Abstract
OBJECTIVE
Patients with type 1 diabetes (T1D) with impaired renal function are at increased risk for end-stage renal disease (ESRD). Although the rate of progression varies, determinants and mechanisms of this variation are unknown.
RESEARCH DESIGN AND METHODS
We examined serum metabolomic profiles associated with variation in renal function decline in participants with T1D (the Joslin Kidney Study prospective cohort). One hundred fifty-eight patients with proteinuria and chronic kidney disease stage 3 were followed for a median of 11 years to determine estimated glomerular filtration rate slopes from serial measurements of serum creatinine and to ascertain time to onset of ESRD. Baseline serum samples were subjected to global metabolomic profiling.
RESULTS
One hundred ten amino acids and purine and pyrimidine metabolites were detected in at least 80% of participants. Serum levels of seven modified metabolites (C-glycosyltryptophan, pseudouridine, O-sulfotyrosine, N-acetylthreonine, N-acetylserine, N6-carbamoylthreonyladenosine, and N6-acetyllysine) were associated with renal function decline and time to ESRD (P < 0.001) independent of the relevant clinical covariates. The significant metabolites correlated with one another and with the indices of tubular injury.
CONCLUSIONS
This prospective cohort study in participants with T1D, proteinuria, and impaired renal function at baseline demonstrated that patients with increased circulating levels of certain modified metabolites experience faster renal function decline, leading to ESRD. Whether some of these candidate metabolites are risk factors or just prognostic biomarkers of progression to ESRD in T1D needs to be determined.
Introduction
Progressive renal decline is the clinical manifestation of the disease process underlying the development of diabetic nephropathy (1,2). Although patients with chronic kidney disease (CKD) are at high risk for end-stage renal disease (ESRD), this risk varies tremendously. Some patients with CKD progress rapidly, whereas others may progress over a decade or more. In the absence of diabetic animal models that develop renal failure, observational studies in humans are valuable to search for new pathways to the development of drugs that would slow renal function decline and postpone ESRD.
The mechanisms responsible for this variation in patients with diabetes are studied with the use of newly developed high throughput omics technologies (3–5). Among these, metabolomic platforms have been used to search for new metabolic pathways (3,6–9). Our global profiling study in type 2 diabetes (T2D) demonstrated that certain metabolites can be predictors of ESRD that occurs 10 years later independent of legacy markers such as albuminuria and baseline renal function (3). To date, no study to our knowledge has examined metabolomic profiles of patients with type 1 diabetes (T1D) who progress to ESRD, the ultimate outcome of diabetic nephropathy.
Research Design and Methods
The Committee on Human Subjects of the Joslin Diabetes Center approved the informed consent and recruitment and examination procedures for this study.
Study Group
Approximately 3,500 adult patients with T1D diagnosed before age 40 receive care at the Joslin clinic. Between 1991 and 2004, we monitored this population for the occurrence of persistent proteinuria by using routinely measured albumin-to-creatinine ratios (ACRs). Patients with persistent proteinuria were invited to participate in a follow-up study on the genetics of diabetic nephropathy (2,10). Eligibility criteria included residence in New England and age at enrollment between 21 and 54 years. In that interval, persistent proteinuria was diagnosed in 790 eligible patients, and 564 (71%) participated in the Joslin Proteinuria Cohort Study. At study enrollment, a standardized baseline examination was performed by trained recruiters who administered a structured interview and brief physical examination. Blood and urine specimens collected at the baseline and follow-up examinations were stored at −85°C until analysis (2,10).
Enrolled patients were followed until 2012. Blood and urine specimens were collected at least every 2 years. Collection of research specimens occurred during routine clinic visits. Patients with less frequent clinic visits were examined at their homes.
Among the patients enrolled in the Joslin Proteinuria Cohort Study, 199 had CKD of whom 158 were included in the current study. Of note, selection of the study participants was based purely on their baseline exposures and was independent from their follow-up characteristics.
Assessment of Abnormalities in Urinary Albumin Excretion
In the Joslin clinical laboratory, albumin concentrations in spot urines are routinely measured at least once a year. Between 1991 and 2009, urinary albumin concentrations were measured by immunonephelometry on a BN ProSpec System (Dade Behring, Newark, DE) with N-albumin kits. Creatinine measurements in urine were assayed by the Jaffe modified picrate method on a Ciba Corning 550 Express Plus Chemistry Analyzer. For each patient, the geometric mean ACR for the preceding 2-year interval of the baseline examination was determined to assign an albuminuria status: normoalbuminuria (ACR <30 μg/g creatinine), microalbuminuria (ACR 30–300 μg/g creatinine), and proteinuria (>300 μg/g creatinine) (11).
