Abstract
Clustered regularly-interspaced short palindromic repeats (CRISPR) is a new and effective genetic editing tool. CRISPR was initially found in bacteria to protect it from virus invasions. In the first step, specific DNA strands of virus are identified by guide RNA that is composed of crRNA and tracrRNA. Then RNAse III is required for producing crRNA from pre-crRNA. In The second step, a crRNA:tracrRNA:Cas9 complex guides RNase III to cleave target DNA. After cleavage of DNA by CRISPR-Cas9, DNA can be fixed by Non-Homologous End Joining (NHEJ) and Homology Directed Repair (HDR). Whereas NHEJ is simple and random, HDR is much more complex and accurate. Gene editing by CRISPR is able to be applied to various biological field such as agriculture and treating genetic diseases in human.
Keywords: CRISPR, Gene editing, Homology directed repair, Nonhomologous end joining
INTRODUCTION
Clustered regularly-interspaced short palindromic repeats (CRISPR) is a new and effective genetic editing tool. Because of the efficiency and affinity, CRISPR has been considered to be a revolutionary invention in the field of genetic-editing and molecular biology. Compared with other genetic-editing techniques, CRISPR has an irreplaceable advantage, which is its accessibility. The application of CRISPR does not require years long training and high laboratory cost. It only demands some basic training and less than 100 dollars for buying the CRISPR molecule, which enables researchers to study genetic editing and process an experiment. By developing CRISPR, numerous applications will be invented in the fields of agriculture, environmental science, biology, and public health care. Of course, there will be huge commercial interest in this field, which will boost the development of those applications. Even though CRISPR has shown us a bright and promising future, it also has its own challenges. One of the biggest challenges is the ethic problem.
HOW DOES CRISPR SYSTEM CORRECTLY CUT TARGET DNA?
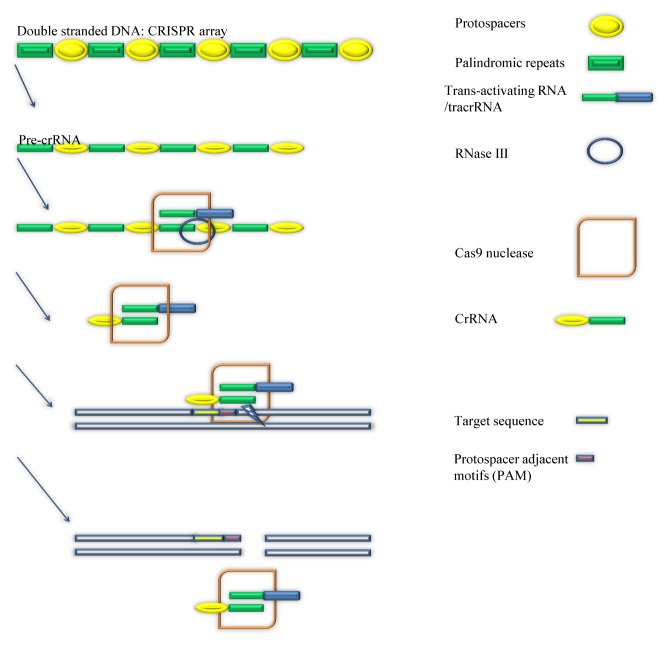
CRISPR is such a strong genetic editing tool is mainly caused by its unique structure and design. But, it might be hard to believe that the creation of CRISPR is based on an immune system of bacteria. At the beginning of the research, scientists found that some genes in the bacteria seem useless for constructing the cells and maintain survival of bacteria (1, 2). But, as scientists discovered that this “useless” gene is commonly existed in bacteria and other micro-organisms, they started to unwind the mystery of the gene. This special gene in bacteria is coding for an interesting function for protecting the bacteria from the invasion by viruses (3). It is also the most primitive form of CRISPR. This defense system of bacteria can achieve elimination of the invading virus by two steps. The first step is to target the invaders and a specific DNA strand of the virus. Then the next step is to cut the target DNA strand of the virus by using a nuclease. As described in Fig. 1, in step one, in order to identify the location of the viruses and target them, the defense system needs a help from the guide RNA. The guide RNA, which is composed of crRNA and tracrRNA, is responsible for locating and targeting the foreign invaders of the bacteria through identification of specific nucleotides of the invading viruses (4). But, guide RNA cannot just automatically find and match the specific nucleotides that bacteria looks for. It has to go through a complex procedure to develop the ability to locate a piece of DNA sequences in the viral genome. When Viruses start to invade the living cells of bacteria, they will release their own invading DNA strands into the bacteria cells for turning the invaded cells into factories that produce more Viruses. At this moment, the immune system of bacteria will make a copy of one DNA strand of the invading virus, and then inserts it into a group of genome of bacteria, which is called CRISPR array (5, 6).
