Abstract
Matrix and tissue rigidity guides many cellular processes, including the differentiation of stem cells and the migration of cells in health and disease. Cells actively and transiently test rigidity using mechanisms limited by inherent physical parameters that include the strength of extracellular attachments, the pulling capacity on these attachments, and the sensitivity of the mechanotransduction system. Here we focus on rigidity sensing mediated through the integrin family of extracellular matrix receptors and linked proteins, and discuss the evidence supporting these proteins as mechanosensors.
What is rigidity and why is it important?
Local rigidities form a landscape within an organism. During development, this landscape acts in concert with chemical cues to shape the organism (Keller et al., 2003). Conversely, recovery from injury can require cells to sense and reshape this landscape, as seen in healing of bones and wounds (Chao et al., 1989; Grinnell, 1994; Hinz, 2006). Numerous pathologies are characterized by disturbance of the rigidity landscape, including atherosclerosis, arthritis, osteoporosis, and fibrosis of the heart, lung, kidney, and liver (Ingber, 2003). There are also a growing number of medical uses for pharmacological drugs that affect the rigidity of tissues, including Botulinum toxin (relaxes smooth muscles) and Rho kinase inhibitors (relaxes cytoskeleton) (Felber, 2006; Hahmann and Schroeter, 2010). Recently, an appreciation for the altered rigidity responses of cancer cells has emerged (Kostic et al., 2009; Wang et al., 2000b). However, rigidity sensing defects are likely to have an important role in classical ‘anchorage independence’ studies (Stoker et al., 1968) that examined the ability of cancerous, but not normal, cells to proliferate on each other when in suspension.
In a broad sense, rigidity is a measure of the relationship between applied forces and the resulting stretch of a material. To quantify it, the elastic (or Young’s) modulus and the shear modulus are commonly employed. Both are defined as the ratio of the stress (applied force per unit area) to the resulting strain. For the elastic modulus (E), the force is applied perpendicular to the material’s surface, whereas for shear modulus (G), the force is applied parallel to the surface (Figure 1A). Because strain is a dimensionless value, the units for rigidity are force per area, with the SI unit being the Pascal (Burdun et al., 1972). However, to ease interpretation of the values, this Review will use the proposed ‘natural’ SI units for pressure of nN/μm2 (equivalent to kPa, Hochmuth, 2000). There are two useful observations that assist the interpretation of elastic and shear moduli values. The first is that they represent the amount of force per unit area required to double the length of a material (elastic modulus) or deflect it by a distance equal to its height (shear modulus). The second is that elastic and shear moduli are related by the following function: E=2G(1+ ν), where v is the Poisson ratio. For materials that do not change volume under stretch, the Poisson ratio equals 0.5. As a consequence, the elastic modulus will be three times its shear modulus.
Figure 1. Rigidity Moduli and the Energy Landscapes of a Slip bond.
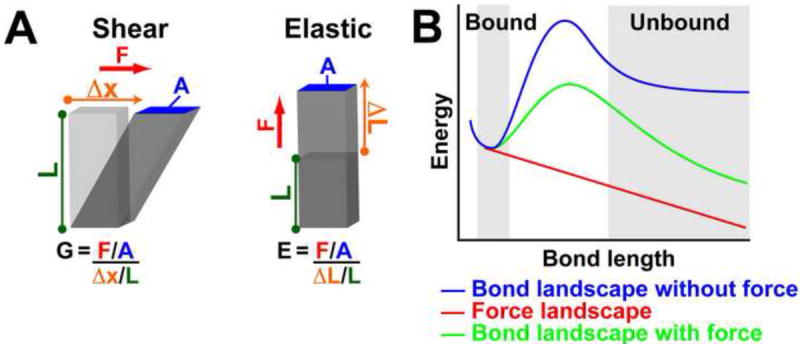
(A) Stress is the amount of force applied per area (F/A) and strain is the displacement in the direction of applied force relative to initial length (Δx/L or ΔL/L). While both elastic and shear moduli are the ratio of stress over strain, there is a difference in the direction of the applied force. (B) The energy landscapes of a slip bond with and without applied force.
There are important limitations to shear and elastic moduli values of biological materials. In particular, they assume a linear relationship between stress and strain, which is not the case for either cells or extracellular matrix (ECM) components (Storm et al., 2005). Rather biological material has a viscoelastic behavior where the relationship between stress and strain depends not only on the magnitude of the force applied but also on the rate of application (Fabry et al., 2001; Puxkandl et al., 2002). This implies that stiffness values reported in the literature will depend on the range of forces applied and on the time scale of force application. This Review will examine the reported physical and motility parameters of tissues, cells, and proteins, and project those constraints onto how cells sense rigidities through integrins.
The Rigidity Landscape
The rigidity landscape of a tissue arises from its constituent cells and ECM. The elastic moduli of tissues range from 0.1 to 30,000,000 nN/μm2 (Table 1). The typical hierarchy of tissue rigidity is that brain (0.1 - 10 nN/μm2) is softer than muscle (10 - 100 nN/μm2), which is softer than areolar connective tissue and arteries (100 - 1,000 nN/μm2), which are softer than bone (15,000,000 - 30,000,000 nN/μm2).
Table 1.
Elasticity of cells and tissues
| Tissue type | Elastic Modulus (nN/μm2) | Key References: |
|---|---|---|
| Brain | 0.1-10 | (Elkin et al., 2007; Gefen et al., 2003; Hirakawa et al., 1981; Metz et al., 1970) |
| Muscle | 12-100 | (Collinsworth et al., 2002; Engler et al., 2006; Mathur et al., 2001) |
| Fat | 20 | (Ophir et al., 1999) |
| Artery | 100 – 3,800 | (Bank and Kaiser, 1998; Intengan et al., 1999) |
| Areolar connective tissue (fibroblasts in collagen) | 600 -1,000 | (Chapuis and Agache, 1992; Wakatsuki et al., 2000) |
| Bone | 17,100,000-28,900,000 | (Reilly et al., 1974; Schaffler and Burr, 1988) |
The rigidity of a tissue is neither static nor uniform. For instance the stiffness of brain decreases with age (Gefen et al., 2003). Local rigidity differences have also been observed in the kidney cortex and hippocampus (Elkin et al., 2007; Kallel et al., 1998). Further, there are local rigidity differences within individual cells (Roca-Cusachs et al., 2008; Yamada et al., 2000).
The Rigidity Preferences of Cells
The rigidity preferences of cells generally reflect their native environments. Neutrophils exist in a broad range of rigidities from the blood to a variety of tissues during inflammation; these cells attach and have similar shapes on glass as on 0.002 nN/μm2 gels (Yeung et al., 2005). From a soft tissue like the brain, neurons are more branched on substrates of 0.05 nN/μm2 compared to 0.55 nN/μm2 and they extend faster on substrates of 0.002 nN/μm2 compared to 0.130 nN/μm2 (Balgude et al., 2001; Flanagan et al., 2002). Fibroblasts and endothelial cells, originating from tissues of ‘midrange’ stiffnesses, do not spread or display actin stress fibers on surfaces softer than 3 nN/μm2 (Pelham and Wang, 1997; Yeung et al., 2005). From the stiff environment of cartilage and bone, chondrocytes do not spread well on substrates softer than 7.4 nN/μm2 (Subramanian and Lin, 2005) and the degree of adhesion protein clustering in pre-osteoblasts is maximized at 60 nN/μm2 (Kong et al., 2005). Cells can also be guided by differences in rigidity, a phenomenon known as durotaxis. Both fibroblasts and leukocytes have been reported to move toward rigid substrates (Lo et al., 2000; Mandeville et al., 1997). Another related cellular process is endothelial cell remodeling to shear stress. Although this process differs from rigidity sensing in that it does not appear to require cells to actively pull on their environment, both processes involve sensing mechanical force in a time- and magnitude-sensitive manner and may therefore share similar mechanosensory mechanisms (Ando and Yamamoto, 2009).
Bond Strengths
Throughout the rigidity sensing process, the key molecular mechanism by which force is detected is through its effect on the binding and conformation of proteins. Two interacting proteins can be described as having a ‘bound’ and an ‘unbound’ state, with an energy barrier necessary to switch between the two. Analogously, the switch between two conformations in a single protein (e.g. folded/unfolded or open/closed channel) can be characterized in the same way. A higher barrier will make it more difficult to change configurations. As first envisioned by Bell (1978), the force required to break a bond can be understood as an input of energy into the system. Force reduces the activation barrier between the two, thereby promoting the transition to the unbound state (Figure 1B). This type of behavior, common to most protein interactions, is called a slip bond. Catch bonds are exceptions to this rule. In response to applied force, the energy landscape of catch bonds changes in such a way as to increase their strength (energy barrier) under certain force regimes (Prezhdo and Pereverzev, 2009; Thomas et al., 2008). The possibility that these bonds have an important role in rigidity sensing will be discussed below.
Quantifying the energy barrier between bound and unbound states is the optimal way to characterize a protein bond. However, this value is difficult to obtain in live cells, which are strongly out of thermodynamic equilibrium (having large local and temporal variations in energy levels, Collin et al., 2005). For this reason, studies usually report the force needed to break a bond in a particular assay. For these assays, the rate at which force is loaded will affect the force measurement (Evans, 2001). This important caveat must be taken into account when comparing force values, and that is why in this Review, we specify the loading rate whenever reported.
Determinants of What Cells Can Sense Through Integrins
Going from the ECM to the inside of the cell, the matrix-integrin bonds are the primary links. Integrins are transmembrane heterodimers composed of an α and a ß chain. In vertebrates, there are 18 α and 8 β subunits that assemble into 24 different heterodimers (Takada et al., 2007). Integrins mediate attachment to a variety of ECM components, including fibronectin, collagen, laminin and vitronectin (Barczyk et al., 2010). In general, the α subunit determines the substrate specificity, while the ß subunit attaches to actin through intermediary proteins (Vicente-Manzanares et al., 2009). Integrins undergo a mechanical cycle in forming adhesive contacts (Puklin-Faucher and Sheetz, 2009) and there are important functional differences between integrin complexes. For instance, less stable αvβ3 integrins initiate signal transduction, while higher matrix forces are supported by α5β1 integrins (Roca-Cusachs et al., 2009).