Assessment of Renal Function
From 2011 to 2014, serum specimens obtained for research purposes at baseline (n = 520) and during follow-up (n = 1,560) in participants in the Joslin Proteinuria Cohort Study were used to measure creatinine concentrations. The measurements were performed in the Advanced Research and Diagnostic Laboratory at the University of Minnesota by using the Roche enzymatic assay (product no. 11775685) on a Roche/Hitache Mod P analyzer. This method is calibrated to be traceable to an isotope dilution mass spectrometry reference assay and was verified by measuring National Institute of Standards and Technology SRM 967.
In addition, we retrieved 2,900 measurements of baseline serum creatinine concentrations (n = 520) from Joslin clinical laboratory records. These measurements were assayed by the Jaffe modified picrate method as described above. For 633 samples, we used duplicate measurements to calibrate the Joslin clinical measurements to the assay method used at the University of Minnesota laboratory. The CKD Epidemiology Collaboration equation was used to determine the estimated glomerular filtration rate (eGFR) (12). For each participant, we used all available serial eGFR determinations performed during follow-up to determine eGFR slopes by general linear model procedure.
Ascertainment of Onset of ESRD
All participants from the Joslin Proteinuria Cohort Study included in the current study were queried against rosters of the U.S. Renal Data System (USRDS) and the National Death Index (NDI), covering all events up to the end of 2012. USRDS maintains a roster of U.S. patients who received renal replacement therapy, which includes dates of dialysis and transplantation (13). The NDI is a comprehensive roster of dates and causes of deaths in the U.S. (14). ESRD was defined by a match with the USRDS roster or a listing of renal failure among the causes of death on an NDI death certificate.
Global Metabolomic Profiling
Baseline serum specimens were subjected to global profiling (Metabolon, Durham, NC). The Metabolon platform uses gas chromatography and liquid chromatography mass spectrometry in positive and negative modes (15,16). The liquid chromatography mass spectrometry portion of the platform incorporates a Waters ACQUITY UPLC system and a Finnigan LTQ mass spectrometer (Thermo Fisher Scientific), including an electrospray ionization source and linear ion trap mass analyzer. The gas chromatography column is 5% phenyl dimethyl silicone, and the temperature ramp is from 40°C to 300°C over 16 min. All samples were then analyzed on a Finnigan TRACE DSQ fast-scanning single-quadrupole mass spectrometer (Thermo Fisher Scientific) using electron impact ionization. After peak identification and quality control filtering, the metabolite-relative concentrations were obtained from median-scaled day-block normalized data for each compound. Samples in this study were run in batches balanced by case status.
Repeated Metabolomic Measurements Over Time
In the study group, we identified 20 participants with a stable diabetic nephropathy phenotype over time (eGFR slope <2.5 mL/min/1.73 m2 per year, ACR ±200 mg/g creatinine per year) for whom we performed global metabolomics measurements in serum samples taken at baseline and 3–8 years later. Global profiling was performed according to the protocol described above. Samples from the same individuals were run on the same batch.
Comparison of Findings in Other Published Reports With Our Global Metabolomics Data
We analyzed amino acids and purine and pyrimidine metabolites reported to be associated with diabetic nephropathy or eGFR decline that were also well detected in our main study by global metabolomic profiling (3,6,8,9,17). We evaluated their associations with eGFR slope and with progression to ESRD in the main study.
Measurements of Glycemic Control and Urinary Glomerular and Tubular Protein Markers
HbA1c was measured in the Joslin clinical laboratory according to the method calibrated to Diabetes Complications and Control standards. In addition to ACR, three other proteins were measured in baseline urine specimens. IgG2 was measured in the Joslin research laboratory as previously described (18). Neutrophil gelatinase-associated lipocalin (NGAL) was measured in the Joslin research laboratory by Quantikine ELISA (DLCN20; R&D Systems, Minneapolis, MN) according to the manufacturer’s protocol. KIM-1 was measured in the laboratory of J.V.B. (Brigham and Women’s Hospital) (19,20). Measurements of protein-based markers (IgG2, KIM-1, and NGAL) were available for 91 study participants. The interassay coefficient of variation was <20% for all analytes. All the urine specimens for marker measurements had the same number of freeze-thaw cycles.
Auxiliary Study of Renal Clearance
Among 365 Joslin Proteinuria Cohort Study participants who had normal renal function at baseline (eGFR >60 mL/min/1.73 m2), 21 who had progressed to ESRD were selected and matched by age, sex, and baseline renal function with 32 participants who had an eGFR <−3.3 mL/min/1.73 m2 (nonprogressors) (2,11). Measurements of the metabolites were performed according to a similar protocol, the global profiling used in the main study. Mass spectra were compared with the library of ∼1,000 purified standards for their analytical identification, whereas measurements performed in the main study were compared with the library of 2,400 standards.