Fig. 1.
How does CRISPR system correctly cut target DNA? (left panel) The Procedures of producing crRNA from pre-crRNA, binding of cr-RNA:tracrRNA: Cas9 complex to the target sequence near the PAM and the cleavage of target DNA are described. (right panel) The components of figures are described at right panel.
The DNA of the virus will be stored in the spacers between two palindromic repeats in the CRISPR array (7). Then bacteria will transcribe a portion of CRISPR array to make a pre-crRNA strand (8) that also includes multiple complementary spacers and palindromic repeats. In the process of turning pre-crRNA to crRNA, a tracrRNA and a double-stranded RNA-specific ribonuclease are required (9). TracrRNA will bind to one of the palindromic repeats of the pre-crRNA (10), and the ribonuclease III will cut the tracrRNA (9, 11). Finally this separated pre-crRNA is matured and called Cr-RNA. Now the crRNA and tracrRNA will bind to the cas9 nuclease to form a crRNA: tracrRNA:Cas9 complex. CrRNA in the complex is completely complementary to a DNA sequence of the invading virus, so that crRNA guide the cas9 nuclease to the viral DNA sequence that are complementary to the crRNA (12). Importantly, a protospacer adjacent motifs (PAM) is required during this process. PAM is a sequence of nucleotides that are located close to the sequences complementary to the crRNA (13), and acts as an initiation of attaching crRNA to its complementary strand in virus (14, 15). After crRNA:tracrRNA: Cas9 complex is launched to the invading virus’s DNA strand, the location of the virus is clearly identified. The tool that kills the virus is the Cas9 nuclease. By binding crRNA:tracrRNA: Cas9 complex to the targeted strand, the Cas9 nuclease will cut the DNA strands of the virus near the PAM (14), so that the virus will have incomplete DNA strands, which will defeat the viruses.
NON-HOMOLOGOUS END JOINING (NHEJ) AND (HOMOLOGY DIRECTED REPAIR) HDR
CRISPR’s capability of cutting not only viruses but also genomes of every living life leads the further development of genetic editing in other organisms. Inside bacteria, CRISPR is able to silence invading viruses by tracing and cutting their DNA strands, due to the existence of Cas9 nuclease and crRNA:tracrRNA:Cas9 complex (12). This tracing and cutting mechanism can also be used in cutting the DNA strand of other organism. Once CRISPR is inserted into a specific site of a DNA strand, it can perform the function of cutting DNA strand by introducing a DNA strands breaks (DSB) to the site, which is just like how the CRISPR kills the invading viruses of the bacteria. But using CRISPR to cut the DNA strand is not the point of doing the CRISPR research. Instead, CRISPR is used to introduce the DSB to those DNA strands for a chance of changing the genetic sequence of the nucleotides on that DNA strand (16). However, CRISPR will not provide such a function of changing the specific nucleotide, because the only function it can provide is cutting the DNA strand. Thus, a method of fixing and editing the broken DNA strands is necessary for the DNA to perform its function again and introducing changes to the targeted DNA sequences. There are two different mechanisms to edit and fix the damaged DNA sequence, which are Non-Homologous End Joining (NHEJ) and Homology Directed Repair (HDR) (17).