Four key parameters determine what rigidities cells can sense through integrins: 1) the strength of integrin binding to extracellular ligands, 2) the force and 3) the speed of cell retractions, and 4) the sensitivity of associated mechanosensors,
Strength of the integrin link to the ECM
Matrix attaches to integrins and integrins attach to actin filaments through several direct and indirect routes. However, talin, filamin, α-actinin and tensin are integrin binding proteins that also bind directly to actin (Delon and Brown, 2007). Of these proteins, talin appears to be the one involved in the initial adhesion events as it is located closer to the leading edge than α-actinin (Izzard, 1988; Zaidel-Bar et al., 2003) while filamins appear to be involved in later adhesion events (Calderwood et al., 2001). As the adhesion matures, additional proteins are recruited to reinforce the linkage, including vinculin, which binds actin and talin (Burridge and Mangeat, 1984). In the absence of talin, vinculin staining is diffuse and traction forces are disrupted (Jiang et al., 2003; Zhang et al., 2008).
Thus, there are five non-covalent interactions in series that are needed to develop the forces on early extracellular linkage: (1) myosin and actin interaction, (2) actin monomer assembly into filaments, (3) actin filament binding to linker protein (e.g. talin), (4) linker binding to integrin, and (5) integrin binding to the ECM component. The strengths of many of these connections are either unknown or poorly characterized (Figure 2A). Actin filaments can withstand forces up to 110pN (Kishino and Yanagida, 1988). In some cases, tropomyosin and crosslinking proteins strengthen actin filaments (Liu and Bretscher, 1989; Tseng et al., 2005). For instance, the elastic modulus of actin filament gels increases from 1.2 to 99 pN/μm2 in the presence of fascin and α-actinin (Tseng et al., 2005). The specific strength of the linker/actin or linker/integrin bond is unclear, but there is an initial talin-mediated slip bond that releases at about 0.7 pN per fibronectin (2pN per fibronectin trimer, Jiang et al., 2003). This likely represents the strength of talin’s initial bond to actin filaments that is subsequently reinforced by additional actin binding proteins like vinculin.
Figure 2. Important Parameters of Rigidity Sensing.

Reported values of the strength (A), rates (B), forces (C) and elasticity (D) of components involved in the coupling of the ECM to the cytoskeleton through integrins. References: (1) (Kishino and Yanagida, 1988), (2) (Jiang et al., 2003), (3) table 2, (4) see text, (5) table 1, (6) (Kovar and Pollard, 2004; Mogilner and Oster, 1996; Peskin et al., 1993), (7) (Tseng et al., 2005).
A number of reports suggest that the weakest link is not within the cell, but rather in the integrin bond to the ECM. The bond strength between α5ß1 integrin and fibronectin has been by far the most studied. Reports have placed its strength as low as 0.1 pN or as high as 69 pN (Table 2). Higher estimates correspond to single molecule measurements, while lower values are average forces per molecule in measurements involving adhesions with multiple molecules (compare values labeled ‘S’ and ‘A’ in Table 2). The large discrepancy between both types of measurements suggests that most integrins in adhesions are not bound and that only a small fraction are sustaining forces in the tens of piconewtons.
Table 2.
Integrin-ECM bond strength
| Integrin/ligand | Method | Loading rate (pN/s) | Strength (pN)* | Reference |
|---|---|---|---|---|
| α5ß1/fibronectin | Magnetic tweezers | - | 0.1-0.65 (A) | (Roca-Cusachs et al., 2009) |
|
| ||||
| Spinning disk | - | 1.5 – 2 (A) | (Friedland et al., 2009) | |
|
| ||||
| Laser tweezers | ~40 | 13 – 28 (S) | (Thoumine et al., 2000) | |
|
| ||||
| AFM | ~10,000 | 39 (S) | (Sun et al., 2005) | |
|
| ||||
| AFM | 100 | ~50 (S) | (Li et al., 2003a) | |
| 10,000 | ~100 (S) | |||
|
| ||||
| αIIbß3 / fibrinogen | Laser tweezers | 20,000 | 60-150 (S) | (Litvinov et al., 2002) |
S: Single molecule measurement, A: Average value calculated per molecule from a measurement on multi-molecular adhesions
Cellular forces
Cells can exert protrusion and retraction forces. Protrusive forces by both filopodia and lamellipodia have been observed (Prass et al., 2006; Sheetz et al., 1992). Recently, it has been reported that the protrusion force of lamellipodial is between 20-80 pN/μm2 (Shahapure et al., 2010). However, protrusion forces are unlikely to mediate rigidity sensing since integrins at the leading edge are not normally attached to ECM ligands (Galbraith et al., 2007; Schmidt et al., 1993). Conversely, retraction forces appear to be the relevant forces since ligand binding causes integrin association to the retrogradely flowing actin (Duband et al., 1988; Felsenfeld et al., 1996; Schmidt et al., 1993). Because microtubules are not involved in retraction forces, the relevant cellular motors are myosin and not kinesin nor dynein (Cai et al., 2010). This agrees with findings that non-muscle myosin II is required for the rigidity sensing of stem cells (Engler et al., 2006).
Contractile forces depend upon bipolar myosin filaments and a single myosin head generates between 1.3 and 5 pN of force (Table 3). During the retraction of cellular structures, the individual forces of myosins are additive and can reach up to 2,000,000 pN for a whole cell (Table 4). However, these larger forces are distributed over numerous integrin-matrix adhesion sites, begging the question of the amount of force felt on individual proteins. Patterned substrates of RGD have demonstrated that a spacing of ~ 60 nm is sufficient for strong binding (Arnold et al., 2004; Cavalcanti-Adam et al., 2007; Koo et al., 2002), with smaller spacing having little additional effect. Thus, the 60 nm spacing (corresponding to a density of approximately 300 integrin receptors per μm2) can be regarded as characterizing the density of bound integrins in cells. The retraction forces on a known integrin substrate can reach up to 10,000 pN/μm2 (Table 4). If we divide this number by the integrin density, this gives an estimate of approximately 30 pN per integrin bond. Complexes of three to five integrins are required in order to couple to the cytoskeleton (Coussen et al., 2002) and the adhesive force per matrix ligand can be 7-fold higher with pentameric complexes (Roca-Cusachs et al., 2009). Thus, an early adhesion complex may sustain on the order of 100-165 pN per adhesion unit. Others have used the maximum density of integrins and thus obtained a much lower force of 1pN per integrin bond (Balaban et al., 2001). Our estimate is based on the idea that only a fraction of integrins are engaged with the ECM and it is consistent with the in vitro single molecule measurements of integrin-ECM strength (Table 2).
Table 3.
Molecular Motor Forces
| Motor | Strength (pN) | Key References: |
|---|---|---|
| Myosin II | 1.3 - 3.7 | (Finer et al., 1994; Guilford et al., 1997; Ishijima et al., 1994; Molloy et al., 1995; Tyska et al., 1999) |
| Myosin V | 3 - 5 | (Clemen et al., 2005; Mehta et al., 1999; Uemura et al., 2004) |
| Myosin VI | 2.8 | (Rock et al., 2001) |
| Kinesin | 1.9 - 6 | (Kuo and Sheetz, 1993; Meyhofer and Howard, 1995; Svoboda and Block, 1994) |
Table 4.
Pulling Strengths of Cells
| Cell type | Method | Substrate | Region | Force (pN) | Force per contact area (pN/μm2) | Maximum loading rate per contact area (pN/s·μm2) | Reference: |
|---|---|---|---|---|---|---|---|
| Macrophage | Optical trap | IgG | Filopodium | 1-16 | 1 - 10b | - | (Kress et al., 2007) |
|
| |||||||
| Spinal neuron | Pillars & optical trap | Polylysine or netrin-1 | Filopodium | 2-60 | 1 - 30a,b | - | (Moore et al., 2009) |
|
| |||||||
| DRG neuron | Micropipette | Polylysine, collagen IV & laminin | Growth cone | 500 | - | - | (Lamoureux et al., 1989) |
|
| |||||||
| SCG neuron | Compliant substrate | Laminin | Filopidium | 970 | - | - | (Bridgman et al., 2001) |
|
| |||||||
| Epithelial | Pillars | Fibronectin | Edge | 5,000 | 1,500 | ~ 2 a | (du Roure et al., 2005) |
|
| |||||||
| Keratocyte | Compliant substrate | Silicone | Whole cell | 16,000 | - | - | (Lee et al., 1994) |
|
| |||||||
| Keratocyte | Compliant substrate | Silicone | Whole cell | 45,000 | 27 - 59 | - | (Oliver et al., 1995) |
|
| |||||||
| Smooth muscle | Pillars | Fibronectin | Edge | 60,000 | 8,500a | 24a | (Tan et al., 2003) |
| Center | 15,000 | 2,100a | |||||
|
| |||||||
| Fibroblast | Microcantilever | Laminin | Leading edge | 7,000 | 870 | - | (Galbraith and Sheetz, 1997) |
| Center | 30,000 | 1,500 | |||||
| Trailing edge | 100,000 | 3,910 | |||||
|
| |||||||
| Keratocytes | Compliant substrate | Silicone | Edge (flank) | 680,000 | 130,000 | 1,250 | (Burton et al., 1999) |
|
| |||||||
| Fibroblast | Compliant substrate | Collagen | Whole cell | 2,000,000 | 10,000 | - | (Dembo and Wang, 1999) |
|
| |||||||
| Smooth muscle | Micropipettes | Anionic bead | Whole cell | 2,100,000 | 26,750b | - | (Warshaw and Fay, 1983) |
Assuming contact is the top surface area of pillar.
Assuming contact is ½ of bead area.
Retraction speeds and loading rates
Cell retraction determines the rate at which force is applied to bonds and that affects how much force is required to break a bond either between or within proteins. Thus, the in vivo retraction rates are important. The reported velocities of non-muscle myosin IIa and IIb are 0.29 and 0.092 μm/s, respectively (Pato et al., 1996; Wang et al., 2000a). In living cells, actin is transported rearward at rates between 0.05-0.21 μm/s (Fisher et al., 1988; Forscher and Smith, 1988; Theriot and Mitchison, 1991). These are very consistent with the reported velocities of integrin rearward movement 0.08-0.23 μm/s (Choquet et al., 1997). In a variety of cases, retraction rates can be as high as 0.6 μm/s when there are no or weak adhesions, but drop to approximately 0.05 μm/s when adhesions occur (Giannone et al., 2004; Kress et al., 2007; Zhang et al., 2008). In cells with adhesions that sense rigidity, the typical values for the loading rate then are in the range of 0.05 μm/s or less.
Although loading rates of force over time have not been addressed directly, rough estimates based on published plots of the traction force over time indicate loading rates between 2 and 1,250 pN/(s·μm2) (Table 4, Burton et al., 1999; du Roure et al., 2005; Tan et al., 2003). Given that there could be up to 300 integrins per square micron (see above), this translates to a loading rate of 0.007 to 4 pN/s on individual integrins.