Data Analysis
Differences in clinical characteristics were tested by ANOVA for continuous variables and χ2 test for categorical variables. Metabolites detected by global profiling in at least 80% of the study participants were considered in further analysis. Correlations of the metabolites with baseline eGFR, other baseline measures, and subsequent eGFR slopes were expressed as Spearman rank correlation coefficients. Adjustment for multiple comparisons was performed with a positive false discovery rate (q-value <0.05 for significance). An independent effect of a metabolite for the progression to ESRD was estimated by using a Cox proportional hazards regression model. After transformation of the metabolite concentration to normally distributed ranks, its effect on risk of progression was expressed as the hazard ratio (HR) per 1 SD difference (3). Discrimination abilities of the risk prediction models were evaluated with C statistics (21). Principal component (PC) analysis was used as a data reduction approach to create a metabolite index. Reproducibility of metabolomic measurements over time was determined by intraclass correlation coefficients (ICCs), which represent ratios of between-person variance to the sum of the within- and between-person variances (modified ICC_SAS macro). Data analysis was performed with SAS 9.4 and JMP Pro 12.0.0 software (SAS Institute, Cary, NC).
Results
Characteristics of the Study Group
Five hundred sixty-four patients attending the Joslin clinic were recruited into the Joslin Proteinuria Cohort Study. Of these patients, 158 with eGFR 30–60 mL/min/1.73 m2 at enrollment were selected for this study. Median follow-up for this study was 11.5 years. Follow-up for participants in whom ESRD did not develop was 4.4–21 years and was understandably shorter (2.4–17.3 years) for participants in whom ESRD did develop. During this time, ESRD developed in 99 participants (63%). Median eGFR slope was −5.0 (−8.4 to −2.5) mL/min/1.73 m2 per year. Participants with more-rapid eGFR decline were younger and had diabetes for a shorter period, worse glycemic control, and higher albuminuria. They differed slightly with regard to their baseline eGFR. Clinical characteristics of the study group according to tertiles of eGFR slope distribution are summarized in Table 1.
Table 1.
Clinical characteristics of the study group stratified by the rate of the renal function decline
| Renal Function Decline |
||||
|---|---|---|---|---|
| Characteristic | Minimal(n = 53) | Moderate(n = 53) | Fast(n = 52) | P value |
| At baseline | ||||
| Male [n (%)] | 25 (47) | 28 (53) | 29 (46) | 0.38 |
| Age (years) | 45 ± 10 | 42 ± 8 | 40 ± 10 | 0.003 |
| Duration of diabetes (years) | 30 ± 8 | 29 ± 8 | 27 ± 9 | 0.007 |
| HbA1c (%) | 8.5 ± 1.3 | 9.3 ± 1.6 | 9.6 ± 1.7 | 0.005 |
| HbA1c (mmol/mol) | 69 ± 14 | 78 ± 18 | 81 ± 19 | 0.005 |
| Serum cholesterol (mg/dL) | 191 ± 43 | 219 ± 63 | 219 ± 71 | 0.03 |
| Systolic blood pressure (mmHg) | 134 ± 20 | 136 ± 19 | 137 ± 24 | 0.57 |
| Diastolic blood pressure (mmHg) | 75 ± 11 | 79 ± 11 | 80 ± 11 | 0.07 |
| Antihypertensive/renoprotective treatment (%) | 93 | 92 | 80 | 0.07 |
| ACR (μg/g creatinine) | 670 (403, 1,009) | 1,007 (680, 1,934) | 1,533 (748, 3,526) | 0.0004 |
| eGFR (mL/min/1.73 m2) | 50 ± 12 | 48 ± 13 | 44 ± 13 | 0.02 |
| Follow-up | ||||
| Length (years) | 14.4 (9.7, 18.4) | 11.8 (7.7, 13.8) | 9.2 (5.6, 12.1) | <0.001 |
| Annual eGFR slope (mL/min/1.73 m2) | −1.9 (−1.0 to −2.5) | −5.0 (−3.9 to −5.9) | −10.9 (−8.5 to −15.6) | By design |
| Incidence of ESRD | ||||
| Cases (n) | 16 | 35 | 48 | <0.001 |
| Rate per 1,000 person-years* | 1.1 (0.6, 1.9) | 3.7 (2.6, 5.1) | 10.5 (7.7, 13.9) | <0.001 |
Data are mean ± SD or median (25th, 75th percentile) unless otherwise indicated.
