Abstract
Background
Side-population (SP) cells that exclude anti-cancer drugs have been found in various tumor cell lines. Moreover, SP cells have a higher proliferative potential and drug resistance than main population cells (Non-SP cells). Also, several ion channels are responsible for the drug resistance and proliferation of SP cells in cancer.
Methods
To confirm the expression and function of voltage-gated potassium (Kv) channels of SP cells, these cells, as well as highly expressed ATP-binding cassette (ABC) transporters and stemness genes, were isolated from a gefitinib-resistant human lung adenocarcinoma cell line (NCI-H460), using Hoechst 33342 efflux.
Results
In the present study, we found that mRNA expression of Kv channels in SP cells was different compared to Non-SP cells, and the resistance of SP cells to gefitinib was weakened with a combination treatment of gefitinib and Kv channel blockers or a Kv7 opener, compared to single-treatment gefitinib, through inhibition of the Ras-Raf signaling pathway.
Conclusions
The findings indicate that Kv channels in SP cells could be new targets for reducing the resistance to gefitinib.
Electronic supplementary material
The online version of this article (doi:10.1186/s40360-017-0118-9) contains supplementary material, which is available to authorized users.
Keywords: Lung cancer cell, Drug resistance, Side population, Voltage-gated potassium channel, Combination therapy
Background
Epidermal growth factor receptor (EGFR) is an oncogene that is involved in the development and progression of several human cancers, including non-small cell lung cancer (NSCLC). Approximately 10–30% of non-small cell lung cancer (NSCLC) patients have EGFR gene mutations. EGFR tyrosine kinase inhibitor (TKI) therapies are effective for NSCLC patients who have EGFR kinase domain mutations that target four exons (exon 18-exon 21) [1, 2]. Gefitinib (ZD 1839, Iressa), a small-molecule EGFR TKI, was approved by United States’ Food and Drug Administration (FDA) in 2003 for NSCLC, which comprises 80% of lung cancers [3]. Gefitinib sensitivity occurs in patients who have EGFR mutations, such as L858R or exon 19 deletion. However, a T790M-like secondary mutation in exon 19 of EGFR was also associated with resistance to gefitinib in NSCLC cells that contain the L858R-EGFR mutation [4]. Moreover, KRAS mutations also demonstrate resistance to gefitinib in vitro and in vivo [4, 5].
Accumulating evidence indicates that cancer stem cells (CSCs) have self-renewal properties in various solid tumors, and play a role in tumor development and progression [6, 7]. Side-population (SP) cells, a fraction of cancer stem cells, can be identified by efflux of Hoechst 33342 dye [8]. Moreover, SP cells have higher clonogenic potential and expression levels of ATP-binding cassette (ABC) transporters than main-population cells (known as Non-SP cells) [9, 10]. Several groups have suggested that SP cells that were dye-excluding cell portion in a tumor including lung cancer were responsible for anti-cancer drug resistance [11, 12]. SP cells in tumors possess phenotypes and signaling pathways similar to those of normal stem cells, which have high efflux of drugs [13, 14]. High expression levels of ABC transporters, especially ABCG2, in normal stem cells and tumor stem cells are considered to be responsible for drug resistance [15–17]. In various types of tumor, SP cells related to drug resistance have been isolated [18–20].
Recently, several reports have proposed that ion channels regulate the survival and growth of cancer stem cells [21, 22]. Silencing of chloride intracellular channel 1 (CLIC1), which is significantly overexpressed in stem/progenitor cells from human glioblastomas, reduced the proliferative and clonogenic capacity of stem/progenitor cells [21]. The transient receptor potential cation channel, subfamily M, member 7 (TRPM7) also leads to increased cancer stem cell proliferation in glioblastoma multiforme (GBM) through activation of the JAK2/STAT3 and/or Notch signaling pathways [22]. Moreover, blockade of CLIC1 induces apoptosis of 1,3-Bis(2-chloroethyl)-1-nitrosourea (BCNU)-resistant cancer stem cells of GBM [23].
However, research regarding the voltage-gated potassium (Kv) channel expression patterns of SP cells, and the involvement of Kv channels in reducing the resistance of SP cells to gefitinib, has not been reported.
Therefore, the present study was performed to compare Kv channel expression between SP cells and Non-SP cells in a gefitinib-resistant NCI-H460 cell line, which had wild-type EGFR and KRAS mutations [4], and to examine the inhibitory effect of combination treatment with gefitinib and Kv channel blockers or a Kv7 opener on the viability of gefitinib-resistant SP cells.
Methods
Cells and reagents
The human lung adenocarcinoma cell line (NCI-H460) was obtained from Korea Cell Line Bank (Seoul, Korea). The cells were maintained in complete growth medium supplemented with 10% fetal bovine serum (WelGene, Korea) and 1% antibiotics (Sigma-Aldrich, St. Louis, MO, USA) in an incubation system at 37 °C with 5% CO2. The cells were harvested using 1% trypsin-EDTA (Sigma-Aldrich) when they were in the logarithmic phase of growth, for SP analysis. Hoechst 33342 and fumitremorgin C (ABCG2 blocker) were purchased from Sigma-Aldrich. The anti-cancer drug gefitinib (Santa Cruz Biotechnology, CA, USA), tetraethylammonium (TEA, Sigma-Aldrich), 4-aminopyridine (4-AP, Sigma-Aldrich), and flupirtine (Tocris Bioscience, Bristol, UK) were used to blockade cell growth.
