Abstract
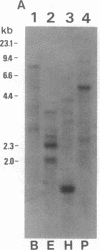
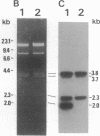
We have characterized a Drosophila glutathione S-transferase (RX:glutathione R-transferase, EC 2.5.1.18) cDNA encoding a protein of 209 amino acids. The cDNA was expressed in Escherichia coli harboring the expression plasmid construct pGTDml-KK. The active enzyme, designated as Drosophila glutathione S-transferase 1-1, had a specific activity toward 1-chloro-2,4-dinitrobenzene comparable to that for the mammalian glutathione S-transferases but did not have as broad a substrate specificity pattern. There is a region of 44 amino acids in this enzyme that shares 66% identity with an analogous region of maize glutathione S-transferase III. Drosophila glutathione S-transferase 1-1 had no obvious homology to any mammalian or parasitic glutathione S-transferases. The gene was found to be a member of a multigene family.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balloul J. M., Sondermeyer P., Dreyer D., Capron M., Grzych J. M., Pierce R. J., Carvallo D., Lecocq J. P., Capron A. Molecular cloning of a protective antigen of schistosomes. Nature. 1987 Mar 12;326(6109):149–153. doi: 10.1038/326149a0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin N., von Gabain A., Bujard H. Isolation of large molecular weight DNA from agarose gels for further digestion by restriction enzymes. FEBS Lett. 1975 Apr 15;53(1):84–86. doi: 10.1016/0014-5793(75)80688-0. [DOI] [PubMed] [Google Scholar]
- Chow N. W., Whang-Peng J., Kao-Shan C. S., Tam M. F., Lai H. C., Tu C. P. Human glutathione S-transferases. The Ha multigene family encodes products of different but overlapping substrate specificities. J Biol Chem. 1988 Sep 15;263(26):12797–12800. [PubMed] [Google Scholar]
- DeJong J. L., Chang C. M., Whang-Peng J., Knutsen T., Tu C. P. The human liver glutathione S-transferase gene superfamily: expression and chromosome mapping of an Hb subunit cDNA. Nucleic Acids Res. 1988 Sep 12;16(17):8541–8554. doi: 10.1093/nar/16.17.8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJong J. L., Morgenstern R., Jörnvall H., DePierre J. W., Tu C. P. Gene expression of rat and human microsomal glutathione S-transferases. J Biol Chem. 1988 Jun 15;263(17):8430–8436. [PubMed] [Google Scholar]
- Echalier G., Ohanessian A. In vitro culture of Drosophila melanogaster embryonic cells. In Vitro. 1970 Nov-Dec;6(3):162–172. doi: 10.1007/BF02617759. [DOI] [PubMed] [Google Scholar]
- Grove G., Zarlengo R. P., Timmerman K. P., Li N. Q., Tam M. F., Tu C. P. Characterization and heterospecific expression of cDNA clones of genes in the maize GSH S-transferase multigene family. Nucleic Acids Res. 1988 Jan 25;16(2):425–438. doi: 10.1093/nar/16.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai H. C., Grove G., Tu C. P. Cloning and sequence analysis of a cDNA for a rat liver glutathione S-transferase Yb subunit. Nucleic Acids Res. 1986 Aug 11;14(15):6101–6114. doi: 10.1093/nar/14.15.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai H. C., Qian B., Grove G., Tu C. P. Gene expression of rat glutathione S-transferases. Evidence for gene conversion in the evolution of the Yb multigene family. J Biol Chem. 1988 Aug 15;263(23):11389–11395. [PubMed] [Google Scholar]
- Lai H. C., Tu C. P. Rat glutathione S-transferases supergene family. Characterization of an anionic Yb subunit cDNA clone. J Biol Chem. 1986 Oct 15;261(29):13793–13799. [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mozer T. J., Tiemeier D. C., Jaworski E. G. Purification and characterization of corn glutathione S-transferase. Biochemistry. 1983 Mar 1;22(5):1068–1072. doi: 10.1021/bi00274a011. [DOI] [PubMed] [Google Scholar]
- Pickett C. B., Lu A. Y. Glutathione S-transferases: gene structure, regulation, and biological function. Annu Rev Biochem. 1989;58:743–764. doi: 10.1146/annurev.bi.58.070189.003523. [DOI] [PubMed] [Google Scholar]
- Reddy C. C., Burgess J. R., Gong Z. Z., Massaro E. J., Tu C. P. Purification and characterization of the individual glutathione S-transferases from sheep liver. Arch Biochem Biophys. 1983 Jul 1;224(1):87–101. doi: 10.1016/0003-9861(83)90192-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimabukuro R. H., Frear D. S., Swanson H. R., Walsh W. C. Glutathione conjugation. An enzymatic basis for atrazine resistance in corn. Plant Physiol. 1971 Jan;47(1):10–14. doi: 10.1104/pp.47.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimabukuro R. H., Swanson H. R., Walsh W. C. Glutathione conjugation: atrazine detoxication mechanism in corn. Plant Physiol. 1970 Jul;46(1):103–107. doi: 10.1104/pp.46.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Davern K. M., Board P. G., Tiu W. U., Garcia E. G., Mitchell G. F. Mr 26,000 antigen of Schistosoma japonicum recognized by resistant WEHI 129/J mice is a parasite glutathione S-transferase. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8703–8707. doi: 10.1073/pnas.83.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Taylor J. B., Vidal A., Torpier G., Meyer D. J., Roitsch C., Balloul J. M., Southan C., Sondermeyer P., Pemble S., Lecocq J. P. The glutathione transferase activity and tissue distribution of a cloned Mr28K protective antigen of Schistosoma mansoni. EMBO J. 1988 Feb;7(2):465–472. doi: 10.1002/j.1460-2075.1988.tb02834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomarev S. I., Zinovieva R. D. Squid major lens polypeptides are homologous to glutathione S-transferases subunits. Nature. 1988 Nov 3;336(6194):86–88. doi: 10.1038/336086a0. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C. P., Matsushima A., Li N. Q., Rhoads D. M., Srikumar K., Reddy A. P., Reddy C. C. Immunological and sequence interrelationships between multiple human liver and rat glutathione S-transferases. J Biol Chem. 1986 Jul 15;261(20):9540–9545. [PubMed] [Google Scholar]
- Tu C. P., Reddy C. C. On the multiplicity of rat liver glutathione S-transferases. J Biol Chem. 1985 Aug 25;260(18):9961–9964. [PubMed] [Google Scholar]
- Tu C. P., Weiss M. J., Li N. Q., Reddy C. C. Tissue-specific expression of the rat glutathione S-transferases. J Biol Chem. 1983 Apr 25;258(8):4659–4662. [PubMed] [Google Scholar]
- Wistow G., Piatigorsky J. Recruitment of enzymes as lens structural proteins. Science. 1987 Jun 19;236(4808):1554–1556. doi: 10.1126/science.3589669. [DOI] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]