*Incidence rate of ESRD is accompanied by 95% CI.
Metabolomic Profiles Associated With Variation in eGFR Slope
Multivariate Analysis
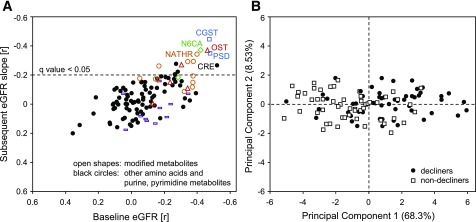
Global metabolomic profiling resulted in the detection of mass spectra of 123 metabolites. Of those, 110 were detected in at least 80% of the participants (87 amino acids and 23 purine and pyrimidine metabolites), and we designated them as commonly detected. Eleven metabolites were significantly associated with eGFR slope in the analysis adjusted for multiple testing, and nine of them (81%) were modified metabolites. Overall, the metabolomic platform’s coverage included 39 modified metabolites featuring five types of modifications: acylation, C-glycosylation, carbamoylation, sulfation, and methylation. Metabolites of all types of modifications, except for methylation, contained numerous biochemicals associated nominally with nephropathy progression. Nine modified metabolites remained significant in the analysis adjusted for multiple testing. C-glycosyltryptophan, N6-carbamoylthreonyladenosine, and O-sulfotyrosine were the top-three metabolites, of which higher levels were associated with a more-rapid eGFR slope. All nine significant modified metabolites correlated with baseline eGFR at a nominal significance level (P < 0.05) (Fig. 1A).
Figure 1.
Associations of metabolites with subsequent renal function decline. A: Global matrix of correlation coefficients of all commonly detected amino acids and nucleotides (n = 114) with baseline eGFR (x-axis) and with a subsequent eGFR slope (y-axis). Each mark represents a correlation coefficient of an individual metabolite. Modified metabolites are represented as open shapes (blue □, C-glycosylated; red △, sulfated; orange ○, acylated; green ◇, carbamylated; purple bars, methylated). All the remaining metabolites are presented as ●. B: Score plot of PC1 and PC2 based on nine significant candidate metabolites identified in the global matrix analysis. Marks represent individual study participants with renal function decline in the bottom tertile of the eGFR slope distribution (decliners) and those in the top tertile of the eGFR slope distribution (nondecliners). The labels of the x- and y-axes include the explained variance. CGST, C-glycosyltryptophan; CRE, creatinine; N6CA, N6-carbamoylthreonyladenosine; NATHR, N-acetylthreonine; OST, O-sulfotyrosine; PSD, pseudouridine.
A PC analysis of all well-detected metabolites (n = 114) explained only a small proportion of the overall variance. PC1all explained 15.5% of the variance, and PC2all explained only 7.1%. Score plots of all PCs with eigenvalues >1 were examined and did not separate well decliners from nondecliners (data not shown). Of note, top loadings contributing to the PC1all corresponded with the nine significant modified metabolites identified in the global matrix analysis. In the further PC analysis that considered only these nine metabolites, PC19 was the only component with an eigenvalue >1 and explained 68.3% of the variance. Contributions to PC19 were comparable among these metabolites, except for N6-acetyllysine and phenol sulfate, for which the loadings were lower. The related PC score plot decently separated decliners from nondecliners (Fig. 1B). PC19 correlated significantly with eGFR slope (r = −0.28).
The multivariate analysis did not reveal a predictive value of essential amino acids or branched chain amino acid–related metabolites for renal function decline. Creatinine was the top metabolite associated with baseline eGFR, but only moderately correlated with the subsequent eGFR slope. Analysis of ratios of modified metabolites (products) to their respective substrates did not improve risk predictions over the effects of the sole modified metabolites (data not shown). Associations of all detected metabolites with eGFR slope and their interrelationships expressed as loadings are listed in Supplementary Table 1.
Modified Metabolites as Candidate Biomarker Predictors of Progression to ESRD
Multivariable Analysis
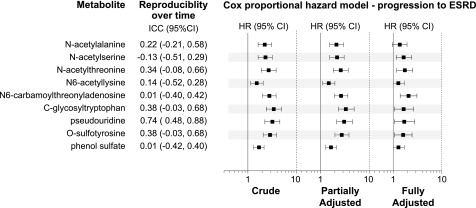
We used a Cox proportional hazards regression model to examine the independence of associations of the modified metabolites with risk of progression to ESRD. All nine modified metabolites associated with eGFR slope were also associated significantly with progression to ESRD in the Cox proportional hazards regression model after controlling for the covariates HbA1c, ACR, and eGFR. Seven of the nine metabolites (except for N-acetylalanine and phenol sulfate) remained significant in the fully adjusted model, controlled for an expanded panel of clinical covariates (Fig. 2). Discrimination ability of the clinical model measured by C-index was 0.706. The addition of each significant metabolite (one at a time) increased the C-index from 0.708 to 0.722. Seven significant metabolites correlated among each, with nonparametric correlation coefficients ranging from r = 0.45 to r = 0.89 (P < 0.0001 for each) (Fig. 3). The C-index of the Cox model containing a metabolite index derived from PC19 and controlled for clinical covariates increased to 0.728, but an improvement in the discrimination ability did not reach statistical significance.