Mechanosensors
How can the generation of force on the matrix be sensed? There are 5 basic mechanisms that have been suggested for mechanosensing through integrins: 1) catch bond formation, 2) channel opening, 3) enzyme regulation, 4) exposure of phosphorylation sites, or 5) exposure of binding sites. All could play significant roles in adhesion-related processes.
Catch bonds
As mentioned above, the lifetime of a bond generally shortens (slips) with applied force, however a subset of interactions ‘catch’ (strengthen) under certain regimes of applied force. Two catch bonds have been described in the ECM connection to the cytoskeleton through integrins: one involving the binding of integrins with the ECM, the other between actin and myosin. Not much is known about the catch bond between myosin II and actin filaments, but it has been reported to have a maximum lifetime at 6 pN (Guo and Guilford, 2006). The catch bond between integrins and the ECM is far more studied. The idea of integrin involvement in the mechanosensing process was proposed originally by Ingber (1991) and several reports have shown that extracellular rigidity causes strengthening of the integrin linkage (Balaban et al., 2001; Choquet et al., 1997; Riveline et al., 2001). Computer simulations have outlined the atomic-level mechanism of how mechanical force could increase the affinity of integrins for their ligand (Jin et al., 2004; Puklin-Faucher et al., 2006). Lateral forces from the cytoskeleton onto integrins may induce structural changes that activate the catch bond (Zhu et al., 2008). However, early work failed to observe a catch bond between α5ß1 integrin and fibronectin (Li et al., 2003a). Recently, using a more sensitive atomic force microscopy (AFM) setup that maintained the force constant at values as low as 4 pN and a retraction rate of 0.2 μm/s (close to physiological rates of ~0.05 μm/s), a catch bond between integrin and fibronectin was observed at forces of 10-30 pN (Kong et al., 2009). Interestingly, this catch bond did not require straightening of the integrin tail region, as previously suspected (Chigaev et al., 2003). The α5s1 integrins bind to fibronectin through two distinct sites: an RGD sequence and a ‘synergy’ site. This catch bond functions, at least in part, through recruitment of the ‘synergy’ binding site (Friedland et al., 2009). Being the primary bond in the rigidity sensing process, the catch bond between the ECM and integrins has an elegant efficiency. However, as discussed below, additional components may be recruited to sense rigidity.
Channel Opening
In hearing, touch, and other mechanical senses, ion channels convert mechanical force into electrical and chemical signals (Sukharev and Corey, 2004). The classical case of force-dependent channel opening is in the hair cells of the auditory system. In such systems, force perpendicular to the membrane opens the channel. Similarly, cytoskeletal forces could pull on a channel associated with the early adhesion complex through lateral associations with the integrins. In endothelial cells, mechanical stress on integrin-bound beads causes calcium entry into cells within 2 to 5 seconds (Matthews et al., 2006). Stretch activated ion-channels have been reported to be important for traction forces at the leading edge, as well as vinculin and tyrosine phosphorylation accumulation at adhesion sites (Munevar et al., 2004). However, the stretch-activated channels were not involved in the initial strengthening of the integrin-mediated link to the ECM, but rather in the later stages of cell repositioning (>60 sec, Matthews et al., 2006). This is consistent with mechanosensitive ion channels requiring filamin recruitment (Glogauer et al., 1998).
Enzyme regulation
A number of enzymes are known to change their kinetics in response to mechanical stimulation; these include kinases, phosphatases, adenylate cyclases and GTPases (Table 5). A key question that has yet to be conclusively answered is whether mechanical force exerted on these proteins alters their function or whether altered kinetics reflects mechanical activation of upstream or downstream effectors. A case for direct activation by mechanical force has been made for the kinase domain in titin (Grater et al., 2005). However, its relevance to integrin-mediated rigidity is unclear. The strongest case for an enzyme regulated by force related to integrin rigidity sensing is focal adhesion kinase (FAK). FAK’s amino FERM domain has been proposed to inhibit its kinase activity (Figure 3A, Cooper et al., 2003). FAK does not bind to integrins directly, but its tyrosine kinase activity increases with mechanical force (Figure 3A, Table 5 and Tomar and Schlaepfer, 2009). This is particularly relevant because loss of FAK inhibits the ability of the cell to sense collagen rigidity (Li et al., 2002a; Wang et al., 2001b).
Table 5.
Mechanical activation of enzymes through integrins
| Enzyme | Type | Function | Regulation by mechanical stimuli | Key references: |
|---|---|---|---|---|
| Adenylate cyclase | Adenylate cyclase | Generates cAMP | Activated by twisting RGD coated beads | (Meyer et al., 2000) |
| FAK | Tyrosine kinase | Focal adhesion turnover and integrin activation | Activated by externally applied force and cell contraction. | (Domingos et al., 2002; Michael et al., 2009; Wang et al., 2001a) |
| Fyn | Tyrosine kinase | Regulates signal transduction. | Activated within ~300 ms of force on fibronectin beads. | (Kostic and Sheetz, 2006; Na et al., 2008; Wang et al., 2005) |
| MAPKs | Serine/threonine kinases | Gene expression | Activated by force on α1 or ß3 integrin | (Schmidt et al., 1998) |
| Rap1 | GTPase | Mitogenic. Activates MAP kinases. | Activated when cell is stretched. cAMP is well established activator of Rap1. | (Sawada et al., 2001) |
| Rho | Rho GTPase | Increases actomyosin contraction | Activated by fluid shear | (Shiu et al., 2004) |
| Rac | Rho GTPase | Promotes lamellipodial and adhesion formation | Inactivated by myosin contraction or cell stretching | (Katsumi et al., 2002) |
| RPTPα | Tyrosine phosphatase | Activates Src family kinases | Activated by restrained fibronectin beads | (von Wichert et al., 2003) |
| Src | Tyrosine kinase | Regulates signal transduction. | Activated by vitronectin. | (Felsenfeld et al., 1999) |
Figure 3. Mechanosensory Proteins in Integrin Mediated Rigidity Sensing.
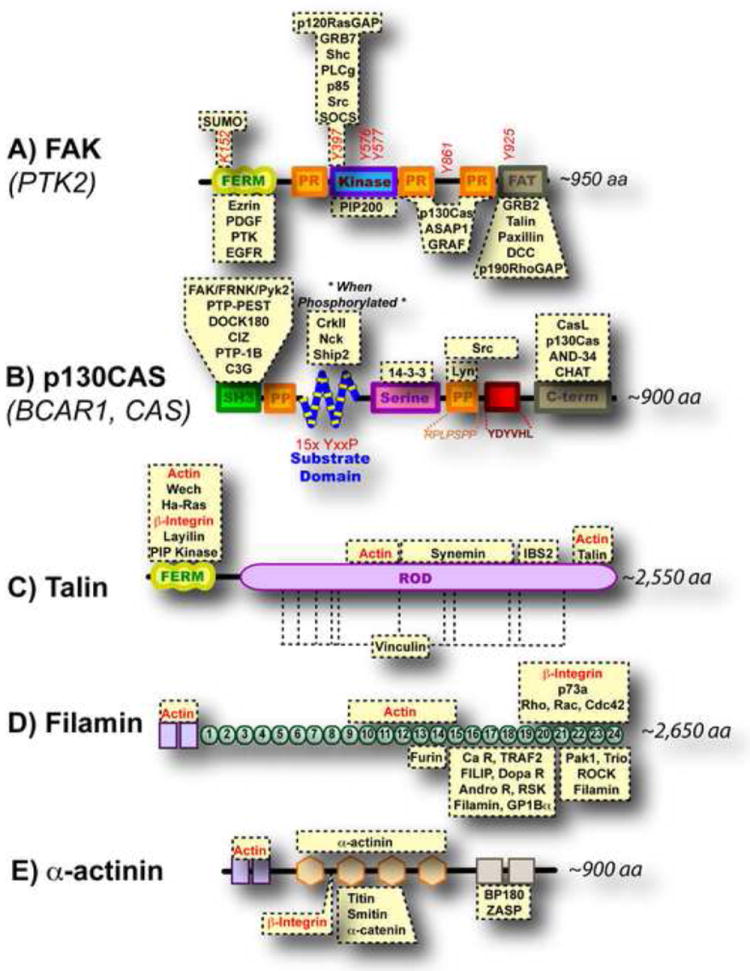
Proteins that bind directly to the depicted domains are highlighted in yellow boxes. (A) FAK does not bind integrins or actin directly but its kinase activity is regulated by mechanical force and it has been hypothesized that removal of the FERM domain from the kinase could play a role (Cooper et al., 2003). (B) The substrate domain of p130Cas contains fifteen tyrosine residues that become exposed upon stretching (Sawada et al., 2006). (C) Stretching of talin’s rod domain exposes vinculin binding sites (del Rio et al., 2009). (D) Extension of filamin immunoglobulin repeats (labeled 1-24) has been shown by AFM (Furuike et al., 2001) and could regulate the binding of proteins. (E) α-actinin forms antiparallel dimers; mechanical force could regulate this dimerization or its association with other proteins.
A local but indirect mechanical activation has been proposed for the Receptor-like Protein Tyrosine Phosphatase-α (RPTPα). This transmembrane phosphatase forms an auto-inhibited dimer in which each of their helix-turn-helix wedges insert into the other’s catalytic cleft (Jiang et al., 2000; Tabernero et al., 2008) that associates with αvß3 integrins and becomes activated by restrained fibronectin (von Wichert et al., 2003). Moreover, it is known to activate Src family kinases (SFKs) which are themselves activated within approximately 300 ms after force application on fibronectin beads (den Hertog et al., 1993; Na et al., 2008; Zheng et al., 2000) and to cause localization of the Src-family kinase fyn near the leading edge (Kostic and Sheetz, 2006).