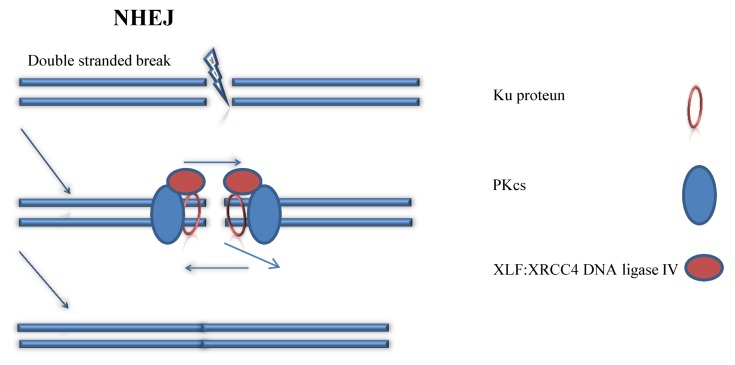
NHEJ is a natural methods of healing damages of DNA strands in most organisms (18). For example, when UV lights damages the DNA strands of the skin cells, our body can use NHEJ to reconnect two broken DNA strands, so they are still functioning. Generally, this method is relatively simple and effective in terms of fixing damages of genes, because this fixing pathway does not require a homologous template to fix the DNA. As described in Fig. 2, in NHEJ, the Ku protein will bind to the two ends of the broken DNA strands and form a Ku DNA end complex (19). This Ku DNA end complex will combine with DNA-PKcs complex to chop away overhangs of nucleotides near the end of the two broken strands (20). Then, with the help from the XLF:XRCC4 DNA ligase IV, the two broken ends of DNA strands will join together and reconnect to each other via ligation (21). Due to the introduction of DSB and some nucleotide deletions during repairing, the original DNA sequence is permanently changed after NHEJ. Thus, eventually, repairing DNA strands via using NHEJ is easy to introduce some unexpected mutations to the targeted DNA strand, because people cannot control what nucleotide is knocked out and what is not. Importantly, due to the simplicity and efficiency of this mechanism, it may have a bright future in the field of non-human genetic editing, like using CRISPR in the field of agriculture and fixing eco-system.
Fig. 2.
Non-Homologous End Joining (NHEJ). (left panel) procedures of NHEJ is described. (right panel) components of the figure is described.
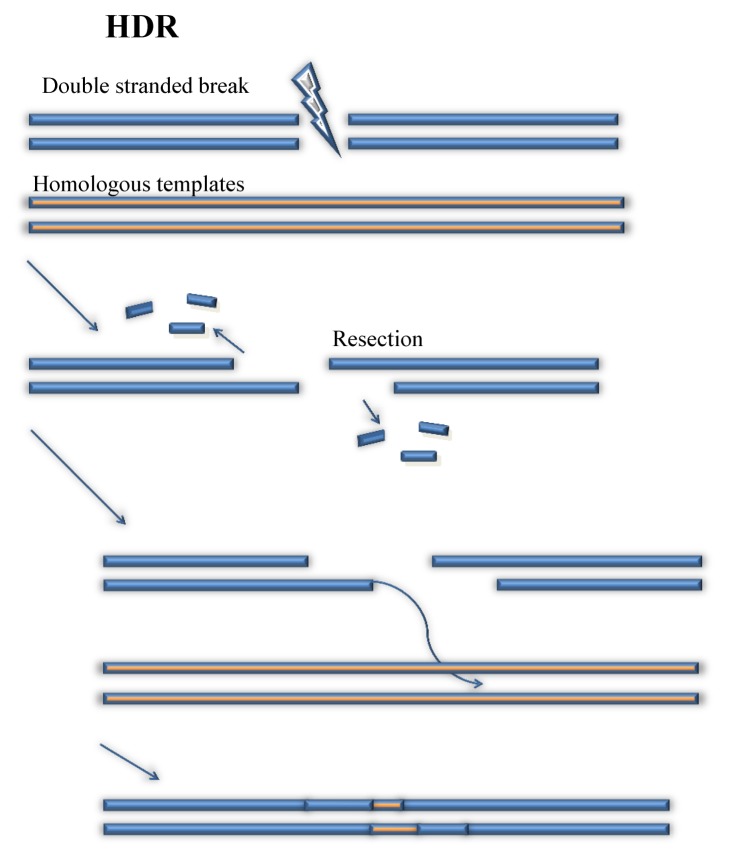
In contrast to the simplicity and randomness of NHEJ, HDR is a much more complex and accurate tool of fixing and editing the genetic sequence of the targeted organism. The main factor that contributes to the greater accuracy of editing is the introduction of a specific template strand (22, 23). NHEJ can repair and edit DNA sequence through deletions of nucleotides to the broken site. Thus, the NHEJ does not have a promising accuracy on processing genetic editing. But HDR is able to edit the DNA sequence with great accuracy by using a prepared DNA strands (Fig. 3) (24). When cr-RNA:tracrRNA:Cas9 complex launches to the PAM site and DSB is introduced, one of the broken DNA strand will lose some nucleotides until it has a 3′ end at the broken site (25). This DNA strand will unzip the prepared DNA strands, and begin to launch at 5′ end of one DNA strand (26, 27). This launching site will become a primer of DNA replication (26). When the invading strand is properly launched, DNA polymerase will pull some nearby nucleotides to create a complementary strand of DNA of the prepared DNA strand from the primer (28). Thus, one of the broken strands successively repairs the broken site and obtains desirable DNA sequences from the prepared strand. Then, as the broken site obtains the desired DNA strand from the provided DNA template, the rest of the missing nucleotides can be reconstructed based on the nucleotides that are available. Eventually, after repairing, the broken site of DNA strands obtain designed genetic sequences from the prepared DNA strands. With assistance of fixing and repairing from NHEJ and HDR, CRISPR is able to edit any gene in any organism.