Isolation of side population
The protocol was based on that of Goodell et al. [24]. Briefly, the NCI-H460 cells were re-suspended at 1 × 106 cells/mL in pre-warmed RPMI 1640 (WelGene) with 2% fetal bovine serum. Hoechst 33342 dye was added at a final concentration of 5 μg/mL in the presence or absence of fumitremorgin C (10 μg/mL), and the cells were incubated in a 37 °C water bath for 90 min with intermittent shaking. At the end of the incubation, the cells were washed with ice-cold Hank’s Balanced Salt Solution (HBSS) (Sigma-Aldrich), centrifuged down at 4 °C, and resuspended in ice-cold HBSS. Propidium iodide (Sigma-Aldrich) at a final concentration of 2 μg/mL was added to the cells to gate the viable cells. The cell preparations were filtered through a 40-μm cell strainer (BD Biosciences, San Jose, CA, USA) to obtain a single cell suspension. Cells were analyzed and sorted into SP and Non-SP using BD FACSAriaIII® (BD Biosciences).
Real-time RT-PCR analysis
Total RNA was extracted using a Hybrid-R prep kit (GeneAll, Korea), according to the manufacturer’s protocol. Reverse transcription was performed using M-MLV reverse transcriptase (Thermo Fisher Scientific, Fremont, CA, USA) and random primer (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Real-time reverse transcription-PCR (real-time RT-PCR) was done with SYBR Green reagents (TAKARA, Japan) on Step-One Plus (Applied Biosystems, Foster City, CA, USA). Primers were designed to generate a PCR product. The relative mRNA expression level of the genes was normalized to GAPDH, and expressed as fold change relative to Non-SP cells. Table 1 represents the list of primers used for real-time RT-PCR.
Table 1.
Primers for real-time RT-PCR
| Gene (Accession number) | Sequences | Product size (base pair) |
|---|---|---|
| ABCG2 (NM_001257386) | F 5′-AGATGGGTTTCCAAGCGTTC-3′ R 5′-TGGTTGGTCGTCAGGAAGAA-3′ |
191 |
| ABCC1 (NM_004996.3) | F 5′- ACTGCCTTGGGATTTTTGCT-3′ R 5′- CATGGTGATGCCCAAGAGAG-3′ |
135 |
| OCT4 (NM_001173531) | F 5′- ATTTTGAGGCTGCTGGGTCT-3′ R 5′- CCTCAGTTTGAATGCATGGG-3′ |
205 |
| NANOG (NM_024865) | F 5′-CAGAAAAACAACTGGCCGAA-3′ R 5′-GGTCTGGTTGCTCCACATTG-3′ |
147 |
| Kv1.3 (NM_002232) | F 5′- TCTCCTTCGAACTGCTGGTG-3′ R 5′- ATGGCCACAATGTCGATCAG-3′ |
95 |
| Kv1.4 (NM_002233.3) | F 5′-ACGAGGGCTTTGTGAGAGAA -3′ R 5′-TAAGATGACCAGGACGGACA- 3′ |
144 |
| Kv4.1 (NM_004979) | F 5′-CTCCCTCAGCTCCTTCTTGG-3′ R 5′-GGGCAATGTTCTGAGGGACT-3′ |
97 |
| Kv7.3 (NM_001204824.1) | F 5′-GGTGCAGGTCACGGAGTATT-3′ R 5′-GGGCTGACTTTGTCAATGGT-3′ |
174 |
| Kv7.5 (NM_001160134.1) | F 5′-CGCTTTCGTTTTTCTCCTTG-3′ R 5′-CGAGCAAACCTCAGTCTTCC-3′ |
207 |
| Kv9.3 (NM_002252.3) | F 5′-CAGTGAGGATGCACCAGAGA-3′ R 5′-TTGCTGTGCAATTCTCCAAG-3′ |
200 |
| Kv10.1 (NM_002238.3) | F 5′-TGACCCCAAACTTATCCGCA-3′ R 5′-CTGCTGATGCCCTCATCCAC-3′ |
116 |
| Kv11.1 (NM_001204798) | F 5′-GACGTGCTGCCTGAGTACAA-3′ R 5′-AGCCGAGTAGGGTGTGAAGA-3′ |
121 |
| GAPDH (NM_002046.4) | F 5′-CTCTGCTCCTCCTGTTCGAC-3′ R 5′-ACGACCAAATCCGTTGACTC-3′ |
112 |
Cell proliferation assay
A cell proliferation assay was performed using the Cell Counting Kit-8 (Dojindo, Japan). Sorted SP cells were plated in 96-well culture plates (SPL, Korea) at 5 × 102 cells per well, and cultured in the complete growth medium. CCK-8 solution was added to each well at 0, 12, 24, 48, and 72 h. After 4 h of incubation, the absorbance was determined through a 450 nm filter in a microplate reader (TECAN, Männedorf, Switzerland), and the growth curve was plotted using optical density (OD) values. Each experiment was performed in triplicate.
Inhibitory effect of gefitinib, TEA, 4-AP, and flupirtine on NCI-H460 cell viability
The cells were seeded in 96-well plates at a concentration of 1 × 103 cells per well. After treatment with gefitinib, Kv channel blockers (TEA, 4-AP), and the Kv7 channel opener (flupirtine), the cell viability was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Briefly, 0.5 mg/mL of thiazolyl blue tetrazolium (Sigma-Aldrich) was added to the cells, and the cells were then incubated for 4 h. The MTT formazan was dissolved with dimethyl sulfoxide (DMSO) (Sigma-Aldrich), and the absorbance at 570 nm was determined using a microplate reader. The results were presented as a percentage of the control values.