Exposing phosphorylation sites
There is considerable evidence that cell stretching and matrix rigidity increases tyrosine phosphorylation of important proteins (Glogauer et al., 1997; Li et al., 2002b; Pelham and Wang, 1997; Schmidt et al., 1998; Wang et al., 2005). An interesting observation can be made when comparing mechanical responses of intact cells versus when their plasma membranes have been stripped away -- intact cells (but not stripped cells) will respond to mechanical stimulus by activating Src family kinases (Na et al., 2008; Sawada et al., 2006; Wang et al., 2005). Importantly, mechanical stimulus in both cases leads to increased tyrosine phosphorylation. This not only reinforces the idea that the transmembrane phosphatase RPTPα is required for Src activation, but also suggests that there is increased availability of phosphorylation sites upon stretching. To date, only the Cas protein family has been proposed to undergo conformational changes that expose phosphorylation sites (Sawada et al., 2006). This protein family contains a substrate domain with multiple Src family kinase tyrosine phosphorylation sites flanked by domains capable of binding a variety of proteins (Figure 3B; Geiger, 2006). When incubated with active Src kinase, increased tyrosine phosphorylation of the substrate domain of p130Cas is observed upon stretch (Sawada et al., 2006). Upon phosphorylation, the substrate domain of Cas becomes a docking site for a variety of proteins including Crk, Nck, and Ship2 (Mayer et al., 1995; Prasad et al., 2001; Schlaepfer et al., 1997) that then activate GTP exchange factors to locally form active small G-proteins such as Rap1 (Tamada et al., 2004). More recent studies are supportive of a role for p130Cas in fibronectin, but not collagen, rigidity sensing (Kostic and Sheetz, 2006).
Exposing protein interaction sites
The other major mechanism for mechanotransduction is the exposure of protein-protein binding sites by stretch. In cells that have been stripped of their plasma membrane, stretching leads to the increased cytoskeleton binding of paxillin, focal adhesion kinase (FAK), p130Cas, PKB/Akt, C3G and CrkII (Sawada and Sheetz, 2002; Tamada et al., 2004). Interestingly, myosin II contractility is required for the recruitment of FAK, zyxin, vinculin, and α-actinin, but not for the recruitment of paxillin, talin, and α1 integrin (Pasapera et al., 2010). Unfortunately, most of the direct protein-protein interactions are unclear. The only case where stretch-dependent binding of one protein to another has been documented is during stretching of talin, which leads to binding of vinculin (del Rio et al., 2009). Talin has 11 potential vinculin binding sites, as well as binding sites for actin filaments, integrins, and a number of other proteins (Figure 3C). Stretching of talin by mechanical forces therefore appears to play an important role in the reinforcement of early adhesions through the recruitment of additional actin binding proteins. Fibronectin is a major ECM protein that has been shown to expose protein interaction sites under mechanical strain (Vogel, 2006). Cellular contraction can expose cryptic sites in fibronectin that are important for its assembly into a matrix (Zhong et al., 1998). Thus, both intracellular and extracellular protein stretching may have important consequences.
Is Protein Stretching Physiological?
A fundamental question in examining protein stretching as a mechanism of mechanotransduction is whether adhesion proteins can be stretched with physiological force. As outlined in the above sections, an estimate of the pulling force on individual integrins is on the order of 30 pN leading to between 100 and 165 pN on a minimal integrin adhesion complex of 3-5 integrins. Although stretching of titin, spectrin and ankyrin have been studied extensively both in vitro and in silico (Sotomayor and Schulten, 2007), their relevance to integrin mediated rigidity sensing is unclear. On the other hand, stretching of talin to expose vinculin binding sites was initially estimated based on computer simulations to require 13 pN (Lee et al., 2007). When AFM was used to measure this value directly, it took only 0.2 seconds for talin to unfold under a constant force of 20 pN (del Rio et al., 2009). Another protein family that bridges integrins to actin is filamin (Zhou et al., 2010). Filamins have an actin-binding domain followed by 24 immunoglobulin repeats and bind integrins through their c-terminal domain (Figure 3D). Using AFM, the immunoglobulin repeats of filamin have been extended with 50 pN of force at a pulling speed of 0.37μm/s (Furuike et al., 2001). Focal adhesion kinase (FAK) is another integrin-associated protein whose elasticity has been addressed. The FAT domain is responsible for targeting FAK to focal adhesions (Hildebrand et al., 1993). Computer simulations have predicted that the FAT domain of FAK will extend with under 75pN of force (Mofrad et al., 2004).
From these data, it seems that the stretching of talin’s vinculin binding domain, filamin’s immunoglobulin domains, and potentially FAK’s FAT domain do occur at physiologically relevant forces. Moreover, direct support has been obtained by looking at the number of exposed cysteines in proteins (Johnson et al., 2007). This technique has shown that in stretched live cells, normally buried cysteines in vimentin and myosin IIa are exposed.
Mechanisms of Rigidity Sensing
Substrate rigidity influences a number of cellular processes including cell adhesion, actin flow, retraction forces, gene expression and cell lineage (Bard and Hay, 1975; Choquet et al., 1997; Engler et al., 2006; Friedl and Brocker, 2000; Giannone et al., 2004; Lo et al., 2000; Opas and Dziak, 1990; Pelham and Wang, 1997; Peyton and Putnam, 2005; Saez et al., 2005; Yeung et al., 2005). However, in its most basic form, integrin-mediated rigidity sensing can be taken as the decision to couple and reinforce the link between an extracellular ligand and the cytoskeleton. For instance, fibroblasts presented with fibronectin coated beads under restraining forces of 0.02 or 0.18 pN/nm will preferentially couple stiff beads to their cytoskeleton (Choquet et al., 1997; Jiang et al., 2006). Whether integrin-cytoskeleton linkages become reinforced depends upon the mechanical properties of the microenvironment and the intracellular components that make up this link. Thus, rigidity responses depend upon both the nature of the matrix and the cell-type specific components involved in the responses.
An early event after ligand binding to integrins involves the activation of Src family kinases (potentially through mechanical effects on integrins that activate RPTPα, as mentioned above). This is supported by their rapid activation (within 300ms) after applied force and the observation that the Src family kinases Fyn and Src are required for rigidity sensing on fibronectin and vitronectin, respectively (Felsenfeld et al., 1999; Kostic and Sheetz, 2006; Na et al., 2008). Although the mechanism is unclear, the activation of Src family kinases leads to the bridging of integrins to the cytoskeleton through talin. This is supported by the observations that ligand binding couples integrins to the cytoskeleton and that talin is not required for SFK activation (Duband et al., 1988; Felsenfeld et al., 1996; Schmidt et al., 1993; Zhang et al., 2008). Once coupled to retrograde flowing actin, mechanical force on integrins could engage the integrin/ECM catch bond. Force on talin could then expose vinculin-binding sites that stabilize and recruit additional links to actin (del Rio et al., 2009). Consistent with this possibility, myosin II contractility is required for vinculin but not for talin recruitment (Pasapera et al., 2010). Finally, activation of FAK could reverse adhesions and restart the process. This is supported by observed roles of FAK in adhesion turnover and the requirement of talin in the activation of FAK (Ilic et al., 1995; Zhang et al., 2008). Thus, rigidity sensing is an active process that is transient and multiple steps could be sensitive to the matrix rigidity (Figure 4A).
Figure 4. The Rigidity Sensing Cycle and Models for Uniform Displacements.
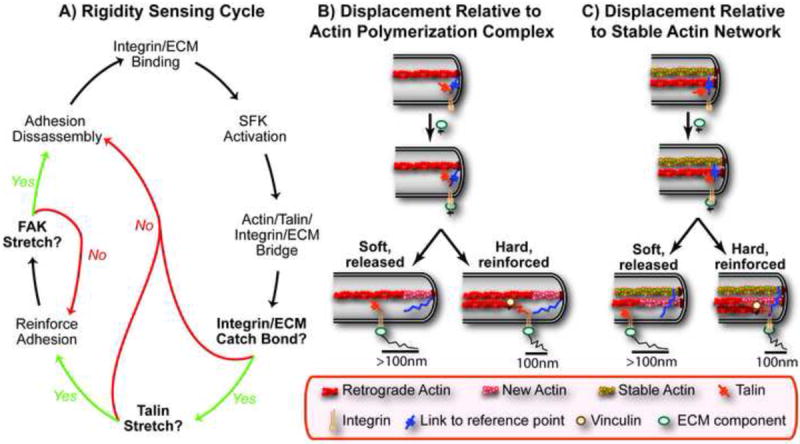
(A) A possible rigidity sensing cycle involves three mechanosensory events: (1) integrin/ECM catch bond formation, (2) stretching of talin that reinforces the adhesion by recruiting vinculin and (3) stretching of FAK that activates its kinase domain leading to the disassembly and recycling of the adhesion. (B,C) Two models to explain the uniform displacements of approximately 100nm, one where the reference structure is the polymerization complex (B) and the other where a stable actin network provides the reference structure (C). In both models the key decision is based on whether the extension of the link to retrograde flowing actin (e.g. talin) occurs before the link to the reference structure is broken.
In such a model, substrate rigidity determines the loading rate felt on the integrin-ECM catch bond. Just as catch bonds have a force providing maximum lifetime in a scenario of constant force application, they will have a corresponding optimal loading rate in scenarios where force is loaded progressively. Thus, at the optimal rigidity, the loading rate will maximize bond lifetime, providing the opportunity for the applied force to trigger subsequent mechanotransduction events. Regardless of the particular model for mechanotransduction, coupling between rearward flowing actin and the substrate is key for rigidity sensing. Detailing the temporal series of mechanosensory events in rigidity sensing will require clever experimental setups that merge high-resolution imaging and sensitive force-sensing techniques.
Rigidity Sensing based on 100nm Displacement Events?
When integrins bind to matrix, there is evidence that rigidity sensing involves uniform displacements of the substrate. If epithelial cells are grown on pillars with spring constants ranging from 1 to 100 pN/nm, they displace the pillars by ~130 nm regardless of their stiffness (Saez et al., 2005). Similarly, when fibroblasts are presented with beads in a laser tweezers force ramp of 0.02 or 0.18 pN/nm, the beads in the softer tweezers are more likely to be released from the cytoskeleton (Jiang et al., 2006). If, however, the softer beads were moved in the tweezers to produce 10-20 pN of force within 100 nm of the initial position, reinforcement occurred as with the stiffer beads. Therefore, cells appear to sense rigidity based on whether a threshold force is obtained within a given displacement of approximately 100-150 nm.
This raises the question of what limits the distance of the sensing event.? If we consider a fibronectin- α5β1 catch bond acting at the force that provides maximum lifetime (20 pN), the substrate rigidity needed to cause a displacement of 100 nm is on the order of 10 nN/μm2 and therefore in the rigidity range of several different tissues (Text Box 1 and Table 1). However, as the substrate rigidity is changed, so will the displacement events, which is not what has been observed experimentally (Saez et al., 2005). Therefore, a mechanism involving more than a catch bond is presumably necessary.
Text Box 1. Estimating optimal rigidity for an integrin-ECM catch bond engagement with an optimal loading of 20 pN.