Figure 2.
Nine modified metabolites significant in the global multivariate analysis as candidate biomarkers for progression to ESRD. Reproducibility over time is expressed as ICCs. HRs are presented per 1 SD of the metabolite. Partially adjusted HRs are controlled for baseline HbA1c, ACR, and eGFR. Fully adjusted HRs are controlled for blood pressure, BMI, smoking status, HbA1c, ACR, eGFR, uric acid levels, treatment with renin-angiotensin system inhibitors, other antihypertensive treatment, and statins.
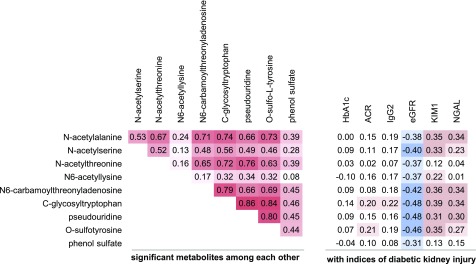
Figure 3.
Matrix of correlation coefficients among and between modified metabolites and baseline indices of diabetic kidney injury: glycemic control (HbA1c), glomerular markers (ACR, IgG2), and tubular markers (KIM-1, NGAL). Cell color indicates strengths and directions of the correlations from red (positive correlation) to white (no correlation) to blue (negative correlation).
Within-Individual Reproducibility Over Time
Repeated measurements of well-detectable metabolites over a median of 5 years demonstrated that 36% of amino acids (31 of 87) and 13% of purine and pyrimidine metabolites (3 of 23) had at least fair reproducibility over time (ICC >0.3) (global data not shown). Among the nine modified metabolites significant in the multivariate analysis, pseudouridine tracked very well over time (ICC 0.74 [95% CI 0.48, 0.88]). Reproducibility of C-glycosyltryptophan, O-sulfotyrosine, and N-acetylthreonine was fairly good (ICC 0.34–0.38) and was poor for the remaining five metabolites (Fig. 2).
Metabolite Levels in the Study Participants Compared With Subjects Without Nephropathy
We compared levels of the nine significant modified metabolites in the multivariate analysis between the study participants with T1D, proteinuria, and CKD (main study) and 20 subjects with T1D, normoalbuminuria, and median diabetes duration of 24 years. Ratios of metabolites in the main study participants compared with that in subjects with normoalbuminuria ranged from 1.21 to 1.94 (P < 0.05 for each comparison) (data not shown).
Candidate Metabolite Biomarkers Across Independent Cohort Studies
We identified 13 metabolites measured in the current study that were formerly reported to be associated with nephropathy phenotype in other independent metabolomics studies (3,6,8,9,17). C-glycosyltryptophan and pseudouridine, O-sulfotyrosine, and tryptophan metabolites were reproducibly associated with renal outcome in this and at least one other independent study (Supplementary Table 2).
Modified Metabolites as Potential Contributors to Diabetic Nephropathy Progression
Modified Metabolites Versus Glycemic Control and Kidney Injury Markers At Baseline
We created a matrix of the correlation coefficients between the nine significant modified metabolites in the multivariate analysis and baseline measurements of HbA1c (glycemic control), markers of glomerular damage (urinary excretion of albumin and IgG2), and markers of tubular damage (urinary excretion of KIM-1 and NGAL). All nine modified metabolites were also correlated, to at least a moderate degree, with baseline eGFR (r = −0.48 to −0.3). The pattern of associations with markers of tubular injury (KIM-1 or NGAL) mirrored associations observed with a baseline eGFR, except for N-acetylthreonine, N6-acetyllysine, and phenol sulfate, whose associations were notably weaker. Associations of the metabolites with ACR and IgG2 were absent or weak (Fig. 3).