Western blotting assay
The western blotting assay was applied to study the proteins related to the EGFR-Ras-Raf signaling pathway. Proteins were extracted and subjected to 10% SDS-PAGE, then transferred to PVDF membranes. The membranes were blocked with 5% skim milk in Tris-buffered saline with Tween™20 (Sigma-Aldrich) for 1 h at room temperature. The membranes were reacted with primary antibodies overnight at 4 °C. The primary antibodies were as follows: phosphorylated EGFR (1:500); Ras (1:500); phosphorylated Raf (1:500); phosphorylated Erk1/2 (1:500, all from Cell Signaling Technology, Danvers, MA, USA); and β-actin (Santa Cruz Biotechnology, 1:1,000). β-actin protein was used as the control.
Statistical analysis
Data were generally presented as the mean ± SE, and the statistical differences between experimental groups were analyzed with Student’s t-test using the statistical software Origin 8.0. A p value of < 0.05 was considered statistically significant in all cases.
Results
Inhibition of viability of NCI-H460 cells by blockade and opening of Kv channels
In order to determine the inhibitory effect of Kv channel blockers (TEA and 4-AP) and Kv7 opener (flupirtine) on the viability of NCI-H460 cells, cell viability was determined using an MTT assay. Cells treated with TEA, 4-AP, and flupirtine at different concentrations were incubated for 24, 48, and 72 h. The results indicated that the viability of NCI-H460 cells treated with TEA, 4-AP, and flupirtine was decreased in a dose-dependent manner (Fig. 1).
Fig. 1.
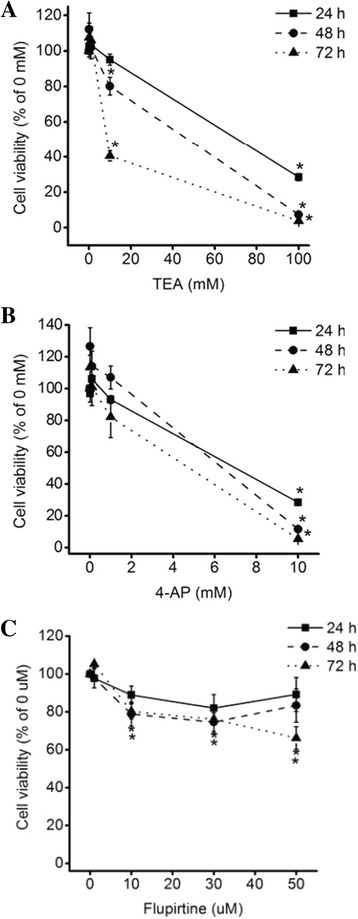
Effect of Kv blockers and an opener on NCI-H460 cell viability. Cell viability of NCI-H460 cells treated with a TEA b 4-AP, or c flupirtine for 24, 48, and 72 h was decreased in a dose-dependent manner. Results are mean ± SE of triplicate experiments, (*) p < 0.05
Isolation of SP cells within the NCI-H460 cell
In order to isolate the SP cells, the NCI-H460 cells were stained with Hoechst 33342, which was extruded by ABC transporters in stem-like cells. Next, we quantified the SP cells with dual-wavelength flow cytometry. The results show that SP cells were detected at 1.75% in multiple independent experiments (n = 34), as illustrated in Fig. 2a. SP cells were also eliminated in the presence of fumitremorgin C, which is specific blocker of ABCG2 (Fig. 2b). According to our results, SP cells, which had dual negative wavelengths of blue and red, were isolated in gefitinib-resistant NCI-H460 cells.
Fig. 2.
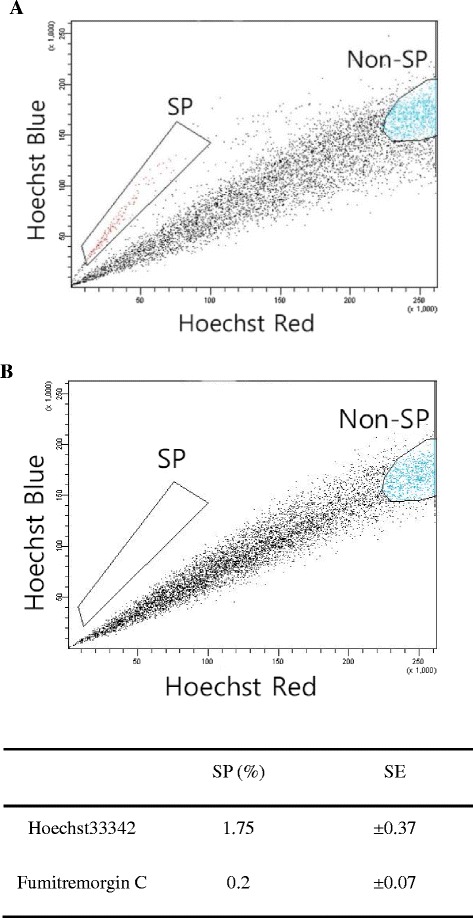
Isolation of SP cells in a cultured NCI-H460 lung cancer cell line. SP cells plotted in the absence (a) or presence (b) of fumitremorgin C. Results are mean ± SE of triplicate experiments, (*) p < 0.05
Characterization of SP cells
In order to confirm the characteristics of the SP cells, the mRNA expression levels of marker genes were identified using real-time RT-PCR, and compared to Non-SP cells. The mRNA expression level of ABCG2 was significantly increased in the SP cells. The mRNA expression levels of OCT4 and NANOG, which are specific markers of self-renewal, were significantly elevated in the SP cells (Fig. 3a). Also, to determine the differences in proliferation potential, the proliferation of SP and Non-SP cells was measured using the CCK-8 assay. After the sorted SP and Non-SP cells were incubated overnight, the CCK-8 assay was performed at 0, 12, 24, 48, and 72 h. The result reveals that the proliferation potential of SP cells was also significantly 2 times higher than Non-SP cells at the 72-h time-point (Fig. 3b).