To evaluate the order of magnitude of the rigidity that would likely result in a displacement of ~100 nm, we did the following calculation. We considered a minimal adhesion complex with three integrins spaced 60 nm from one another, each exerting a force of 20 pN. We thus assumed a total force (F) of 60 pN, which for simplicity we modeled as a single point force exerted in the middle of three integrins located at a distance (x) of 30 nm from the source. The displacement d in a material of shear modulus G caused by such a point force as a function of the distance to the source x (along the direction of force application) is d = F/(2πGx) (Landau et al., 1986). If we consider a displacement (d) of 100 nm, we can calculate the corresponding shear modulus as G = F/(2πdx). By assuming an incompressible material with a Poisson ratio of 0.5 where the elastic modulus is 3 times the shear modulus, then E = 3G = 3F/(2πdx) ≈ 10 nN/μm2. Given the assumptions and approximations, this value should be taken only as an estimate of the order of magnitude involved, but it does provide a useful guide to the range of rigidities for which such a configuration would be tuned.
One possibility is that this distance represents the stretching of a mechanosensor linking the integrin complex to a reference structure within the cell. Proteins are typically folded into domains of 2.5 nm containing 200 amino acids (Bao, 2009). If we assume that the maximum extension per amino acid is 0.4nm (Ainavarapu et al., 2007), a domain of only 200 amino acids could be stretched to 80 nm – in others words by 32 times its original length and close to the observed displacement of 100 nm. A portion of 407 amino acids from Talin’s rod domain (containing five of its eleven potential vinculin binding sites) increases its length by 140 nm upon the application of force (del Rio et al., 2009). Thus extension of intracellular proteins is within the range of the observed displacements of approximately 100nm.
A final point that emerges if protein stretching underlies the uniform displacement events is that it requires attachment to a reference structure. Potential reference structures within the cell include the actin polymerization complex or a rigid actin network (Figures 4B and 4C). In regard to the latter possibility, two overlapping actin networks are seen at the periphery of cells (Giannone et al., 2007; Ponti et al., 2004). For simplicity, these rigidity-sensing models are based on only single integrin receptors. As discussed above, complexes of three to five integrins act synergistically (Coussen et al., 2002). Integrin multimerization may initiate downstream signaling and be an integral part of the rigidity sensing machinery (Li et al., 2003b; Paszek et al., 2005). Thus, more complicated models involving integrin multimerizaiton and potentially mechanosensory proteins that bridge integrins may be necessary.
Conclusion
In summary, the mechanism of integrin-mediated rigidity sensing is constrained by physical characteristics of the cytoskeleton-integrin-matrix link. Relevant factors include the force and loading rate that cells employ to probe the substrate, the sensitivity of their mechanotransduction system and the strength of their attachment to the ECM. Current measurements indicate that the liganded integrins move rearward at about 50 nm/s and can support on the order of 30 pN per bond. Although integrin-matrix catch bonds can explain reduced adhesion to soft surfaces and cell rounding, they cannot explain the observed uniform displacements over a wide range of rigidity. If displacements of approximately 100nm are indeed important elements of the rigidity sensing process of cells, a key challenge will be to identify the structure that provides the reference point.
We focused here on rigidity sensing through integrins. However, tissue rigidity can influence cellular behavior through cadherin-mediated adhesions (Tsai and Kam, 2009). Little is known of the rigidity sensing mediated through cell-cell or other ECM receptors; although, recently it has been shown that mechanical forces regulate adherens cell-cell junction size (Liu et al., 2010). Because other transmembrane receptors are bridged to the cytoskeleton through many of the same intracellular proteins, it is likely that similar rigidity sensing mechanisms exist.
Finally, an important gap in our knowledge is how the short-term rigidity sensing described here is translated into the long-term rigidity sensing involved in cell differentiation. Although much may be shared between the two processes, they may have radically different time constants and frequencies. In fibroblasts, there are periodic pulls on the matrix that produce early adhesions every 24 seconds (Giannone et al., 2004). However, it is not evident that stem cells test the rigidity of the environment in the same way and with the same frequency over the full period of differentiation that can take 7 days or more. Thus, it remains a key challenge to link the early rigidity mechanisms described here to the long-term processes involved in cellular differentiation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ainavarapu SR, Brujic J, Huang HH, Wiita AP, Lu H, Li L, Walther KA, Carrion-Vazquez M, Li H, Fernandez JM. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys J. 2007;92:225–33. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando J, Yamamoto K. Vascular mechanobiology: endothelial cell responses to fluid shear stress. Circ J. 2009;73:1983–92. doi: 10.1253/circj.cj-09-0583. [DOI] [PubMed] [Google Scholar]
- Arnold M, Cavalcanti-Adam EA, Glass R, Blummel J, Eck W, Kantlehner M, Kessler H, Spatz JP. Activation of integrin function by nanopatterned adhesive interfaces. Chemphyschem. 2004;5:383–8. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, Geiger B. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3:466–72. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- Balgude AP, Yu X, Szymanski A, Bellamkonda RV. Agarose gel stiffness determines rate of DRG neurite extension in 3D cultures. Biomaterials. 2001;22:1077–84. doi: 10.1016/s0142-9612(00)00350-1. [DOI] [PubMed] [Google Scholar]
- Bank AJ, Kaiser DR. Smooth muscle relaxation: effects on arterial compliance, distensibility, elastic modulus, and pulse wave velocity. Hypertension. 1998;32:356–9. doi: 10.1161/01.hyp.32.2.356. [DOI] [PubMed] [Google Scholar]
- Bao G. Protein Mechanics: A New Frontier in Biomechanics. Exp Mech. 2009;49:153–164. doi: 10.1007/s11340-008-9154-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barczyk M, Carracedo S, Gullberg D. Integrins. Cell Tissue Res. 2010;339:269–80. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard JB, Hay ED. The behavior of fibroblasts from the developing avian cornea. Morphology and movement in situ and in vitro. J Cell Biol. 1975;67:400–18. doi: 10.1083/jcb.67.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–27. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Bridgman PC, Dave S, Asnes CF, Tullio AN, Adelstein RS. Myosin IIB is required for growth cone motility. J Neurosci. 2001;21:6159–69. doi: 10.1523/JNEUROSCI.21-16-06159.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdun GD, Kolchin OM, Kogan AI, Gorbunkov AV. Application of the International (SI) System of Units to mechanics, strength of materials, and mechanical engineering. Measurement Techniques. 1972;15:860–864. [Google Scholar]
- Burridge K, Mangeat P. An interaction between vinculin and talin. Nature. 1984;308:744–6. doi: 10.1038/308744a0. [DOI] [PubMed] [Google Scholar]
- Burton K, Park JH, Taylor DL. Keratocytes generate traction forces in two phases. Mol Biol Cell. 1999;10:3745–69. doi: 10.1091/mbc.10.11.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Rossier O, Gauthier NC, Biais N, Fardin MA, Zhang X, Miller LW, Ladoux B, Cornish VW, Sheetz MP. Cytoskeletal coherence requires myosin-IIA contractility. J Cell Sci. 2010 doi: 10.1242/jcs.058297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood DA, Huttenlocher A, Kiosses WB, Rose DM, Woodside DG, Schwartz MA, Ginsberg MH. Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat Cell Biol. 2001;3:1060–8. doi: 10.1038/ncb1201-1060. [DOI] [PubMed] [Google Scholar]
- Cavalcanti-Adam EA, Volberg T, Micoulet A, Kessler H, Geiger B, Spatz JP. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys J. 2007;92:2964–74. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao EY, Aro HT, Lewallen DG, Kelly PJ. The effect of rigidity on fracture healing in external fixation. Clin Orthop Relat Res. 1989:24–35. [PubMed] [Google Scholar]
- Chapuis JF, Agache P. A new technique to study the mechanical properties of collagen lattices. J Biomech. 1992;25:115–20. doi: 10.1016/0021-9290(92)90250-5. [DOI] [PubMed] [Google Scholar]
- Chigaev A, Buranda T, Dwyer DC, Prossnitz ER, Sklar LA. FRET detection of cellular alpha4-integrin conformational activation. Biophys J. 2003;85:3951–62. doi: 10.1016/S0006-3495(03)74809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet D, Felsenfeld DP, Sheetz MP. Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell. 1997;88:39–48. doi: 10.1016/s0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- Clemen AE, Vilfan M, Jaud J, Zhang J, Barmann M, Rief M. Force-dependent stepping kinetics of myosin-V. Biophys J. 2005;88:4402–10. doi: 10.1529/biophysj.104.053504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin D, Ritort F, Jarzynski C, Smith SB, Tinoco I, Jr, Bustamante C. Verification of the Crooks fluctuation theorem and recovery of RNA folding free energies. Nature. 2005;437:231–4. doi: 10.1038/nature04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinsworth AM, Zhang S, Kraus WE, Truskey GA. Apparent elastic modulus and hysteresis of skeletal muscle cells throughout differentiation. Am J Physiol Cell Physiol. 2002;283:C1219–27. doi: 10.1152/ajpcell.00502.2001. [DOI] [PubMed] [Google Scholar]
- Cooper LA, Shen TL, Guan JL. Regulation of focal adhesion kinase by its amino-terminal domain through an autoinhibitory interaction. Mol Cell Biol. 2003;23:8030–41. doi: 10.1128/MCB.23.22.8030-8041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussen F, Choquet D, Sheetz MP, Erickson HP. Trimers of the fibronectin cell adhesion domain localize to actin filament bundles and undergo rearward translocation. J Cell Sci. 2002;115:2581–90. doi: 10.1242/jcs.115.12.2581. [DOI] [PubMed] [Google Scholar]