Renal Clearance of the Modified Metabolites and Risk of Progression to ESRD
To elucidate whether an increase in metabolites in participants with proteinuria and CKD is not due to impaired clearance, we conducted an auxiliary study of subjects with comparable proteinuria but normal renal function for whom we evaluated metabolite levels in plasma and urine according to the nephropathy risk. Clinical characteristics of the auxiliary study subjects are presented in Supplementary Table 3. Median follow-up for the auxiliary study subjects was 12.3 years. Follow-up for nonprogressors was 6.9–21 years and 2.4–19 years for ESRD progressors. The library of the metabolomic platform in the auxiliary study had less comprehensive coverage than the main study because only four significant modified metabolites were measured. Risk patterns remained similar to that observed in the main study. The auxiliary study was small; therefore, confidence intervals around HRs were understandably broader. Of note, in the excretion patterns analysis, metabolites measured in urine or measured as fractional excretions were not significantly associated with progression to ESRD (Supplementary Fig. 1).
We also compared free acetyllysine with circulating proteomic content of acetyllysine in a subset of subjects. We did not observe a significant correlation between the two. These results suggest that circulating acetyllysine residues do not likely reflect proteolysis of circulating proteins (Supplementary Data).
Conclusions
This study was conducted in a cohort of patients with T1D, proteinuria, and CKD stage 3 at baseline who were followed for approximately a decade to assess eGFR trajectories and identify time to onset of ESRD. To our knowledge, this and our former study in T2D (3) are the only to date that evaluated the associations of circulating metabolomic profiles with progression to the ultimate outcome of diabetic nephropathy: ESRD.
In the current study, global metabolomic profiling revealed nine candidate metabolite biomarkers as potential risk factors of progression to ESRD in T1D. These metaboites were C-glycosyltryptophan, pseudouridine, O-sulfotyrosine, N-acetylthreonine, N-acetylserine, N6-carbamoylthreonyladenosine, N6-acetyllysine, N-acetylalanine, and phenol sulfate. The effect of these metabolites on risk of ESRD remained independent after adjustment for glycemic control, renal function, and albuminuria. Seven of these nine (except for N-acetylalanine and phenol sulfate) remained significant in multivariable models adjusted for a comprehensive selection of clinical covariates. C-indices of models containing added biomarkers were higher than the C-index of the clinical model alone, but improvements in the discrimination abilities of models were not significant, likely reflecting that the study may not have had sufficient power because of the number of applied adjustments.
Do these findings have the potential for generalization? This report and two others (Joslin Kidney Study in T2D [3] and Cooperative Health Research in the Region of Augsburg [KORA] F4 [9]) have reported that C-glycosyltryptophan, pseudouridine, and O-sulfotyrosine are among the top risk metabolites that predict GFR-based renal outcome. KORA F4 evaluated incident CKD in the general population, and all three studies used the same metabolomic platform. The Framingham cohort study used a different platform (6) wherein the authors identified tryptophan derivatives as determinants of incident CKD. Tryptophan-derived metabolites also reached nominal significance in the current study.
Do the candidate metabolites actually contribute to disease progression? This study demonstrates that significant modified metabolites markedly correlate with one another and further with baseline indices of kidney tubular injury. We hypothesize that a common, currently unknown mechanism leads to an increase in these modified metabolites in circulation and that these metabolites may be toxic to kidney tubules, contributing to subsequent eGFR loss. Functional studies are needed to validate such a hypothesis.
The presence of impaired renal clearance at baseline in the current study raises concerns about whether retention can account for increased serum levels of metabolites (9,22–24). Nevertheless, effects of metabolites were independent from baseline renal function in the multivariable models examined. Second, we provide limited evidence that renal clearances of the selected modified metabolites were unaltered with regard to ESRD risk.
One may speculate that an increase in significant metabolites may also be due to the increased posttranslational modifications in tissues of patients at risk. Increased glycosylation, sulfation, acetylation, and carbamoylation of amino acids and proteins have been reported in diabetic and uremic tissues (25–28). The importance of the circulating metabolites rather than their urinary levels suggests sources other than the kidney, such as liver and muscle. No studies to our knowledge have systematically examined the content of modified metabolites across tissues in the presence of diabetes. However, basic research studies have demonstrated that the modification of metabolites identified in the current study have been determined in many tissues of subjects without diabetes, including kidney, liver, and muscle (27–35). A number of studies also have highlighted the importance of modifications of metabolites in the presence of chronic diabetic complications (25–28,34,36–38). Different modifications are involved in a variety of biological processes, such as inflammation (36,37), glucose metabolism (34,35), and epigenetic regulation (38), among others.