Fig. 3.
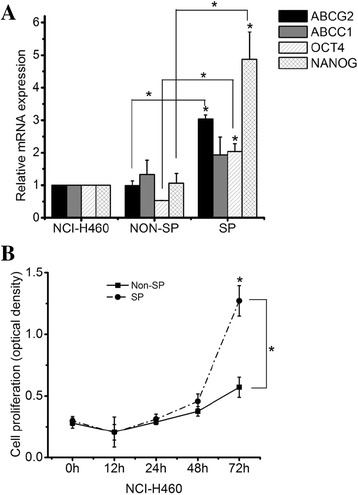
Characterization of SP cells in NCI-H460 lung cancer cells. a Real-time RT-PCR analysis of cancer stem cell marker genes b showing proliferation of sorted SP cells and Non-SP cells. Results are mean ± SE of triplicate experiments, (*) p < 0.05
Comparison of mRNA expression level of Kv channels between SP- and Non-SP cells
To determine the mRNA expression level of Kv channel subtypes in SP cells, mRNA expression levels were measured with a real-time RT-PCR. The result shows that the mRNA expression levels of Kv channel subtypes in SP cells were significantly different compared to Non-SP cells. The expression levels of Kv1.4, Kv7.3, Kv7.5, Kv10.1, and Kv11.1 mRNA in SP cells were less than in Non-SP cells. In other types of Kv channels, mRNA expression levels of Kv4.1 and Kv9.3 in SP cells were 1.58- and 2.03-fold higher, respectively, than in Non-SP cells (Fig. 4). The mRNA expression level of Kv1.3 between the SP and Non-SP cells was not significantly different (p = 0.06). This result indicates that the expression patterns of Kv channel subtypes are different between SP cells and Non-SP cells.
Fig. 4.
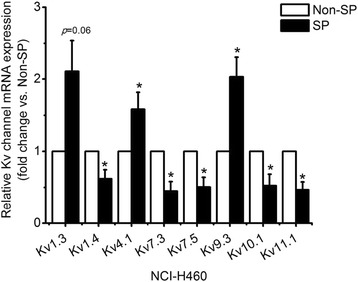
Comparison of Kv channel expression between sorted SP cells and Non-SP cells. Real-time RT-PCR analysis of Kv channel subtypes. Results are mean ± SE of triplicate experiments, (*) p < 0.05
Reduction of resistance to gefitinib with Kv channel blockers and opener in SP cells
To evaluate their inhibitory effects of gefitinib, Kv channel blockers and Kv7 opener on viability of the sorted SP cells, the viability of sorted cells treated with Kv channel blockers, Kv7 opener, and gefitinib for 72 h was estimated. The results showed that the viability of SP cells treated with gefitinib alone at an insensitive concentration (2 μM) [4] was not decreased. Compared to the SP cells, the viability of Non-SP cells treated with gefitinib alone at a concentration of 2 μM was decreased by 40.3% (Fig. 5). In cells treated with gefitinib at a concentration of 1 μM, viability was not decreased in the SP and Non-SP cells. An additional file shows this in detail (see Additional file 1).
Fig. 5.
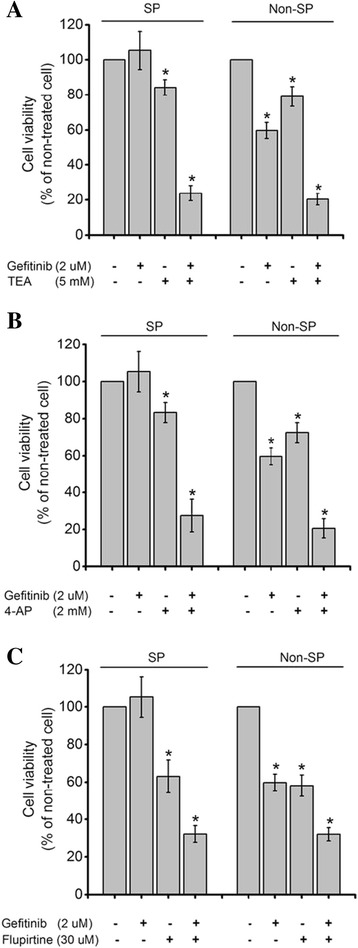
Inhibitory effect of combination treatment with gefitinib and Kv blockers (TEA and 4-AP) or an opener (flupirtine) on sorted SP cells and Non-SP cell viability. Cell viability of SP cells and Non-SP cells treated with a TEA 5 mM b 4-AP 2 mM, or c flupirtine 30 μM together gefitinib 2 μM. Results are mean ± SE of triplicate experiments, (*) p < 0.05
In addition, the viability of SP cells treated with TEA alone at a concentration of 5 mM or with a combination treatment of TEA and gefitinib at concentrations of 5 mM and 2 μM, respectively, was decreased by 15.8 and 76.16 (Fig. 5a). After treatment with 4-AP alone at a concentration of 2 mM, or a combination treatment of 4-AP and gefitinib at concentrations of 2 mM and 2 μM, the viability of the SP cells was decreased by 16.7 and 72.6%, respectively (Fig. 5b). The viability of the SP cells treated with flupirtine alone at a concentration of 30 μM, or with a combination treatment of flupirtine and gefitinib at concentrations of 30 μM and 2 μM, was decreased 37 and 69.4%, respectively (Fig. 5c). According to the results, SP cells were less sensitive to gefitinib than Non-SP cells, and the viability of SP cells with combination treatments was decreased more than with gefitinib as a single treatment. However, the degree of reduction between SP and Non-SP cells in each group was not different. Table 2 summarizes the percentage of decreased viability of SP and Non-SP cells from combination and single treatments.