Fig. 3.
Homology Directed Repair (HDR). (left panel) procedures of HDR is described.
APPLICATIONS OF CRISPR IN GENE EDITING
Theoretically, human obtains the ability to change the genome sequences using CRISPR. This possibility sounds very attractive to the fields that are related to biology, because there are numerous opportunities that can be offered by CRISPR.
For agriculture, CRISPR may be used to edit the genome of crops and plants to offer them phenotypes that provide resistance to bacteria, so that less pesticide will be used to treat it, which will possibly save money for farmers and lower the damages from overuse pesticides. CRISPR can also be used in treating genetic diseases that are caused by some mutations in DNA strands. For example, Huntington’s disease is caused by the mutation on HTT genes (29), which can be seen as having incorrect nucleotide pairs on the genome. If CRISPR can be used to replace the incorrect nucleotides on HTT gene with correct ones, Huntington’s disease can be easily treated (30). Although CRISPR has infinite applications, it still faces two challenges. The first challenge is the off-target problem. Recently, a Chinese team tried to use CRISPR to edit the genetic information of human embryos to code for a mutation on CCR5 gene for getting resistance to HIV. HIV needs a very specific launching spot on normal white blood cells to invade them, but a mutation on CCR5 gene can cause a change on that spot and HIV will lose the ability to invade those cells (31). Thus, by introducing a mutation to CCR5 gene, there is a hope to develop resistance to HIV. However, when scientists tried to introduce this mutation, they discovered a high off-target rate for CRISPR (32). These mismatched nucleotides or unexpected mutations can all lead to serious problems.
Another problem that CRISPR has to face before real applications is the ethics. The ethical problems of CRISPR does not come from editing the DNA sequences of organism like what happened last century when people still concerned about side effect of genetic editing. Most of countries now offer permissions for processing genetic editing on organisms for improving agriculture or other biological use. However, even if permission of editing organisms for agricultural or biological use is granted, editing human genome is still very controversial. There are two main reasons that cause this ethical problem. First, CRISPR is not a completely developed and mature method yet to process precise genetic editing, especially when the target is human genome. If mistakes or off-target effect takes place during genetic editing, patients may result in serious genetic mutation, which will not only affect the health condition of patients but also the future generations of patients. Thus, before scientists develop more advanced version of CRISPR, it is better not to use it on human. Another ethical problem for using CRISPR is not related to the technical problem. CRISPR can be considered as the strongest genetic editing tool that is invented in human history, because it enables scientists to edit any genome in any organisms. Thus, some ethicists concern about the consequences of CRISPR overuse in editing human genome. If CRISPR is commonly used to modify human genome, huge social conflicts can be possibly generated. For example, people may be able to design their children based on their interests and some people can possibly have a much longer life span than the others due to the use of CRISPR. These possible applications sound very attractive to us, but they may cause very serious social conflicts in terms of the increasing social inequality and other problems. Thus, ethicists think that CRISPR still needs a long period of time to be allowed to edit human genome, because society has to gradually adapt to the changes that are brought by CRISPR.
As a summary, CRISPR is a young and very powerful genetic editing tool. Its mechanism allows scientists to edit any DNA sequences in any organisms, which is historical for genetic editing. The breakthrough of CRISPR will possibly lead to a golden age of genetic editing. In the future, CRISPR and genetic editing will be applied to almost every corner of our life.