Finally, we must consider the strengths and limitations of the current study. The study was conducted in Caucasians, so it may not be generalizable to populations of other ethnic ancestry. Our study is novel because the modified metabolites were studied as risk factors for subsequent progression of diabetic nephropathy; on the contrary, in past cross-sectional comparisons, posttranslational modifications in diabetic complications were evaluated as features associated with an already-present disease. The current study includes a well-characterized population with a long follow-up and ascertainment of ESRD together with eGFR slopes. Associations of the metabolites also were concordant between two renal outcomes and in other studies in T2D and the general population (3,6,9). Future replication in independent prospective cohorts of T1D is needed to validate these predictive associations in patients with this disease phenotype. An important advantage of the nontargeted metabolomic approach utilized in our study is that it examines metabolites comprehensively and assigns a ranking to identified metabolic alterations. For example, not all modifications were associated with disease progression (acetylation was, whereas methylation was not).
In summary, the metabolomic approach is an attractive line of investigation in diabetes because of the metabolic nature of the disease. It is relevant in kidney disease because the kidney is involved in handling numerous metabolites, and the metabolites themselves may contribute further to kidney injury. We report associations of metabolomic determinants with risk of ESRD in T1D. Future directions should involve elucidating mechanisms that underlie increased acetylation or C-glycosylation of metabolites in patients at increased risk for renal disease, whereas independent replication studies will expand our knowledge about the potential generalizability of the current findings.
Supplementary Material
Article Information
Funding. This study was supported by grants from JDRF (career development award 5-CDA-2015-89-A-B to M.A.N. and 17-2013-311 to A.S.K.) and from the National Institute of Diabetes and Digestive and Kidney Diseases (DK-072381 to J.V.B., DK-082841 to S.P., and DK-41526 to A.S.K.).
Duality of Interest. J.V.B. is co-inventor on KIM-1 patents, which have been licensed by Partners Healthcare to several companies. He has received royalties from Partners Healthcare and grant funding from Novo Nordisk. No other potential conflicts relevant to this article were reported.
Author Contributions. M.A.N. designed the study, supervised experimental data collection, analyzed and interpreted data, and wrote the manuscript. A.V.M. supervised measurements of acetyllysine, analyzed the data, and edited the manuscript. S.C. contributed to the data collection and performed the relevant laboratory measurements. J.B. performed the measurements of acetyllysine and analyzed the data. M.M. examined patients and contributed to the data collection. V.S.S. performed measurements of urinary KIM-1 and analyzed the data. A.S. supervised phenotypic data collection and contributed to the data analysis. J.V.B. contributed to the data interpretation and edited the manuscript. S.P. contributed to the study design and data interpretation and edited the manuscript. A.S.K. designed the study, contributed to the data interpretation, and edited the manuscript. M.A.N. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc16-0173/-/DC1.
References
- 1.Krolewski AS. Progressive renal decline: the new paradigm of diabetic nephropathy in type 1 diabetes. Diabetes Care 2015;38:954–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skupien J, Warram JH, Smiles AM, et al. The early decline in renal function in patients with type 1 diabetes and proteinuria predicts the risk of end-stage renal disease. Kidney Int 2012;82:589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niewczas MA, Sirich TL, Mathew AV, et al. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: metabolomic study. Kidney Int 2014;85:1214–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merchant ML, Perkins BA, Boratyn GM, et al. Urinary peptidome may predict renal function decline in type 1 diabetes and microalbuminuria. J Am Soc Nephrol 2009;20:2065–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pezzolesi MG, Satake E, McDonnell KP, Major M, Smiles AM, Krolewski AS. Circulating TGF-β1-regulated miRNAs and the risk of rapid progression to ESRD in type 1 diabetes. Diabetes 2015;64:3285–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhee EP, Clish CB, Ghorbani A, et al. A combined epidemiologic and metabolomic approach improves CKD prediction. J Am Soc Nephrol 2013;24:1330–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sas KM, Karnovsky A, Michailidis G, Pennathur S. Metabolomics and diabetes: analytical and computational approaches. Diabetes 2015;64:718–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma K, Karl B, Mathew AV, et al. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J Am Soc Nephrol 2013;24:1901–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekula P, Goek ON, Quaye L, et al. A metabolome-wide association study of kidney function and disease in the general population. J Am Soc Nephrol 2015;27:1175–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosolowsky ET, Skupien J, Smiles AM, et al. Risk for ESRD in type 1 diabetes remains high despite renoprotection. J Am Soc Nephrol 2011;22:545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warram JH, Gearin G, Laffel L, Krolewski AS. Effect of duration of type I diabetes on the prevalence of stages of diabetic nephropathy defined by urinary albumin/creatinine ratio. J Am Soc Nephrol 1996;7:930–937 [DOI] [PubMed] [Google Scholar]