Table 2.
Viability of SP- and Non-SP cells treated with gefitinib and Kv channels blocker or Kv7 opener
| Cell viability (%) | Gefitinib (2 μM) | TEA (5 mM) | 4-AP (2 mM) | Flupirtine (30 μM) | TEA (5 mM) | 4-AP (2 mM) | Flupirtine (30 μM) |
|---|---|---|---|---|---|---|---|
| (Gef 2 μM) | |||||||
| Non-SP | 59.68 (±4.54) | 79.09 (±5.62) | 72.46 (±5.45) | 58.08 (±5.67) | 20.39 (±3.27) | 20.54 (±5.22) | 32.02 (±3.48) |
| SP | 105.32 (±10.92) | 84.22 (±4.40) | 83.32 (±5.43) | 63.00 (±8.81) | 23.84 (±4.27) | 27.44 (±8.87) | 30.63 (±3.95) |
Inhibition of activated EGFR-Ras-Raf-Erk signaling pathway with combination treatment
To determine the signaling pathway involved in gefitinib resistance in cells with the combination treatment, western blotting to detect proteins of the EGFR-Ras-Raf signaling pathway was performed. The result showed that phosphorylated EGFR (p-EGFR) protein in cells with the combination treatment was decreased, compared to single-treatment gefitinib or blockers (Fig. 6). Also, total Ras protein in cells with the combination treatment was decreased, compared to single treatment with a gefitinib or blockers. Phosphorylated Erk (p-Erk) protein in cells with the combination treatment was decreased as compared to single treatments with 4-AP and flupirtine. However, the expression level of phosphorylated Raf (p-Raf) protein was not different between the combination treatment and the single treatment. According to the results, combination treatment with gefitinib and Kv channel blockers or the Kv7 opener reduced the viability of gefitinib-resistant NCI-H460 cells through inhibition of the EGFR-Ras-Raf-ERK pathway.
Fig. 6.
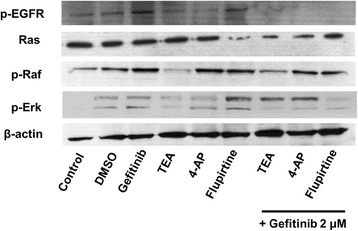
Inhibition of the EGFR-Ras-Raf signaling pathway on NCI-H460 cells with combination treatment. The protein expression level of p-EGFR, total Ras, p-Raf, and p-ERK1/2 was inhibited in cells with gefitinib and Kv blockers (TEA and 4-AP) or an opener (flupirtine)
Discussion
In the present study, isolated SP cells responsible for resistance to gefitinib in the NCI-H460 cell line demonstrated different Kv channel expression patterns compared to Non-SP cells. Our results also indicated that the viability of SP cells treated with gefitinib and Kv channel blockers or the Kv7 opener was more significantly inhibited than with single treatments through inhibition of the activated EGFR-Ras-Raf-Erk signaling pathway.
Several research groups have suggested that SP cells (known as cancer stem cells) were isolated from various cancer cell lines, including lung cancer, at various frequencies [12, 25–27]. The frequency of isolated cancer stem cells from human cancers was infrequent [28]. In our study, SP cells in gefitinib-resistant lung cancer cells were isolated (1.87%). Moreover, SP cells had characteristics similar to those of normal stem cells that were responsible for drug resistance [13, 15]. SP cells in tumor had high expression of ABC transporters and a high proliferative capacity, similar to normal stem cells [29, 30]. Some reports suggest that ABCG2 regulates self-renewal and stem cell marker expression in radiation-resistant glioma cells and NSCLC cell lines [31, 32]. Furthermore, other studies have proposed that gefitinib is a substrate extruded by ABCG2 and that a high expression of ABCG2 is responsible for acquired resistance to gefitinib [33–35]. Corresponding to previous studies, we isolated SP cells with high mRNA expression of ABCG2, OCT4, and NANOG genes and high proliferative potential from gefitinib-resistant NCI-H460 lung cancer cells. Moreover, SP cells in the NCI-H460 cell line demonstrate less sensitivity to gefitinib than Non-SP cells.