ACKNOWLEDGEMENTS
This work was supported by the NRF-2015R1A2A1A15054247 grant to Haihong Shen, the NRF-2016R1A2B1007135 grant to Xuexiu Zheng and Cell Logistics Research center (2016R1A 5A1007318) funded by the National Research Foundation (NRF) of Korea, and an integrative aging research grant at the Gwangju Institute of Science and Technology (GIST).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim KI. Recent advances in developing molecular tools for targeted genome engineering of mammalian cells. BMB Rep. 2015;48:6–12. doi: 10.5483/BMBRep.2015.48.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrangou R, Fremaux C, Deveau H, et al. Crispr provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 4.Hwang WY, Fu Y, Reyon D, et al. Efficient genome editing in zebrafish using a crispr-cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deveau H, Garneau JE, Moineau S. Crispr/cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 6.Deveau H, Barrangou R, Garneau JE, et al. Phage response to crispr-encoded resistance in streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhaya D, Davison M, Barrangou R. Crispr-cas systems in bacteria and archaea: Versatile small rnas for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 8.Brouns SJ, Jore MM, Lundgren M, et al. Small crispr rnas guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V. Crrna and tracrrna guide cas9-mediated DNA interference in streptococcus thermophilus. RNA Biol. 2013;10:841–851. doi: 10.4161/rna.24203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carte J, Christopher RT, Smith JT, et al. The three major types of crispr-cas systems function independently in crispr rna biogenesis in streptococcus thermophilus. Mol Microbiol. 2014;93:98–112. doi: 10.1111/mmi.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deltcheva E, Chylinski K, Sharma CM, et al. Crispr rna maturation by trans-encoded small rna and host factor rnase iii. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crrna ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swarts DC, Mosterd C, van Passel MW, Brouns SJ. Crispr interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cencic R, Miura H, Malina A, et al. Protospacer adjacent motif (pam)-distal sequences engage crispr cas9 DNA target cleavage. PLoS One. 2014;9:e109213. doi: 10.1371/journal.pone.0109213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SH, Bae S. Structural and dynamic views of the CRISPR-Cas system at the single-molecule level. BMB Rep. 2016;49:201–207. doi: 10.5483/BMBRep.2016.49.4.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. Rna-guided editing of bacterial genomes using crispr-cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iliakis G, Wang H, Perrault AR, et al. Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet Genome Res. 2004;104:14–20. doi: 10.1159/000077461. [DOI] [PubMed] [Google Scholar]
- 18.Gu J, Lieber MR. Mechanistic flexibility as a conserved theme across 3 billion years of nonhomologous DNA end-joining. Genes Dev. 2008;22:411–415. doi: 10.1101/gad.1646608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein ku, novel domains in the ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu S, Peng A. Non-homologous end joining repair in xenopus egg extract. Sci Rep. 2016;6:27797. doi: 10.1038/srep27797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riballo E, Woodbine L, Stiff T, Walker SA, Goodarzi AA, Jeggo PA. Xlf-cernunnos promotes DNA ligase iv-xrcc4 re-adenylation following ligation. Nucleic Acids Res. 2009;37:482–492. doi: 10.1093/nar/gkn957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using crispr/cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zu Y, Tong X, Wang Z, et al. Talen-mediated precise genome modification by homologous recombination in zebrafish. Nat Methods. 2013;10:329–331. doi: 10.1038/nmeth.2374. [DOI] [PubMed] [Google Scholar]
- 24.Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res. 2001;477:131–153. doi: 10.1016/S0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 25.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the mre11/rad50/xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motamedi MR, Szigety SK, Rosenberg SM. Double-strand-break repair recombination in escherichia coli: Physical evidence for a DNA replication mechanism in vivo. Genes Dev. 1999;13:2889–2903. doi: 10.1101/gad.13.21.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 29.Walker FO. Huntington’s disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 30.Trager U, Andre R, Lahiri N, et al. Htt-lowering reverses huntington’s disease immune dysfunction caused by nfkappab pathway dysregulation. Brain. 2014;137:819–833. doi: 10.1093/brain/awt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopalco L. Ccr5: From natural resistance to a new anti-hiv strategy. Viruses. 2010;2:574–600. doi: 10.3390/v2020574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang X, He W, Huang Y, et al. Introducing precise genetic modifications into human 3pn embryos by crispr/cas-mediated genome editing. J Assist Reprod Genet. 2016;33:581–588. doi: 10.1007/s10815-016-0710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]