- del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–41. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delon I, Brown NH. Integrins and the actin cytoskeleton. Curr Opin Cell Biol. 2007;19:43–50. doi: 10.1016/j.ceb.2006.12.013. [DOI] [PubMed] [Google Scholar]
- Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76:2307–16. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hertog J, Pals CE, Peppelenbosch MP, Tertoolen LG, de Laat SW, Kruijer W. Receptor protein tyrosine phosphatase alpha activates pp60c-src and is involved in neuronal differentiation. EMBO J. 1993;12:3789–98. doi: 10.1002/j.1460-2075.1993.tb06057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingos PP, Fonseca PM, Nadruz W, Jr, Franchini KG. Load-induced focal adhesion kinase activation in the myocardium: role of stretch and contractile activity. Am J Physiol Heart Circ Physiol. 2002;282:H556–64. doi: 10.1152/ajpheart.00534.2001. [DOI] [PubMed] [Google Scholar]
- du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Silberzan P, Ladoux B. Force mapping in epithelial cell migration. Proc Natl Acad Sci U S A. 2005;102:2390–5. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duband JL, Nuckolls GH, Ishihara A, Hasegawa T, Yamada KM, Thiery JP, Jacobson K. Fibronectin receptor exhibits high lateral mobility in embryonic locomoting cells but is immobile in focal contacts and fibrillar streaks in stationary cells. J Cell Biol. 1988;107:1385–96. doi: 10.1083/jcb.107.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkin BS, Azeloglu EU, Costa KD, Morrison B., 3rd Mechanical heterogeneity of the rat hippocampus measured by atomic force microscope indentation. J Neurotrauma. 2007;24:812–22. doi: 10.1089/neu.2006.0169. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Evans E. Probing the relation between force--lifetime--and chemistry in single molecular bonds. Annu Rev Biophys Biomol Struct. 2001;30:105–28. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- Fabry B, Maksym GN, Butler JP, Glogauer M, Navajas D, Fredberg JJ. Scaling the microrheology of living cells. Phys Rev Lett. 2001;87:148102. doi: 10.1103/PhysRevLett.87.148102. [DOI] [PubMed] [Google Scholar]
- Felber ES. Botulinum toxin in primary care medicine. J Am Osteopath Assoc. 2006;106:609–14. [PubMed] [Google Scholar]
- Felsenfeld DP, Choquet D, Sheetz MP. Ligand binding regulates the directed movement of beta1 integrins on fibroblasts. Nature. 1996;383:438–40. doi: 10.1038/383438a0. [DOI] [PubMed] [Google Scholar]
- Felsenfeld DP, Schwartzberg PL, Venegas A, Tse R, Sheetz MP. Selective regulation of integrin--cytoskeleton interactions by the tyrosine kinase Src. Nat Cell Biol. 1999;1:200–6. doi: 10.1038/12021. [DOI] [PubMed] [Google Scholar]
- Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–9. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- Fisher GW, Conrad PA, DeBiasio RL, Taylor DL. Centripetal transport of cytoplasm, actin, and the cell surface in lamellipodia of fibroblasts. Cell Motil Cytoskeleton. 1988;11:235–47. doi: 10.1002/cm.970110403. [DOI] [PubMed] [Google Scholar]
- Flanagan LA, Ju YE, Marg B, Osterfield M, Janmey PA. Neurite branching on deformable substrates. Neuroreport. 2002;13:2411–5. doi: 10.1097/01.wnr.0000048003.96487.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forscher P, Smith SJ. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J Cell Biol. 1988;107:1505–16. doi: 10.1083/jcb.107.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Brocker EB. T cell migration in three-dimensional extracellular matrix: guidance by polarity and sensations. Dev Immunol. 2000;7:249–66. doi: 10.1155/2000/56473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323:642–4. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- Furuike S, Ito T, Yamazaki M. Mechanical unfolding of single filamin A (ABP-280) molecules detected by atomic force microscopy. FEBS Lett. 2001;498:72–5. doi: 10.1016/s0014-5793(01)02497-8. [DOI] [PubMed] [Google Scholar]
- Galbraith CG, Sheetz MP. A micromachined device provides a new bend on fibroblast traction forces. Proc Natl Acad Sci U S A. 1997;94:9114–8. doi: 10.1073/pnas.94.17.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith CG, Yamada KM, Galbraith JA. Polymerizing actin fibers position integrins primed to probe for adhesion sites. Science. 2007;315:992–5. doi: 10.1126/science.1137904. [DOI] [PubMed] [Google Scholar]
- Gefen A, Gefen N, Zhu Q, Raghupathi R, Margulies SS. Age-dependent changes in material properties of the brain and braincase of the rat. J Neurotrauma. 2003;20:1163–77. doi: 10.1089/089771503770802853. [DOI] [PubMed] [Google Scholar]
- Geiger B. A role for p130Cas in mechanotransduction. Cell. 2006;127:879–81. doi: 10.1016/j.cell.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Giannone G, Dubin-Thaler BJ, Dobereiner HG, Kieffer N, Bresnick AR, Sheetz MP. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116:431–43. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- Giannone G, Dubin-Thaler BJ, Rossier O, Cai Y, Chaga O, Jiang G, Beaver W, Dobereiner HG, Freund Y, Borisy G, Sheetz MP. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128:561–75. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glogauer M, Arora P, Chou D, Janmey PA, Downey GP, McCulloch CA. The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J Biol Chem. 1998;273:1689–98. doi: 10.1074/jbc.273.3.1689. [DOI] [PubMed] [Google Scholar]
- Glogauer M, Arora P, Yao G, Sokholov I, Ferrier J, McCulloch CA. Calcium ions and tyrosine phosphorylation interact coordinately with actin to regulate cytoprotective responses to stretching. J Cell Sci. 1997;110(Pt 1):11–21. doi: 10.1242/jcs.110.1.11. [DOI] [PubMed] [Google Scholar]
- Grater F, Shen J, Jiang H, Gautel M, Grubmuller H. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys J. 2005;88:790–804. doi: 10.1529/biophysj.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinnell F. Fibroblasts, myofibroblasts, and wound contraction. J Cell Biol. 1994;124:401–4. doi: 10.1083/jcb.124.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilford WH, Dupuis DE, Kennedy G, Wu J, Patlak JB, Warshaw DM. Smooth muscle and skeletal muscle myosins produce similar unitary forces and displacements in the laser trap. Biophys J. 1997;72:1006–21. doi: 10.1016/S0006-3495(97)78753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Guilford WH. Mechanics of actomyosin bonds in different nucleotide states are tuned to muscle contraction. Proc Natl Acad Sci U S A. 2006;103:9844–9. doi: 10.1073/pnas.0601255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahmann C, Schroeter T. Rho-kinase inhibitors as therapeutics: from pan inhibition to isoform selectivity. Cell Mol Life Sci. 2010;67:171–7. doi: 10.1007/s00018-009-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand JD, Schaller MD, Parsons JT. Identification of sequences required for the efficient localization of the focal adhesion kinase, pp125FAK, to cellular focal adhesions. J Cell Biol. 1993;123:993–1005. doi: 10.1083/jcb.123.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B. Masters and servants of the force: the role of matrix adhesions in myofibroblast force perception and transmission. Eur J Cell Biol. 2006;85:175–81. doi: 10.1016/j.ejcb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Hirakawa K, Hashizume K, Hayashi T. Viscoelastic property of human brain -for the analysis of impact injury (author’s transl) No To Shinkei. 1981;33:1057–65. [PubMed] [Google Scholar]
- Hochmuth RM. Micropipette aspiration of living cells. J Biomech. 2000;33:15–22. doi: 10.1016/s0021-9290(99)00175-x. [DOI] [PubMed] [Google Scholar]
- Ilic D, Furuta Y, Kanazawa S, Takeda N, Sobue K, Nakatsuji N, Nomura S, Fujimoto J, Okada M, Yamamoto T. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature. 1995;377:539–44. doi: 10.1038/377539a0. [DOI] [PubMed] [Google Scholar]
- Ingber D. Integrins as mechanochemical transducers. Curr Opin Cell Biol. 1991;3:841–8. doi: 10.1016/0955-0674(91)90058-7. [DOI] [PubMed] [Google Scholar]
- Ingber DE. Mechanobiology and diseases of mechanotransduction. Ann Med. 2003;35:564–77. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- Intengan HD, Thibault G, Li JS, Schiffrin EL. Resistance artery mechanics, structure, and extracellular components in spontaneously hypertensive rats : effects of angiotensin receptor antagonism and converting enzyme inhibition. Circulation. 1999;100:2267–75. doi: 10.1161/01.cir.100.22.2267. [DOI] [PubMed] [Google Scholar]
- Ishijima A, Harada Y, Kojima H, Funatsu T, Higuchi H, Yanagida T. Single-molecule analysis of the actomyosin motor using nano-manipulation. Biochem Biophys Res Commun. 1994;199:1057–63. doi: 10.1006/bbrc.1994.1336. [DOI] [PubMed] [Google Scholar]
- Izzard CS. A precursor of the focal contact in cultured fibroblasts. Cell Motil Cytoskeleton. 1988;10:137–42. doi: 10.1002/cm.970100118. [DOI] [PubMed] [Google Scholar]
- Jiang G, den Hertog J, Hunter T. Receptor-like protein tyrosine phosphatase alpha homodimerizes on the cell surface. Mol Cell Biol. 2000;20:5917–29. doi: 10.1128/mcb.20.16.5917-5929.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature. 2003;424:334–7. doi: 10.1038/nature01805. [DOI] [PubMed] [Google Scholar]
- Jiang G, Huang AH, Cai Y, Tanase M, Sheetz MP. Rigidity sensing at the leading edge through alphavbeta3 integrins and RPTPalpha. Biophys J. 2006;90:1804–9. doi: 10.1529/biophysj.105.072462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Andricioaei I, Springer TA. Conversion between three conformational states of integrin I domains with a C-terminal pull spring studied with molecular dynamics. Structure. 2004;12:2137–47. doi: 10.1016/j.str.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317:663–6. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallel F, Ophir J, Magee K, Krouskop T. Elastographic imaging of low-contrast elastic modulus distributions in tissue. Ultrasound Med Biol. 1998;24:409–25. doi: 10.1016/s0301-5629(97)00287-1. [DOI] [PubMed] [Google Scholar]
- Katsumi A, Milanini J, Kiosses WB, del Pozo MA, Kaunas R, Chien S, Hahn KM, Schwartz MA. Effects of cell tension on the small GTPase Rac. J Cell Biol. 2002;158:153–64. doi: 10.1083/jcb.200201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R, Davidson LA, Shook DR. How we are shaped: the biomechanics of gastrulation. Differentiation. 2003;71:171–205. doi: 10.1046/j.1432-0436.2003.710301.x. [DOI] [PubMed] [Google Scholar]
- Kishino A, Yanagida T. Force measurements by micromanipulation of a single actin filament by glass needles. Nature. 1988;334:74–6. doi: 10.1038/334074a0. [DOI] [PubMed] [Google Scholar]