Potassium channels are involved in the regulation of anti-cancer-drug resistance. The up regulation of Kv1.5 increased the sensitivity of human gastric cancer (SGC7901) cells to chemotherapeutic drugs [36], and Kv1.1 specific blocker reduced gefitinib-resistant H460 cell viability [37]. Furthermore, inhibition of intermediate conductance calcium-activated potassium (KCa3.1) channels in a stem-like subpopulation from primary GBM cells induced reducing motility of stem-like subpopulation [38]. Kv channels are also involved in cell differentiation [39]. In particular, the expression of Kv1.3 was altered in poorly differentiated breast cancer [40], while Kv1.1, Kv1.2, Kv1.3, Kv1.4, Kv4.2, Kv4.3 and Kv9.3 decreased, as undifferentiated human mesenchymal stem cells (MSCs) differentiated into adipocytes [41]. In addition, the expression of neural Kv7 genes was increased during murine myoblast cell differentiation [42]. Likewise, our results also suggested that SP cells which were less sensitive to gefitinib have different mRNA expression patterns of Kv channel subtypes compared to Non-SP cells.
Over the last several years, in order to overcome the resistance of TKIs, many molecular targets have been investigated in cancer cells [43–45]. Kv channels as targets for suppression of cancer cell growth have been proposed [46–48]. Moreover, the inhibitory effect of combination therapies with EGFR TKIs and potassium channel blockers on the viability of cancer cells has been measured [37, 49]. However, the function of Kv channel blockers and a Kv7 opener on gefitinib-resistant SP cells has not been reported. According to our results, the combination treatment of gefitinib and Kv channel blockers or the Kv7 opener further attenuated the resistance of SP cells to gefitinib compared to single treatments. Therefore, our results demonstrate the synergic effect of gefitinib and Kv channel blockers (TEA and 4-AP) or the Kv7 opener (flupirtine) on the viability of gefitinib-resistant SP cells.
In addition, the signaling pathway related to reducing the resistance of SP cells to gefitinib with combination treatment of gefitinib and Kv channel blockers or the Kv7 opener has not been characterized. Recently, several reports have demonstrated that K+ channels are regulated by EGFR [50, 51]. Moreover, K+ currents have been induced by the Ras-Raf cascade [52]. Our study also suggests that the blockade or opening of Kv channels is associated with the EGFR-Ras-Raf signaling pathway.
Conclusion
The SP cells in the NCI-H460 cell line have different Kv channel expressions compared to Non-SP cells, and the resistance of SP cells to gefitinib, with a highly expressed ABC transporter and a stemness gene, was attenuated through combination treatment with gefitinib and Kv channel blockers (TEA and 4-AP) or Kv7 opener (flupirtine). Therefore, the Kv channels of SP cells are useful targets for overcoming gefitinib resistance in lung cancer patients.
Acknowledgments
Funding
This research was supported by Basic Science Research Program through National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2012-R1A2A2A01047151,NRF-2014R1A1A3A04052757) and by the Bio & Medical Technology Development Program of the National Research Foundation funded by the Ministry of Science, ICT & Future Planning (2016M3A9B6026771).
Availability of data and materials
All datasets generated or analyzed during this study are included in this published article and its supplementary information files.
Author’s contributions
SYC carried out experiments, analyzed data and wrote manuscript. HRK contributed to isolation and discussion of SP cells. PDR contributed to discussions about the manuscript. SYL designed the study and wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
According to the guidelines of Seoul National University Institutional Review Board (SNUIRB), the research involving human cancer cell lines obtained from Korea Cell Line Bank was not target for research ethics review.
Abbreviations
- 4-AP
4-aminopyridine
- ABC
ATP-binding cassette
- BCNU
1,3-Bis(2-chloroethyl)-1-nitrosourea
- CLIC1
Chloride intracellular channel 1
- CSCs
Cancer stem cells
- EGFR
Epidermal growth factor receptor
- GBM
Glioblastoma multiforme
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NSCLC
Non-small cell lung cancer
- SP
Side-population
- TEA
Tetraethylammonium
- TKIs
Tyrosine kinase inhibitors
- TRPM7
Transient receptor potential cation channel, subfamily M, member 7
Additional file
Effect of gefitinb 1 μM on sorted SP cells and Non-SP cell viability. Sorted SP and Non-SP cells were seeded at 1 × 103 cells in 96 well culture plates. After 24 h, cells were treated with gefitinib 1 μM. Cell viability was measured after 72 h. Results are mean ± SE of triplicate experiments. (PPTX 100 kb)
Contributor Information
Seon Young Choi, Email: rainbowbbang@snu.ac.kr.
Hang-Rae Kim, Email: hangrae2@snu.ac.kr.
Pan Dong Ryu, Email: pdryu@snu.ac.kr.
So Yeong Lee, Phone: +82 2 8801283, Email: leeso@snu.ac.kr.