- 12.Inker LA, Schmid CH, Tighiouart H, et al.; CKD-EPI Investigators . Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med 2012;367:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.U.S. Renal Data System USRDS 2010 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. Bethesda, MD, National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, 2013 [Google Scholar]
- 14.Center for Disease Control and Prevention; National Center for Health Statistics. National Death Index [Internet]. Available from https://www.cdc.gov/nchs/ndi. Accessed 1 September 2013
- 15.Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform 2010;2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohta T, Masutomi N, Tsutsui N, et al. Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicol Pathol 2009;37:521–535 [DOI] [PubMed] [Google Scholar]
- 17.Looker HC, Colombo M, Hess S, et al.; SUMMIT Investigators . Biomarkers of rapid chronic kidney disease progression in type 2 diabetes. Kidney Int 2015;88:888–896 [DOI] [PubMed] [Google Scholar]
- 18.Gohda T, Walker WH, Wolkow P, et al. Elevated urinary excretion of immunoglobulins in nonproteinuric patients with type 1 diabetes. Am J Physiol Renal Physiol 2012;303:F157–F162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabbisetti VS, Waikar SS, Antoine DJ, et al. Blood kidney injury molecule-1 is a biomarker of acute and chronic kidney injury and predicts progression to ESRD in type I diabetes. J Am Soc Nephrol 2014;25:2177–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaidya VS, Niewczas MA, Ficociello LH, et al. Regression of microalbuminuria in type 1 diabetes is associated with lower levels of urinary tubular injury biomarkers, kidney injury molecule-1, and N-acetyl-β-D-glucosaminidase. Kidney Int 2011;79:464–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pencina MJ, D’Agostino RB Sr, D’Agostino RB Jr, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 2008;27:157–172; discussion 207–212 [DOI] [PubMed] [Google Scholar]
- 22.Duranton F, Cohen G, De Smet R, et al.; European Uremic Toxin Work Group . Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol 2012;23:1258–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhee EP, Souza A, Farrell L, et al. Metabolite profiling identifies markers of uremia. J Am Soc Nephrol 2010;21:1041–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka H, Sirich TL, Plummer NS, Weaver DS, Meyer TW. An enlarged profile of uremic solutes. PLoS One 2015;10:e0135657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ihara Y, Manabe S, Kanda M, et al. Increased expression of protein C-mannosylation in the aortic vessels of diabetic Zucker rats. Glycobiology 2005;15:383–392 [DOI] [PubMed] [Google Scholar]
- 26.Kanan Y, Brobst D, Han Z, Naash MI, Al-Ubaidi MR. Fibulin 2, a tyrosine O-sulfated protein, is up-regulated following retinal detachment. J Biol Chem 2014;289:13419–13433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosanam H, Thai K, Zhang Y, et al. Diabetes induces lysine acetylation of intermediary metabolism enzymes in the kidney. Diabetes 2014;63:2432–2439 [DOI] [PubMed] [Google Scholar]
- 28.Kalim S, Karumanchi SA, Thadhani RI, Berg AH. Protein carbamylation in kidney disease: pathogenesis and clinical implications. Am J Kidney Dis 2014;64:793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hille A, Rosa P, Huttner WB. Tyrosine sulfation: a post-translational modification of proteins destined for secretion? FEBS Lett 1984;177:129–134 [DOI] [PubMed] [Google Scholar]
- 30.Moore KL. The biology and enzymology of protein tyrosine O-sulfation. J Biol Chem 2003;278:24243–24246 [DOI] [PubMed] [Google Scholar]
- 31.Furmanek A, Hofsteenge J. Protein C-mannosylation: facts and questions. Acta Biochim Pol 2000;47:781–789 [PubMed] [Google Scholar]
- 32.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 2000;49:341–351 [DOI] [PubMed] [Google Scholar]
- 33.Chow CS, Lamichhane TN, Mahto SK. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem Biol 2007;2:610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu R, Zhong Y, Li X, et al. Role of transcription factor acetylation in diabetic kidney disease. Diabetes 2014;63:2440–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao S, Xu W, Jiang W, et al. Regulation of cellular metabolism by protein lysine acetylation. Science 2010;327:1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Nicholls SJ, Rodriguez ER, et al. Protein carbamylation links inflammation, smoking, uremia and atherogenesis. Nat Med 2007;13:1176–1184 [DOI] [PubMed] [Google Scholar]
- 37.Farzan M, Mirzabekov T, Kolchinsky P, et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 1999;96:667–676 [DOI] [PubMed] [Google Scholar]
- 38.Kato M, Natarajan R. Diabetic nephropathy—emerging epigenetic mechanisms. Nat Rev Nephrol 2014;10:517–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.