric deconstruction”. J Mol Biol. 2011;407:744–763. doi: 10.1016/j.jmb.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Xia X, Longo LM, Blaber M. Mutation choice to eliminate buried free cysteines in protein therapeutics. J Pharm Sci. 2015;104:566–576. doi: 10.1002/jps.24188. [DOI] [PubMed] [Google Scholar]
- 22.Copeland RA, Ji H, Halfpenny AJ, et al. The structure of human acidic fibroblast growth factor and its interaction with heparin. Arch Biochem Biophys. 1991;289(1):53–61. doi: 10.1016/0003-9861(91)90441-k. [DOI] [PubMed] [Google Scholar]
- 23.Blaber SI, Culajay JF, Khurana A, Blaber M. Reversible thermal denaturation of human FGF-1 induced by low concentrations of guanidine hydrochloride. Biophys J. 1999;77:470–477. doi: 10.1016/S0006-3495(99)76904-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Culajay JF, Blaber SI, Khurana A, Blaber M. Thermodynamic characterization of mutants of human fibroblast growth factor 1 with an increased physiological half-life. Biochemistry. 2000;39(24):7153–7158. doi: 10.1021/bi9927742. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, Blaber M. Structural basis for conserved cysteine in the fibroblast growth factor family: evidence for a vestigial half-cystine. J Mol Biol. 2009;393:128–139. doi: 10.1016/j.jmb.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Brych SR, Kim J, Logan TM, Blaber M. Accommodation of a highly symmetric core within a symmetric protein superfold. Protein Sci. 2003;12:2704–2718. doi: 10.1110/ps.03374903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubey VK, Lee J, Somasundaram T, Blaber S, Blaber M. Spackling the crack: stabilizing human fibroblast growth factor-1 by targeting the N and C terminus beta-strand interactions. J Mol Biol. 2007;371(1):256–268. doi: 10.1016/j.jmb.2007.05.065. [DOI] [PubMed] [Google Scholar]
- 28.Gasteiger E, Hoogland C, Gattiker A, et al. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handbook. New York, NY: Humana Press; 2005. [Google Scholar]
- 29.Kim JJ, Iyer V, Joshi SB, Volkin DB, Middaugh CR. Improved data visualization techniques for analyzing macromolecule structural changes. Protein Sci. 2012;21:1540–1553. doi: 10.1002/pro.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zambrano R, Jamroz M, Szczasiuk A, Pujols J, Kmiecik S, Ventura S. AGGRESCAN3D (A3D): server for prediction of aggregation properties of protein structures. Nucleic Acids Res. 2015;43:W306–W313. doi: 10.1093/nar/gkv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brych SR, Blaber SI, Logan TM, Blaber M. Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core region of a β-trefoil. Protein Sci. 2001;10:2587–2599. doi: 10.1110/ps.ps.34701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eftink MR. The use of fluorescence methods to monitor unfolding transitions in proteins. Biophys J. 1994;66:482–501. doi: 10.1016/s0006-3495(94)80799-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pace CN, Scholtz JM. Measuring the conformational stability of a protein. In: Creighton TE, editor. Protein Structure: A Practical Approach. Oxford: Oxford University Press; 1997. pp. 299–321. [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Zwart PH, Afonine PV, Grosse-Kunstleve RW, et al. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 38.Ornitz DM, Xu J, Colvin JS, et al. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271(25):15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 39.Dubey VK, Lee J, Blaber M. Redesigning symmetry-related “mini-core” regions of FGF-1 to increase primary structure symmetry: thermodynamic and functional consequences of structural symmetry. Protein Sci. 2005;14(9):2315–2323. doi: 10.1110/ps.051494405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linemeyer DL, Menke JG, Kelly LJ, et al. Disulfide bonds are neither required, present, nor compatible with full activity of human recombinant acidic fibroblast growth factor. Growth Factors. 1990;3:287–298. doi: 10.3109/08977199009003671. [DOI] [PubMed] [Google Scholar]
- 41.Seitz CS, Brocker EB, Trautman A. Management of allergy to heparins in postoperative care: subcutaneous allergy and intravenous tolerance. Dermatology. 2008;14(9):4. [PubMed] [Google Scholar]
- 42.Prechel M, Walenga JM. Heparin-induced thrombocytopenia: an update. Semin Thromb Hemost. 2012;38:483–496. doi: 10.1055/s-0032-1306432. [DOI] [PubMed] [Google Scholar]
- 43.Beenken A, Eliseenkova AV, Ibrahimi OA, Olsen SK, Mohammadi M. Plasticity in interactions of fibroblast growth factor 1 (FGF1) N terminus with FGF receptors underlies promiscuity of FGF1. J Biol Chem. 2012;287(5):3067–3078. doi: 10.1074/jbc.M111.275891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ornitz DM. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4(3):215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mamonova TB, Glyakina AV, Galzitskaya OV, Kurnikova MG. Stability and rigidity/flexibility—Two sides of the same coin? Biochim Biophys Acta. 2013;1834(5):854–866. doi: 10.1016/j.bbapap.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 46.Karshikoff A, Nilsson L, Ladenstein R. Rigidity versus flexibility: the dilemma of understanding protein thermal stability. FEBS J. 2015;282(20):3899–3917. doi: 10.1111/febs.13343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.