References
- 1.Melosky B. Review of EGFR TKIs in metastatic NSCLC, including ongoing trials. Front Oncol. 2014;4:244. doi: 10.3389/fonc.2014.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin Y, Wang X, Jin H. EGFR-TKI resistance in NSCLC patients: mechanisms and strategies. Am J Cancer Res. 2014;4:411–35. [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen MH, Williams GA, Sridhara R, Chen G, Pazdur R. FDA drug approval summary: gefitinib (ZD1839) (Iressa) tablets. Oncologist. 2003;8:303–6. doi: 10.1634/theoncologist.8-4-303. [DOI] [PubMed] [Google Scholar]
- 4.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 5.Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7:493–507. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K, Zeng J, Luo L, Yang J, Chen J, Li B, et al. Identification of a cancer stem cell-like side population in the HeLa human cervical carcinoma cell line. Oncol Lett. 2013;6:1673–80. doi: 10.3892/ol.2013.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richard V, Nair MG, Santhosh Kumar TR, Pillai MR. Side population cells as prototype of chemoresistant, tumor-initiating cells. Biomed Res Int. 2013;2013:517237. doi: 10.1155/2013/517237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moserle L, Ghisi M, Amadori A, Indraccolo S. Side population and cancer stem cells: therapeutic implications. Cancer Lett. 2010;288:1–9. doi: 10.1016/j.canlet.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 9.Britton KM, Kirby JA, Lennard TW, Meeson AP. Cancer stem cells and side population cells in breast cancer and metastasis. Cancers (Basel) 2011;3:2106–30. doi: 10.3390/cancers3022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang B, Huang YJ, Yao ZJ, Chen X, Guo SJ, Mao XP, et al. Cancer stem cell-like side population cells in clear cell renal cell carcinoma cell line 769P. PLoS One. 2013;8:e68293. doi: 10.1371/journal.pone.0068293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–43. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 12.Leon G, MacDonagh L, Finn SP, Cuffe S, Barr MP. Cancer stem cells in drug resistant lung cancer: targeting cell surface markers and signaling pathways. Pharmacol Ther. 2016;158:71–90. doi: 10.1016/j.pharmthera.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 14.Zhou S, Schuetz JD, Bunting KD, Colapietro AM, Sampath J, Morris JJ, et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 2001;7:1028–34. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 15.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–84. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 16.Kathawala RJ, Gupta P, Ashby CR, Jr, Chen ZS. The modulation of ABC transporter-mediated multidrug resistance in cancer: a review of the past decade. Drug Resist Updat. 2015;18:1–17. doi: 10.1016/j.drup.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Singh A, Wu H, Zhang P, Happel C, Ma J, Biswal S. Expression of ABCG2 (BCRP) is regulated by Nrf2 in cancer cells that confers side population and chemoresistance phenotype. Mol Cancer Ther. 2010;9:2365–76. doi: 10.1158/1535-7163.MCT-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu L, McArthur C, Jaffe RB. Ovarian cancer stem-like side-population cells are tumourigenic and chemoresistant. Br J Cancer. 2010;102:1276–83. doi: 10.1038/sj.bjc.6605626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown MD, Gilmore PE, Hart CA, Samuel JD, Ramani VA, George NJ, et al. Characterization of benign and malignant prostate epithelial Hoechst 33342 side populations. Prostate. 2007;67:1384–96. doi: 10.1002/pros.20620. [DOI] [PubMed] [Google Scholar]
- 20.Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–33. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 21.Setti M, Savalli N, Osti D, Richichi C, Angelini M, Brescia P, et al. Functional role of CLIC1 ion channel in glioblastoma-derived stem/progenitor cells. J Natl Cancer Inst. 2013;105:1644–55. doi: 10.1093/jnci/djt278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu M, Inoue K, Leng T, Guo S, Xiong ZG. TRPM7 channels regulate glioma stem cell through STAT3 and Notch signaling pathways. Cell Signal. 2014;26:2773–81. doi: 10.1016/j.cellsig.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang MK, Kang SK. Pharmacologic blockade of chloride channel synergistically enhances apoptosis of chemotherapeutic drug-resistant cancer stem cells. Biochem Biophys Res Commun. 2008;373:539–44. doi: 10.1016/j.bbrc.2008.06.070. [DOI] [PubMed] [Google Scholar]
- 24.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, et al. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A. 2004;101:14228–33. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiang L, Yang Y, Ma YJ, Chen FH, Zhang LB, Liu W, et al. Isolation and characterization of cancer stem like cells in human glioblastoma cell lines. Cancer Lett. 2009;279:13–21. doi: 10.1016/j.canlet.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Fang DD, Kim YJ, Lee CN, Aggarwal S, McKinnon K, Mesmer D, et al. Expansion of CD133(+) colon cancer cultures retaining stem cell properties to enable cancer stem cell target discovery. Br J Cancer. 2010;102:1265–75. doi: 10.1038/sj.bjc.6605610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishizawa K, Rasheed ZA, Karisch R, Wang Q, Kowalski J, Susky E, et al. Tumor-initiating cells are rare in many human tumors. Cell Stem Cell. 2010;7:279–82. doi: 10.1016/j.stem.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Neaves WB. Normal stem cells and cancer stem cells: the niche matters. Cancer Res. 2006;66:4553–7. doi: 10.1158/0008-5472.CAN-05-3986. [DOI] [PubMed] [Google Scholar]