- Kong F, Garcia AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185:1275–84. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HJ, Polte TR, Alsberg E, Mooney DJ. FRET measurements of cell-traction forces and nano-scale clustering of adhesion ligands varied by substrate stiffness. Proc Natl Acad Sci U S A. 2005;102:4300–5. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo LY, Irvine DJ, Mayes AM, Lauffenburger DA, Griffith LG. Co-regulation of cell adhesion by nanoscale RGD organization and mechanical stimulus. J Cell Sci. 2002;115:1423–33. doi: 10.1242/jcs.115.7.1423. [DOI] [PubMed] [Google Scholar]
- Kostic A, Lynch CD, Sheetz MP. Differential matrix rigidity response in breast cancer cell lines correlates with the tissue tropism. PLoS One. 2009;4:e6361. doi: 10.1371/journal.pone.0006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A, Sheetz MP. Fibronectin rigidity response through Fyn and p130Cas recruitment to the leading edge. Mol Biol Cell. 2006;17:2684–95. doi: 10.1091/mbc.E05-12-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci U S A. 2004;101:14725–30. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress H, Stelzer EH, Holzer D, Buss F, Griffiths G, Rohrbach A. Filopodia act as phagocytic tentacles and pull with discrete steps and a load-dependent velocity. Proc Natl Acad Sci U S A. 2007;104:11633–8. doi: 10.1073/pnas.0702449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo SC, Sheetz MP. Force of single kinesin molecules measured with optical tweezers. Science. 1993;260:232–4. doi: 10.1126/science.8469975. [DOI] [PubMed] [Google Scholar]
- Lamoureux P, Buxbaum RE, Heidemann SR. Direct evidence that growth cones pull. Nature. 1989;340:159–62. doi: 10.1038/340159a0. [DOI] [PubMed] [Google Scholar]
- Landau LD, Pitaevskii LP, Lifshitz EM, Kosevich AM. Theory of elasticity. viii. Pergamon Press; Oxford [Oxfordshire] ; New York: 1986. p. 187. [Google Scholar]
- Lee J, Leonard M, Oliver T, Ishihara A, Jacobson K. Traction forces generated by locomoting keratocytes. J Cell Biol. 1994;127:1957–64. doi: 10.1083/jcb.127.6.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Kamm RD, Mofrad MR. Force-induced activation of talin and its possible role in focal adhesion mechanotransduction. J Biomech. 2007;40:2096–106. doi: 10.1016/j.jbiomech.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Li F, Redick SD, Erickson HP, Moy VT. Force measurements of the alpha5beta1 integrin-fibronectin interaction. Biophys J. 2003a;84:1252–62. doi: 10.1016/S0006-3495(03)74940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Okura M, Imamoto A. Focal adhesions require catalytic activity of Src family kinases to mediate integrin-matrix adhesion. Mol Cell Biol. 2002a;22:1203–17. doi: 10.1128/MCB.22.4.1203-1217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Mitra N, Gratkowski H, Vilaire G, Litvinov R, Nagasami C, Weisel JW, Lear JD, DeGrado WF, Bennett JS. Activation of integrin alphaIIbbeta3 by modulation of transmembrane helix associations. Science. 2003b;300:795–8. doi: 10.1126/science.1079441. [DOI] [PubMed] [Google Scholar]
- Li S, Butler P, Wang Y, Hu Y, Han DC, Usami S, Guan JL, Chien S. The role of the dynamics of focal adhesion kinase in the mechanotaxis of endothelial cells. Proc Natl Acad Sci U S A. 2002b;99:3546–51. doi: 10.1073/pnas.052018099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinov RI, Shuman H, Bennett JS, Weisel JW. Binding strength and activation state of single fibrinogen-integrin pairs on living cells. Proc Natl Acad Sci U S A. 2002;99:7426–31. doi: 10.1073/pnas.112194999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HP, Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989;57:233–42. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- Liu Z, Tan JL, Cohen DM, Yang MT, Sniadecki NJ, Ruiz SA, Nelson CM, Chen CS. Mechanical tugging force regulates the size of cell-cell junctions. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0914547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–52. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandeville JT, Lawson MA, Maxfield FR. Dynamic imaging of neutrophil migration in three dimensions: mechanical interactions between cells and matrix. J Leukoc Biol. 1997;61:188–200. doi: 10.1002/jlb.61.2.188. [DOI] [PubMed] [Google Scholar]
- Mathur AB, Collinsworth AM, Reichert WM, Kraus WE, Truskey GA. Endothelial, cardiac muscle and skeletal muscle exhibit different viscous and elastic properties as determined by atomic force microscopy. J Biomech. 2001;34:1545–53. doi: 10.1016/s0021-9290(01)00149-x. [DOI] [PubMed] [Google Scholar]
- Matthews BD, Overby DR, Mannix R, Ingber DE. Cellular adaptation to mechanical stress: role of integrins, Rho, cytoskeletal tension and mechanosensitive ion channels. J Cell Sci. 2006;119:508–18. doi: 10.1242/jcs.02760. [DOI] [PubMed] [Google Scholar]
- Mayer BJ, Hirai H, Sakai R. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr Biol. 1995;5:296–305. doi: 10.1016/s0960-9822(95)00060-1. [DOI] [PubMed] [Google Scholar]
- Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–3. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- Metz H, McElhaney J, Ommaya AK. A comparison of the elasticity of live, dead, and fixed brain tissue. J Biomech. 1970;3:453–8. doi: 10.1016/0021-9290(70)90017-5. [DOI] [PubMed] [Google Scholar]
- Meyer CJ, Alenghat FJ, Rim P, Fong JH, Fabry B, Ingber DE. Mechanical control of cyclic AMP signalling and gene transcription through integrins. Nat Cell Biol. 2000;2:666–8. doi: 10.1038/35023621. [DOI] [PubMed] [Google Scholar]
- Meyhofer E, Howard J. The force generated by a single kinesin molecule against an elastic load. Proc Natl Acad Sci U S A. 1995;92:574–8. doi: 10.1073/pnas.92.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael KE, Dumbauld DW, Burns KL, Hanks SK, Garcia AJ. Focal adhesion kinase modulates cell adhesion strengthening via integrin activation. Mol Biol Cell. 2009;20:2508–19. doi: 10.1091/mbc.E08-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mofrad MR, Golji J, Abdul Rahim NA, Kamm RD. Force-induced unfolding of the focal adhesion targeting domain and the influence of paxillin binding. Mech Chem Biosyst. 2004;1:253–65. [PubMed] [Google Scholar]
- Mogilner A, Oster G. Cell motility driven by actin polymerization. Biophys J. 1996;71:3030–45. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy JE, Burns JE, Sparrow JC, Tregear RT, Kendrick-Jones J, White DC. Single-molecule mechanics of heavy meromyosin and S1 interacting with rabbit or Drosophila actins using optical tweezers. Biophys J. 1995;68:298S–303S. 303S–305S. [PMC free article] [PubMed] [Google Scholar]
- Moore SW, Biais N, Sheetz MP. Traction on immobilized netrin-1 is sufficient to reorient axons. Science. 2009;325:166. doi: 10.1126/science.1173851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munevar S, Wang YL, Dembo M. Regulation of mechanical interactions between fibroblasts and the substratum by stretch-activated Ca2+ entry. J Cell Sci. 2004;117:85–92. doi: 10.1242/jcs.00795. [DOI] [PubMed] [Google Scholar]
- Na S, Collin O, Chowdhury F, Tay B, Ouyang M, Wang Y, Wang N. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc Natl Acad Sci U S A. 2008;105:6626–31. doi: 10.1073/pnas.0711704105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver T, Dembo M, Jacobson K. Traction forces in locomoting cells. Cell Motil Cytoskeleton. 1995;31:225–40. doi: 10.1002/cm.970310306. [DOI] [PubMed] [Google Scholar]
- Opas M, Dziak E. Effects of a tumour promoter, 12-0-tetradecanoyl-phorbol-13-acetate (TPA), on expression of differentiated phenotype in the chick retinal pigmented epithelial cells and on their interactions with the native basement membrane and with artificial substrata. Differentiation. 1990;43:20–8. doi: 10.1111/j.1432-0436.1990.tb00426.x. [DOI] [PubMed] [Google Scholar]
- Ophir J, Alam SK, Garra B, Kallel F, Konofagou E, Krouskop T, Varghese T. Elastography: ultrasonic estimation and imaging of the elastic properties of tissues. Proc Inst Mech Eng H. 1999;213:203–33. doi: 10.1243/0954411991534933. [DOI] [PubMed] [Google Scholar]
- Pasapera AM, I, Schneider C, Rericha E, Schlaepfer DD, Waterman CM. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J Cell Biol. 2010;188:877–90. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Pato MD, Sellers JR, Preston YA, Harvey EV, Adelstein RS. Baculovirus expression of chicken nonmuscle heavy meromyosin II-B. Characterization of alternatively spliced isoforms. J Biol Chem. 1996;271:2689–95. doi: 10.1074/jbc.271.5.2689. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–5. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskin CS, Odell GM, Oster GF. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys J. 1993;65:316–24. doi: 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol. 2005;204:198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–6. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- Prasad N, Topping RS, Decker SJ. SH2-containing inositol 5’-phosphatase SHIP2 associates with the p130(Cas) adapter protein and regulates cellular adhesion and spreading. Mol Cell Biol. 2001;21:1416–28. doi: 10.1128/MCB.21.4.1416-1428.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prass M, Jacobson K, Mogilner A, Radmacher M. Direct measurement of the lamellipodial protrusive force in a migrating cell. J Cell Biol. 2006;174:767–72. doi: 10.1083/jcb.200601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prezhdo OV, Pereverzev YV. Theoretical aspects of the biological catch bond. Acc Chem Res. 2009;42:693–703. doi: 10.1021/ar800202z. [DOI] [PubMed] [Google Scholar]
- Puklin-Faucher E, Gao M, Schulten K, Vogel V. How the headpiece hinge angle is opened: New insights into the dynamics of integrin activation. J Cell Biol. 2006;175:349–60. doi: 10.1083/jcb.200602071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puklin-Faucher E, Sheetz MP. The mechanical integrin cycle. J Cell Sci. 2009;122:179–86. doi: 10.1242/jcs.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puxkandl R, Zizak I, Paris O, Keckes J, Tesch W, Bernstorff S, Purslow P, Fratzl P. Viscoelastic properties of collagen: synchrotron radiation investigations and structural model. Philos Trans R Soc Lond B Biol Sci. 2002;357:191–7. doi: 10.1098/rstb.2001.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly DT, Burstein AH, Frankel VH. The elastic modulus for bone. J Biomech. 1974;7:271–5. doi: 10.1016/0021-9290(74)90018-9. [DOI] [PubMed] [Google Scholar]
- Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 2001;153:1175–86. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P, Alcaraz J, Sunyer R, Samitier J, Farre R, Navajas D. Micropatterning of single endothelial cell shape reveals a tight coupling between nuclear volume in G1 and proliferation. Biophys J. 2008;94:4984–95. doi: 10.1529/biophysj.107.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P, Gauthier NC, Del Rio A, Sheetz MP. Clustering of alpha(5)beta(1) integrins determines adhesion strength whereas alpha(v)beta(3) and talin enable mechanotransduction. Proc Natl Acad Sci U S A. 2009;106:16245–50. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock RS, Rice SE, Wells AL, Purcell TJ, Spudich JA, Sweeney HL. Myosin VI is a processive motor with a large step size. Proc Natl Acad Sci U S A. 2001;98:13655–9. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Buguin A, Silberzan P, Ladoux B. Is the mechanical activity of epithelial cells controlled by deformations or forces? Biophys J. 2005;89:L52–4. doi: 10.1529/biophysj.105.071217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, Nakamura K, Doi K, Takeda K, Tobiume K, Saitoh M, Morita K, Komuro I, De Vos K, Sheetz M, Ichijo H. Rap1 is involved in cell stretching modulation of p38 but not ERK or JNK MAP kinase. J Cell Sci. 2001;114:1221–7. doi: 10.1242/jcs.114.6.1221. [DOI] [PubMed] [Google Scholar]
- Sawada Y, Sheetz MP. Force transduction by Triton cytoskeletons. J Cell Biol. 2002;156:609–15. doi: 10.1083/jcb.200110068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–26. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffler MB, Burr DB. Stiffness of compact bone: effects of porosity and density. J Biomech. 1988;21:13–6. doi: 10.1016/0021-9290(88)90186-8. [DOI] [PubMed] [Google Scholar]
- Schlaepfer DD, Broome MA, Hunter T. Fibronectin-stimulated signaling from a focal adhesion kinase-c-Src complex: involvement of the Grb2, p130cas, and Nck adaptor proteins. Mol Cell Biol. 1997;17:1702–13. doi: 10.1128/mcb.17.3.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Pommerenke H, Durr F, Nebe B, Rychly J. Mechanical stressing of integrin receptors induces enhanced tyrosine phosphorylation of cytoskeletally anchored proteins. J Biol Chem. 1998;273:5081–5. doi: 10.1074/jbc.273.9.5081. [DOI] [PubMed] [Google Scholar]
- Schmidt CE, Horwitz AF, Lauffenburger DA, Sheetz MP. Integrin-cytoskeletal interactions in migrating fibroblasts are dynamic, asymmetric, and regulated. J Cell Biol. 1993;123:977–91. doi: 10.1083/jcb.123.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahapure R, Difato F, Laio A, Bisson G, Ercolini E, Amin L, Ferrari E, Torre V. Force generation in lamellipodia is a probabilistic process with fast growth and retraction events. Biophys J. 2010;98:979–88. doi: 10.1016/j.bpj.2009.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz MP, Wayne DB, Pearlman AL. Extension of filopodia by motor-dependent actin assembly. Cell Motil Cytoskeleton. 1992;22:160–9. doi: 10.1002/cm.970220303. [DOI] [PubMed] [Google Scholar]
- Shiu YT, Li S, Marganski WA, Usami S, Schwartz MA, Wang YL, Dembo M, Chien S. Rho mediates the shear-enhancement of endothelial cell migration and traction force generation. Biophys J. 2004;86:2558–65. doi: 10.1016/S0006-3495(04)74311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotomayor M, Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316:1144–8. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- Stoker M, O’Neill C, Berryman S, Waxman V. Anchorage and growth regulation in normal and virus-transformed cells. Int J Cancer. 1968;3:683–93. doi: 10.1002/ijc.2910030517. [DOI] [PubMed] [Google Scholar]
- Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nonlinear elasticity in biological gels. Nature. 2005;435:191–4. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Lin HY. Crosslinked chitosan: its physical properties and the effects of matrix stiffness on chondrocyte cell morphology and proliferation. J Biomed Mater Res A. 2005;75:742–53. doi: 10.1002/jbm.a.30489. [DOI] [PubMed] [Google Scholar]
- Sukharev S, Corey DP. Mechanosensitive channels: multiplicity of families and gating paradigms. Sci STKE. 2004;2004:re4. doi: 10.1126/stke.2192004re4. [DOI] [PubMed] [Google Scholar]
- Sun Z, Martinez-Lemus LA, Trache A, Trzeciakowski JP, Davis GE, Pohl U, Meininger GA. Mechanical properties of the interaction between fibronectin and alpha5beta1-integrin on vascular smooth muscle cells studied using atomic force microscopy. Am J Physiol Heart Circ Physiol. 2005;289:H2526–35. doi: 10.1152/ajpheart.00658.2004. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–84. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Tabernero L, Aricescu AR, Jones EY, Szedlacsek SE. Protein tyrosine phosphatases: structure-function relationships. FEBS J. 2008;275:867–82. doi: 10.1111/j.1742-4658.2008.06251.x. [DOI] [PubMed] [Google Scholar]
- Takada Y, Ye X, Simon S. The integrins. Genome Biol. 2007;8:215. doi: 10.1186/gb-2007-8-5-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada M, Sheetz MP, Sawada Y. Activation of a signaling cascade by cytoskeleton stretch. Dev Cell. 2004;7:709–18. doi: 10.1016/j.devcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci U S A. 2003;100:1484–9. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ. Actin microfilament dynamics in locomoting cells. Nature. 1991;352:126–31. doi: 10.1038/352126a0. [DOI] [PubMed] [Google Scholar]
- Thomas WE, Vogel V, Sokurenko E. Biophysics of catch bonds. Annu Rev Biophys. 2008;37:399–416. doi: 10.1146/annurev.biophys.37.032807.125804. [DOI] [PubMed] [Google Scholar]
- Thoumine O, Kocian P, Kottelat A, Meister JJ. Short-term binding of fibroblasts to fibronectin: optical tweezers experiments and probabilistic analysis. Eur Biophys J. 2000;29:398–408. doi: 10.1007/s002490000087. [DOI] [PubMed] [Google Scholar]
- Tomar A, Schlaepfer DD. Focal adhesion kinase: switching between GAPs and GEFs in the regulation of cell motility. Curr Opin Cell Biol. 2009;21:676–83. doi: 10.1016/j.ceb.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Kam L. Rigidity-dependent cross talk between integrin and cadherin signaling. Biophys J. 2009;96:L39–41. doi: 10.1016/j.bpj.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y, Kole TP, Lee JS, Fedorov E, Almo SC, Schafer BW, Wirtz D. How actin crosslinking and bundling proteins cooperate to generate an enhanced cell mechanical response. Biochem Biophys Res Commun. 2005;334:183–92. doi: 10.1016/j.bbrc.2005.05.205. [DOI] [PubMed] [Google Scholar]
- Tyska MJ, Dupuis DE, Guilford WH, Patlak JB, Waller GS, Trybus KM, Warshaw DM, Lowey S. Two heads of myosin are better than one for generating force and motion. Proc Natl Acad Sci U S A. 1999;96:4402–7. doi: 10.1073/pnas.96.8.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Higuchi H, Olivares AO, De La Cruz EM, Ishiwata S. Mechanochemical coupling of two substeps in a single myosin V motor. Nat Struct Mol Biol. 2004;11:877–83. doi: 10.1038/nsmb806. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Choi CK, Horwitz AR. Integrins in cell migration--the actin connection. J Cell Sci. 2009;122:199–206. doi: 10.1242/jcs.018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel V. Mechanotransduction involving multimodular proteins: converting force into biochemical signals. Annu Rev Biophys Biomol Struct. 2006;35:459–88. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- von Wichert G, Jiang G, Kostic A, De Vos K, Sap J, Sheetz MP. RPTP-alpha acts as a transducer of mechanical force on alphav/beta3-integrin-cytoskeleton linkages. J Cell Biol. 2003;161:143–53. doi: 10.1083/jcb.200211061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakatsuki T, Kolodney MS, Zahalak GI, Elson EL. Cell mechanics studied by a reconstituted model tissue. Biophys J. 2000;79:2353–68. doi: 10.1016/S0006-3495(00)76481-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Harvey EV, Conti MA, Wei D, Sellers JR. A conserved negatively charged amino acid modulates function in human nonmuscle myosin IIA. Biochemistry. 2000a;39:5555–60. doi: 10.1021/bi000133x. [DOI] [PubMed] [Google Scholar]
- Wang HB, Dembo M, Hanks SK, Wang Y. Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc Natl Acad Sci U S A. 2001a;98:11295–300. doi: 10.1073/pnas.201201198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HB, Dembo M, Wang YL. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am J Physiol Cell Physiol. 2000b;279:C1345–50. doi: 10.1152/ajpcell.2000.279.5.C1345. [DOI] [PubMed] [Google Scholar]
- Wang SC, Makino K, Xia W, Kim JS, Im SA, Peng H, Mok SC, Singletary SE, Hung MC. DOC-2/hDab-2 inhibits ILK activity and induces anoikis in breast cancer cells through an Akt-independent pathway. Oncogene. 2001b;20:6960–4. doi: 10.1038/sj.onc.1204873. [DOI] [PubMed] [Google Scholar]
- Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–5. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- Warshaw DM, Fay FS. Tension transients in single isolated smooth muscle cells. Science. 1983;219:1438–41. doi: 10.1126/science.6828870. [DOI] [PubMed] [Google Scholar]
- Yamada S, Wirtz D, Kuo SC. Mechanics of living cells measured by laser tracking microrheology. Biophys J. 2000;78:1736–47. doi: 10.1016/S0006-3495(00)76725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R, Ballestrem C, Kam Z, Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J Cell Sci. 2003;116:4605–13. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- Zhang X, Jiang G, Cai Y, Monkley SJ, Critchley DR, Sheetz MP. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat Cell Biol. 2008;10:1062–8. doi: 10.1038/ncb1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XM, Resnick RJ, Shalloway D. A phosphotyrosine displacement mechanism for activation of Src by PTPalpha. EMBO J. 2000;19:964–78. doi: 10.1093/emboj/19.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C, Chrzanowska-Wodnicka M, Brown J, Shaub A, Belkin AM, Burridge K. Rho-mediated contractility exposes a cryptic site in fibronectin and induces fibronectin matrix assembly. J Cell Biol. 1998;141:539–51. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou AX, Hartwig JH, Akyurek LM. Filamins in cell signaling, transcription and organ development. Trends Cell Biol. 2010;20:113–23. doi: 10.1016/j.tcb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Zhu J, Luo BH, Xiao T, Zhang C, Nishida N, Springer TA. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol Cell. 2008;32:849–61. doi: 10.1016/j.molcel.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]