- 30.Chen K, Huang YH, Chen JL. Understanding and targeting cancer stem cells: therapeutic implications and challenges. Acta Pharmacol Sin. 2013;34:732–40. doi: 10.1038/aps.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wee B, Pietras A, Ozawa T, Bazzoli E, Podlaha O, Antczak C, et al. ABCG2 regulates self-renewal and stem cell marker expression but not tumorigenicity or radiation resistance of glioma cells. Sci Rep. 2016;6:25956. doi: 10.1038/srep25956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galetti M, Petronini PG, Fumarola C, Cretella D, La Monica S, Bonelli M, et al. Effect of ABCG2/BCRP expression on efflux and uptake of gefitinib in NSCLC cell lines. PLoS One. 2015;10:e0141795. doi: 10.1371/journal.pone.0141795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozvegy-Laczka C, Hegedus T, Varady G, Ujhelly O, Schuetz JD, Varadi A, et al. High-affinity interaction of tyrosine kinase inhibitors with the ABCG2 multidrug transporter. Mol Pharmacol. 2004;65:1485–95. doi: 10.1124/mol.65.6.1485. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Cusatis G, Brahmer J, Sparreboom A, Robey RW, Bates SE, et al. Association of variant ABCG2 and the pharmacokinetics of epidermal growth factor receptor tyrosine kinase inhibitors in cancer patients. Cancer Biol Ther. 2007;6:432–8. doi: 10.4161/cbt.6.3.3763. [DOI] [PubMed] [Google Scholar]
- 35.Chen YJ, Huang WC, Wei YL, Hsu SC, Yuan P, Lin HY, et al. Elevated BCRP/ABCG2 expression confers acquired resistance to gefitinib in wild-type EGFR-expressing cells. PLoS One. 2011;6:e21428. doi: 10.1371/journal.pone.0021428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han Y, Shi Y, Han Z, Sun L, Fan D. Detection of potassium currents and regulation of multidrug resistance by potassium channels in human gastric cancer cells. Cell Biol Int. 2007;31:741–7. doi: 10.1016/j.cellbi.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 37.Jeon WI, Ryu PD, Lee SY. Effects of voltage-gated K+ channel blockers in gefitinib-resistant H460 non-small cell lung cancer cells. Anticancer Res. 2012;32:5279–84. [PubMed] [Google Scholar]
- 38.Ruggieri P, Mangino G, Fioretti B, Catacuzzeno L, Puca R, Ponti D, et al. The inhibition of KCa3.1 channels activity reduces cell motility in glioblastoma derived cancer stem cells. PLoS One. 2012;7:e47825. doi: 10.1371/journal.pone.0047825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li GR, Deng XL. Functional ion channels in stem cells. World J Stem Cells. 2011;3:19–24. doi: 10.4252/wjsc.v3.i3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pardo LA, Stuhmer W. The roles of K(+) channels in cancer. Nat Rev Cancer. 2014;14:39–48. doi: 10.1038/nrc3635. [DOI] [PubMed] [Google Scholar]
- 41.You MH, Song MS, Lee SK, Ryu PD, Lee SY, Kim DY. Voltage-gated K+ channels in adipogenic differentiation of bone marrow-derived human mesenchymal stem cells. Acta Pharmacol Sin. 2013;34:129–36. doi: 10.1038/aps.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roura-Ferrer M, Sole L, Martinez-Marmol R, Villalonga N, Felipe A. Skeletal muscle Kv7 (KCNQ) channels in myoblast differentiation and proliferation. Biochem Biophys Res Commun. 2008;369:1094–7. doi: 10.1016/j.bbrc.2008.02.152. [DOI] [PubMed] [Google Scholar]
- 43.Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol. 2012;30:679–92. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- 44.Eleftheria T, Allison ME, Younsoo B. Combination effects of docetaxel and doxorubicin in hormone-refractory prostate cancer cells. Biochem Res Int. 2012;2012:10. doi: 10.1155/2012/832059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wulff H, Castle NA, Pardo LA. Voltage-gated potassium channels as therapeutic targets. Nat Rev Drug Discov. 2009;8:982–1001. doi: 10.1038/nrd2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jang SH, Choi SY, Ryu PD, Lee SY. Anti-proliferative effect of Kv1.3 blockers in A549 human lung adenocarcinoma in vitro and in vivo. Eur J Pharmacol. 2011;651:26–32. doi: 10.1016/j.ejphar.2010.10.066. [DOI] [PubMed] [Google Scholar]
- 47.Lee BH, Ryu PD, Lee SY. Serum starvation-induced voltage-gated potassium channel Kv7.5 expression and its regulation by Sp1 in canine osteosarcoma cells. Int J Mol Sci. 2014;15:977–93. doi: 10.3390/ijms15010977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ru Q, Tian X, Wu YX, Wu RH, Pi MS, Li CY. Voltage-gated and ATP-sensitive K+ channels are associated with cell proliferation and tumorigenesis of human glioma. Oncol Rep. 2014;31:842–8. doi: 10.3892/or.2013.2875. [DOI] [PubMed] [Google Scholar]
- 49.Zhang DY, Zhang YH, Sun HY, Lau CP, Li GR. Epidermal growth factor receptor tyrosine kinase regulates the human inward rectifier potassium K(IR)2.3 channel, stably expressed in HEK 293 cells. Br J Pharmacol. 2011;164:1469–78. doi: 10.1111/j.1476-5381.2011.01424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowlby MR, Fadool DA, Holmes TC, Levitan IB. Modulation of the Kv1.3 potassium channel by receptor tyrosine kinases. J Gen Physiol. 1997;110:601–10. doi: 10.1085/jgp.110.5.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L, Zhang C, Su X, Lin D. Kcnj10 is a major type of K+ channel in mouse corneal epithelial cells and plays a role in initiating EGFR signaling. Am J Physiol Cell Physiol. 2014;307:C710–7. doi: 10.1152/ajpcell.00040.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y, Rane SG. Potassium channel induction by the Ras/Raf signal transduction cascade. J Biol Chem. 1994;269:31183–9. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated or analyzed during this study are included in this published article and its supplementary information files.


